Hyperspectral Image Classification Based on Fusing S3-PCA, 2D-SSA and Random Patch Network
Abstract
1. Introduction
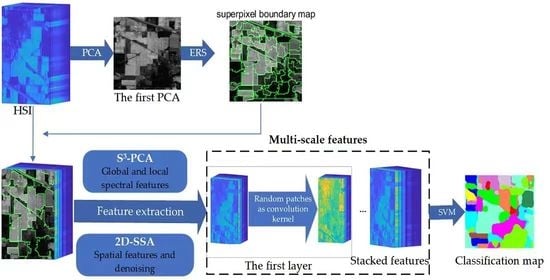
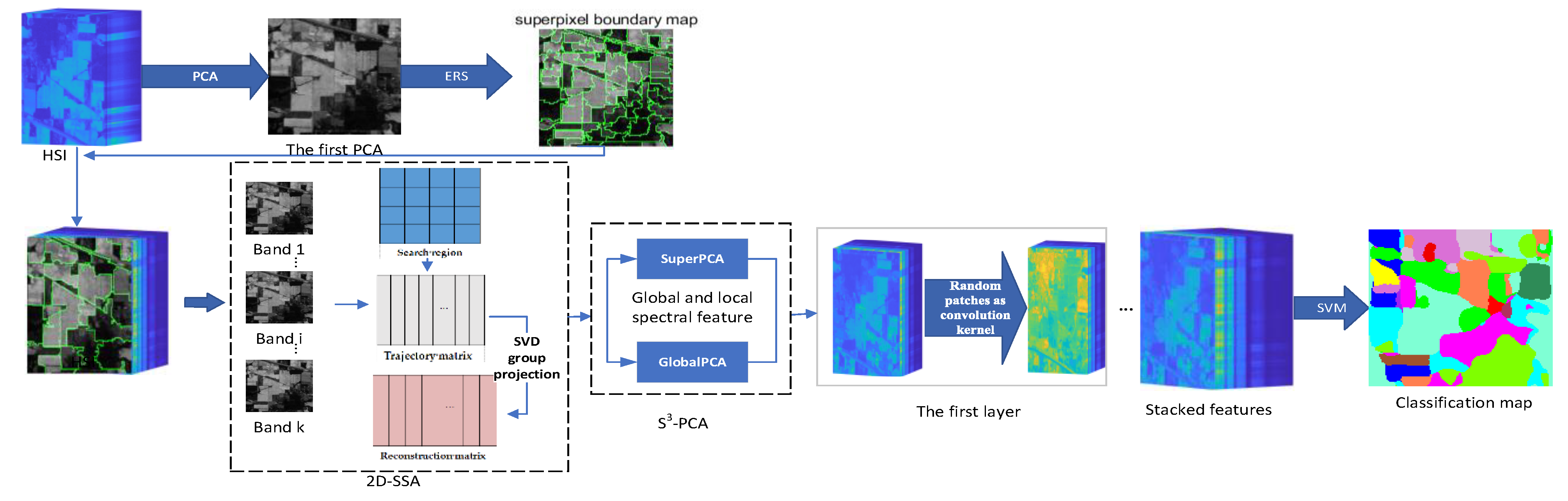
2. Methods
2.1. Spectral–Spatial and SuperPCA (S3-PCA)
2.2. Two-Dimensional Singular Spectrum Analysis (2D-SSA)
2.3. Random Patches Network (RPNet)
2.4. Proposed MS-RPNet Model
2.4.1. S3-PCA Domain Feature Extraction and Fusion with 2D-SSA
2.4.2. Convolution with Random Patches
| Algorithm 1. The proposed hyperspectral image classification algorithm. |
| Input: HSI image D, principal component number (PC_num), superpixel number (Pixel_num), layer number (Layernum). The first layer:
|
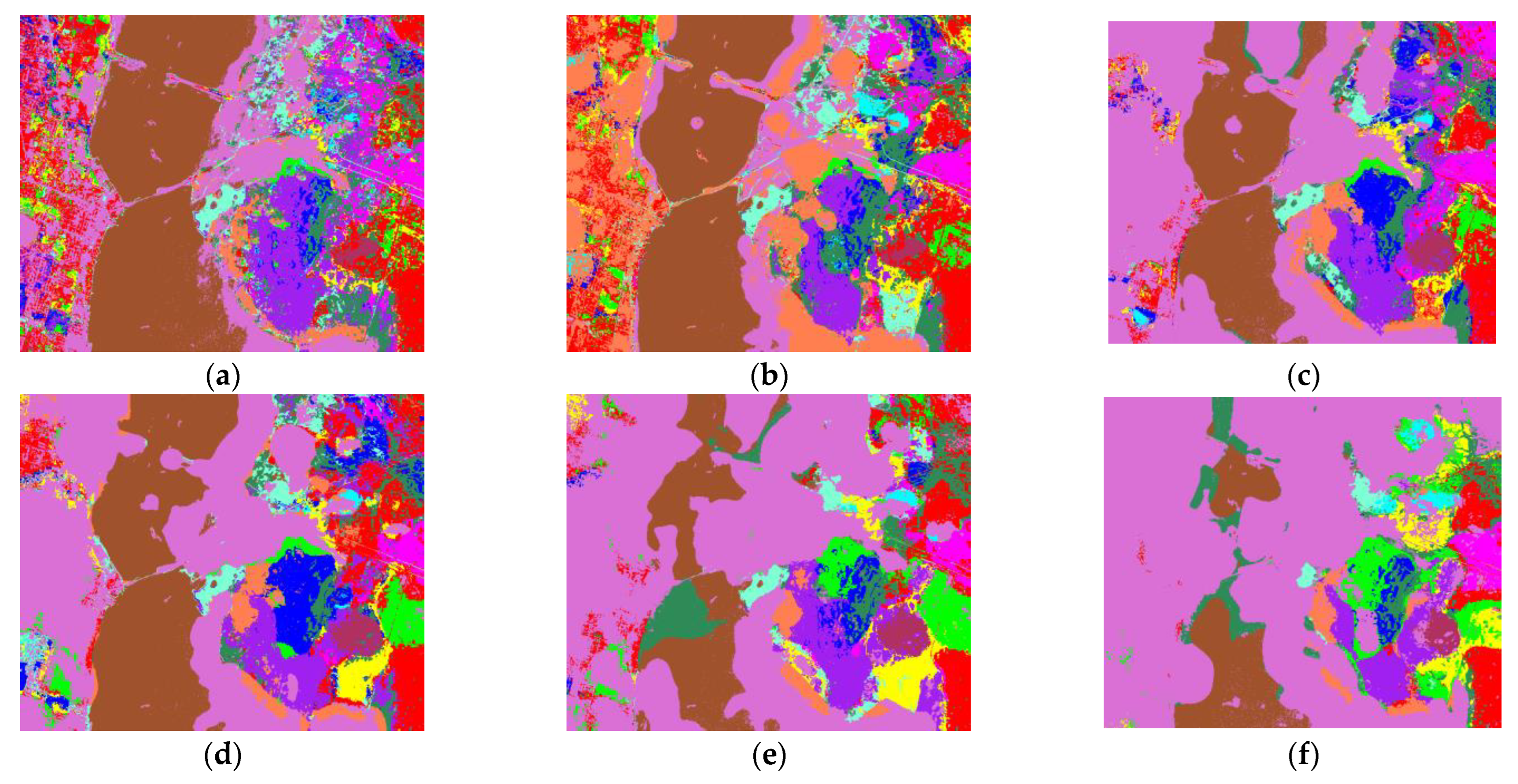
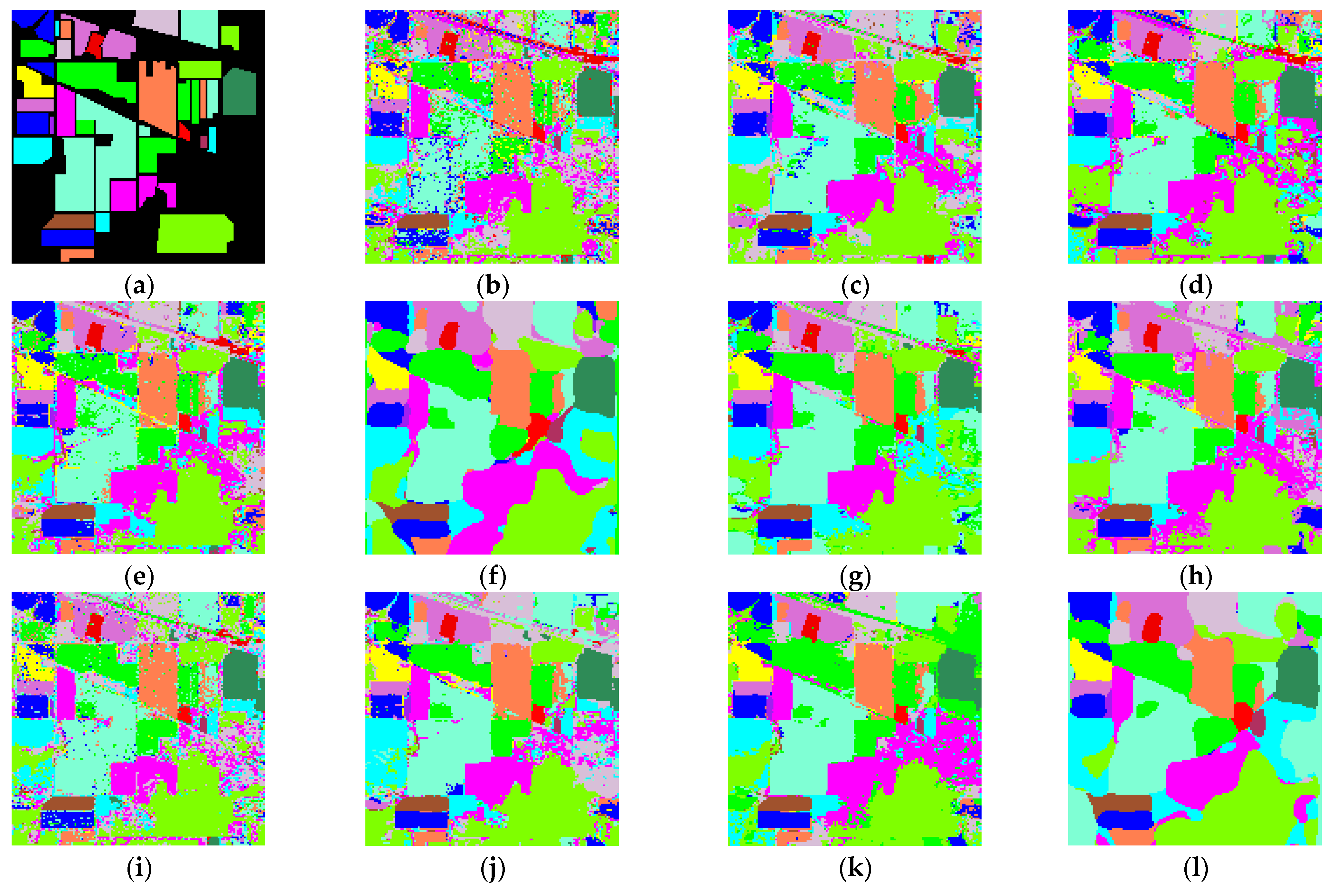
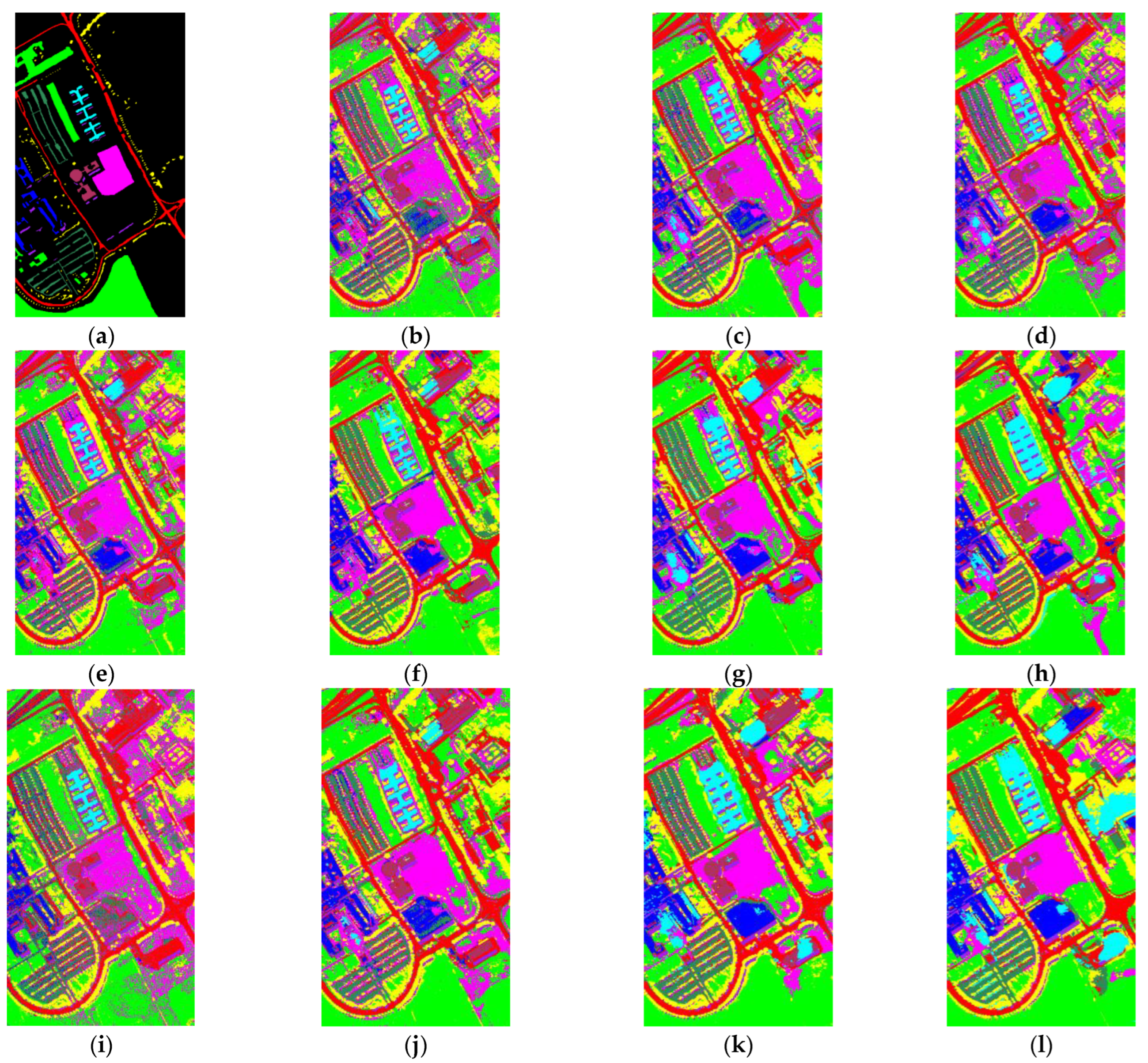
3. Experiments
3.1. Introduction of Datasets
3.2. Parameter Analysis
- (1)
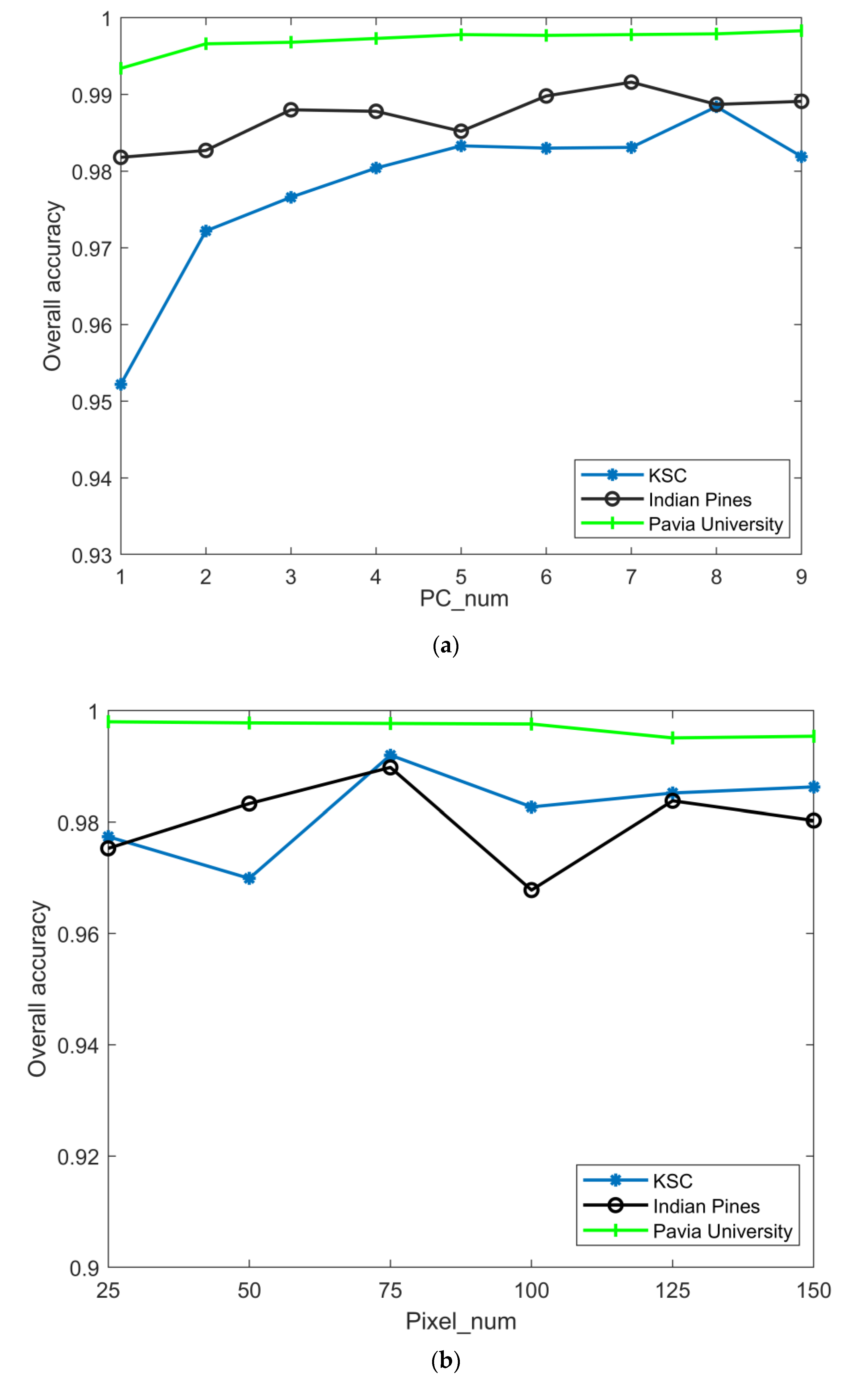
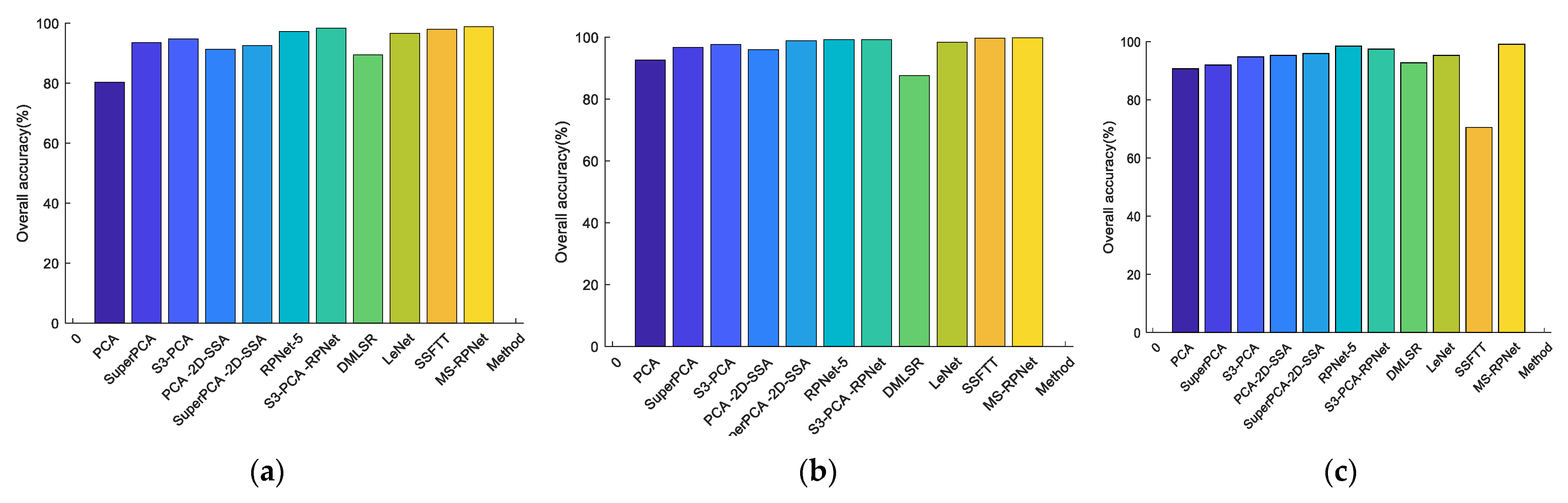
- Analyze the effect of the parameter PC_num (number of principal components) on the experiment. The values of PC_num were divided into 9 cases (PC_num = 1, 2, 3, 4, 5, 6, 7, 8, 9), the effect of parameter PC_num on classification accuracy was observed in 9 cases, and the specific classification accuracy is plotted in Figure 5. From the figure, it can be seen that for the Indian Pines dataset, the variation in PCA does not affect the overall precision of classification, and the principal component dimension is taken as 7; for the Pavia University dataset, the change in the principal component dimension has a smaller impact on the overall accuracy, and the low-dimensional matrix is considered to be more beneficial to the subsequent calculation of the model, and PC_num = 5. For the KSC dataset, the change in the principal component dimension causes the overall accuracy to fluctuate, and the curves show a tendency to increase and then level, and PC_num = 8.
- (2)
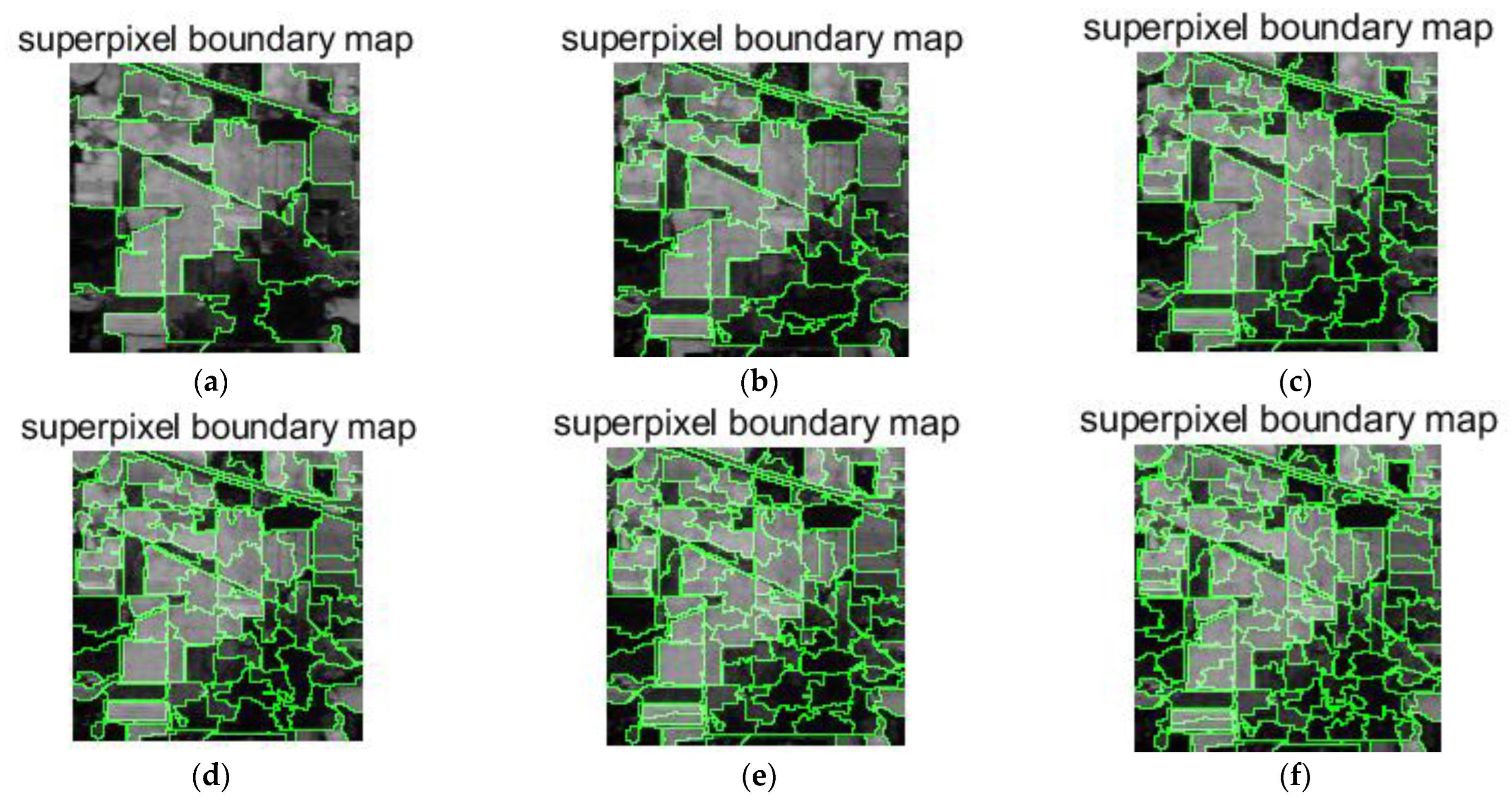
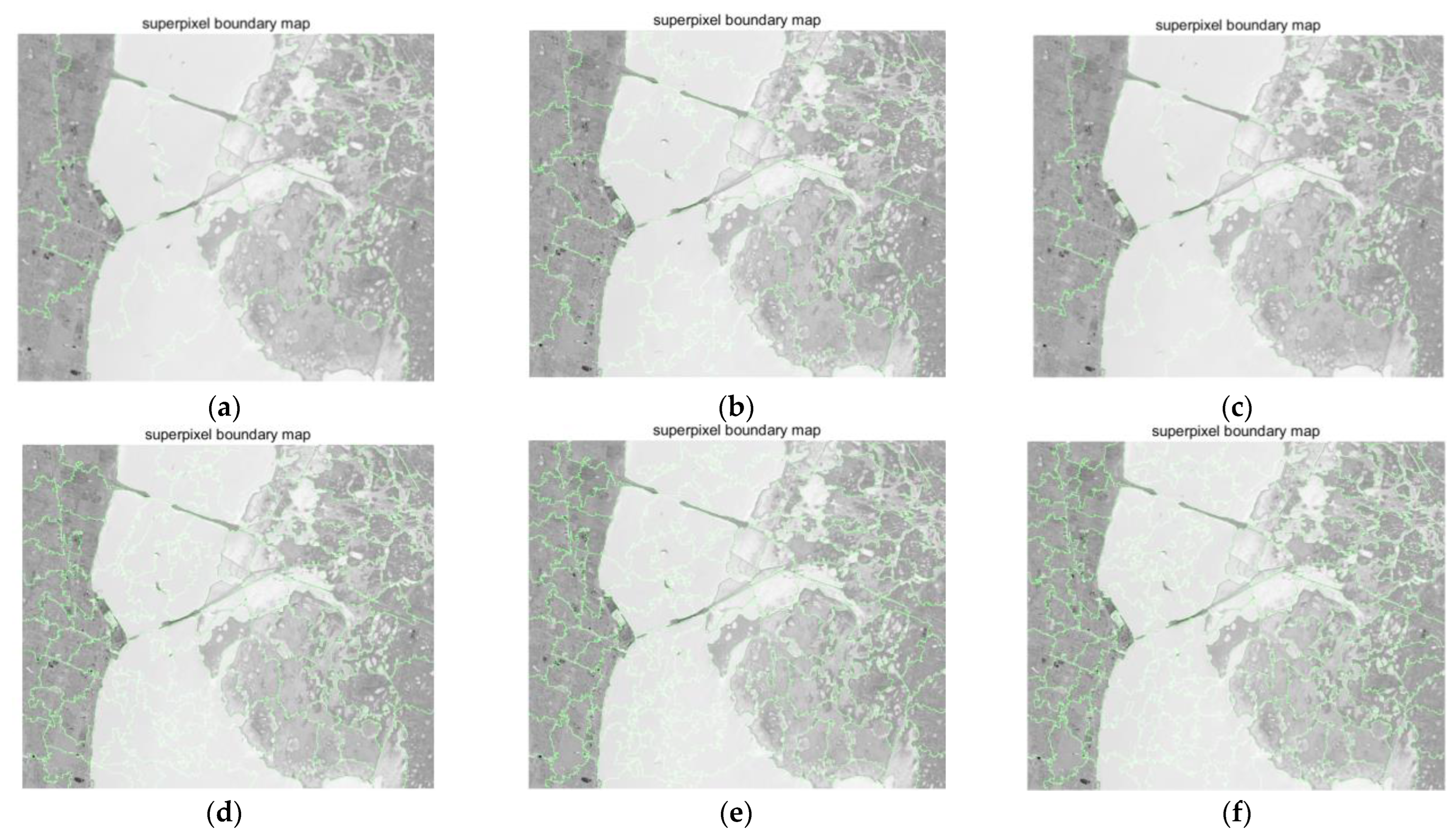
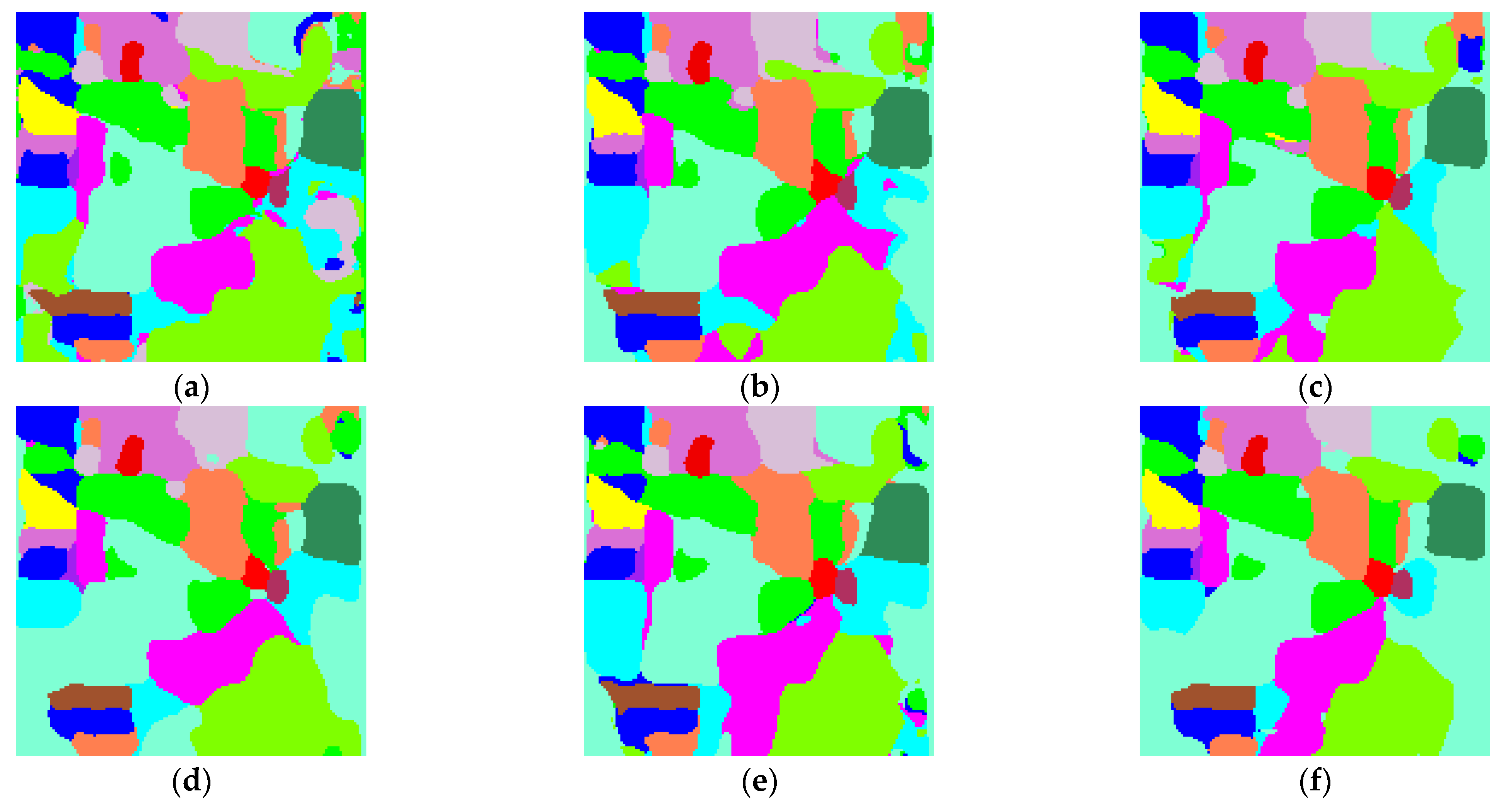
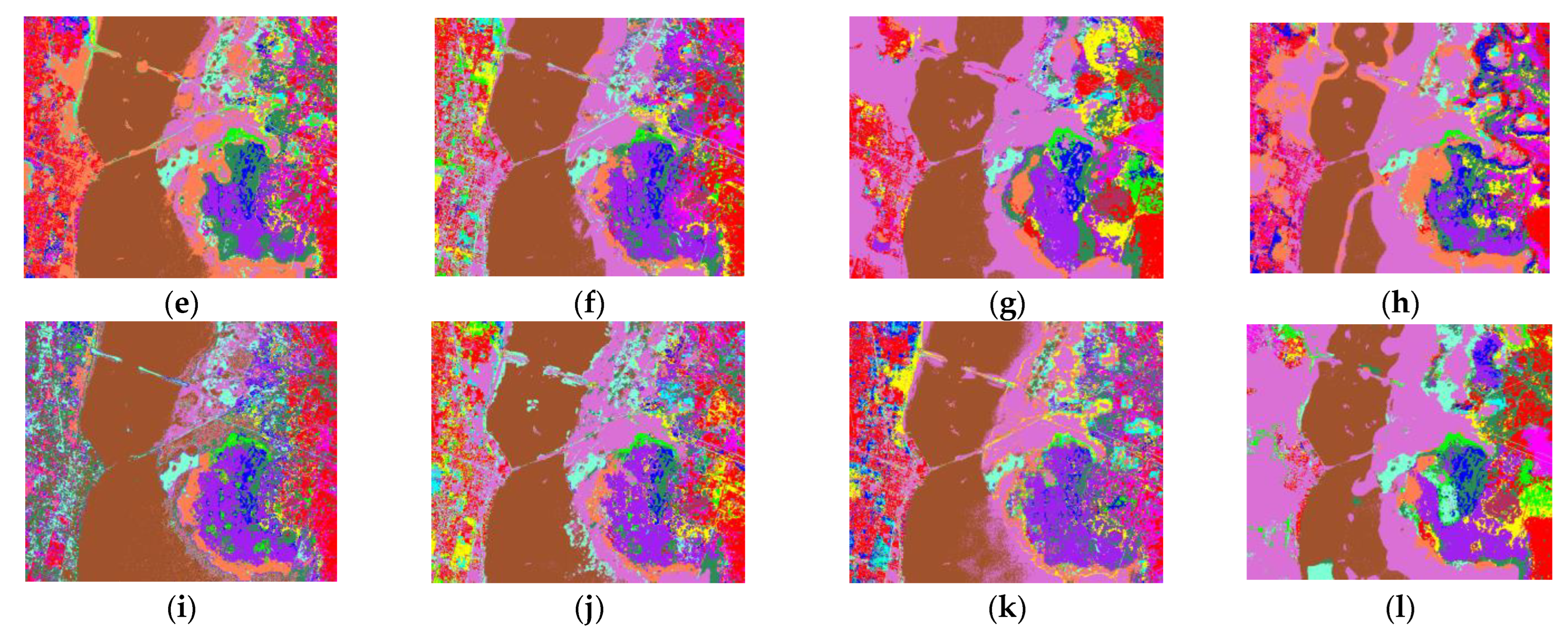
- Analyze the effect of the parameter Pixel_num (superpixel number) on the experiment. The PC_nums are fixed according to the optimal number in (1). The specific superpixel segmentation graphs obtained by dividing the Pixel_num values into six cases (Pixel_num = 25, 50, 75, 100, 125, 150 are shown in Figure 6, Figure 7 and Figure 8. The number of superpixels determines the granularity of the segmentation result and various classification results. A larger number of superpixels produces finer-grained segmentation results that can better preserve the detailed image information, but may retain redundant information, while a smaller number of superpixels produces coarser segmentation results, but may lose some details. Thus, we need to choose this according to the specific application requirements and image characteristics. From Figure 5, it is obvious that different superpixel numbers make a greater difference to the classification accuracy for the first two datasets, which further indicates that the introduction of superpixel segmentation helps to improve classification accuracy. For the Pavia University dataset, the effect of the change in the number of superpixels on the overall accuracy is also not significant. An increase in the number of superpixels leads to an increase in the computational complexity of the algorithm. Considering the computational complexity, the parameter Pixel_num is set to Pixel_num = 75, and a high overall accuracy is achieved on the validation set of each dataset.
- (3)
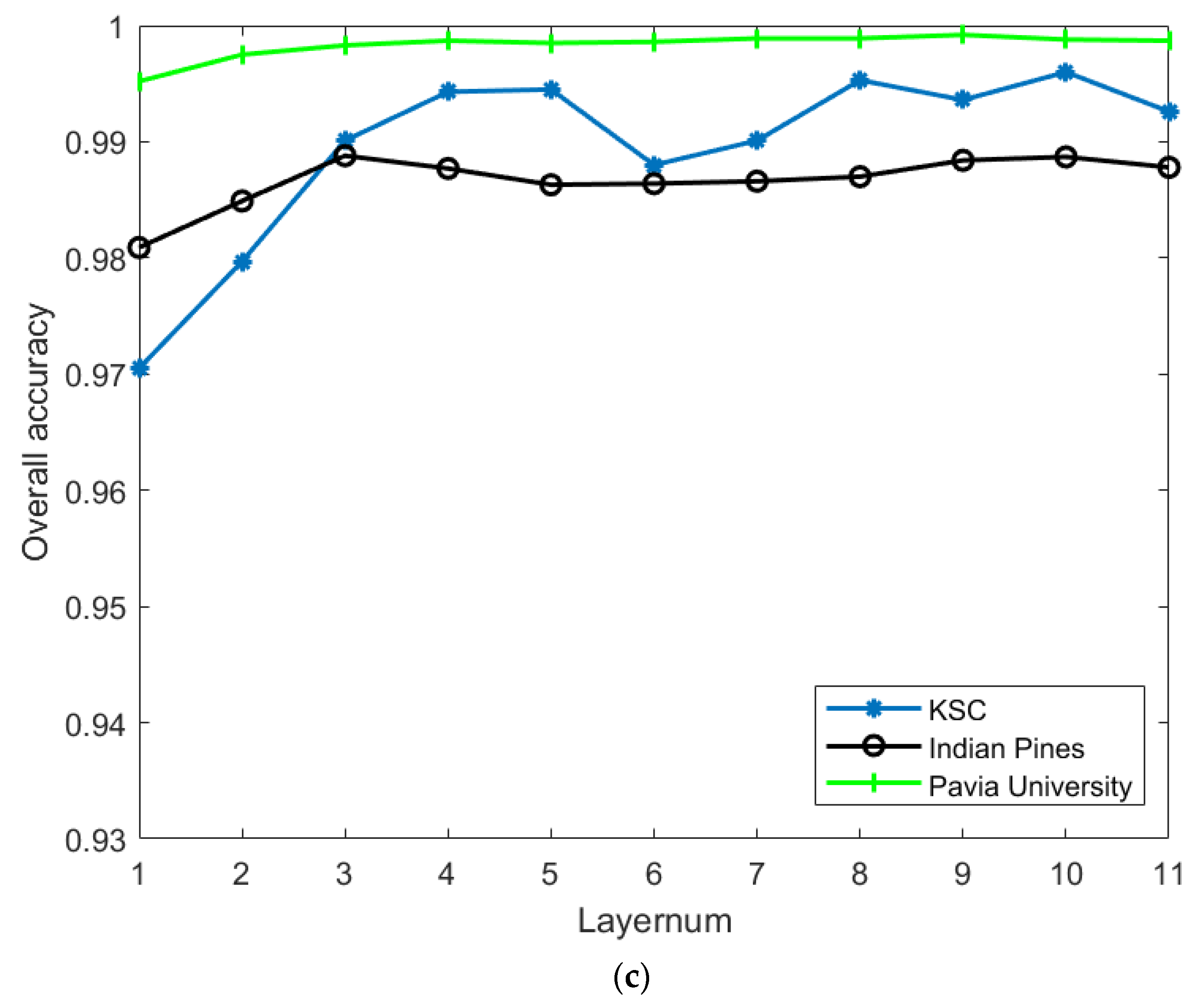
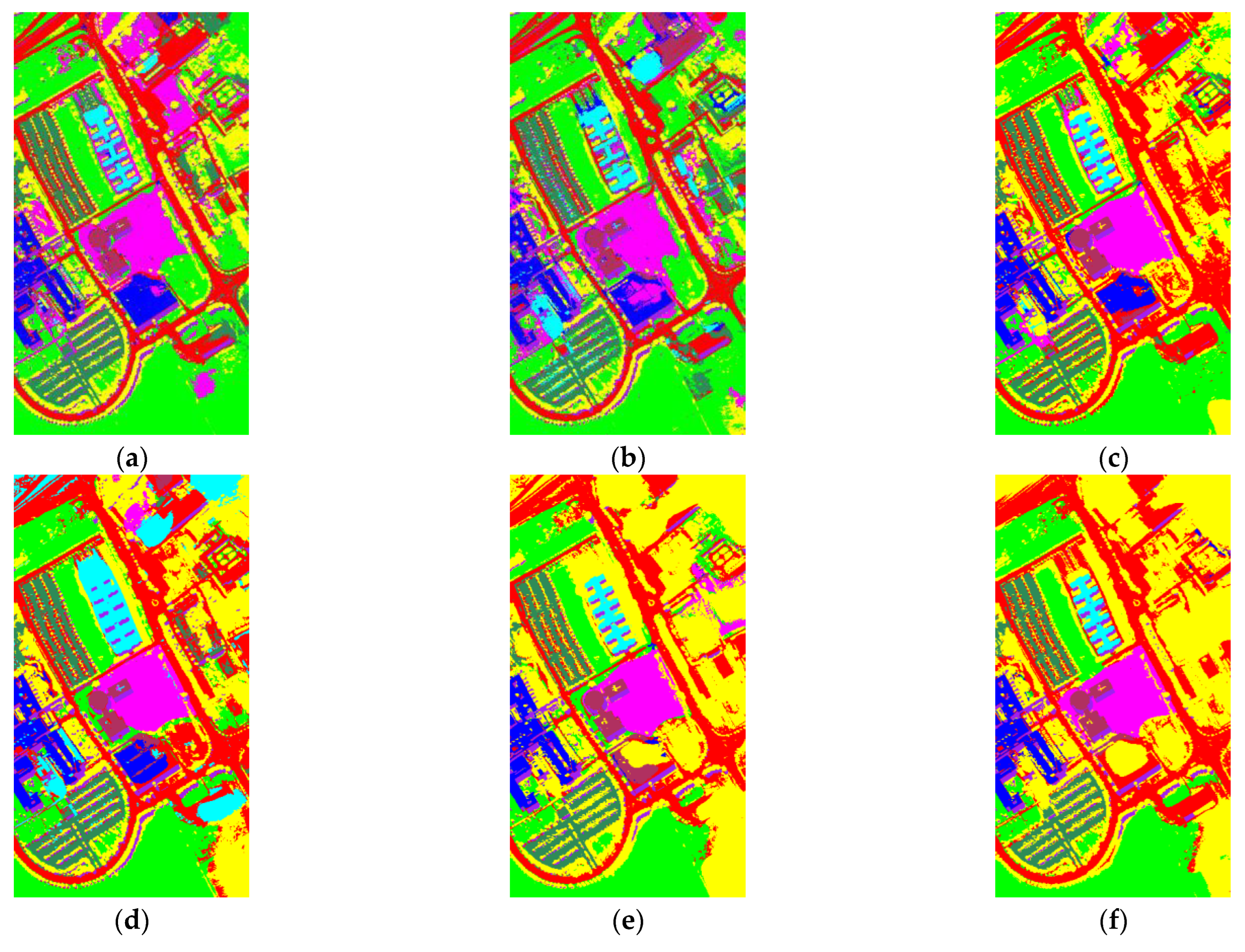
- Analyze the effect of the parameter Layernum on the experiment. The PC_nums and pixel_nums are fixed according to the optimal number in (1). The classification results are shown in Figure 9, Figure 10 and Figure 11. The overall accuracy gradually increases and then stabilizes when the layer depth increases, which indicates that the random blocks extracted from the HSI contain useful information. However, an architecture that is too deep not only does not improve the accuracy, but also increases the computational complexity. According to Figure 5., the number of layers is taken as 3, 3, 5 respectively.
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Camps-Valls, G.; Tuia, D.; Bruzzone, L.; Benediktsson, J.A. Advances in Hyperspectral Image Classification: Earth Monitoring with Statistical Learning Methods. IEEE Signal Process. Mag. 2013, 31, 45–54. [Google Scholar] [CrossRef]
- Matthews, M.W.; Bernard, S.; Evers-King, H.; Lain, L.R. Distinguishing cyanobacteria from algae in optically complex inland waters using a hyperspectral radiative transfer inversion algorithm. Remote Sens. Environ. 2020, 248, 111981. [Google Scholar] [CrossRef]
- Carrino, T.A.; Crósta, A.P.; Toledo, C.L.B.; Silva, A.M. Hyperspectral remote sensing applied to mineral exploration in southern Peru: A multiple data integration approach in the Chapi Chiara gold prospect. Int. J. Appl. Earth Obs. Geoinf. 2018, 64, 287–300. [Google Scholar] [CrossRef]
- Tong, X.Y.; Xia, G.S.; Lu, Q.; Shen, H.; Li, S.; You, S.; Zhang, L. Land-cover classification with high-resolution remote sensing images using transferable deep models. Remote Sens. Environ. 2020, 237, 111322. [Google Scholar] [CrossRef]
- Ma, W.; Gong, C.; Hu, Y.; Meng, P.; Xu, F. The Hughes phenomenon in hyperspectral classification based on the ground spectrum of grasslands in the region around Qinghai Lake. In International Symposium on Photoelectronic Detection and Imaging 2013: Imaging Spectrometer Technologies and Applications; SPIE: Bellingham, WA, USA, 2013; Volume 8910, pp. 363–373. [Google Scholar]
- Du, Q.; Yang, H. Similarity-Based Unsupervised Band Selection for Hyperspectral Image Analysis. IEEE Geosci. Remote Sens. Lett. 2008, 5, 564–568. [Google Scholar] [CrossRef]
- Yang, H.; Du, Q.; Chen, G. Unsupervised hyperspectral band selection using graphics processing units. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2011, 4, 660–668. [Google Scholar] [CrossRef]
- Wang, Q.; Lin, J.; Yuan, Y. Salient Band Selection for Hyperspectral Image Classification via Manifold Ranking. IEEE Trans. Neural Netw. Learn. Syst. 2016, 27, 1279–1289. [Google Scholar] [CrossRef]
- Jia, S.; Tang, G.; Zhu, J.; Li, Q. A Novel Ranking-Based Clustering Approach for Hyperspectral Band Selection. IEEE Trans. Geosci. Remote Sens. 2015, 54, 88–102. [Google Scholar] [CrossRef]
- Bruce, L.; Koger, C.; Li, J. Dimensionality reduction of hyperspectral data using discrete wavelet transform feature extraction. IEEE Trans. Geosci. Remote Sens. 2002, 40, 2331–2338. [Google Scholar] [CrossRef]
- Zhao, W.; Du, S. Spectral–Spatial Feature Extraction for Hyperspectral Image Classification: A Dimension Reduction and Deep Learning Approach. IEEE Trans. Geosci. Remote Sens. 2016, 54, 4544–4554. [Google Scholar] [CrossRef]
- Prasad, S.; Bruce, L.M. Limitations of Principal Components Analysis for Hyperspectral Target Recognition. IEEE Geosci. Remote Sens. Lett. 2008, 5, 625–629. [Google Scholar] [CrossRef]
- Bandos, T.V.; Bruzzone, L.; Camps-Valls, G. Classification of Hyperspectral Images With Regularized Linear Discriminant Analysis. IEEE Trans. Geosci. Remote Sens. 2009, 47, 862–873. [Google Scholar] [CrossRef]
- Lixin, G.; Weixin, X.; Jihong, P. Segmented minimum noise fraction transformation for efficient feature extraction of hyperspectral images. Pattern Recognit. 2015, 48, 3216–3226. [Google Scholar] [CrossRef]
- Zabalza, J.; Ren, J.; Ren, J.; Liu, Z.; Marshall, S. Structured covariance principal component analysis for real-time onsite feature extraction and dimensionality reduction in hyperspectral imaging. Appl. Opt. 2014, 53, 4440–4449. [Google Scholar] [CrossRef]
- Jia, X.; Richards, J.A. Segmented principal components transformation for efficient hyperspectral remote-sensing image display and classification. IEEE Trans. Geosci. Remote Sens. 1999, 37, 538–542. [Google Scholar] [CrossRef]
- Zabalza, J.; Ren, J.; Yang, M.; Zhang, Y.; Wang, J.; Marshall, S.; Han, J. Novel Folded-PCA for improved feature extraction and data reduction with hyperspectral imaging and SAR in remote sensing. ISPRS J. Photogramm. Remote Sens. 2014, 93, 112–122. [Google Scholar] [CrossRef]
- Zhang, H.; Li, J.; Huang, Y.; Zhang, L. A Nonlocal Weighted Joint Sparse Representation Classification Method for Hyperspectral Imagery. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2013, 7, 2056–2065. [Google Scholar] [CrossRef]
- Jiang, J.; Ma, J.; Chen, C.; Wang, Z.; Cai, Z.; Wang, L. SuperPCA: A Superpixelwise PCA Approach for Unsupervised Feature Extraction of Hyperspectral Imagery. IEEE Trans. Geosci. Remote Sens. 2018, 56, 4581–4593. [Google Scholar] [CrossRef]
- Zhang, X.; Jiang, X.; Jiang, J.; Zhang, Y.; Liu, X.; Cai, Z. Spectral–Spatial and Superpixelwise PCA for Unsupervised Feature Extraction of Hyperspectral Imagery. IEEE Trans. Geosci. Remote Sens. 2021, 60, 5502210. [Google Scholar] [CrossRef]
- Donoho, D.L. High-dimensional data analysis: The curses and blessings of dimensionality. AMS Math Chall. Lect. 2000, 1, 32. [Google Scholar]
- Zhang, L.; Zhang, L.; Tao, D.; Huang, X. On combining multiple features for hyperspectral remote sensing image classification. IEEE Trans. Geosci. Remote Sens. 2011, 50, 879–893. [Google Scholar] [CrossRef]
- Fauvel, M.; Tarabalka, Y.; Benediktsson, J.A.; Chanussot, J.; Tilton, J.C. Advances in spectral-spatial classification of hyperspectral images. Proc. IEEE 2013, 101, 652–675. [Google Scholar] [CrossRef]
- Bao, R.; Xia, J.; Mura, M.D.; Du, P.; Chanussot, J.; Ren, J. Combining Morphological Attribute Profiles via an Ensemble Method for Hyperspectral Image Classification. IEEE Geosci. Remote Sens. Lett. 2016, 13, 359–363. [Google Scholar] [CrossRef]
- Gu, Y.; Liu, T.; Jia, X.; Benediktsson, J.A.; Chanussot, J. Nonlinear Multiple Kernel Learning With Multiple-Structure-Element Extended Morphological Profiles for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2016, 54, 3235–3247. [Google Scholar] [CrossRef]
- Xia, J.; Mura, M.D.; Chanussot, J.; Du, P.; He, X. Random Subspace Ensembles for Hyperspectral Image Classification With Extended Morphological Attribute Profiles. IEEE Trans. Geosci. Remote Sens. 2015, 53, 4768–4786. [Google Scholar] [CrossRef]
- Zabalza, J.; Ren, J.; Wang, Z.; Marshall, S.; Wang, J. Singular Spectrum Analysis for Effective Feature Extraction in Hyperspectral Imaging. IEEE Geosci. Remote Sens. Lett. 2014, 11, 1886–1890. [Google Scholar] [CrossRef]
- Qiao, T.; Ren, J.; Wang, Z.; Zabalza, J.; Sun, M.; Zhao, H.; Li, S.; Benediktsson, J.A.; Dai, Q.; Marshall, S. Effective Denoising and Classification of Hyperspectral Images Using Curvelet Transform and Singular Spectrum Analysis. IEEE Trans. Geosci. Remote Sens. 2016, 55, 119–133. [Google Scholar] [CrossRef]
- Zabalza, J.; Ren, J.; Zheng, J.; Han, J.; Zhao, H.; Li, S.; Marshall, S. Novel Two-Dimensional Singular Spectrum Analysis for Effective Feature Extraction and Data Classification in Hyperspectral Imaging. IEEE Trans. Geosci. Remote Sens. 2015, 53, 4418–4433. [Google Scholar] [CrossRef]
- Yan, Y.; Ren, J.; Liu, Q.; Zhao, H.; Sun, H.; Zabalza, J. PCA-Domain Fused Singular Spectral Analysis for Fast and Noise-Robust Spectral–Spatial Feature Mining in Hyperspectral Classification. IEEE Geosci. Remote Sens. Lett. 2021, 20, 5505405. [Google Scholar] [CrossRef]
- Fukushima, K. Neocognitron: A self-organizing neural network model for a mechanism of pattern recognition unaffected by shift in position. Biol. Cybern. 1980, 36, 193–202. [Google Scholar] [CrossRef]
- Bengio, Y.; Lamblin, P.; Popovici, D.; Larochelle, H. Greedy layer-wise training of deep networks. In Proceedings of the Advances in Neural Information Processing Systems 19 (NIPS 2006), Vancouver, BC, Canada, 4–7 December 2006. [Google Scholar]
- Hinton, G.E.; Osindero, S.; Teh, Y.-W. A Fast Learning Algorithm for Deep Belief Nets. Neural Comput. 2006, 18, 1527–1554. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Xiao, L.; Zhao, Y.Q.; Chan, J.C. Variational regularization network with attentive deep prior for hyperspectral–multispectral image fusion. IEEE Trans. Geosci. Remote Sens. 2021, 60, 5508817. [Google Scholar] [CrossRef]
- Li, X.; Ding, M.; Gu, Y.; Pižurica, A. An End-to-End Framework for Joint Denoising and Classification of Hyperspectral Images. IEEE Trans. Neural Netw. Learn. Syst. 2023, 1–15. [Google Scholar] [CrossRef]
- Xue, J.; Zhao, Y.; Huang, S.; Liao, W.; Chan, J.C.-W.; Kong, S.G. Multilayer Sparsity-Based Tensor Decomposition for Low-Rank Tensor Completion. IEEE Trans. Neural Netw. Learn. Syst. 2021, 33, 6916–6930. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Xue, J.; Luong, H.Q.; Philips, W. Multimodal Core Tensor Factorization and Its Applications to Low-Rank Tensor Completion. IEEE Trans. Multimed. 2022, 1–15. [Google Scholar] [CrossRef]
- Yang, J.; Xiao, L.; Zhao, Y.-Q.; Chan, J.C.-W. Unsupervised Deep Tensor Network for Hyperspectral–Multispectral Image Fusion. IEEE Trans. Neural Netw. Learn. Syst. 2023, 1–15. [Google Scholar] [CrossRef]
- Chen, Y.; Zhao, X.; Jia, X. Spectral–Spatial Classification of Hyperspectral Data Based on Deep Belief Network. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 8, 2381–2392. [Google Scholar] [CrossRef]
- Xu, Y.; Du, B.; Zhang, F.; Zhang, L. Hyperspectral image classification via a random patches network. ISPRS J. Photogramm. Remote Sens. 2018, 142, 344–357. [Google Scholar] [CrossRef]
- Hong, D.; Han, Z.; Yao, J.; Gao, L.; Zhang, B.; Plaza, A.; Chanussot, J. SpectralFormer: Rethinking Hyperspectral Image Classification With Transformers. IEEE Trans. Geosci. Remote Sens. 2021, 60, 5518615. [Google Scholar] [CrossRef]
- Zhou, X.; Cai, X.; Zhang, H.; Zhang, Z.; Jin, T.; Chen, H.; Deng, W. Multi-strategy competitive-cooperative co-evolutionary algorithm and its application. Inf. Sci. 2023, 635, 328–344. [Google Scholar] [CrossRef]
- Li, M.; Zhang, W.; Hu, B.; Kang, J.; Wang, Y.; Lu, S. Automatic Assessment of Depression and Anxiety through Encoding Pupil-wave from HCI in VR Scenes. ACM Trans. Multimed. Comput. Commun. Appl. 2022. [Google Scholar] [CrossRef]
- Sun, Q.; Zhang, M.; Zhou, L.; Garme, K.; Burman, M. A machine learning-based method for prediction of ship performance in ice: Part I. ice resistance. Mar. Struct. 2022, 83, 103181. [Google Scholar] [CrossRef]
- Duan, Z.; Song, P.; Yang, C.; Deng, L.; Jiang, Y.; Deng, F.; Jiang, X.; Chen, Y.; Yang, G.; Ma, Y.; et al. The impact of hyperglycaemic crisis episodes on long-term outcomes for inpatients presenting with acute organ injury: A prospective, multicentre follow-up study. Front. Endocrinol. 2022, 13, 1057089. [Google Scholar] [CrossRef]
- Xie, C.; Zhou, L.; Ding, S.; Liu, R.; Zheng, S. Experimental and numerical investigation on self-propulsion performance of polar merchant ship in brash ice channel. Ocean Eng. 2023, 269, 113424. [Google Scholar] [CrossRef]
- Chen, T.; Song, P.; He, M.; Rui, S.; Duan, X.; Ma, Y.; Armstrong, D.G.; Deng, W. Sphingosine-1-phosphate derived from PRP-Exos promotes angiogenesis in diabetic wound healing via the S1PR1/AKT/FN1 signalling pathway. Burn. Trauma 2023, 11, tkad003. [Google Scholar] [CrossRef]
- Ren, Z.; Zhen, X.; Jiang, Z.; Gao, Z.; Li, Y.; Shi, W. Underactuated control and analysis of single blade installation using a jackup installation vessel and active tugger line force control. Mar. Struct. 2023, 88, 103338. [Google Scholar] [CrossRef]
- Chen, M.; Shao, H.; Dou, H.; Li, W.; Liu, B. Data Augmentation and Intelligent Fault Diagnosis of Planetary Gearbox Using ILoFGAN Under Extremely Limited Samples. IEEE Trans. Reliab. 2022, 1–9. [Google Scholar] [CrossRef]
- Song, Y.; Zhao, G.; Zhang, B.; Chen, H.; Deng, W.; Deng, W. An enhanced distributed differential evolution algorithm for portfolio optimization problems. Eng. Appl. Artif. Intell. 2023, 121, 106004. [Google Scholar] [CrossRef]
- Yan, S.; Shao, H.; Min, Z.; Peng, J.; Cai, B.; Liu, B. FGDAE: A new machinery anomaly detection method towards complex operating conditions. Reliab. Eng. Syst. Saf. 2023, 236, 109319. [Google Scholar] [CrossRef]
- Li, M.; Zhang, J.; Song, J.; Li, Z.; Lu, S. A Clinical-Oriented Non-Severe Depression Diagnosis Method Based on Cognitive Behavior of Emotional Conflict. IEEE Trans. Comput. Soc. Syst. 2022, 10, 131–141. [Google Scholar] [CrossRef]
- Cai, J.; Ding, S.; Zhang, Q.; Liu, R.; Zeng, D.; Zhou, L. Broken ice circumferential crack estimation via image techniques. Ocean Eng. 2022, 259, 111735. [Google Scholar] [CrossRef]
- Yu, Y.; Tang, K.; Liu, Y. A Fine-Tuning Based Approach for Daily Activity Recognition between Smart Homes. Appl. Sci. 2023, 13, 5706. [Google Scholar] [CrossRef]
- Lin, J.; Shao, H.; Zhou, X.; Cai, B.; Liu, B. Generalized MAML for few-shot cross-domain fault diagnosis of bearing driven by heterogeneous signals. Expert Syst. Appl. 2023, 230, 120696. [Google Scholar] [CrossRef]
- Huang, C.; Zhou, X.; Ran, X.; Wang, J.; Chen, H.; Deng, W. Adaptive cylinder vector particle swarm optimization with differential evolution for UAV path planning. Eng. Appl. Artif. Intell. 2023, 121, 105942. [Google Scholar] [CrossRef]
- Chen, X.; Shao, H.; Xiao, Y.; Yan, S.; Cai, B.; Liu, B. Collaborative fault diagnosis of rotating machinery via dual adversarial guided unsupervised multi-domain adaptation network. Mech. Syst. Signal Process. 2023, 198, 1104270. [Google Scholar] [CrossRef]
- Caywood, M.S.; Willmore, B.; Tolhurst, D.J. Independent Components of Color Natural Scenes Resemble V1 Neurons in Their Spatial and Color Tuning. J. Neurophysiol. 2004, 91, 2859–2873. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, W.; Li, H.-C.; Tao, R.; Du, Q. Discriminative Marginalized Least-Squares Regression for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2019, 58, 3148–3161. [Google Scholar] [CrossRef]
- Sun, L.; Zhao, G.; Zheng, Y.; Wu, Z. Spectral–spatial feature tokenization transformer for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2022, 60, 5522214. [Google Scholar] [CrossRef]






| Related Information | Data Sets | ||
|---|---|---|---|
| Indian Pines | Pavia University | KSC | |
| Sensor | AVIRIS | ROSIS | AVIRIS |
| Size (pixels) | 145 × 145 | 610 × 340 | 512 × 614 |
| Bands | 200 | 103 | 176 |
| Class | 16 | 9 | 13 |
| Spatial resolution (m) | 20 | 1.3 | 18 |
| Spectral wavelength (µm) | 0.4–2.45 | 0.43–0.86 | 0.4–2.5 |
| Class Number | Class Name | Training | Test |
|---|---|---|---|
| 1 | Alfalfa | 30 | 16 |
| 2 | Corn–notill | 150 | 1278 |
| 3 | Corn–mintill | 150 | 680 |
| 4 | Corn | 100 | 137 |
| 5 | Grass–pasture | 150 | 333 |
| 6 | Grass–trees | 150 | 580 |
| 7 | Grass–pasture–mowed | 20 | 8 |
| 8 | Hay–windrowed | 150 | 328 |
| 9 | Oats | 15 | 5 |
| 10 | Soybean–notill | 150 | 822 |
| 11 | Soybean–mintill | 150 | 2305 |
| 12 | Soybean–clean | 150 | 443 |
| 13 | Wheat | 150 | 55 |
| 14 | Woods | 150 | 1115 |
| 15 | Buildings–Grass–Trees–Drivers | 50 | 336 |
| 16 | Stone–Steel–Towers | 50 | 43 |
| Total | 1765 | 8484 |
| Class Number | Class Name | Training | Test |
|---|---|---|---|
| 1 | Asphalt | 548 | 6083 |
| 2 | Meadows | 540 | 18,109 |
| 3 | Gravel | 392 | 1707 |
| 4 | Trees | 542 | 2522 |
| 5 | Metal sheets | 256 | 1089 |
| 6 | Bare soil | 532 | 4497 |
| 7 | Bitumen | 375 | 955 |
| 8 | Bricks | 514 | 3168 |
| 9 | Shadows | 231 | 716 |
| Total | 3930 | 38,846 |
| Class Number | Class Name | Training | Test |
|---|---|---|---|
| 1 | Scrub | 33 | 728 |
| 2 | Willow swamp | 23 | 220 |
| 3 | CP hammock | 24 | 232 |
| 4 | CP/Oak | 24 | 228 |
| 5 | Slash pine | 15 | 146 |
| 6 | Oak/Broadleaf | 22 | 207 |
| 7 | Hardwood swamp | 9 | 96 |
| 8 | Graminoid marsh | 38 | 393 |
| 9 | Spartina marsh | 51 | 469 |
| 10 | Catiail marsh | 39 | 365 |
| 11 | Salt marsh | 41 | 378 |
| 12 | Mud flats | 49 | 454 |
| 13 | Water | 91 | 836 |
| Total | 459 | 4752 |
| Class | PCA | SuperPCA | S3-PCA | PCA -2D-SSA | SuperPCA -2D-SSA | RPNet-5 | S3-PCA -RPNet | DMLSR | LeNet | SSFTT | Proposed |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 87.50 | 87.50 | 100 | 93.75 | 100 | 100 | 100 | 82.14 | 100 | 95.12 | 87.50 |
| 2 | 75.04 | 89.36 | 92.49 | 87.87 | 94.91 | 96.48 | 97.97 | 85.98 | 92.78 | 97.57 | 97.10 |
| 3 | 79.85 | 93.24 | 96.18 | 93.53 | 88.09 | 98.38 | 97.94 | 82.93 | 97.98 | 96.94 | 98.53 |
| 4 | 78.10 | 92.70 | 97.81 | 94.89 | 97.08 | 97.82 | 100 | 77.62 | 99.48 | 97.10 | 100 |
| 5 | 94.59 | 98.80 | 99.70 | 99.40 | 97.30 | 99.40 | 98.80 | 93.77 | 98.31 | 99.04 | 99.10 |
| 6 | 97.07 | 99.66 | 100 | 98.97 | 100 | 99.66 | 99.83 | 99.32 | 98.87 | 99.20 | 99.83 |
| 7 | 100 | 100 | 87.50 | 87.50 | 87.50 | 100 | 87.50 | 93.75 | 100 | 95.65 | 100 |
| 8 | 99.09 | 100 | 100 | 100 | 99.70 | 100 | 100 | 99.66 | 99.74 | 99.51 | 100 |
| 9 | 100 | 100 | 100 | 100 | 80.00 | 100 | 100 | 83.33 | 100 | 92.86 | 100 |
| 10 | 80.41 | 94.89 | 90.63 | 93.55 | 93.43 | 93.43 | 98.66 | 87.84 | 94.35 | 97.56 | 99.51 |
| 11 | 70.07 | 91.02 | 91.71 | 84.21 | 86.20 | 95.70 | 97.74 | 88.53 | 93.45 | 96.88 | 98.05 |
| 12 | 86.23 | 95.49 | 96.39 | 94.36 | 95.26 | 99.32 | 99.10 | 91.83 | 98.26 | 98.97 | 99.32 |
| 13 | 98.18 | 100 | 100 | 100 | 100 | 100 | 100 | 99.19 | 100 | 100 | 100 |
| 14 | 91.93 | 99.55 | 98.83 | 95.87 | 97.76 | 99.91 | 99.91 | 94.33 | 99.05 | 99.91 | 99.91 |
| 15 | 58.63 | 94.35 | 94.64 | 90.77 | 86.31 | 96.13 | 91.67 | 72.41 | 98.13 | 98.78 | 97.32 |
| 16 | 97.69 | 97.67 | 97.67 | 100 | 90.70 | 100 | 95.35 | 94.55 | 99.00 | 87.50 | 97.67 |
| OA (%) | 80.33 | 93.58 | 94.78 | 91.34 | 92.57 | 97.21 | 98.30 | 89.49 | 96.67 | 97.94 | 98.87 |
| Kappa (%) | 77.37 | 92.57 | 93.94 | 89.99 | 91.41 | 96.87 | 98.02 | 88.02 | 95.84 | 97.65 | 98.05 |
| Class | PCA | SuperPCA | S3-PCA | PCA -2D-SSA | SuperPCA -2D-SSA | RPNet-5 | S3-PCA -RPNet | DMLSR | LeNet | SSFTT | Proposed |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 90.79 | 93.69 | 97.32 | 93.84 | 98.75 | 98.29 | 98.83 | 86.51 | 98.32 | 99.78 | 99.64 |
| 2 | 92.94 | 97.58 | 97.97 | 96.36 | 98.35 | 99.37 | 99.32 | 88.31 | 97.05 | 99.99 | 99.83 |
| 3 | 84.07 | 94.49 | 95.84 | 94.32 | 99.82 | 99.36 | 99.18 | 78.53 | 98.00 | 99.90 | 99.82 |
| 4 | 98.06 | 98.02 | 97.22 | 98.89 | 99.33 | 99.41 | 98.69 | 96.76 | 99.00 | 98.77 | 99.37 |
| 5 | 99.54 | 99.82 | 99.45 | 99.45 | 99.54 | 100 | 99.82 | 100 | 99.73 | 100 | 99.63 |
| 6 | 94.62 | 97.80 | 97.98 | 96.22 | 99.69 | 99.91 | 99.29 | 85.65 | 97.51 | 99.87 | 99.91 |
| 7 | 93.30 | 95.92 | 98.53 | 96.54 | 99.90 | 99.79 | 99.16 | 92.61 | 99.31 | 99.84 | 99.79 |
| 8 | 87.31 | 95.04 | 96.62 | 93.75 | 99.34 | 99.34 | 99.34 | 80.54 | 98.05 | 98.02 | 99.21 |
| 9 | 100 | 99.86 | 99.86 | 100 | 100 | 100 | 99.86 | 100 | 99.88 | 98.43 | 99.86 |
| OA (%) | 92.60 | 96.74 | 97.71 | 95.97 | 98.88 | 99.27 | 99.21 | 87.54 | 98.34 | 99.64 | 99.76 |
| Kappa (%) | 90.02 | 95.66 | 96.87 | 94.52 | 98.77 | 99.18 | 98.93 | 80.15 | 97.52 | 99.52 | 99.68 |
| Class | PCA | SuperPCA | S3-PCA | PCA -2D-SSA | SuperPCA -2D-SSA | RPNet-5 | S3-PCA -RPNet | DMLSR | LeNet | SSFTT | Proposed |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 90.80 | 91.07 | 90.38 | 94.92 | 92.72 | 97.12 | 96.84 | 89.35 | 91.74 | 53.04 | 99.73 |
| 2 | 82.27 | 90.45 | 98.18 | 97.73 | 98.64 | 98.64 | 94.55 | 92.89 | 90.09 | 60.66 | 100 |
| 3 | 88.79 | 95.26 | 93.10 | 94.40 | 97.41 | 99.14 | 95.26 | 93.75 | 86.34 | 33.33 | 97.84 |
| 4 | 67.98 | 78.07 | 85.53 | 90.35 | 92.54 | 90.79 | 95.61 | 75.98 | 76.80 | 39.33 | 92.98 |
| 5 | 63.70 | 58.90 | 80.82 | 84.25 | 90.41 | 98.63 | 93.15 | 79.39 | 92.40 | 93.94 | 97.26 |
| 6 | 69.57 | 57.00 | 87.44 | 61.35 | 90.82 | 99.03 | 87.44 | 77.30 | 90.34 | 0 | 99.52 |
| 7 | 92.71 | 100 | 96.88 | 80.21 | 100 | 100 | 92.71 | 78.16 | 90.94 | 50.00 | 96.88 |
| 8 | 90.59 | 96.95 | 99.24 | 98.47 | 92.11 | 98.22 | 97.96 | 93.24 | 94.35 | 57.54 | 98.73 |
| 9 | 97.87 | 97.01 | 97.44 | 98.72 | 98.29 | 100 | 100 | 99.28 | 97.85 | 85.92 | 100 |
| 10 | 89.32 | 86.03 | 96.16 | 98.08 | 97.81 | 98.63 | 96.71 | 98.77 | 99.48 | 66.75 | 98.90 |
| 11 | 97.35 | 93.92 | 97.35 | 98.41 | 94.18 | 98.94 | 99.74 | 99.11 | 99.89 | 94.34 | 99.47 |
| 12 | 98.46 | 95.37 | 94.05 | 99.78 | 96.92 | 100 | 99.56 | 90.86 | 98.55 | 79.34 | 100 |
| 13 | 99.28 | 99.88 | 99.88 | 99.88 | 99.88 | 99.28 | 100 | 99.60 | 100 | 93.72 | 99.76 |
| OA (%) | 90.72 | 91.98 | 94.80 | 95.24 | 95.88 | 98.46 | 97.43 | 92.73 | 95.29 | 70.51 | 99.11 |
| Kappa (%) | 89.78 | 91.07 | 94.22 | 94.70 | 95.41 | 98.29 | 96.25 | 91.90 | 94.97 | 53.91 | 99.02 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, H.; Wang, T.; Chen, T.; Deng, W. Hyperspectral Image Classification Based on Fusing S3-PCA, 2D-SSA and Random Patch Network. Remote Sens. 2023, 15, 3402. https://doi.org/10.3390/rs15133402
Chen H, Wang T, Chen T, Deng W. Hyperspectral Image Classification Based on Fusing S3-PCA, 2D-SSA and Random Patch Network. Remote Sensing. 2023; 15(13):3402. https://doi.org/10.3390/rs15133402
Chicago/Turabian StyleChen, Huayue, Tingting Wang, Tao Chen, and Wu Deng. 2023. "Hyperspectral Image Classification Based on Fusing S3-PCA, 2D-SSA and Random Patch Network" Remote Sensing 15, no. 13: 3402. https://doi.org/10.3390/rs15133402
APA StyleChen, H., Wang, T., Chen, T., & Deng, W. (2023). Hyperspectral Image Classification Based on Fusing S3-PCA, 2D-SSA and Random Patch Network. Remote Sensing, 15(13), 3402. https://doi.org/10.3390/rs15133402