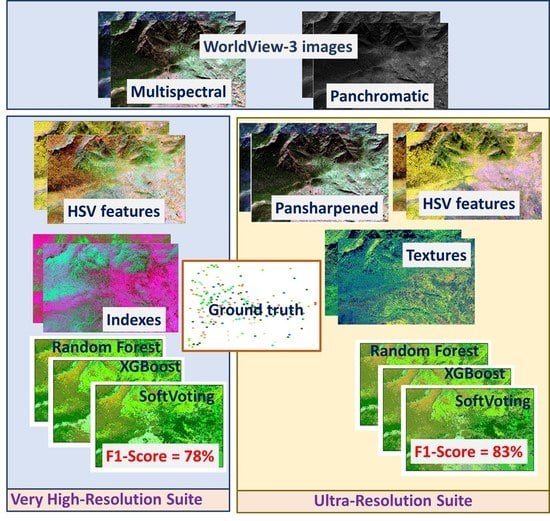
An Ultra-Resolution Features Extraction Suite for Community-Level Vegetation Differentiation and Mapping at a Sub-Meter Resolution
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area
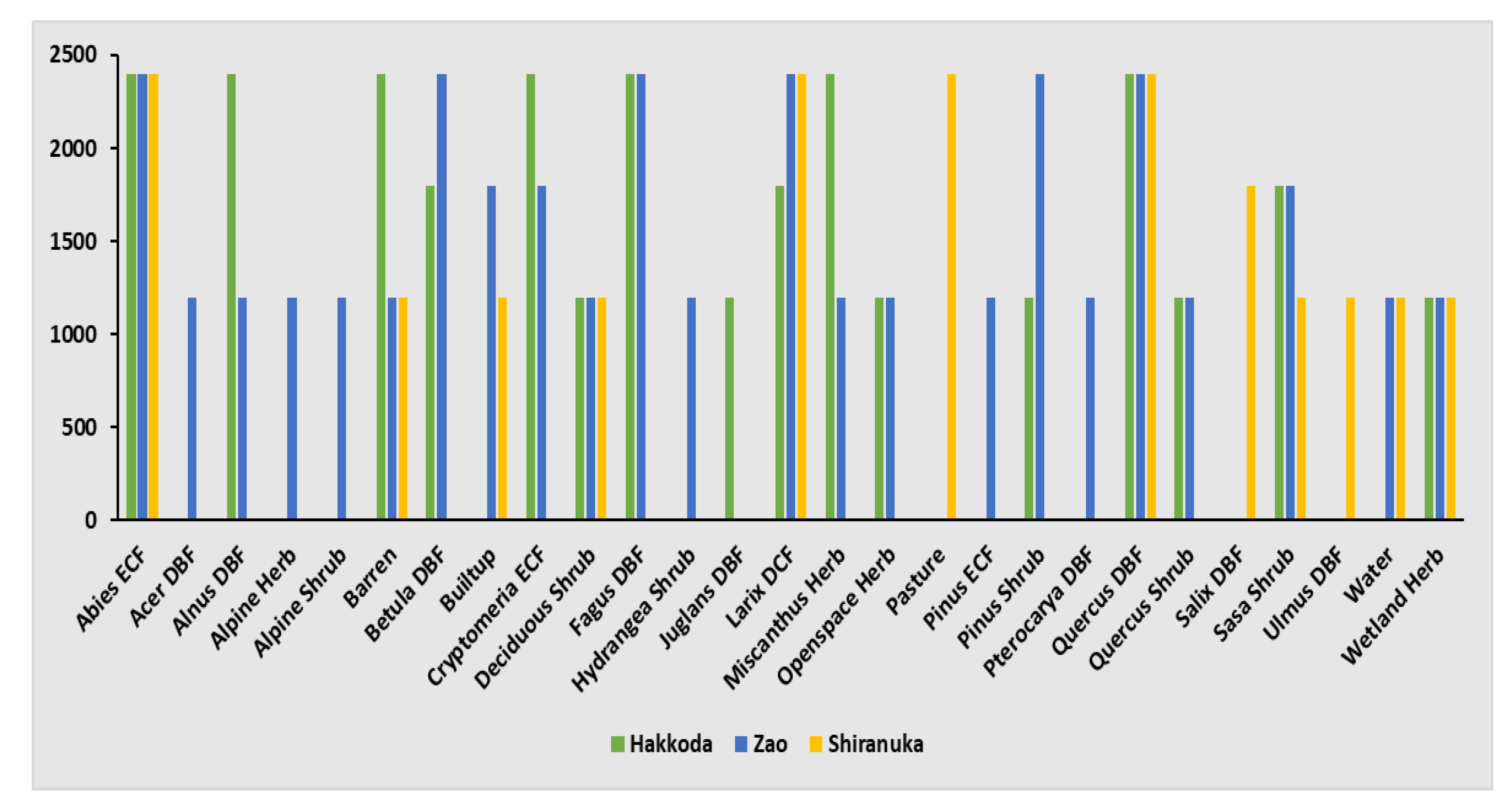
2.2. Collection of Ground Truth Data
2.3. Processing of WorldView-3 Images
2.4. Features Extraction
2.5. Machine Learning and Mapping
3. Results
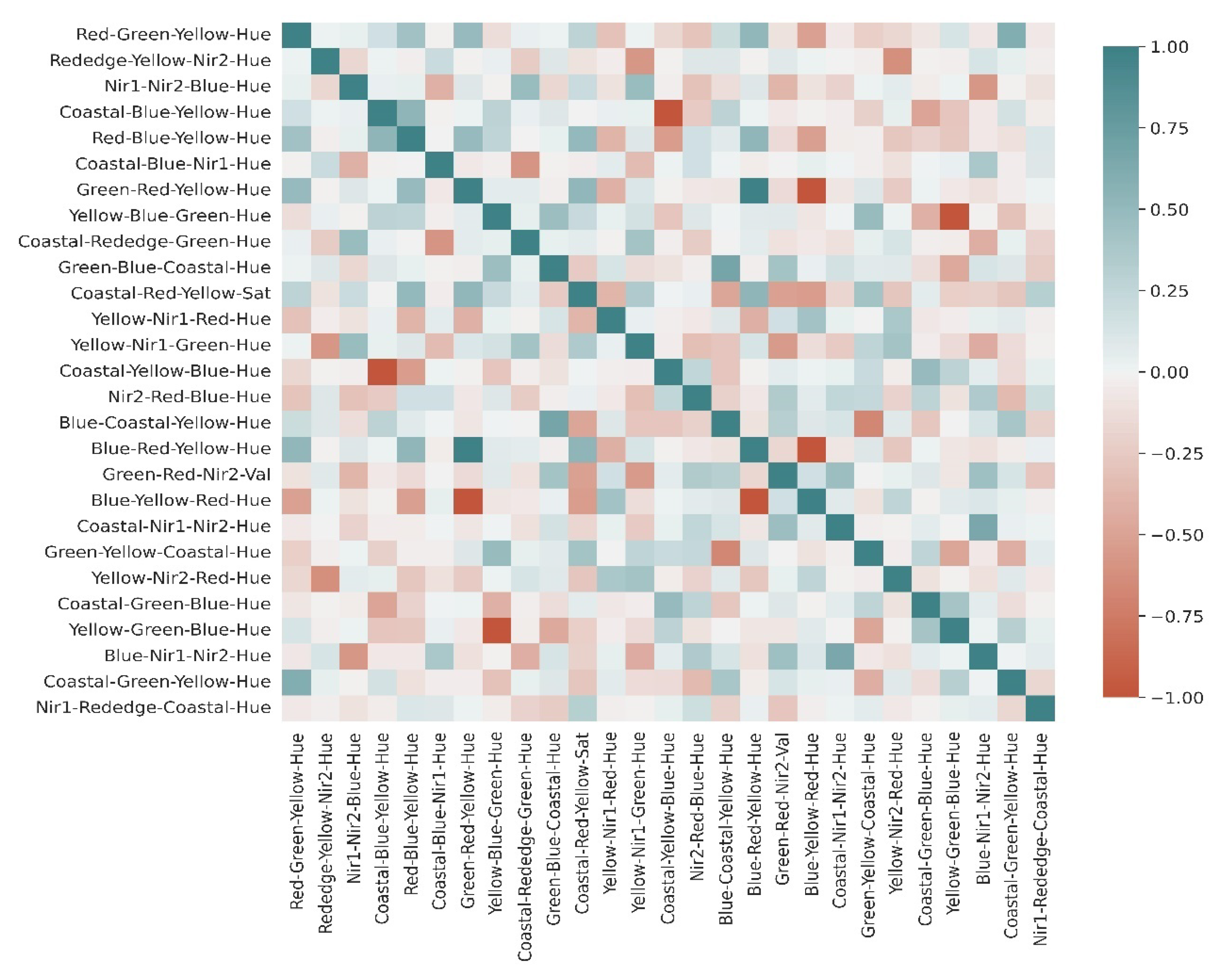
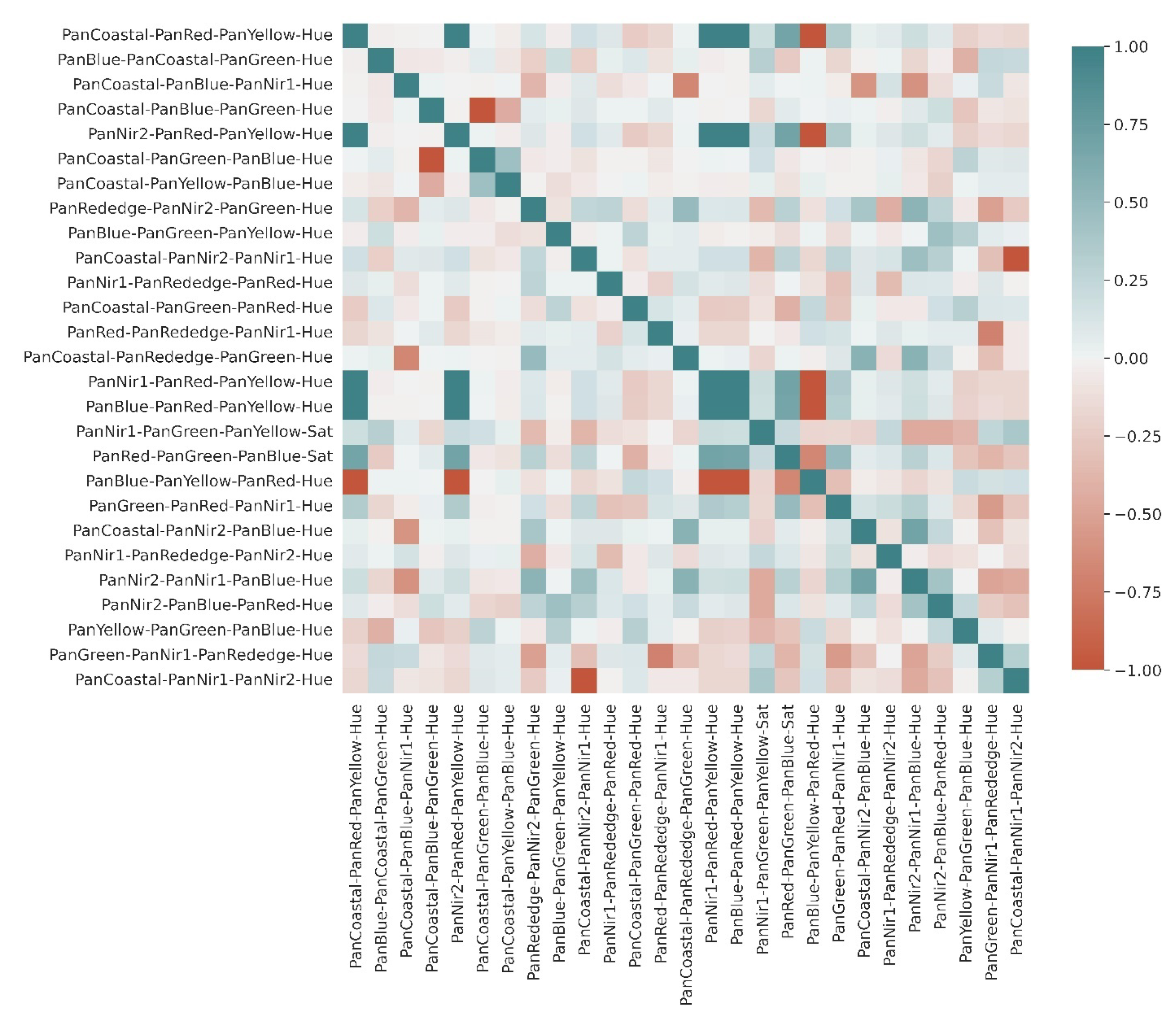
3.1. Extraction of Least-Correlated Features
3.2. Pan-Sharpened versus Multi-Spectral Images
3.3. Effect of Classifiers in the Case of Single-Date Autumn Images
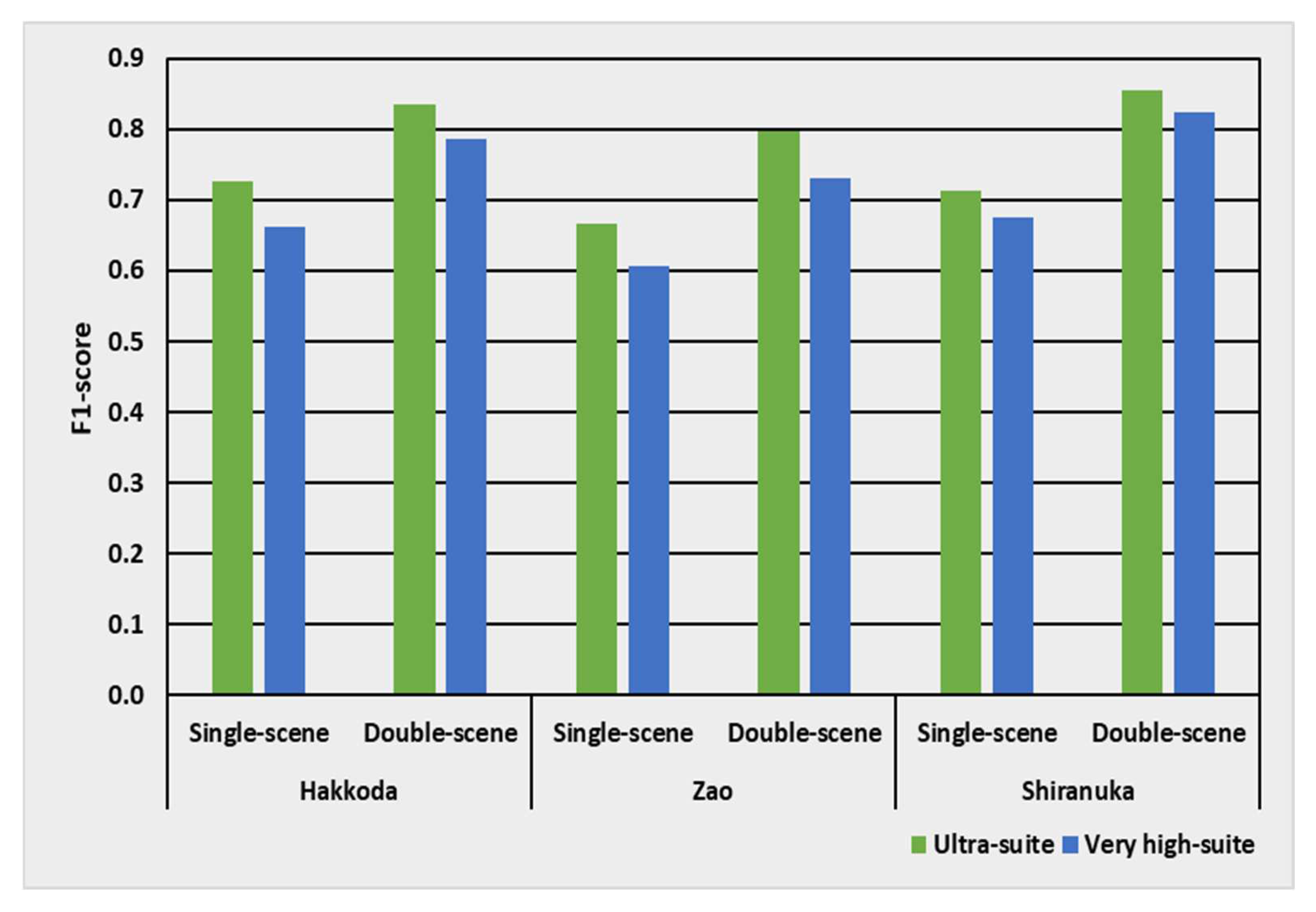
3.4. Effect of Classifiers in the Case of Bi-Seasonal Images
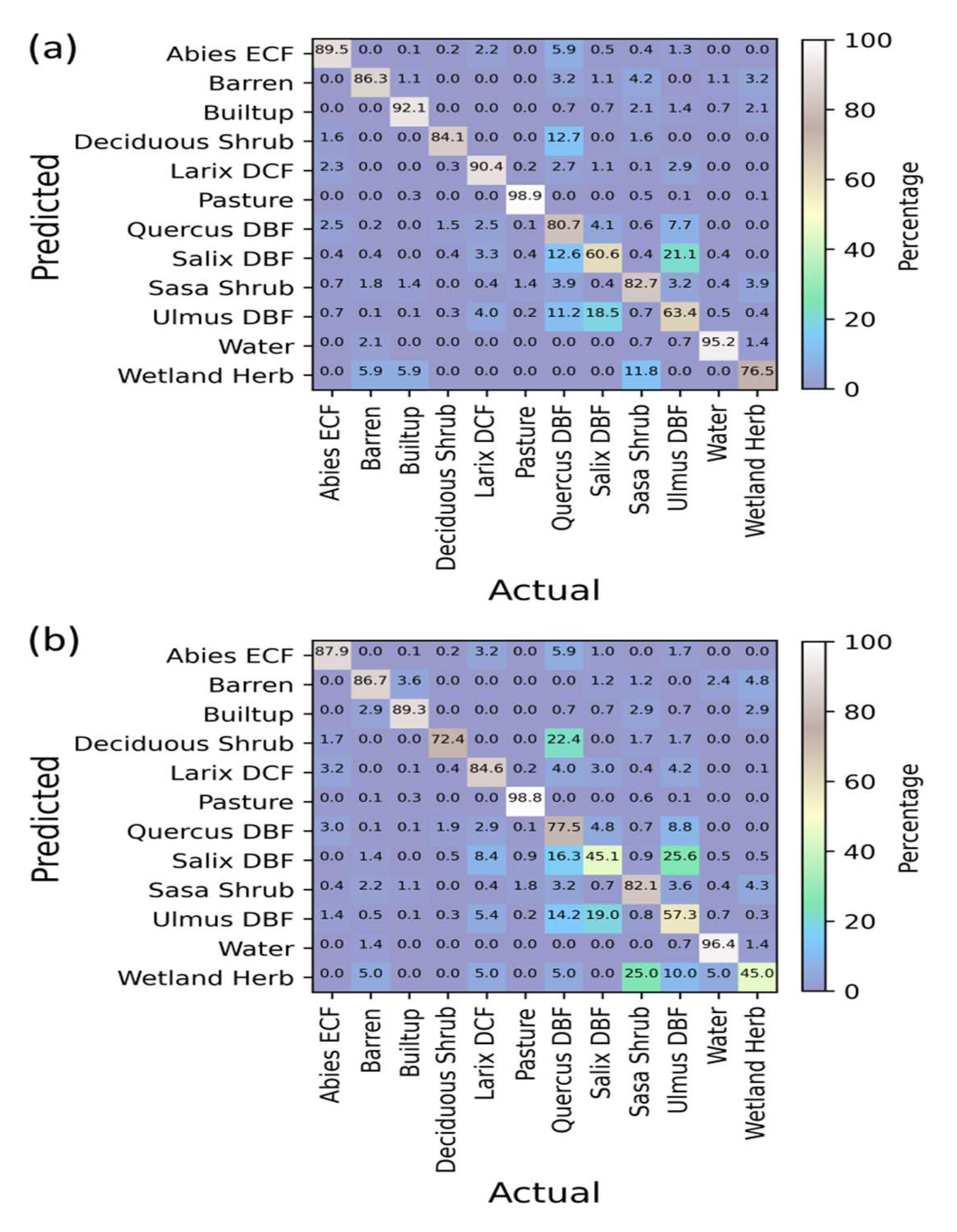
3.5. Confusion Matrices Using Bi-Seasonal Images
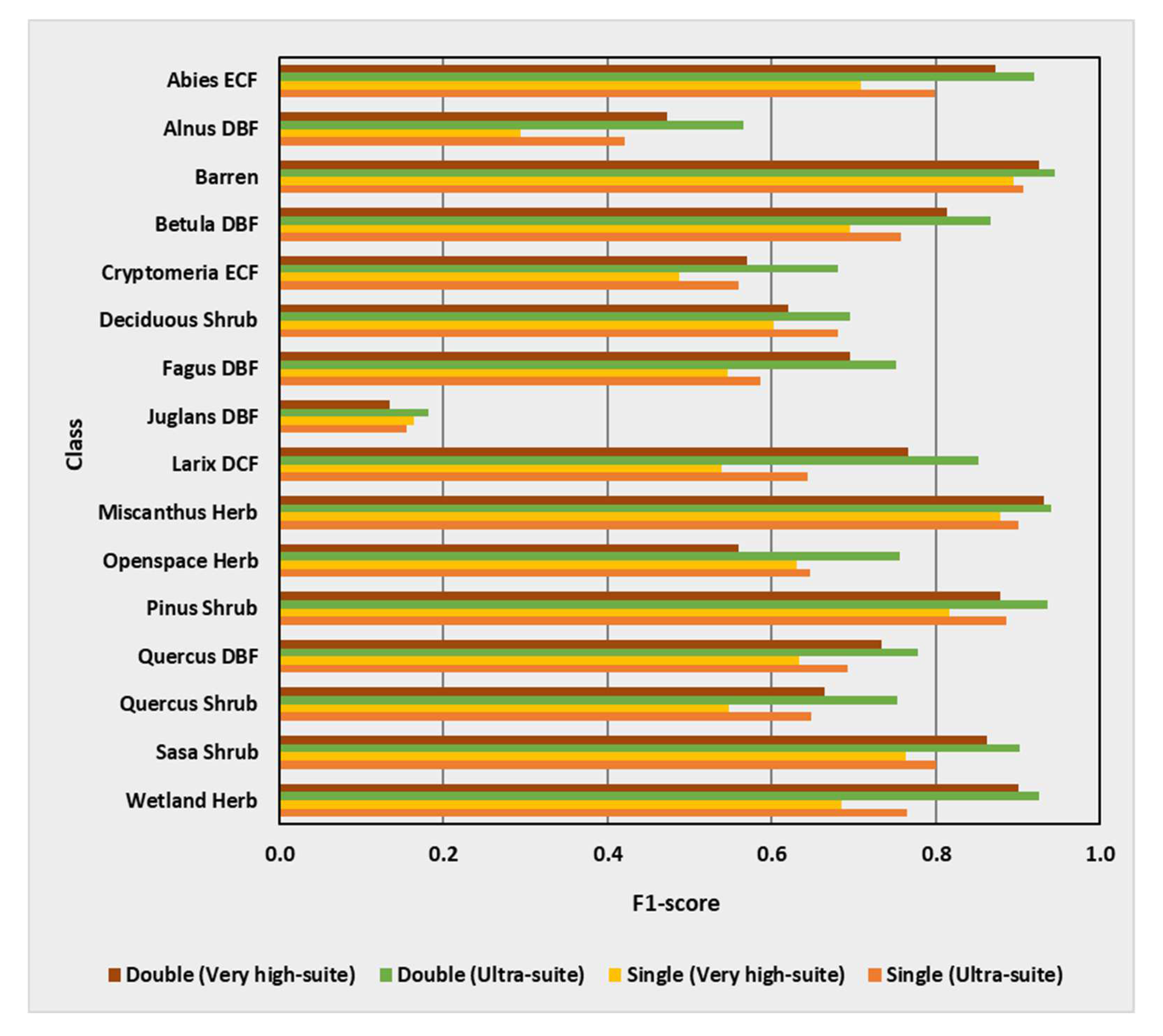
3.6. Class-Wise Changes
3.7. Performance Summary
3.8. Ultra-Resolution Maps
4. Discussion
5. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Lauer, D.T.; Morain, S.A.; Salomonson, V.V. The Landsat Program: Its Origins, Evolution, and Impacts. Photogramm. Eng. Remote Sens. 1997, 63, 831–838. [Google Scholar]
- Woodcock, C.E.; Allen, R.; Anderson, M.; Belward, A.; Bindschadler, R.; Cohen, W.; Gao, F.; Goward, S.N.; Helder, D.; Helmer, E.; et al. Free Access to Landsat Imagery. Science 2008, 320, 1011. [Google Scholar] [CrossRef] [PubMed]
- Wulder, M.A.; Masek, J.G.; Cohen, W.B.; Loveland, T.R.; Woodcock, C.E. Opening the Archive: How Free Data Has Enabled the Science and Monitoring Promise of Landsat. Remote Sens. Environ. 2012, 122, 2–10. [Google Scholar] [CrossRef]
- Irons, J.R.; Dwyer, J.L.; Barsi, J.A. The next Landsat Satellite: The Landsat Data Continuity Mission. Remote Sens. Environ. 2012, 122, 11–21. [Google Scholar] [CrossRef]
- Zhu, Z.; Wulder, M.A.; Roy, D.P.; Woodcock, C.E.; Hansen, M.C.; Radeloff, V.C.; Healey, S.P.; Schaaf, C.; Hostert, P.; Strobl, P.; et al. Benefits of the Free and Open Landsat Data Policy. Remote Sens. Environ. 2019, 224, 382–385. [Google Scholar] [CrossRef]
- Drusch, M.; Del Bello, U.; Carlier, S.; Colin, O.; Fernandez, V.; Gascon, F.; Hoersch, B.; Isola, C.; Laberinti, P.; Martimort, P.; et al. Sentinel-2: ESA’s Optical High-Resolution Mission for GMES Operational Services. Remote Sens. Environ. 2012, 120, 25–36. [Google Scholar] [CrossRef]
- Li, J.; Roy, D. A Global Analysis of Sentinel-2A, Sentinel-2B and Landsat-8 Data Revisit Intervals and Implications for Terrestrial Monitoring. Remote Sens. 2017, 9, 902. [Google Scholar] [CrossRef]
- Anderson, N.T.; Marchisio, G.B. WorldView-2 and the Evolution of the DigitalGlobe Remote Sensing Satellite Constellation: Introductory Paper for the Special Session on WorldView-2. In Proceedings of the Algorithms and Technologies for Multispectral, Hyperspectral, and Ultraspectral Imagery XVIII, Baltimore, MD, USA, 23–27 April 2012; Shen, S.S., Lewis, P.E., Eds.; SPIE: Bellingham, WA, USA, 2012; pp. 166–180. [Google Scholar]
- Ustin, S.L.; Middleton, E.M. Current and Near-Term Advances in Earth Observation for Ecological Applications. Ecol. Process 2021, 10, 1. [Google Scholar] [CrossRef]
- Qin, R.; Liu, T. A Review of Landcover Classification with Very-High Resolution Remotely Sensed Optical Images—Analysis Unit, Model Scalability and Transferability. Remote Sens. 2022, 14, 646. [Google Scholar] [CrossRef]
- Ponzoni, F.J.; Galvão, L.S.; Epiphanio, J.C.N. Spatial Resolution Influence on the Identification of Land Cover Classes in the Amazon Environment. An. Acad. Bras. Ciênc. 2002, 74, 717–725. [Google Scholar] [CrossRef][Green Version]
- Achard, F.; Eva, H.; Mayaux, P. Tropical Forest Mapping from Coarse Spatial Resolution Satellite Data: Production and Accuracy Assessment Issues. Int. J. Remote Sens. 2001, 22, 2741–2762. [Google Scholar] [CrossRef]
- Xu, K.; Zhang, Z.; Yu, W.; Zhao, P.; Yue, J.; Deng, Y.; Geng, J. How Spatial Resolution Affects Forest Phenology and Tree-Species Classification Based on Satellite and Up-Scaled Time-Series Images. Remote Sens. 2021, 13, 2716. [Google Scholar] [CrossRef]
- Liu, M.; Popescu, S.; Malambo, L. Effects of Spatial Resolution on Burned Forest Classification with ICESat-2 Photon Counting Data. Front. Remote Sens. 2021, 2, 666251. [Google Scholar] [CrossRef]
- Wulder, M.A.; Hall, R.J.; Coops, N.C.; Franklin, S.E. High Spatial Resolution Remotely Sensed Data for Ecosystem Characterization. BioScience 2004, 54, 511. [Google Scholar] [CrossRef]
- Wickham, J.; Riitters, K.H. Influence of High-Resolution Data on the Assessment of Forest Fragmentation. Landsc. Ecol. 2019, 34, 2169–2182. [Google Scholar] [CrossRef]
- Durgun, Y.Ö.; Gobin, A.; Duveiller, G.; Tychon, B. A Study on Trade-Offs between Spatial Resolution and Temporal Sampling Density for Wheat Yield Estimation Using Both Thermal and Calendar Time. Int. J. Appl. Earth Obs. Geoinf. 2020, 86, 101988. [Google Scholar] [CrossRef]
- Monasterio, M.; Sarmiento, G. Phenological Strategies of Plant Species in the Tropical Savanna and the Semi-Deciduous Forest of the Venezuelan Llanos. J. Biogeogr. 1976, 3, 325. [Google Scholar] [CrossRef]
- de L. Brooke, M.; Jones, P.J.; Vickery, J.A.; Waldren, S. Seasonal Patterns of Leaf Growth and Loss, Flowering and Fruiting on a Subtropical Central Pacific Island. Biotropica 1996, 28, 164. [Google Scholar] [CrossRef]
- Xue, J.; Su, B. Significant Remote Sensing Vegetation Indices: A Review of Developments and Applications. J. Sens. 2017, 2017, 1–17. [Google Scholar] [CrossRef]
- Lee, J.-H.; Chang, B.-H.; Kim, S.-D. Comparison of Colour Transformations for Image Segmentation. Electron. Lett. 1994, 30, 1660–1661. [Google Scholar] [CrossRef]
- Reed, T.R.; Dubuf, J.M.H. A Review of Recent Texture Segmentation and Feature Extraction Techniques. CVGIP Image Underst. 1993, 57, 359–372. [Google Scholar] [CrossRef]
- Humeau-Heurtier, A. Texture Feature Extraction Methods: A Survey. IEEE Access 2019, 7, 8975–9000. [Google Scholar] [CrossRef]
- Smith, A.R. Color Gamut Transform Pairs. In Proceedings of the 5th Annual Conference on Computer Graphics and Interactive techniques—SIGGRAPH ’78, Atlanta, GA, USA, 23–25 August 1978; ACM Press: New York, NY, USA, 1978; pp. 12–19. [Google Scholar]
- Hamuda, E.; Mc Ginley, B.; Glavin, M.; Jones, E. Automatic Crop Detection under Field Conditions Using the HSV Colour Space and Morphological Operations. Comput. Electron. Agric. 2017, 133, 97–107. [Google Scholar] [CrossRef]
- Chang, C.-L.; Lin, K.-M. Smart Agricultural Machine with a Computer Vision-Based Weeding and Variable-Rate Irrigation Scheme. Robotics 2018, 7, 38. [Google Scholar] [CrossRef]
- Wang, D.; Fang, S.; Yang, Z.; Wang, L.; Tang, W.; Li, Y.; Tong, C. A Regional Mapping Method for Oilseed Rape Based on HSV Transformation and Spectral Features. Int. J. Geo-Inf. 2018, 7, 224. [Google Scholar] [CrossRef]
- Iovan, C.; Boldo, D.; Cord, M. Detection, Characterization, and Modeling Vegetation in Urban Areas from High-Resolution Aerial Imagery. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2008, 1, 206–213. [Google Scholar] [CrossRef]
- Liu, H.; An, H. Urban Greening Tree Species Classification Based on HSV Colour Space of WorldView-2. J. Indian Soc. Remote Sens. 2019, 47, 1959–1967. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef]
- Mishra, V.N.; Prasad, R.; Rai, P.K.; Vishwakarma, A.K.; Arora, A. Performance Evaluation of Textural Features in Improving Land Use/Land Cover Classification Accuracy of Heterogeneous Landscape Using Multi-Sensor Remote Sensing Data. Earth Sci. Inform. 2019, 12, 71–86. [Google Scholar] [CrossRef]
- Farwell, L.S.; Gudex-Cross, D.; Anise, I.E.; Bosch, M.J.; Olah, A.M.; Radeloff, V.C.; Razenkova, E.; Rogova, N.; Silveira, E.M.O.; Smith, M.M.; et al. Satellite Image Texture Captures Vegetation Heterogeneity and Explains Patterns of Bird Richness. Remote Sens. Environ. 2021, 253, 112175. [Google Scholar] [CrossRef]
- Guo, W.; Rees, G.; Hofgaard, A. Delineation of the Forest-Tundra Ecotone Using Texture-Based Classification of Satellite Imagery. Int. J. Remote Sens. 2020, 41, 6384–6408. [Google Scholar] [CrossRef]
- Ireland, A.W.; Smith, F.G.F.; Jaffe, B.D.; Palandro, D.A.; Mercer, S.M.; Liu, L.; Renton, J. Field Experiment Demonstrates the Potential Utility of Satellite-Derived Reflectance Indices for Monitoring Regeneration of Boreal Forest Communities. Trees For. People 2021, 6, 100145. [Google Scholar] [CrossRef]
- Wang, M.; Fei, X.; Zhang, Y.; Chen, Z.; Wang, X.; Tsou, J.Y.; Liu, D.; Lu, X. Assessing Texture Features to Classify Coastal Wetland Vegetation from High Spatial Resolution Imagery Using Completed Local Binary Patterns (CLBP). Remote Sens. 2018, 10, 778. [Google Scholar] [CrossRef]
- Zhang, X.; Cui, J.; Wang, W.; Lin, C. A Study for Texture Feature Extraction of High-Resolution Satellite Images Based on a Direction Measure and Gray Level Co-Occurrence Matrix Fusion Algorithm. Sensors 2017, 17, 1474. [Google Scholar] [CrossRef]
- Ferreira, M.P.; Wagner, F.H.; Aragão, L.E.O.C.; Shimabukuro, Y.E.; de Souza Filho, C.R. Tree Species Classification in Tropical Forests Using Visible to Shortwave Infrared WorldView-3 Images and Texture Analysis. ISPRS J. Photogramm. Remote Sens. 2019, 149, 119–131. [Google Scholar] [CrossRef]
- Tamondong, A.M.; Blanco, A.C.; Fortes, M.D.; Nadaoka, K. Mapping of Seagrass and Other Benthic Habitats in Bolinao, Pangasinan Using Worldview-2 Satellite Image. In Proceedings of the 2013 IEEE International Geoscience and Remote Sensing Symposium—IGARSS, Melbourne, Australia, 21–26 July 2013; IEEE: Piscataway, NJ, USA, 2013; pp. 1579–1582. [Google Scholar]
- Carle, M.V.; Wang, L.; Sasser, C.E. Mapping Freshwater Marsh Species Distributions Using WorldView-2 High-Resolution Multispectral Satellite Imagery. Int. J. Remote Sens. 2014, 35, 4698–4716. [Google Scholar] [CrossRef]
- Collin, A.; Lambert, N.; Etienne, S. Satellite-Based Salt Marsh Elevation, Vegetation Height, and Species Composition Mapping Using the Superspectral WorldView-3 Imagery. Int. J. Remote Sens. 2018, 39, 5619–5637. [Google Scholar] [CrossRef]
- Immitzer, M.; Atzberger, C.; Koukal, T. Tree Species Classification with Random Forest Using Very High Spatial Resolution 8-Band WorldView-2 Satellite Data. Remote Sens. 2012, 4, 2661–2693. [Google Scholar] [CrossRef]
- Pu, R.; Landry, S. A Comparative Analysis of High Spatial Resolution IKONOS and WorldView-2 Imagery for Mapping Urban Tree Species. Remote Sens. Environ. 2012, 124, 516–533. [Google Scholar] [CrossRef]
- Waser, L.; Küchler, M.; Jütte, K.; Stampfer, T. Evaluating the Potential of WorldView-2 Data to Classify Tree Species and Different Levels of Ash Mortality. Remote Sens. 2014, 6, 4515–4545. [Google Scholar] [CrossRef]
- Jombo, S.; Adam, E.; Odindi, J. Classification of Tree Species in a Heterogeneous Urban Environment Using Object-Based Ensemble Analysis and World View-2 Satellite Imagery. Appl. Geomat. 2021, 13, 373–387. [Google Scholar] [CrossRef]
- Ngubane, Z.; Odindi, J.; Mutanga, O.; Slotow, R. Assessment of the Contribution of WorldView-2 Strategically Positioned Bands in Bracken Fern (Pteridium aquilinum (L.) Kuhn) Mapping. S. Afr. J. Geomat. 2014, 3, 210. [Google Scholar] [CrossRef]
- Adam, E.; Mureriwa, N.; Newete, S. Mapping Prosopis Glandulosa (Mesquite) in the Semi-Arid Environment of South Africa Using High-Resolution WorldView-2 Imagery and Machine Learning Classifiers. J. Arid Environ. 2017, 145, 43–51. [Google Scholar] [CrossRef]
- Mureriwa, N.F.; Adam, E.; Adelabu, S. Cost Effective Approach for Mapping Prosopis Invasion in Arid South Africa Using SPOT-6 Imagery and Two Machine Learning Classifiers. In Proceedings of the IGARSS 2019—2019 IEEE International Geoscience and Remote Sensing Symposium, Yokohama, Japan, 28 July–2 August 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 3724–3727. [Google Scholar]
- Schulze-Brüninghoff, D.; Wachendorf, M.; Astor, T. Potentials and Limitations of WorldView-3 Data for the Detection of Invasive Lupinus Polyphyllus Lindl. in Semi-Natural Grasslands. Remote Sens. 2021, 13, 4333. [Google Scholar] [CrossRef]
- Novack, T.; Esch, T.; Kux, H.; Stilla, U. Machine Learning Comparison between WorldView-2 and QuickBird-2-Simulated Imagery Regarding Object-Based Urban Land Cover Classification. Remote Sens. 2011, 3, 2263–2282. [Google Scholar] [CrossRef]
- Heumann, B.W. An Object-Based Classification of Mangroves Using a Hybrid Decision Tree—Support Vector Machine Approach. Remote Sens. 2011, 3, 2440–2460. [Google Scholar] [CrossRef]
- Kavzoglu, T.; Colkesen, I.; Yomralioglu, T. Object-Based Classification with Rotation Forest Ensemble Learning Algorithm Using Very-High-Resolution WorldView-2 Image. Remote Sens. Lett. 2015, 6, 834–843. [Google Scholar] [CrossRef]
- Chuang, Y.-C.; Shiu, Y.-S. A Comparative Analysis of Machine Learning with WorldView-2 Pan-Sharpened Imagery for Tea Crop Mapping. Sensors 2016, 16, 594. [Google Scholar] [CrossRef]
- Jackson, C.M.; Adam, E. Machine Learning Classification of Endangered Tree Species in a Tropical Submontane Forest Using WorldView-2 Multispectral Satellite Imagery and Imbalanced Dataset. Remote Sens. 2021, 13, 4970. [Google Scholar] [CrossRef]
- Sharma, R.C.; Hirayama, H.; Yasuda, M.; Asai, M.; Hara, K. Classification and Mapping of Plant Communities Using Multi-Temporal and Multi-Spectral Satellite Images. J. Geogr. Geol. 2022, 14, 43. [Google Scholar] [CrossRef]
- Sharma, R.C. Genus-Physiognomy-Ecosystem (GPE) System for Satellite-Based Classification of Plant Communities. Ecologies 2021, 2, 203–213. [Google Scholar] [CrossRef]
- Farr, T.G.; Rosen, P.A.; Caro, E.; Crippen, R.; Duren, R.; Hensley, S.; Kobrick, M.; Paller, M.; Rodriguez, E.; Roth, L.; et al. The Shuttle Radar Topography Mission. Rev. Geophys. 2007, 45, RG2004. [Google Scholar] [CrossRef]
- Aguilar, M.A.; del Saldaña, M.M.; Aguilar, F.J. Assessing Geometric Accuracy of the Orthorectification Process from GeoEye-1 and WorldView-2 Panchromatic Images. Int. J. Appl. Earth Obs. Geoinf. 2013, 21, 427–435. [Google Scholar] [CrossRef]
- Belfiore, O.; Parente, C. Comparison of Different Algorithms to Orthorectify WorldView-2 Satellite Imagery. Algorithms 2016, 9, 67. [Google Scholar] [CrossRef]
- Soenen, S.A.; Peddle, D.R.; Coburn, C.A. SCS+C: A Modified Sun-Canopy-Sensor Topographic Correction in Forested Terrain. IEEE Trans. Geosci. Remote Sens. 2005, 43, 2148–2159. [Google Scholar] [CrossRef]
- Richter, R.; Kellenberger, T.; Kaufmann, H. Comparison of Topographic Correction Methods. Remote Sens. 2009, 1, 184–196. [Google Scholar] [CrossRef]
- Updike, T.; Comp, C. Radiometric Use of WorldView-2 Imagery; Technical Note; DigitalGlobe: Longmont, CO, USA, 2010; pp. 1–17. [Google Scholar]
- Rouse, J.W.; Haas, R.H.; Schell, J.A.; Deering, D.W. Monitoring Vegetation Systems in the Great Plains with ERTS. NASA Spec. Publ. 1974, 351, 309. [Google Scholar]
- Falkowski, M.J.; Gessler, P.E.; Morgan, P.; Hudak, A.T.; Smith, A.M.S. Characterizing and Mapping Forest Fire Fuels Using ASTER Imagery and Gradient Modeling. For. Ecol. Manag. 2005, 217, 129–146. [Google Scholar] [CrossRef]
- Huete, A.R. A Soil-Adjusted Vegetation Index (SAVI). Remote Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- Qi, J.; Chehbouni, A.; Huete, A.R.; Kerr, Y.H.; Sorooshian, S. A Modified Soil Adjusted Vegetation Index. Remote Sens. Environ. 1994, 48, 119–126. [Google Scholar] [CrossRef]
- Kaufman, Y.J.; Tanre, D. Atmospherically Resistant Vegetation Index (ARVI) for EOS-MODIS. IEEE Trans. Geosci. Remote Sens. 1992, 30, 261–270. [Google Scholar] [CrossRef]
- Daughtry, C. Estimating Corn Leaf Chlorophyll Concentration from Leaf and Canopy Reflectance. Remote Sens. Environ. 2000, 74, 229–239. [Google Scholar] [CrossRef]
- Wolf, A.F. Using WorldView-2 Vis-NIR Multispectral Imagery to Support Land Mapping and Feature Extraction Using Normalized Difference Index Ratios. In Proceedings of the Algorithms and Technologies for Multispectral, Hyperspectral, and Ultraspectral Imagery XVIII, Baltimore, MD, USA, 23–27 April 2012; Shen, S.S., Lewis, P.E., Eds.; SPIE: Bellingham, WA, USA, 2012; pp. 188–195. [Google Scholar]
- Penuelas, J.; Frederic, B.; Filella, I. Semi-Empirical Indices to Assess Carotenoids/Chlorophyll-a Ratio from Leaf Spectral Reflectance. Photosynthetica 1995, 31, 221–230. [Google Scholar]
- Huete, A.; Didan, K.; Miura, T.; Rodriguez, E.P.; Gao, X.; Ferreira, L.G. Overview of the Radiometric and Biophysical Performance of the MODIS Vegetation Indices. Remote Sens. Environ. 2002, 83, 195–213. [Google Scholar] [CrossRef]
- Mhangara, P.; Mapurisa, W.; Mudau, N. Comparison of Image Fusion Techniques Using Satellite Pour l’Observation de La Terre (SPOT) 6 Satellite Imagery. Appl. Sci. 2020, 10, 1881. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Friedman, J.H. Greedy Function Approximation: A Gradient Boosting Machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; ACM: New York, NY, USA, 2016; pp. 785–794. [Google Scholar]
- Mitchell, R.; Frank, E. Accelerating the XGBoost Algorithm Using GPU Computing. PeerJ Comput. Sci. 2017, 3, e127. [Google Scholar] [CrossRef]
- Bhagwat, R.U.; Uma Shankar, B. A Novel Multilabel Classification of Remote Sensing Images Using XGBoost. In Proceedings of the 2019 IEEE 5th International Conference for Convergence in Technology (I2CT), Bombay, India, 29–31 March 2019; IEEE: Bombay, India, 2019; pp. 1–5. [Google Scholar]
- Zhang, H.; Eziz, A.; Xiao, J.; Tao, S.; Wang, S.; Tang, Z.; Zhu, J.; Fang, J. High-Resolution Vegetation Mapping Using EXtreme Gradient Boosting Based on Extensive Features. Remote Sens. 2019, 11, 1505. [Google Scholar] [CrossRef]
- Muthoka, J.M.; Salakpi, E.E.; Ouko, E.; Yi, Z.-F.; Antonarakis, A.S.; Rowhani, P. Mapping Opuntia Stricta in the Arid and Semi-Arid Environment of Kenya Using Sentinel-2 Imagery and Ensemble Machine Learning Classifiers. Remote Sens. 2021, 13, 1494. [Google Scholar] [CrossRef]
- Zhang, T.; Su, J.; Xu, Z.; Luo, Y.; Li, J. Sentinel-2 Satellite Imagery for Urban Land Cover Classification by Optimized Random Forest Classifier. Appl. Sci. 2021, 11, 543. [Google Scholar] [CrossRef]
- Huang, L.; Liu, Y.; Huang, W.; Dong, Y.; Ma, H.; Wu, K.; Guo, A. Combining Random Forest and XGBoost Methods in Detecting and Mid-Term Winter Wheat Stripe Rust Using Canopy Level Hyperspectral Measurements. Agriculture 2022, 12, 74. [Google Scholar] [CrossRef]
- Amani, M.; Salehi, B.; Mahdavi, S.; Brisco, B.; Shehata, M. A Multiple Classifier System to Improve Mapping Complex Land Covers: A Case Study of Wetland Classification Using SAR Data in Newfoundland, Canada. Int. J. Remote Sens. 2018, 39, 7370–7383. [Google Scholar] [CrossRef]
- Hanson, C.C.; Brabyn, L.; Gurung, S.B. Diversity-Accuracy Assessment of Multiple Classifier Systems for the Land Cover Classification of the Khumbu Region in the Himalayas. J. Mt. Sci. 2022, 19, 365–387. [Google Scholar] [CrossRef]
- Mugiraneza; Nascetti; Ban WorldView-2 Data for Hierarchical Object-Based Urban Land Cover Classification in Kigali: Integrating Rule-Based Approach with Urban Density and Greenness Indices. Remote Sens. 2019, 11, 2128. [CrossRef]
- Wilson, K.L.; Wong, M.C.; Devred, E. Comparing Sentinel-2 and WorldView-3 Imagery for Coastal Bottom Habitat Mapping in Atlantic Canada. Remote Sens. 2022, 14, 1254. [Google Scholar] [CrossRef]
- Du, P.; Xia, J.; Zhang, W.; Tan, K.; Liu, Y.; Liu, S. Multiple Classifier System for Remote Sensing Image Classification: A Review. Sensors 2012, 12, 4764–4792. [Google Scholar] [CrossRef]
- Shi, D.; Yang, X. Mapping Vegetation and Land Cover in a Large Urban Area Using a Multiple Classifier System. Int. J. Remote Sens. 2017, 38, 4700–4721. [Google Scholar] [CrossRef]
- Talukdar, S.; Singha, P.; Mahato, S.; Shahfahad; Pal, S.; Liou, Y.-A.; Rahman, A. Land-Use Land-Cover Classification by Machine Learning Classifiers for Satellite Observations—A Review. Remote Sens. 2020, 12, 1135. [Google Scholar] [CrossRef]
- Rommel, E.; Giese, L.; Fricke, K.; Kathöfer, F.; Heuner, M.; Mölter, T.; Deffert, P.; Asgari, M.; Näthe, P.; Dzunic, F.; et al. Very High-Resolution Imagery and Machine Learning for Detailed Mapping of Riparian Vegetation and Substrate Types. Remote Sens. 2022, 14, 954. [Google Scholar] [CrossRef]
- Coulter, L.; Stow, D.; Hope, A.; O’Leary, J.; Turner, D.; Longmire, P.; Peterson, S.; Kaiser, J. Comparison of High Spatial Resolution Imagery for Efficient Generation of GIS Vegetation Layers. Photogramm. Eng. Remote Sens. 2000, 66, 1329–1336. [Google Scholar]
- Belward, A.S.; Skøien, J.O. Who Launched What, When and Why; Trends in Global Land-Cover Observation Capacity from Civilian Earth Observation Satellites. ISPRS J. Photogramm. Remote Sens. 2015, 103, 115–128. [Google Scholar] [CrossRef]
- Fisher, J.R.B.; Acosta, E.A.; Dennedy-Frank, P.J.; Kroeger, T.; Boucher, T.M. Impact of Satellite Imagery Spatial Resolution on Land Use Classification Accuracy and Modeled Water Quality. Remote Sens. Ecol. Conserv. 2018, 4, 137–149. [Google Scholar] [CrossRef]
- Joshi, P.K.K.; Roy, P.S.; Singh, S.; Agrawal, S.; Yadav, D. Vegetation Cover Mapping in India Using Multi-Temporal IRS Wide Field Sensor (WiFS) Data. Remote Sens. Environ. 2006, 103, 190–202. [Google Scholar] [CrossRef]
- Jawak, S.D.; Luis, A.J. Improved Land Cover Mapping Using High Resolution Multiangle 8-Band WorldView-2 Satellite Remote Sensing Data. J. Appl. Remote Sens. 2013, 7, 073573. [Google Scholar] [CrossRef]
- Rapinel, S.; Clément, B.; Magnanon, S.; Sellin, V.; Hubert-Moy, L. Identification and Mapping of Natural Vegetation on a Coastal Site Using a Worldview-2 Satellite Image. J. Environ. Manag. 2014, 144, 236–246. [Google Scholar] [CrossRef]
- Odindi, J.; Adam, E.; Ngubane, Z.; Mutanga, O.; Slotow, R. Comparison between WorldView-2 and SPOT-5 Images in Mapping the Bracken Fern Using the Random Forest Algorithm. J. Appl. Remote Sens 2014, 8, 083527. [Google Scholar] [CrossRef]
- Lewis, K.; de V. Barros, F.; Cure, M.B.; Davies, C.A.; Furtado, M.N.; Hill, T.C.; Hirota, M.; Martins, D.L.; Mazzochini, G.G.; Mitchard, E.T.A.; et al. Mapping Native and Non-Native Vegetation in the Brazilian Cerrado Using Freely Available Satellite Products. Sci. Rep. 2022, 12, 1588. [Google Scholar] [CrossRef]
- Wuttke, S.; Middelmann, W.; Stilla, U. Improving the Efficiency of Land Cover Classification by Combining Segmentation, Hierarchical Clustering, and Active Learning. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2018, 11, 4016–4031. [Google Scholar] [CrossRef]
- Lassiter, A.; Darbari, M. Assessing Alternative Methods for Unsupervised Segmentation of Urban Vegetation in Very High-Resolution Multispectral Aerial Imagery. PLoS ONE 2020, 15, e0230856. [Google Scholar] [CrossRef]
- Maxwell, A.E.; Warner, T.A.; Fang, F. Implementation of Machine-Learning Classification in Remote Sensing: An Applied Review. Int. J. Remote Sens. 2018, 39, 2784–2817. [Google Scholar] [CrossRef]
- Gašparović, M.; Dobrinić, D. Comparative Assessment of Machine Learning Methods for Urban Vegetation Mapping Using Multitemporal Sentinel-1 Imagery. Remote Sens. 2020, 12, 1952. [Google Scholar] [CrossRef]
- Omer, G.; Mutanga, O.; Abdel-Rahman, E.M.; Adam, E. Performance of Support Vector Machines and Artificial Neural Network for Mapping Endangered Tree Species Using WorldView-2 Data in Dukuduku Forest, South Africa. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 8, 4825–4840. [Google Scholar] [CrossRef]
- Tang, Y.; Jing, L.; Li, H.; Liu, Q.; Yan, Q.; Li, X. Bamboo Classification Using WorldView-2 Imagery of Giant Panda Habitat in a Large Shaded Area in Wolong, Sichuan Province, China. Sensors 2016, 16, 1957. [Google Scholar] [CrossRef] [PubMed]
- Saad, F.; Biswas, S.; Huang, Q.; Corte, A.P.D.; Coraiola, M.; Macey, S.; Carlucci, M.B.; Leimgruber, P. Detectability of the Critically Endangered Araucaria Angustifolia Tree Using Worldview-2 Images, Google Earth Engine and UAV-LiDAR. Land 2021, 10, 1316. [Google Scholar] [CrossRef]
- Bransky, N.; Sankey, T.; Sankey, B.J.; Johnson, M.; Jamison, L. Monitoring Tamarix Changes Using WorldView-2 Satellite Imagery in Grand Canyon National Park, Arizona. Remote Sens. 2021, 13, 958. [Google Scholar] [CrossRef]
- Ghosh, A.; Joshi, P.K. A Comparison of Selected Classification Algorithms for Mapping Bamboo Patches in Lower Gangetic Plains Using Very High Resolution WorldView 2 Imagery. Int. J. Appl. Earth Obs. Geoinf. 2014, 26, 298–311. [Google Scholar] [CrossRef]
- Abutaleb, K.; Newete, S.W.; Mangwanya, S.; Adam, E.; Byrne, M.J. Mapping Eucalypts Trees Using High Resolution Multispectral Images: A Study Comparing WorldView 2 vs. SPOT 7. Egypt. J. Remote Sens. Space Sci. 2021, 24, 333–342. [Google Scholar] [CrossRef]
- Deval, K.; Joshi, P.K. Vegetation Type and Land Cover Mapping in a Semi-Arid Heterogeneous Forested Wetland of India: Comparing Image Classification Algorithms. Environ. Dev. Sustain. 2022, 24, 3947–3966. [Google Scholar] [CrossRef]
- Li, D.; Ke, Y.; Gong, H.; Li, X. Object-Based Urban Tree Species Classification Using Bi-Temporal WorldView-2 and WorldView-3 Images. Remote Sens. 2015, 7, 16917–16937. [Google Scholar] [CrossRef]
- Wendelberger, K.; Gann, D.; Richards, J. Using Bi-Seasonal WorldView-2 Multi-Spectral Data and Supervised Random Forest Classification to Map Coastal Plant Communities in Everglades National Park. Sensors 2018, 18, 829. [Google Scholar] [CrossRef]
- Ibarrola-Ulzurrun, E.; Gonzalo-Martín, C.; Marcello, J. Influence of Pansharpening in Obtaining Accurate Vegetation Maps. Can. J. Remote Sens. 2017, 43, 528–544. [Google Scholar] [CrossRef]
- Castillejo-González, I. Mapping of Olive Trees Using Pansharpened QuickBird Images: An Evaluation of Pixel- and Object-Based Analyses. Agronomy 2018, 8, 288. [Google Scholar] [CrossRef]
- Karlson, M.; Ostwald, M.; Reese, H.; Bazié, H.R.; Tankoano, B. Assessing the Potential of Multi-Seasonal WorldView-2 Imagery for Mapping West African Agroforestry Tree Species. Int. J. Appl. Earth Obs. Geoinf. 2016, 50, 80–88. [Google Scholar] [CrossRef]
- Adamo, M.; Tomaselli, V.; Tarantino, C.; Vicario, S.; Veronico, G.; Lucas, R.; Blonda, P. Knowledge-Based Classification of Grassland Ecosystem Based on Multi-Temporal WorldView-2 Data and FAO-LCCS Taxonomy. Remote Sens. 2020, 12, 1447. [Google Scholar] [CrossRef]

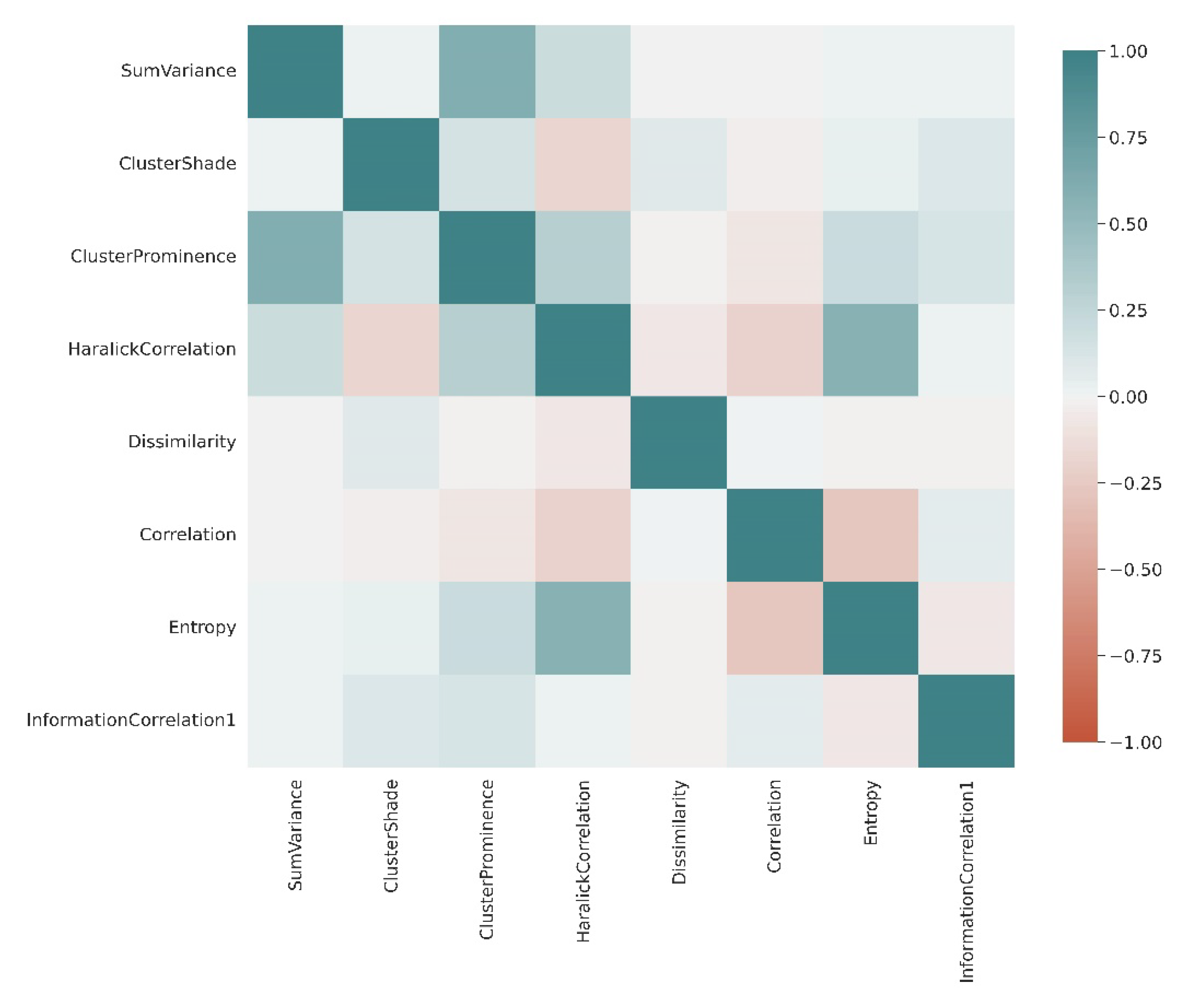

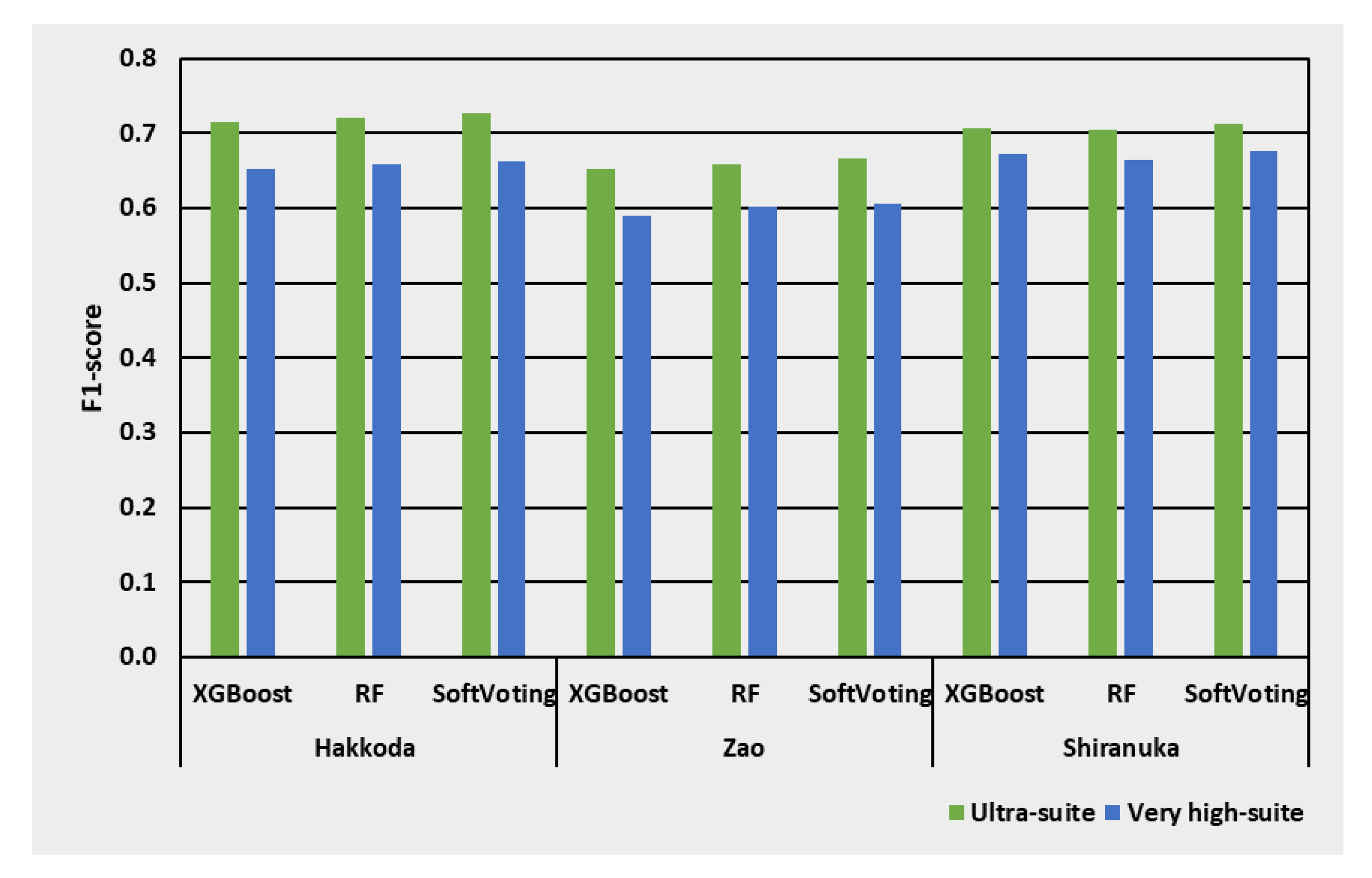

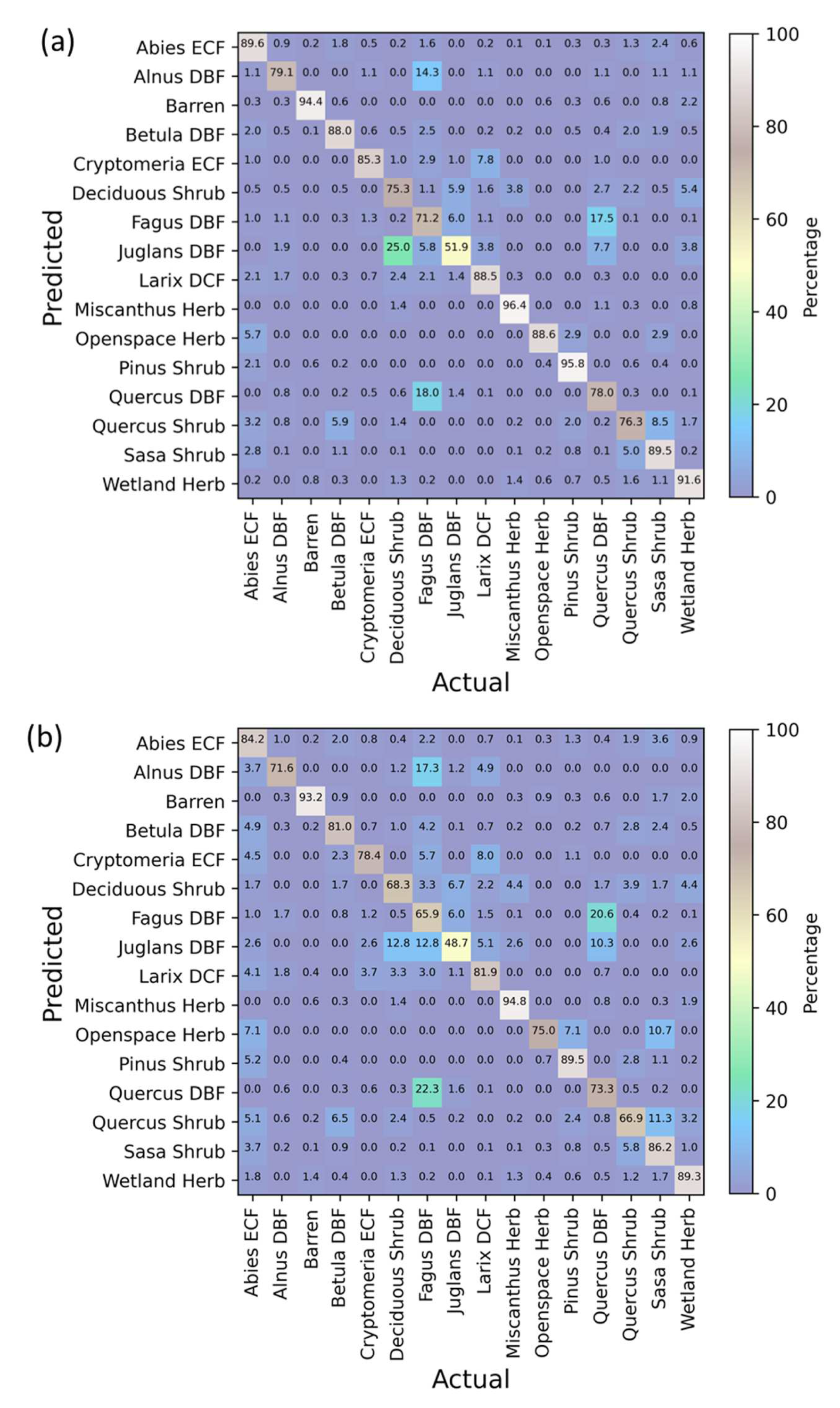
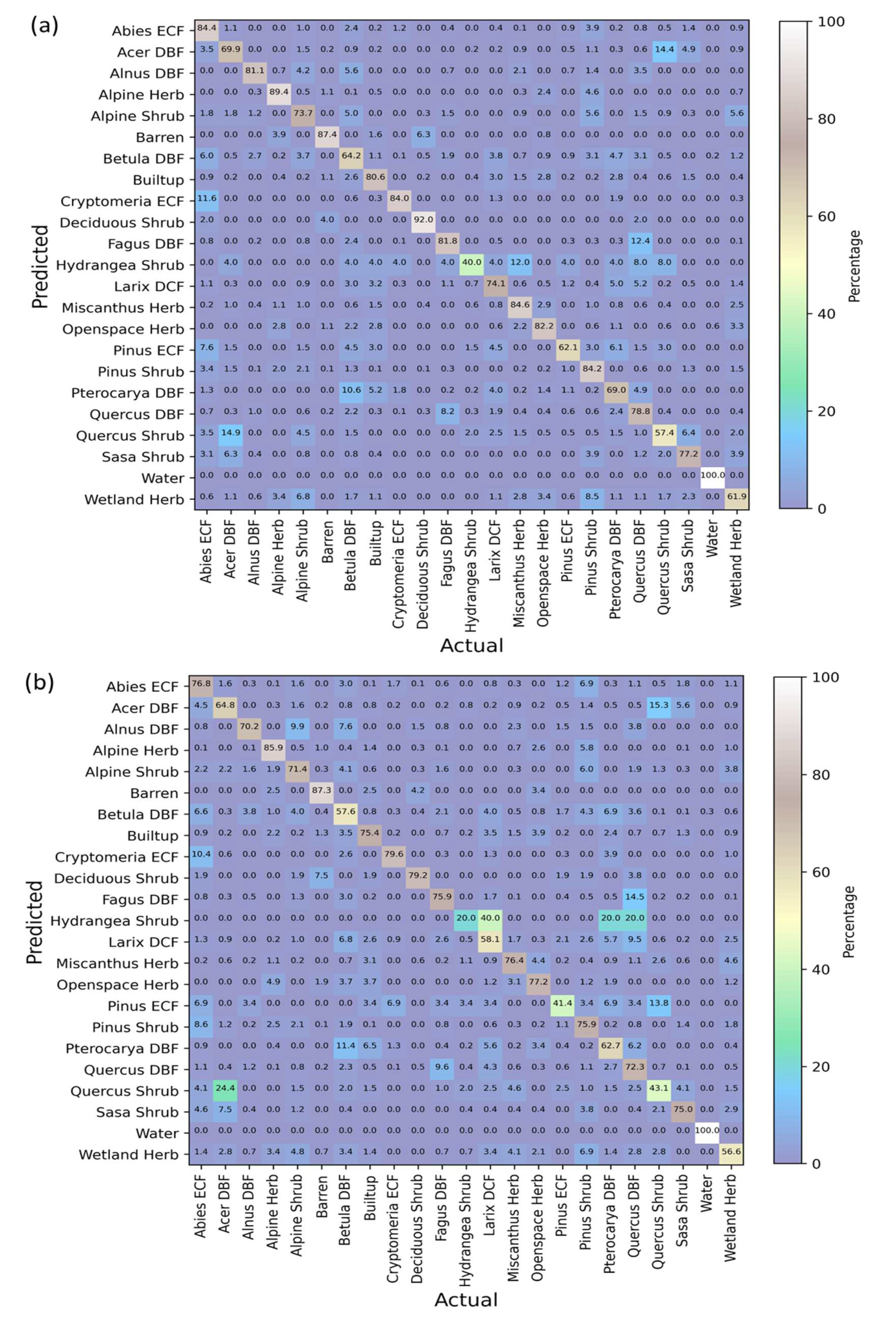





| Indices | References |
|---|---|
| Normalized Difference Vegetation Index | Rouse et al. [62] |
| Green–Red Vegetation Index | Falkowski et al. [63] |
| Soil-Adjusted Vegetation Index | Huete [64] |
| Modified Soil-Adjusted Vegetation Index | Qi et al. [65] |
| Atmospherically Resistant Vegetation Index | Kaufman and Tanre [66] |
| Modified Chlorophyll Absorption Ratio Index | Daughtry et al. [67] |
| Non-Homogeneous Feature Difference | Wolf [68] |
| Structure-Insensitive Pigment Index | Penuelas et al. [69] |
| Enhanced Vegetation Index | Huete et al. [70] |
| Suite | Features | Total Features | |
|---|---|---|---|
| Very high-resolution suite (2 m) | Multi-spectral bands | 8 | 44 |
| Multi-spectral indices | 9 | ||
| Color-transformation (multi-spectral) | 27 | ||
| Ultra-resolution suite (0.5 m) | Panchromatic band | 1 | 44 |
| Pan-sharpened bands | 8 | ||
| Textural features | 8 | ||
| Color-transformation (pan-sharpened) | 27 | ||
| Site | Model | Overall Accuracy | Kappa Coefficient | F1-Score | Recall | Precision |
|---|---|---|---|---|---|---|
| Hakkoda | XGBoost | 0.653 | 0.622 | 0.653 | 0.653 | 0.653 |
| RF | 0.659 | 0.629 | 0.659 | 0.659 | 0.659 | |
| SoftVoting | 0.663 | 0.634 | 0.663 | 0.663 | 0.663 | |
| Zao | XGBoost | 0.590 | 0.566 | 0.590 | 0.590 | 0.590 |
| RF | 0.601 | 0.577 | 0.601 | 0.601 | 0.601 | |
| SoftVoting | 0.606 | 0.582 | 0.606 | 0.606 | 0.606 | |
| Shiranuka | XGBoost | 0.673 | 0.627 | 0.673 | 0.673 | 0.673 |
| RF | 0.665 | 0.619 | 0.665 | 0.665 | 0.665 | |
| SoftVoting | 0.676 | 0.631 | 0.676 | 0.676 | 0.676 |
| Site | Model | Overall Accuracy | Kappa Coefficient | F1-Score | Recall | Precision |
|---|---|---|---|---|---|---|
| Hakkoda | XGBoost | 0.715 | 0.690 | 0.715 | 0.715 | 0.715 |
| RF | 0.720 | 0.696 | 0.720 | 0.720 | 0.720 | |
| SoftVoting | 0.726 | 0.703 | 0.726 | 0.726 | 0.726 | |
| Zao | XGBoost | 0.652 | 0.631 | 0.652 | 0.652 | 0.652 |
| RF | 0.659 | 0.638 | 0.659 | 0.659 | 0.659 | |
| SoftVoting | 0.666 | 0.645 | 0.666 | 0.666 | 0.666 | |
| Shiranuka | XGBoost | 0.707 | 0.666 | 0.707 | 0.707 | 0.707 |
| RF | 0.704 | 0.662 | 0.704 | 0.704 | 0.704 | |
| SoftVoting | 0.712 | 0.672 | 0.712 | 0.712 | 0.712 |
| Site | Model | Overall Accuracy | Kappa Coefficient | F1-Score | Recall | Precision |
|---|---|---|---|---|---|---|
| Hakkoda | XGBoost | 0.783 | 0.752 | 0.783 | 0.783 | 0.783 |
| RF | 0.779 | 0.746 | 0.779 | 0.779 | 0.779 | |
| SoftVoting | 0.786 | 0.755 | 0.786 | 0.786 | 0.786 | |
| Zao | XGBoost | 0.729 | 0.700 | 0.729 | 0.729 | 0.729 |
| RF | 0.728 | 0.699 | 0.728 | 0.728 | 0.728 | |
| SoftVoting | 0.730 | 0.701 | 0.730 | 0.730 | 0.730 | |
| Shiranuka | XGBoost | 0.818 | 0.776 | 0.818 | 0.818 | 0.818 |
| RF | 0.818 | 0.776 | 0.818 | 0.818 | 0.818 | |
| SoftVoting | 0.825 | 0.784 | 0.825 | 0.825 | 0.825 |
| Site | Model | Overall Accuracy | Kappa Coefficient | F1-Score | Recall | Precision |
|---|---|---|---|---|---|---|
| Hakkoda | XGBoost | 0.832 | 0.807 | 0.832 | 0.832 | 0.832 |
| RF | 0.828 | 0.803 | 0.828 | 0.828 | 0.828 | |
| SoftVoting | 0.835 | 0.811 | 0.835 | 0.835 | 0.835 | |
| Zao | XGBoost | 0.797 | 0.776 | 0.797 | 0.797 | 0.797 |
| RF | 0.794 | 0.773 | 0.794 | 0.794 | 0.794 | |
| SoftVoting | 0.798 | 0.777 | 0.798 | 0.798 | 0.798 | |
| Shiranuka | XGBoost | 0.852 | 0.818 | 0.852 | 0.852 | 0.852 |
| RF | 0.849 | 0.815 | 0.849 | 0.849 | 0.849 | |
| SoftVoting | 0.855 | 0.822 | 0.855 | 0.855 | 0.855 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, R.C. An Ultra-Resolution Features Extraction Suite for Community-Level Vegetation Differentiation and Mapping at a Sub-Meter Resolution. Remote Sens. 2022, 14, 3145. https://doi.org/10.3390/rs14133145
Sharma RC. An Ultra-Resolution Features Extraction Suite for Community-Level Vegetation Differentiation and Mapping at a Sub-Meter Resolution. Remote Sensing. 2022; 14(13):3145. https://doi.org/10.3390/rs14133145
Chicago/Turabian StyleSharma, Ram C. 2022. "An Ultra-Resolution Features Extraction Suite for Community-Level Vegetation Differentiation and Mapping at a Sub-Meter Resolution" Remote Sensing 14, no. 13: 3145. https://doi.org/10.3390/rs14133145
APA StyleSharma, R. C. (2022). An Ultra-Resolution Features Extraction Suite for Community-Level Vegetation Differentiation and Mapping at a Sub-Meter Resolution. Remote Sensing, 14(13), 3145. https://doi.org/10.3390/rs14133145