Abstract
The ability to measure and monitor wildlife populations is important for species management and conservation. The use of near-infrared spectroscopy (NIRS) to rapidly detect physiological traits from wildlife scat and other body materials could play an important role in the conservation of species. Previous research has demonstrated the potential for NIRS to detect diseases such as the novel COVID-19 from saliva, parasites from feces, and numerous other traits from animal skin, hair, and scat, such as cortisol metabolites, diet quality, sex, and reproductive status, that may be useful for population monitoring. Models developed from NIRS data use light reflected from a sample to relate the variation in the sample’s spectra to variation in a trait, which can then be used to predict that trait in unknown samples based on their spectra. The modelling process involves calibration, validation, and evaluation. Data sampling, pre-treatments, and the selection of training and testing datasets can impact model performance. We review the use of NIRS for measuring physiological traits in animals that may be useful for wildlife management and conservation and suggest future research to advance the application of NIRS for this purpose.
Keywords:
spectroscopy; near-infrared; wildlife; conservation; management; health; disease; diet; nutrition; reproductive status 1. Introduction
Accurate and timely information about wildlife populations is necessary to make appropriate decisions about their management and conservation. A combination of invasive and non-invasive methods is typically used to obtain this information [1,2]. However, management decisions are often made without all of the information needed due to the difficulty, time, and/or cost associated with acquiring it [3]. Invasive methods for collecting data on animals usually require them to be captured, which can occasionally cause injury or even death [1,4]. In addition, it may not be possible to locate or capture cryptic, rare, dangerous, or easily excitable species [5]. Non-invasive field sampling techniques such as the collection of feces, hair, footprints, and urine samples can provide useful information about wildlife, without needing to see or interact with the animal [2,6]. For many species, scats are easy to find in the environment and can provide in-depth information about diet [7], stress and reproductive hormones [8,9], sex [10], and parasite load [11]. However, extracting this information from scat can be time-consuming, expensive, and require multiple procedures [12]. For example, DNA sequencing has been used to identify diet composition from scat [13], but these analyses require highly skilled personnel and costly analytical equipment.
Near-infrared spectroscopy (NIRS) is a non-invasive analytical technique that can provide a variety of qualitative and quantitative information rapidly and inexpensively [14]. A detailed description of this technique is provided in the Section 2. NIRS is widely used in livestock industries, where it provides information about diet selection and the nutritional properties of pasture and feed to help farmers increase yields [15,16,17,18,19]. NIRS is also used extensively to quantify the concentrations of chemical compounds in dry goods [20], dairy milk, and other liquid consumables [21,22], as well as to rapidly detect the presence of hazardous chemicals for security screening [23]. In addition, NIRS has been used to measure the quality of plant browse for wild herbivores to investigate plant–animal interactions and the relationships between browse quality and habitat quality [24,25,26,27,28,29,30].
Recently, NIRS has demonstrated the potential to detect biophysical traits from animal excrement, saliva, and tissue samples that may be useful for a number of applications in wildlife management and conservation [31]. These include disease and parasite status [32,33], sex [10,34], species [35], age class [10,36], reproductive status [10,37,38,39], stress levels [40,41], body composition [42,43], and nutritional state [44]. The development of portable field sensors and lower-cost NIRS instruments, particularly in comparison with the equipment required for conventional assays, is increasing interest in a wide range of ecological applications [31,45,46]. Another major benefit of NIRS is that, after calibration, a practitioner can measure several traits simultaneously from one sample with little preparation [14]. In this review, we provide a brief overview of NIRS and discuss the application of NIRS for determining disease, population demographics, and diet quality using spectral data collected from animal scat, urine, saliva, hair, skin, and scales. We also highlight areas that should be the focus of future research, such as how the pre-treatment of sample material could improve NIRS models, whether the aging of samples impacts the information obtained, and other considerations such as sample size, validation, data variability, and lastly the potential for the shareability of NIRS calibration models.
2. NIRS in Practice
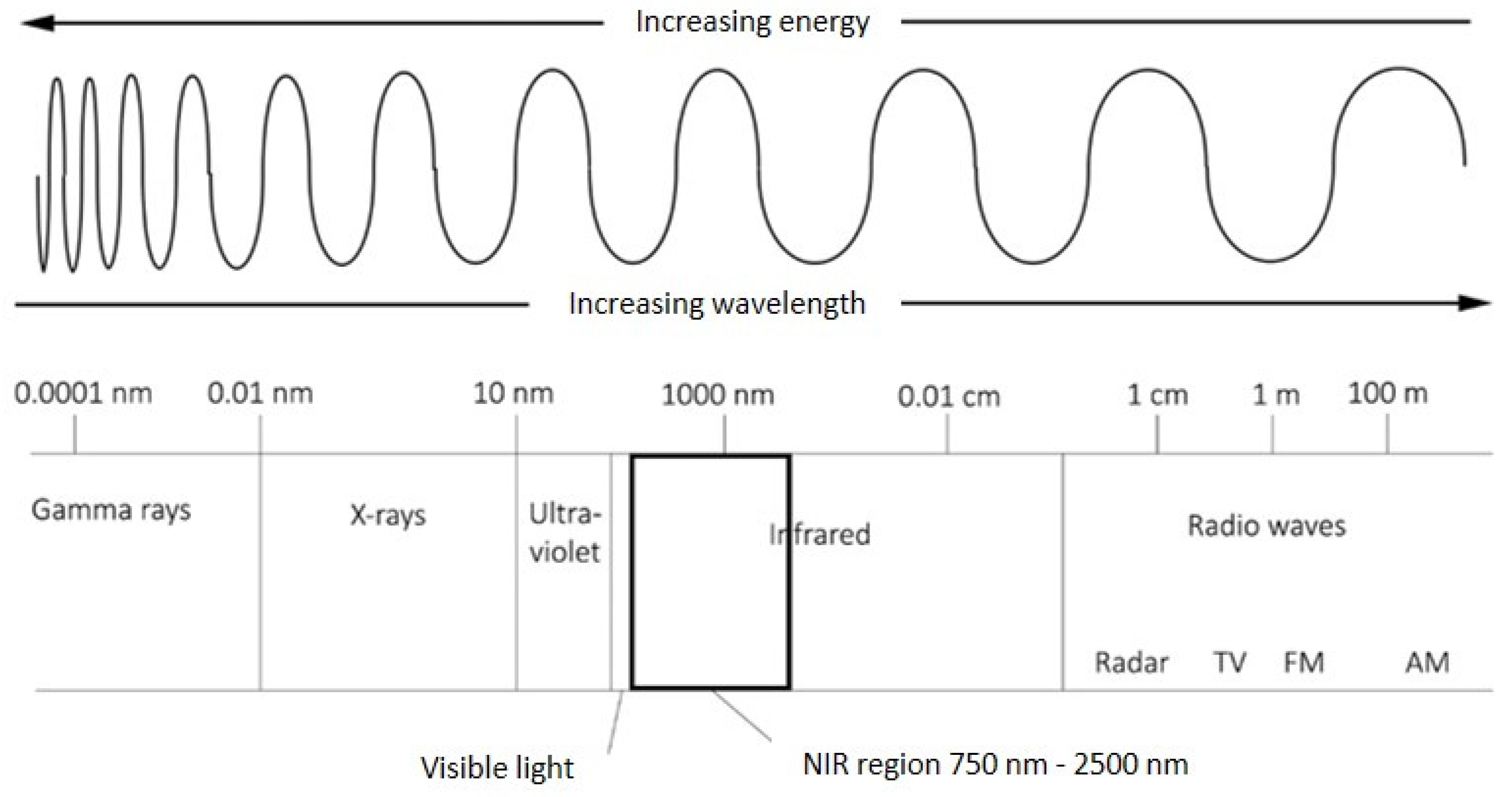
NIRS involves collecting the light reflected from or transmitted through a material across the near-infrared (NIR, 750 nm to 1000 nm) and short-wave infrared (SWIR, 1000 nm to 2500 nm) regions of the electromagnetic spectrum, often collectively referred to as near-infrared (Figure 1, [45]). Chemical bonds vibrate as they rotate, bend, and stretch in response to their atomic interactions and energy level. These vibrations absorb electromagnetic radiation at wavelengths that correspond to their energy state and create measurable overtones and harmonics in the NIR wavelengths [47]. Given that different materials contain specific combinations of chemical constituents, and thus the patterns of reflected and absorbed light are also different between materials, NIRS can be used to obtain information about the chemical composition of a sample [14,48]. Notably, near-infrared radiation is absorbed primarily by C-H, N-H, and O-H chemical bonds, which make up most organic compounds, so it is particularly useful for the quantitative analysis of organic material [14]. In addition to measuring some chemical constituents directly from specific wavelengths associated with chemical bonds, NIRS and its associated statistical methodology—chemometrics—can also identify relationships between complex traits that are not determined by any one chemical compound but by numerous interacting effects and multiple wavelengths within the electromagnetic spectrum [14,29,49].
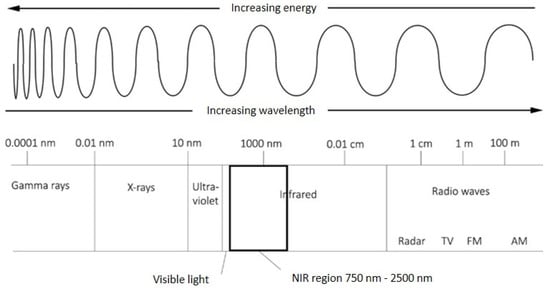
Figure 1.
A visualisation of wavelengths in the electromagnetic spectrum, highlighting the near-infrared region. Adapted from proccesssensors.com blog from 2013 (https://www.processsensors.com/, accessed on 8 September 2021).
NIRS allows for the rapid collection of thousands of data points across the electromagnetic spectrum. These data can be collected using a variety of different imaging modes (i.e., reflectance, transmittance, or interactance) and instrument types, which are reviewed elsewhere (for example, see [50]). Although there are some simple models that associate a single wavelength or band of wavelengths with a constituent of interest, such as water and ethanol, most variables require reflectance values from tens or hundreds of wavelengths for accurate measurements [45]. Models developed from NIRS result in accurate predictions when consistent associations between the measured variable and variations in the reflectance of particular wavelengths are found; however, the underlying mechanism driving those relationships is not always known [51]. For example, the ability of NIRS to discriminate males from females in some animals may not be due to wavelengths associated with the chemical bonds of sex hormones but possibly sex-based differences in diet selection that result in samples from males and females having a different chemical composition [46]. Although understanding the scientific basis for model success can help in the assessment of its applicability and limitations, few studies address this issue. Most research focuses on developing and testing spectral calibration models to predict values in new samples using chemometric methods. Chemometrics evolved alongside NIRS as a collection of mathematical and statistical tools to extract relevant information from the large, multidimensional datasets that are common with NIRS and other forms of spectroscopy. A complete review of chemometrics is beyond the scope of this paper and has already been addressed in the existing literature [52,53]. However, we provide an overview of some common mathematical pre-treatments and statistical methods for the qualitative and quantitative analysis of NIRS data below.
Principal Component Regression (PCR) and Partial Least Squares Regression (PLSR) are the most common statistical methods for developing calibration models with NIRS data because they are particularly useful for reducing the large, multivariate, and often highly correlated datasets to focus on relevant variables [53,54,55]. Both methods allow models to be developed using a number of variables that is larger than the number of samples, and they combine independent variables into factors to reduce data dimensionality and avoid co-linearity issues. PCR and PLSR typically provide similar model results, but PLSR usually requires fewer principal components (PCs) than PCR-based models. A high number of PCs reduces the variability not accounted for by the model and a model may be overfit, meaning it can predict the training data well but not independent data [56]. For this reason, PLSR is often the favoured method for identifying the linear combination of wavebands to explain the response variable, although overfitting can still be an issue for any of these models. Principle component and partial least squares approaches also have variations that can be used when the response variable is categorical rather than continuous, and these are called discriminate analyses (DA). Machine learning techniques such as Artificial Neural Networks (ANN), Random Forests (RF), and Cubists approaches are other methods to develop non-linear models that are gaining in popularity, but these are still relatively new tools for developing NIRS calibrations [57].
The process of developing NIRS calibrations to predict traits in unknown spectra involves a number of steps. First, NIR spectral data are collected from the samples of interest with a spectrometer. The way that samples are prepared can have a great influence on the quality and consistency of the spectral information that can be obtained. The most appropriate methods to prepare samples prior to collecting spectral data are discussed in a separate section at the end of the manuscript because this requires specific considerations and additional research for some of the applications we review later. Once spectral data have been collected, they are pre-processed to reduce noise, or non-useful information. Pre-processing treatments such as standardisation (i.e., centering and scaling) [58], multiplicative scatter correction, and standard normal variate are often applied to reduce this noise and “clean” the spectral data prior to modelling [59,60]. In addition, first and second derivatives of the spectral data are generally created to accentuate features of interest [61]. After pre-treatment, the spectra can be used in model development.
Constituent values must be determined using other methods in a subset of the samples from which spectra were collected. “Constituent” here refers to the trait of interest, and that can be the amount or presence of a certain chemical compound or a trait such as sex (i.e., male or female) that may be linked to the presence or absence of a number of different compounds within a sample. Constituent values for model training and testing data should be obtained using appropriate and robust methods because the predictive models ultimately will be constrained by the quality of input data for the training and testing datasets [46].
The spectral data with known constituent values are then divided randomly into training and testing datasets, and the model is usually developed using one of the methods outlined above. The number of samples required for training depends on many factors, including the spectral variability of the material used for modelling and the potential range of values (e.g., chemical concentrations) and variability in materials (e.g., different species, sample collection localities, etc.) across which predictions are going to be made [53]. Pasquini [45] recommended that 50 to 100 samples be used for natural systems depending on the complexity and variability of the sample matrix. However, training datasets can easily require several hundred samples if there is substantial variability in the material being analysed [62]. If the sources for the variability in the data are not present in the training data, the resulting model may not be robust when faced with variability beyond which it was trained. Having a training dataset that includes all the potential sources of variability in a population is ideal but usually unachievable. Efforts should be made to identify and include potentially large sources of variability, and models should be used with an awareness of potential limitations. A subset of samples should be verified using alternative analyses when models are applied to data that may have additional variability beyond the training dataset to further test model performance.
Sometimes, the number of samples available for training and testing is constrained by the number that it is possible to obtain. Where it has been determined that there are insufficient data for model training and independent validation, cross-validation can be used to estimate model error based on data resampling, where the model is both trained and tested on all available data [63,64]. Although cross-validation is commonly used in NIRS model development, the ability of the model to fit new data depends on whether the training data are spectrally representative of the entire population of the data. Cross-validation alone is appropriate for feasibility studies, but independent data should be used to verify model performance before using a model for a specific application in the real world [53]. Cross-validation can also provide some indication of a model’s performance on future datasets by comparing training and cross-validation testing standard errors, which should be similar. If they are not similar, this indicates that the amount of spectral variability in the population may be much larger than the training dataset can accommodate.
3. Measuring Animal Traits with NIRS
The following sections review the use of NIRS in the existing literature to provide information on animal health and disease, population demographics, and diet quality.
3.1. Animal Health and Disease
Disease and parasitism can have substantial impacts on animal populations [65,66]. Likewise, stress can have negative effects on animal reproduction, body condition, and immune function [67]. Assessing animal health and disease is traditionally determined through clinical evaluation by a qualified individual, which is often a veterinarian [68]. For instance, cortisol, a hormone associated with stress, is usually detected through the saliva or blood of animals, using hormone extraction and quantification methods such as radioimmunoassay [69]. Similarly, diagnosing diseases, such as chlamydia in koalas (Phascolarctos cinereus), often involves collecting a sample from the affected area on the animal for antigen, antibody, DNA, or microbial testing [70]. These investigations may require a thorough clinical evaluation and the capture and sometimes sedation of an individual animal, or many individuals, to understand the disease and health status of the population [1,4].
Indicators of stress and disease are found in many bodily materials, including hair [41,71] and feces [72,73], that can be collected without interacting with an animal. A recent study found that NIRS of cattle (Bos taurus) hair was able to accurately predict cortisol levels [41], and similar success was reported for measuring cortisol in gorillas (Gorilla gorilla gorilla) from fecal cortisol metabolites in scat [40]. NIRS has also been used for the non-invasive detection of parasitic infection and bacterial pathogens from scat [33,65,74].
Even when animals must be caught, assessment methods that do not require extracting fluid or tissue may be preferable. Vance et al. [66] were able to predict disease status and sex in Chinese giant salamanders (Andrias davidianus) using NIR spectral data collected from the animals’ skin without causing harm. The study provided valuable information for aquaculture managers to mitigate disease outbreaks in the study population and reduce intrasex aggression during reintroductions [66]. Other applications of NIRS of animal skin for monitoring health include identifying differences in burn scars at various stages of healing in cattle [75]. NIRS has also been used to investigate differences in body tissue composition between animals eating different diets [42,76,77,78].
Malaria and many other zoonotic diseases, such as COVID-19, are also potential threats to human populations. The ability to rapidly detect them could help to control disease outbreaks across species. For example, Esperança et al. [32] used NIRS of insect bodies to identify malaria (Plasmodium berghei) infected mosquitoes (Anopheles stephensi), which is essential for monitoring the transmission potential of this parasite to humans and the efficacy of control interventions. Recent research has found that the novel COVID-19 virus can be detected from serum and pharynx exudate samples in humans using NIRS [79,80]. This method also may be able to provide a rapid screening tool for coronaviruses in bats and other animals that could threaten human populations.
3.2. Population Demographics
Information on population demographics, such as sex ratios, reproductive status, and age classes within a population, is essential for population management and conservation. These data can be used to inform estimates of population growth and decline rates [10,66,81]. A number of studies have used NIRS to successfully predict demographic traits in a range of different species, including giant pandas (Ailuropoda melanoleuca) (sex, age class, and reproductive status; [10,38]), snow leopards (Panthera uncia) (sex and species; [82]), red deer (Cervus elaphus), fallow deer (Dama dama) (sex and species; [35]), giant salamanders (Andrias davidianus) (sex; [66]), red snapper (Lutjanus campechanus) (age, growth; [83]), sharks (Sphyrna mokarran, Carcharhinus sorrah) (age; [84]), fruit flies (Drosophila melanogaster) (species, sex, and age class; [85]), mosquitoes (Aedes aegypti) (age; [86]), tsetse flies (Glossina spp.) (sex; [87]), sheep (Ovis aries) (pregnancy status; [88]), and cattle (Bos taurus taurus) (reproductive status and sex; [34,37,89]).
The ability to detect fertility and reproductive cycles can be a valuable tool in wildlife conservation. In pandas, the NIRS-derived reproductive status from urine has been used to successfully monitor and manage populations for captive breeding programs [10,38]. Similarly, Kinoshita et al. [39] used NIRS to determine the estrogen and creatine concentrations in the urine of Bornean orangutans (Pongo pygmaeus) to better understand their reproductive physiology. Prior to this study, there was no easy monitoring method for reproductive physiology in this critically endangered species [39]. Although NIRS has proven to be useful in this capacity for some species, it is unclear whether this method is broadly applicable for a wide range of species or using NIRS data collected from other bodily materials. Some studies have found that animal age and the stage of the reproductive cycle may impact the ability of NIRS models to determine sex and reproductive status [34,89].
NIRS can be used to differentiate species using chemotaxonomy, which identifies species based on metabolomics (i.e., the chemical compounds that an organism produces) [90]. This methodology has proven particularly successful in entomology, demonstrating the potential of NIRS to identify multiple different species (from insect bodies) within the same family across a diverse order of taxa including Blattodea, Orthoptera, Psocodea, Hemiptera, Coleoptera, Diptera, Lepidoptera, and Hymenoptera (reviewed in [90]). Similarly, NIRS also has been used to identify different origins of individuals from a single species, such as sea bass (Dicentrarchus labrax) from body tissue, which can be particularly useful for monitoring trade from both farmed and free-ranging wildlife [91].
The ability to age individuals within populations is essential for understanding and predicting population growth dynamics. Aging is particularly important for managing fisheries, and the application of NIRS to aging larval, juvenile, and adult fish is revolutionising fish aging by replacing traditional, labor-intensive methods with the comparably rapid and inexpensive Fourier transform NIRS [92]. Growing fish accumulate layers of calcium carbonate and a protein matrix on their otoliths, and scanning these otoliths with a focused beam from a spectrophotometer has provided rapid and accurate age estimations for a wide range of commercially valuable fish species [83,92,93,94].
Despite many successful studies involving the detection of population demographics in wildlife using NIRS, some studies have been unable to link spectra with traits of interest. Although Aw et al. [85] were able to quantify the sex and species in Drosophila flies, they were unable to accurately predict fly ages. Godfrey et al. [88] successfully predicted the pregnancy status of sheep from scat, yet were unable to determine sex. In this case, diet composition appeared to be a confounding factor [88]. In other situations, differences in diets between sexes can aid in sex discrimination and may be the basis for the success of some NIRS predictive models [46]; however, it is important to recognize the limitations of such models because they may not be discriminating sex per se, but diet.
3.3. Diet Quality
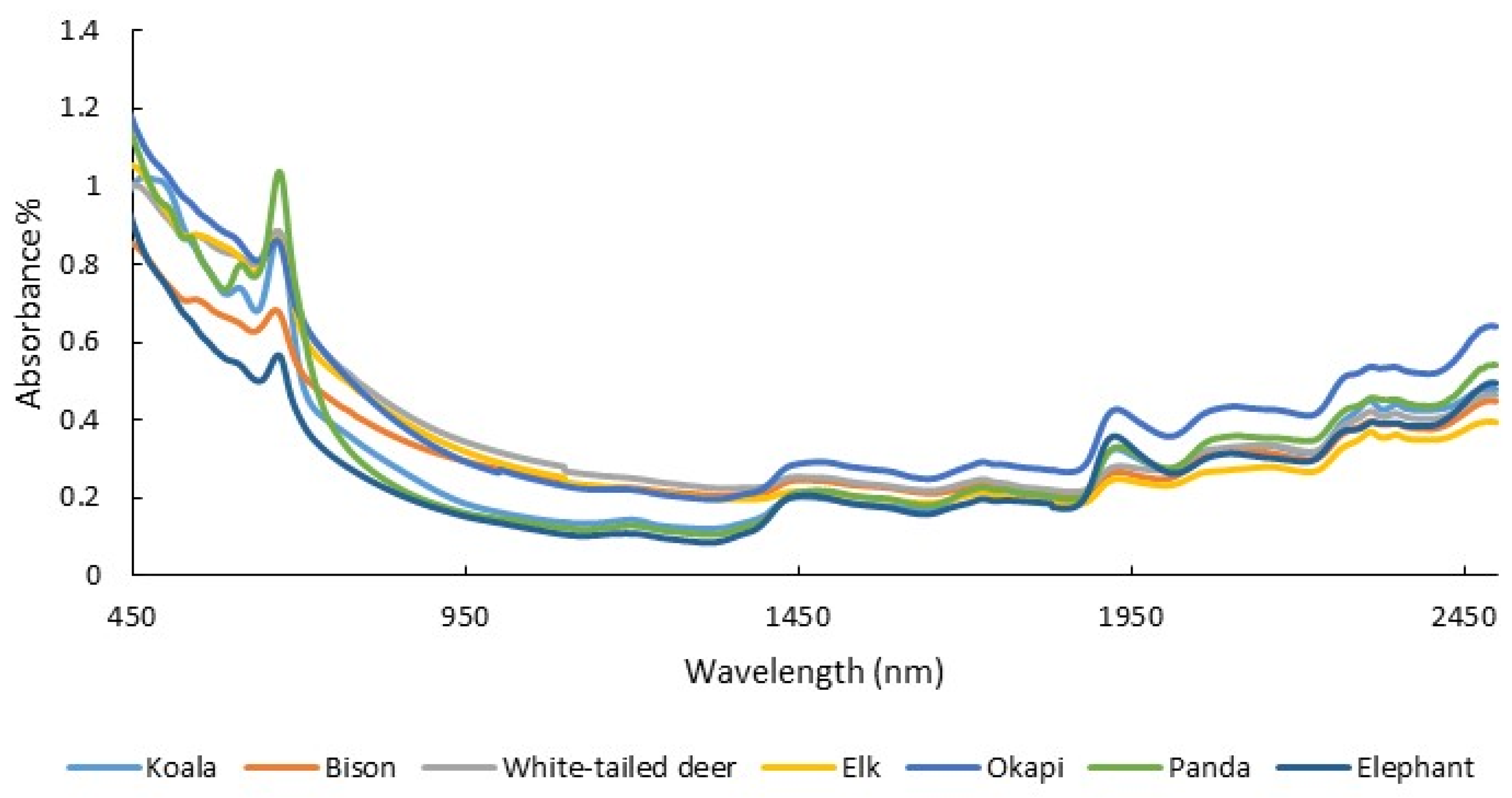
The amount of energy, protein, minerals, and various other dietary constituents that animals consume and digest influences their nutritional state [95,96]. This, in turn, affects animal health, growth rate, and reproduction. As a consequence, the capacity to measure diet quality can be useful for population management [18,46]. Diet quality can be detected through the measurement of the diet itself (e.g., foliar nitrogen) but also indirectly through the chemical composition of bodily material, often feces. NIRS has been used extensively to measure diet quality and intake in agricultural animals [17,97,98]. The use of feces in conjunction with diet, as opposed to using diet alone to measure diet quality, has been especially successful. For example, Lyons and Stuth [97] found a strong relationship between diet quality reference chemistry (crude protein and digestible organic matter) and fecal NIRS-predicted estimates of these constituents. Since then, multiple studies have related the NIR spectra of feces (Figure 2) to many different aspects of diet, including crude protein, digestible organic matter, phosphorous, and plant secondary metabolites that can adversely affect nutritional quality, such as dietary tannins [24,26,99,100,101,102].
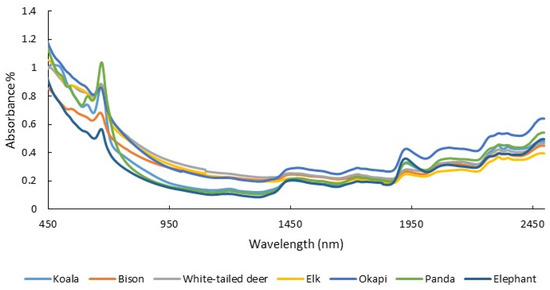
Figure 2.
Absorbance spectra (450–2500 nm) from the dried, ground scat of several herbivorous species collected with a laboratory NIR spectrometer (unpublished data).
One limitation of the above approach is the need for accurate measures of diet composition, intake and/or digestibility parameters to pair with fecal spectra, which may be difficult to obtain for many wildlife species. An alternative approach is to develop calibrations that directly measure constituents in feces that indicate diet quality. In some animals, for example, fecal nitrogen concentrations increase with dietary protein intake [103,104]. Due to the low protein concentrations in many plants, protein can be a limiting nutrient for many herbivores. As a consequence, protein, or nitrogen as a proxy for protein, is often a key measure of diet quality [46,104,105]. However, for browsing herbivores and some grazers, total fecal nitrogen may not correlate with the protein that is available to the animal, since the tannins found in some herbivorous diets can bind to nitrogen and make it unavailable for digestion, inflating the amount of nitrogen excreted in scats [104,106]. In this instance, the amount of unbound nitrogen in scats may be a better indicator of diet quality [44]. In recognition of this, Windley et al. [44] were able to use NIRS to determine the concentration of in vitro digestible nitrogen in scats of female common brushtail possums (Trichosurus vulpecula) and showed that these measurements were correlated with their reproductive success. Digestible nitrogen has also been measured using NIRS of vegetation from airborne and satellite sensors in addition to laboratory spectrophotometers, opening the door to relatively rapid, landscape-scale assessments of forage quality [64,107].
The accuracy of NIRS models for estimating diet quality from scat can be affected by factors such as high levels of moisture in samples and contamination from other substances [97,108,109]. It may be necessary to dry fecal samples prior to diet quality analyses, and care should be taken to avoid contaminants [14,110]. Additional considerations for sample pre-treatment that may impact model performance, such as grinding to homogenize samples, are discussed in the next section. The variation in diet between sampled individuals is a potential issue with all NIRS models developed from scat and should be considered when testing the effectiveness of NIR models. Diet represents a considerable portion of the spectra of excrement, and diet can be chemically variable in itself. However, several studies have demonstrated that multispecies calibrations for fecal N can be developed [111,112], which may be particularly useful when trying to assess diet quality from rare species that forage and deposit scat in the same landscapes as more abundant grazers.
4. Considerations for the Application of NIRS to Wildlife Health, Population, and Diet Assessments
The following section discusses aspects of data collection, sample preparation, and data sharing that can impact the accuracy and applicability of NIRS models for assessing animal physiological traits.
4.1. Data Collection
A problem with the applicability of some NIRS models is that data collection takes place under a narrow set of conditions, often due to limitations in the capacity to sample widely, and thus cannot accurately predict the same trait in samples collected from another location or set of conditions. For example, Tolleson et al. [35] looked at sex and species in red and fallow deer, but all individuals in the study lived in a small region and consumed a monoculture of ryegrass (Lolium perenne) pastures. Wild deer consume a much larger range of foods, and thus the NIRS calibrations may not adequately predict traits in wild deer feeding on different forage in other locations. Similarly, calibrations developed from the bodily materials of wild animals in captive or otherwise unnatural conditions, may only be relevant for animals under those conditions [10].
Sample condition (e.g., age, ground particle size, drying temperatures, storage conditions, potential contamination, and other environmental factors) can also impact model development and performance [66]. Studies that seek to use samples of different ages should first determine how the aging process influences the capacity to determine the trait(s) of interest [113]. For example, it may be particularly important to collect fresh fecal samples when looking at stress levels, given the fact that cortisol metabolites may degrade in feces over time [114]. Even if aging does not result in the loss of the trait of interest, calibrations should incorporate samples of all ages to which the final models will be applied, because other external factors such as microbial activity may change over time and impact spectral signatures [46]. Although it is acceptable to conduct feasibility studies on smaller, non-representative samples, it is critical that the diversity of conditions under which the final NIRS model will be applied is considered during sample collection and model development, or the model will not be widely applicable [10,115].
The independence of the training samples on which a calibration is built is another important consideration. Pseudoreplication is a process whereby samples from the same individual are treated as independent data points. Models from these data can be very good at predicting traits from the individuals used for model development but may be limited in their ability to predict the same traits from other individuals [116]. A number of studies utilizing NIRS to determine traits from wildlife have had extensive pseudoreplication in their datasets [10,82]. For example, Wiedower et al. [10] collected 239 fecal samples over 2 years from 12 giant pandas. The resulting models were very good at determining traits in those 12 individuals but may have performed poorly in correctly determining traits in new individuals. The best way to overcome limitations that can be associated with pseudoreplication is to develop models with truly independent samples (e.g., different individuals rather than multiple samples from the same individual). In addition, it is important to consider the sources of variability that might occur in the population for which prediction is desired and attempt to account for that variability through the sample selection.
4.2. Pre-Treatment of Samples
Numerous factors besides the traits of interest can influence the NIR spectral profile of samples. For example, Fernandez-Cabanas et al. [117] found that particle size caused the greatest spectral variation in ground plant samples. The same is likely to be true for scat samples. Fine and homogeneous milling can reduce the effect of heterogenous particle size on light scattering detected by the NIRS sensor [117]. Grinding also can help to homogenize a sample so that the collected spectra is more representative of the whole.
Water interacts with light in the same areas on the spectrum that other compounds interact, which causes the information from some traits to be obscured [118,119]. A study on the moisture content of cattle manure found that feces containing high moisture levels generally reduced the calibration’s predictive capacity [109]. For these reasons, for some traits, drying samples before collecting spectral data may be desirable. However, drying may also degrade some chemicals or dissipate volatile compounds that could be important for determining other traits with NIRS. Freeze-drying is an alternative to oven drying that avoids the breakdown of some chemicals from heating [120]. In some situations, it might be necessary to use fresh samples because volatile compounds can be lost during freeze-drying as well, and this should be determined on a case-by-case basis.
4.3. Data Sharing and Building Widely Applicable Calibration Models
Collecting spatial data in the location where samples were collected is particularly useful for NIRS data sharing and research purposes. Moore et al. [49] used NIRS together with spatial data to map tree palatability for koalas in a 7.6 ha fenced reserve. This allowed a subsequent study to demonstrate relationships among foliage choice by koalas, tree nutritional quality, and tree location, providing novel information about koala dietary choices [29]. Data sharing could also aid in the development of more robust calibration models. If multiple labs collaborate to compile large spectral datasets and calibration models that could be continuously expanded, it would likely result in very robust and widely applicable models [31]. However, standardisation across equipment, sample collection, processing, and storage currently prevent this occurring on a global scale [31].
One of the main constraints on using data sharing to develop widely applicable models is the difficulty in cross-calibrating instruments. Machines—even spectrometers produced by the same manufacturer—often do not produce the exact sample spectrum from the same sample [121]. Cross-calibrating machines is theoretically possible but rarely performed in practice because most instrument manufacturers do not provide affordable or accessible pathways to achieve this. It may be possible to overcome this by developing calibrations with spectra collected on several instruments by different operators to build models that could account for variability across machines and users. Developing and sharing standard methods for sample collection, preparation, and storage would also help to reduce these sources of variability to improve calibration models developed across labs. Identifying ways to overcome limitations such as these is important to advance the application of NIRS for wildlife management and conservation.
5. Conclusions
Species traits such as sex, reproductive status, diet quality, stress levels, and disease status shape population management decisions, reintroduction plans, and conservation priorities. Many of these traits have been measured in a variety of animals using NIRS, which has numerous advantages over more traditional methods for assessing population demographics, health, and diet quality [10,26,33,35,36,40,82]. NIRS allows for the collection of data quickly, inexpensively, and often non-invasively, which makes this approach ideal for measuring animal traits, particularly in rare or cryptic species [14,31]. However, it is important that studies attempt to represent or otherwise account for variability in their populations, use independent validation datasets for testing model accuracy, and avoid pseudoreplication if models are intended to be applicable beyond the group used for model development. Sample pre-treatment can influence model performance, and it is necessary to determine the most appropriate methodology for specific traits and applications at the outset. Overcoming issues that can impede data sharing and collaborative calibration model development will help us to build more robust and widely applicable models.
Author Contributions
Conceptualisation, K.N.Y., K.J.M., D.R.T.; literature curation, L.R.M., K.N.Y., D.R.T.; writing—original draft preparation, L.R.M.; writing—review and editing, L.R.M., K.N.Y., K.J.M., D.R.T.; supervision, K.J.M., K.N.Y.; funding acquisition, K.N.Y., K.J.M. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Victorian Government Department of Environment, Land, Water, and Planning (grant 345963), Australian Research Council, (grant DE150101870), and The Minderoo Foundation, (grant TI_ANU001).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
We thank William Foley, Carrie Vance, David Lindenmayer and James Skewes for helpful comments on early versions of this manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Arnemo, J.M.; Ahlqvist, P.; Andersen, R.; Berntsen, F.; Ericsson, G.; Odden, J.; Brunberg, S.; Segerström, P.; Swenson, J.E. Risk of capture-related mortality in large free-ranging mammals: Experiences from Scandinavia. Wildl. Biol. 2006, 12, 109–113. [Google Scholar] [CrossRef]
- Beja-Pereira, A.; Oliveira, R.; Alves, P.C.; Schwartz, M.; Luikart, G. Advancing ecological understandings through technological transformations in noninvasive genetics. Mol. Ecol. Resour. 2009, 9, 1279–1301. [Google Scholar] [CrossRef]
- Eyvindson, K.; Hakanen, J.; Mönkkönen, M.; Juutinen, A.; Karvanen, J. Value of information in multiple criteria decision making: An application to forest conservation. Stoch. Environ. Res. Risk Assess. 2019, 33, 2007–2018. [Google Scholar] [CrossRef]
- McMahon, C.R.; Hindell, M.; Harcourt, R. Publish or perish: Why it’s important to publicise how, and if, research activities affect animals. Wildl. Res. 2012, 39, 375–377. [Google Scholar] [CrossRef]
- Alacs, E.; Alpers, D.; De Tores, P.J.; Dillon, M.; Spencer, P.B.S. Identifying the presence of quokkas (Setonix brachyurus) and other macropods using cytochrome b analyses from faeces. Wildl. Res. 2003, 30, 41–47. [Google Scholar] [CrossRef]
- Taberlet, P.; Waits, L.P.; Luikart, G. Noninvasive genetic sampling: Look before you leap. Trends Ecol. Evol. 1999, 14, 323–327. [Google Scholar] [CrossRef]
- Kohn, M.H.; Wayne, R.K. Facts from feces revisited. Trends Ecol. Evol. 1997, 12, 223–227. [Google Scholar] [CrossRef]
- Li, C.; Jiang, Z.; Jiang, G.; Fang, J. Seasonal Changes of Reproductive Behavior and Fecal Steroid Concentrations in Père David’s Deer. Horm. Behav. 2001, 40, 518–525. [Google Scholar] [CrossRef] [PubMed]
- Goymann, W. Noninvasive Monitoring of Hormones in Bird Droppings: Physiological Validation, Sampling, Extraction, Sex Differences, and the Influence of Diet on Hormone Metabolite Levels. Ann. N. Y. Acad. Sci. 2005, 1046, 35–53. [Google Scholar] [CrossRef]
- Wiedower, E.E.; Kouba, A.J.; Vance, C.K.; Hansen, R.L.; Stuth, J.W.; Tolleson, D.R. Fecal Near Infrared Spectroscopy to Discriminate Physiological Status in Giant Pandas. PLoS ONE 2012, 7, e38908. [Google Scholar] [CrossRef] [PubMed]
- Pilotte, N.; Zaky, W.I.; Abrams, B.P.; Chadee, D.D.; Williams, S. A Novel Xenomonitoring Technique Using Mosquito Excreta/Feces for the Detection of Filarial Parasites and Malaria. PLOS Negl. Trop. Dis. 2016, 10, e0004641. [Google Scholar] [CrossRef]
- Jean, P.-O.; Bradley, R.L.; Giroux, M.-A.; Tremblay, J.-P.; Côté, S.D. Near Infrared Spectroscopy and Fecal Chemistry as Predictors of the Diet Composition of White-Tailed Deer. Rangel. Ecol. Manag. 2014, 67, 154–159. [Google Scholar] [CrossRef]
- Valentini, A.; Miquel, C.; Nawaz, M.A.; Bellemain, E.; Coissac, E.; Pompanon, F.; Gielly, L.; Cruaud, C.; Nascetti, G.; Wincker, P.; et al. New perspectives in diet analysis based on DNA barcoding and parallel pyrosequencing: ThetrnL approach. Mol. Ecol. Resour. 2009, 9, 51–60. [Google Scholar] [CrossRef]
- Foley, W.J.; McIlwee, A.; Lawler, I.; Aragones, L.; Woolnough, A.P.; Berding, N. Ecological applications of near infrared reflectance spectroscopy-a tool for rapid, cost-effective prediction of the composition of plant and animal tissues and aspects of animal performance. Oecologia 1998, 116, 293–305. [Google Scholar] [CrossRef]
- Bailey, D.W.; Thomas, M.G.; Walker, J.W.; Witmore, B.K.; Tolleson, D. Effect of Previous Experience on Grazing Patterns and Diet Selection of Brangus Cows in the Chihuahuan Desert. Rangel. Ecol. Manag. 2010, 63, 223–232. [Google Scholar] [CrossRef]
- Dixon, R.; Coates, D.; Jackson, D. Utilizing faecal near infrared spectroscopy to improve nutritional management of grazing cattle in the tropics of northern Australia. Adv. Anim. Biosci. 2010, 1, 432–433. [Google Scholar] [CrossRef]
- Landau, S.; Dvash, L.; Roudman, M.; Muklada, H.; Barkai, D.; Yehuda, Y.; Ungar, E. Faecal near-IR spectroscopy to determine the nutritional value of diets consumed by beef cattle in east Mediterranean rangelands. Animal 2016, 10, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Jancewicz, L.J.; Penner, G.; Swift, M.L.; Waldner, C.; Gibb, D.; McAllister, T.A. Predictability of growth performance in feedlot cattle using fecal near infrared spectroscopy. Can. J. Anim. Sci. 2017, 97, 701–720. [Google Scholar] [CrossRef]
- Johnson, J.R.; Carstens, G.E.; Prince, S.D.; Ominski, K.H.; Wittenberg, K.M.; Undi, M.; Forbes, T.A.; Hafla, A.N.; Tolleson, D.R.; Basarab, J.A. Application of fecal near-infrared reflectance spectroscopy profiling for the prediction of diet nutritional characteristics and voluntary intake in beef cattle. J. Anim. Sci. 2017, 95, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Hu, W.; Zhou, Y.; Zhang, X.; Xu, S.; Guo, Q.; Qi, P.; Chen, L.; Yang, X.; Zhang, F.; et al. Use of near-infrared spectroscopy for the rapid evaluation of soybean [Glycine max (L.) Merri.] water soluble protein content. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 224, 117400. [Google Scholar] [CrossRef]
- Tsenkova, R.; Atanassova, S.; Toyoda, K.; Ozaki, Y.; Itoh, K.; Fearn, T. Near-Infrared Spectroscopy for Dairy Management: Measurement of Unhomogenized Milk Composition. J. Dairy Sci. 1999, 82, 2344–2351. [Google Scholar] [CrossRef]
- Engelhard, S.; Löhmannsröben, H.-G.; Schael, F. Quantifying Ethanol Content of Beer Using Interpretive Near-Infrared Spectroscopy. Appl. Spectrosc. 2004, 58, 1205–1209. [Google Scholar] [CrossRef] [PubMed]
- Correia, R.M.; Domingos, E.; Tosato, F.; dos Santos, N.A.; Leite, J.D.A.; da Silva, M.; Marcelo, M.C.A.; Ortiz, R.S.; Filgueiras, P.; Romão, W. Portable near infrared spectroscopy applied to abuse drugs and medicine analyses. Anal. Methods 2018, 10, 593–603. [Google Scholar] [CrossRef]
- Leite, E.; Stuth, J. Fecal NIRS equations to assess diet quality of free-ranging goats. Small Rumin. Res. 1995, 15, 223–230. [Google Scholar] [CrossRef]
- Landau, S.; Nitzan, R.; Barkai, D.; Dvash, L. Excretal Near Infrared Reflectance Spectrometry to Monitor the Nutrient Content of Diets of Grazing Young Ostriches (Struthio Camelus). S. Afr. J. Anim. Sci. 2006, 36, 248–256. [Google Scholar]
- Showers, S.E.; Tolleson, D.R.; Stuth, J.W.; Kroll, J.C.; Koerth, B.H. Predicting Diet Quality of White-Tailed Deer via NIRS Fecal Profiling. Rangel. Ecol. Manag. 2006, 59, 300–307. [Google Scholar] [CrossRef]
- Glasser, T.; Landau, S.; Ungar, E.D.; Perevolotsky, A.; Dvash, L.; Muklada, H.; Kababya, D.; Walker, J.W. A fecal near-infrared reflectance spectroscopy-aided methodology to determine goat dietary composition in a Mediterranean shrubland1. J. Anim. Sci. 2008, 86, 1345–1356. [Google Scholar] [CrossRef]
- Rothman, J.M.; Chapman, C.A.; Hansen, J.L.; Cherney, D.J.R.; Pell, A.N. Rapid Assessment of the Nutritional Value of Foods Eaten by Mountain Gorillas: Applying Near-Infrared Reflectance Spectroscopy to Primatology. Int. J. Primatol. 2009, 30, 729–742. [Google Scholar] [CrossRef]
- Marsh, K.J.; Moore, B.D.; Wallis, I.R.; Foley, W.J. Feeding rates of a mammalian browser confirm the predictions of a ‘foodscape’ model of its habitat. Oecologia 2013, 174, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Au, J.; Clark, R.G.; Allen, C.; Marsh, K.; Foley, W.J.; Youngentob, K.N. A nutritional mechanism underpinning folivore occurrence in disturbed forests. For. Ecol. Manag. 2019, 453, 117585. [Google Scholar] [CrossRef]
- Vance, C.K.; Tolleson, D.R.; Kinoshita, K.; Rodriguez, J.; Foley, W.J. Near Infrared Spectroscopy in Wildlife and Biodiversity. J. Near Infrared Spectrosc. 2016, 24, 1–25. [Google Scholar] [CrossRef]
- Esperança, P.M.; Blagborough, A.M.; Da, D.F.; Dowell, F.E.; Churcher, T.S. Detection of Plasmodium berghei infected Anopheles stephensi using near-infrared spectroscopy. Parasites Vectors 2018, 11, 377. [Google Scholar] [CrossRef]
- Tolleson, D.; Teel, P.; Stuth, J.; Strey, O.; Welsh, T.; Carstens, G. Fecal NIRS: Detection of tick infestations in cattle and horses. Vet. Parasitol. 2007, 144, 146–152. [Google Scholar] [CrossRef]
- Tolleson, D.R. Determination of Sex in Ungulate Herbivores via near Infrared Spectroscopy of Hair: Growing Cattle as a Surrogate Model. J. Anim. Sci. 2021, 99, 23–24. [Google Scholar] [CrossRef]
- Tolleson, D.; Randel, R.; Stuth, J.; Neuendorff, D. Determination of sex and species in red and fallow deer by near infrared reflectance spectroscopy of the faeces. Small Rumin. Res. 2005, 57, 141–150. [Google Scholar] [CrossRef]
- Perez-Mendoza, J.; Dowell, F.E.; Broce, A.B.; Throne, J.E.; Wirtz, R.A.; Xie, F.; Fabrick, J.A.; Baker, J.E. Chronological age-grading of house flies by using near-infrared spectroscopy. J. Med. Èntomol. 2002, 39, 499–508. [Google Scholar] [CrossRef][Green Version]
- Tolleson, D.R.; Schafer, D.W. Detection of Pregnancy in Arizona Range Cattle Using near Infrared Spectroscopy of Feces. J. Anim. Sci. 2012, 90, 3442–3450. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, K.; Morita, H.; Miyazaki, M.; Hama, N.; Kanemitsu, H.; Kawakami, H.; Wang, P.; Ishikawa, O.; Kusunoki, H.; Tsenkova, R. Near infrared spectroscopy of urine proves useful for estimating ovulation in giant panda (Ailuropoda melanoleuca). Anal. Methods 2010, 2, 1671–1675. [Google Scholar] [CrossRef]
- Kinoshita, K.; Kuze, N.; Kobayashi, T.; Miyakawa, E.; Narita, H.; Inoue-Murayama, M.; Idani, G.; Tsenkova, R. Detection of urinary estrogen conjugates and creatinine using near infrared spectroscopy in Bornean orangutans (Pongo pygmaeus). Primates 2015, 57, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Tallo-Parra, O.; Albanell, E.; Carbajal, A.; Monclús, L.; Sabes-Alsina, M.; Riba, C.; Martin, M.; Abelló, M.; Lopez-Bejar, M. Prediction of Faecal Cortisol Metabolites from Western Lowland Gorilla (Gorilla Gorilla Gorilla) by near Infrared Reflectance Spectroscopy (NIRS). In Proceedings of the 5th ISWE CONFERENCE, Berlin, Germany, 12–14 October 2015; p. 61. [Google Scholar]
- Tallo-Parra, O.; Albanell, E.; Carbajal, A.; Monclús, L.; Manteca, X.; Lopez-Bejar, M. Prediction of Cortisol and Progesterone Concentrations in Cow Hair Using Near-Infrared Reflectance Spectroscopy (NIRS). Appl. Spectrosc. 2017, 71, 1954–1961. [Google Scholar] [CrossRef] [PubMed]
- Ancin-Murguzur, F.J.; Tarroux, A.; Bråthen, K.A.; Bustamante, P.; Descamps, S. Using near-infrared reflectance spectroscopy (NIRS) to estimate carbon and nitrogen stable isotope composition in animal tissues. Ecol. Evol. 2021, 11, 10483–10488. [Google Scholar] [CrossRef]
- Peiretti, P.G.; Meineri, G.; Masoero, G. NIRS of body and tissues in growing rabbits fed diets with different fat sources and supplemented with Curcuma longa. World Rabbit Sci. 2013, 21, 85–90. [Google Scholar] [CrossRef][Green Version]
- Windley, H.R.; Wallis, I.R.; DeGabriel, J.L.; Moore, B.D.; Johnson, C.N.; Foley, W.J. A faecal index of diet quality that predicts reproductive success in a marsupial folivore. Oecologia 2013, 173, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Pasquini, C. Near Infrared Spectroscopy: Fundamentals, practical aspects and analytical applications. J. Braz. Chem. Soc. 2003, 14, 198–219. [Google Scholar] [CrossRef]
- Dixon, R.; Coates, D. Review: Near Infrared Spectroscopy of Faeces to Evaluate the Nutrition and Physiology of Herbivores. J. Near Infrared Spectrosc. 2009, 17, 1–31. [Google Scholar] [CrossRef]
- Kokaly, R. Spectroscopic Determination of Leaf Biochemistry Using Band-Depth Analysis of Absorption Features and Stepwise Multiple Linear Regression. Remote Sens. Environ. 1999, 67, 267–287. [Google Scholar] [CrossRef]
- Shenk, J.S.; Workman, J.J., Jr.; Westerhaus, M.O. Application of NIR Spectroscopy to Agricultural Products. In Handbook of Near-Infrared Analysis; CRC Press: Boca Raton, FL, USA, 2020; pp. 365–404. [Google Scholar]
- Moore, B.D.; Lawler, I.R.; Wallis, I.R.; Beale, C.M.; Foley, W. Palatability mapping: A koala’s eye view of spatial variation in habitat quality. Ecology 2010, 91, 3165–3176. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Sun, D.-W. Advanced applications of hyperspectral imaging technology for food quality and safety analysis and assessment: A review—Part I: Fundamentals. Innov. Food Sci. Emerg. Technol. 2013, 19, 1–14. [Google Scholar] [CrossRef]
- Tolleson, D.R.; Teel, P.D.; Carstens, G.E.; Welsh, T.H. The physiology of tick-induced stress in grazing animals. In Ticks: Disease, Management and Control; Nova Science Publishers, Inc: Hauppauge, NY, USA, 2012; pp. 1–18. ISBN 9781620811368. [Google Scholar]
- Ziegel, E.R. Chemometrics: Statistics and Computer Application in Analytical Chemistry. Technometrics 2001, 43, 240. [Google Scholar] [CrossRef]
- Næs, T.; Isaksson, T.; Fearn, T.; Davies, T. A User-Friendly Guide to Multivariate Calibration and Classification; IM Publications Open LLP: Chichester, UK, 2017; Volume 6. [Google Scholar]
- Wold, H. Soft Modelling by Latent Variables: The Non-Linear Iterative Partial Least Squares (NIPALS) Approach. J. Appl. Probab. 1975, 12, 117–142. [Google Scholar] [CrossRef]
- Shenk, J.S.; Westerhaus, M.O. Populations Structuring of Near Infrared Spectra and Modified Partial Least Squares Regression. Crop. Sci. 1991, 31, 1548–1555. [Google Scholar] [CrossRef]
- Lawler, I.R.; Aragones, L.; Berding, N.; Marsh, H.; Foley, W. Near-Infrared Reflectance Spectroscopy is a Rapid, Cost-Effective Predictor of Seagrass Nutrients. J. Chem. Ecol. 2006, 32, 1353–1365. [Google Scholar] [CrossRef]
- Pérez-Marín, D.; Garrido-Varo, A.; Guerrero, J.E.; Gutiérrez-Estrada, J.C. Use of Artificial Neural Networks in Near-Infrared Reflectance Spectroscopy Calibrations for Predicting the Inclusion Percentages of Wheat and Sunflower Meal in Compound Feedingstuffs. Appl. Spectrosc. 2006, 60, 1062–1069. [Google Scholar] [CrossRef]
- Bro, R.; Smilde, A.K. Centering and scaling in component analysis. J. Chemom. 2003, 17, 16–33. [Google Scholar] [CrossRef]
- Geladi, P.; MacDougall, D.; Martens, H. Linearization and Scatter-Correction for Near-Infrared Reflectance Spectra of Meat. Appl. Spectrosc. 1985, 39, 491–500. [Google Scholar] [CrossRef]
- Barnes, R.J.; Dhanoa, M.S.; Lister, S.J. Standard Normal Variate Transformation and De-Trending of Near-Infrared Diffuse Reflectance Spectra. Appl. Spectrosc. 1989, 43, 772–777. [Google Scholar] [CrossRef]
- Savitzky, A.; Golay, M.J.E. Smoothing and Differentiation of Data by Simplified Least Squares Procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Au, J.; Youngentob, K.N.; Foley, W.J.; Moore, B.; Fearn, T. Sample selection, calibration and validation of models developed from a large dataset of near infrared spectra of tree leaves. J. Near Infrared Spectrosc. 2020, 28, 186–203. [Google Scholar] [CrossRef]
- Elisseeff, A.; Pontil, M. Leave-one-out error and stability of learning algorithms with applications. In Advances in Learning Theory: Methods, Models, and Applications; Suykens, J.A.K., Ed.; IOS Press: Leuven, Belgium, 2002; p. 415. [Google Scholar]
- Youngentob, K.N.; Renzullo, L.J.; Held, A.A.; Jia, X.; Lindenmayer, D.B.; Foley, W. Using imaging spectroscopy to estimate integrated measures of foliage nutritional quality. Methods Ecol. Evol. 2011, 3, 416–426. [Google Scholar] [CrossRef]
- Anderson, K.; Norby, B.; Prince, S.; Ball, G.; Tolleson, D. Detection of Paratuberculosis in Dairy Cattle via near Infrared Reflectance Spectroscopy of Feces. J. Anim. Sci. 2007, 85, 42. [Google Scholar]
- Vance, C.K.; Kouba, A.J.; Zhang, H.-X.; Zhao, H.; Wang, Q.; Willard, S.T. Near Infrared Reflectance Spectroscopy Studies of Chinese Giant Salamanders in Aquaculture Production. NIR News 2015, 26, 4–7. [Google Scholar] [CrossRef]
- Hing, S.; Narayan, E.; Thompson, R.C.A.; Godfrey, S. The relationship between physiological stress and wildlife disease: Consequences for health and conservation. Wildl. Res. 2016, 43, 51–60. [Google Scholar] [CrossRef]
- Scott, M.E. The Impact of Infection and Disease on Animal Populations: Implications for Conservation Biology. Conserv. Biol. 1988, 2, 40–56. [Google Scholar] [CrossRef]
- Andrew, A.; Edward, D.; Solomon, A.; Isaac, A.; Ibrahim, Y. The Cortisol Steroid Levels as a Determinant of Health Status in Animals. J. Proteomics Bioinform. 2017, 10, 277–283. [Google Scholar] [CrossRef]
- Hanger, J.; Loader, J.; Wan, C.; Beagley, K.W.; Timms, P.; Polkinghorne, A. Comparison of antigen detection and quantitative PCR in the detection of chlamydial infection in koalas (Phascolarctos cinereus). Vet. J. 2013, 195, 391–393. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bennett, A.; Hayssen, V. Measuring cortisol in hair and saliva from dogs: Coat color and pigment differences. Domest. Anim. Endocrinol. 2010, 39, 171–180. [Google Scholar] [CrossRef]
- Sachsenröder, J.; Twardziok, S.; Hammerl, J.A.; Janczyk, P.; Wrede, P.; Hertwig, S.; Johne, R. Simultaneous Identification of DNA and RNA Viruses Present in Pig Faeces Using Process-Controlled Deep Sequencing. PLoS ONE 2012, 7, e34631. [Google Scholar] [CrossRef]
- Yamazaki, W.; Makino, R.; Nagao, K.; Mekata, H.; Tsukamoto, K. New Micro-amount of Virion Enrichment Technique (Mi VET ) to detect influenza A virus in the duck faeces. Transbound. Emerg. Dis. 2019, 66, 341–348. [Google Scholar] [CrossRef]
- Norby, B.; Tolleson, D.; Ball, G.; Jordan, E.; Stuth, J. Near Infrared Spectroscopy: A New Approach to Diagnosis of Paratuberculosis in Cattle. In Proceedings of the International Epidemiology Conference, Cairns, Australia; 2006; pp. 2003–2005. Available online: http://www.sciquest.org.nz/node/64094 (accessed on 8 September 2021).
- Tolleson, D.R.; Schafer, D.W. Evaluation of non-invasive bioforensic techniques for determining the age of hot-iron brand burn scars in cattle. Transl. Anim. Sci. 2021, 5, txab108. [Google Scholar] [CrossRef]
- Masoero, G.; Sala, G.; Meineri, G.; Cornale, P.; Tassone, S.; Peiretti, P.G. Nir Spectroscopy and Electronic Nose Evaluation on Live Rabbits and on the Meat of Rabbits Fed Increasing Levels of Chia (Salvia hispanica L.) Seeds. J. Anim. Vet. Adv. 2008, 7, 1394–1399. [Google Scholar]
- Tolleson, D.R.; Hollingsworth, K.; Barboza, P.S. Determination of Species and Sex in Deer via near Infrared Spectroscopy of Liver Tissue. In Proceedings of the Society for Range Management Annual Meetings, Denver, CO, USA, 20 January 2020. [Google Scholar]
- Tolleson, D.R.; Garza, N.; Hollingsworth, K.; Diaz, J.M.; Welsh, T.H. 157 Spectroscopic analysis of tissues collected from male goats differing in genetic propensity to consume juniper. J. Anim. Sci. 2020, 98, 119–120. [Google Scholar] [CrossRef]
- Zhang, L.; Xiao, M.; Wang, Y.; Peng, S.; Chen, Y.; Zhang, D.; Zhang, D.; Guo, Y.; Wang, X.; Luo, H.; et al. Fast Screening and Primary Diagnosis of COVID-19 by ATR–FT-IR. Anal. Chem. 2021, 93, 2191–2199. [Google Scholar] [CrossRef] [PubMed]
- Wood, B.R.; Kochan, K.; Bedolla, D.E.; Salazar-Quiroz, N.; Grimley, S.L.; Perez-Guaita, D.; Baker, M.J.; Vongsvivut, J.; Tobin, M.J.; Bambery, K.R.; et al. Infrared Based Saliva Screening Test for COVID-19. Angew. Chem. 2021, 133, 17239–17244. [Google Scholar] [CrossRef]
- Kimura, M. Introduction to Population Genetics. Evol. Hum. Genome I 2020, 3, 85–101. [Google Scholar] [CrossRef]
- Johnson-Ulrich, L.; Vance, C.K.; Kouga, A.J.; Willard, S.T. Fecal Near Infrared Reflectance FNIR Spectroscopy for Discrimination of Species and Gender for Amur Leopards and Snow Leopards. NIR-2013 Proc. 2013, 1438, 495–505. [Google Scholar]
- Passerotti, M.S.; E Helser, T.; Benson, I.M.; Barnett, B.K.; Ballenger, J.C.; Bubley, W.J.; Reichert, M.J.M.; Quattro, J.M. Age estimation of red snapper (Lutjanus campechanus) using FT-NIR spectroscopy: Feasibility of application to production ageing for management. ICES J. Mar. Sci. 2020, 77, 2144–2156. [Google Scholar] [CrossRef]
- Rigby, C.L.; Wedding, B.B.; Grauf, S.; Simpfendorfer, C.A. Novel method for shark age estimation using near infrared spectroscopy. Mar. Freshw. Res. 2016, 67, 537. [Google Scholar] [CrossRef]
- Aw, W.C.; E Dowell, F.; O Ballard, J.W. Using Near-Infrared Spectroscopy to Resolve the Species, Gender, Age, and the Presence of Wolbachia Infection in Laboratory-Reared Drosophila. G3 Genes Genomes Genet. 2012, 2, 1057–1065. [Google Scholar] [CrossRef] [PubMed]
- Lambert, B.; Sikulu-Lord, M.T.; Mayagaya, V.S.; Devine, G.; Dowell, F.; Churcher, T.S. Monitoring the Age of Mosquito Populations Using Near-Infrared Spectroscopy. Sci. Rep. 2018, 8, 5274. [Google Scholar] [CrossRef]
- Dowell, F.; Parker, A.; Benedict, M.; Robinson, A.; Broce, A.; Wirtz, R. Sex separation of tsetse fly pupae using near-infrared spectroscopy. Bull. Èntomol. Res. 2005, 95, 249–257. [Google Scholar] [CrossRef]
- Peberdy, J.F.; Moore, P.M. Chitin Synthase in Mortierella vinacea: Properties, Cellular Location and Synthesis in Growing Cultures. J. Gen. Microbiol. 1975, 90, 228–236. [Google Scholar] [CrossRef][Green Version]
- Tolleson, D.; Willard, S.; Gandy, B.; Stuth, J. Determination of Reproductive Status in Dairy Cattle Using near Infrared Reflectance Spectroscopy of Feces. J. Anim. Sci. 2001, 79, 21. [Google Scholar]
- Johnson, J. Near-infrared spectroscopy (NIRS) for taxonomic entomology: A brief review. J. Appl. Èntomol. 2020, 144, 241–250. [Google Scholar] [CrossRef]
- Xiccato, G.; Trocino, A.; Tulli, F.; Tibaldi, E. Prediction of chemical composition and origin identification of european sea bass (Dicentrarchus labrax L.) by near infrared reflectance spectroscopy (NIRS). Food Chem. 2004, 86, 275–281. [Google Scholar] [CrossRef]
- Benson, I.M.; Barnett, B.K. In Proceedings of the Research Workshop on the Rapid Estimation of Fish Age Using Fourier Transform Near-Infrared Spec-Troscopy (FT-NIRS) Age and Growth Program, September 2019. Available online: https://www.fisheries.noaa.gov/alaska/science-data/alaska-fisheries-science-center-publications (accessed on 12 September 2021).
- Wedding, B.B.; Forrest, A.J.; Wright, C.; Grauf, S.; Exley, P.; Poole, S.E. A novel method for the age estimation of Saddletail snapper (Lutjanus malabaricus) using Fourier Transform-near infrared (FT-NIR) spectroscopy. Mar. Freshw. Res. 2014, 65, 894–900. [Google Scholar] [CrossRef]
- Robins, J.; Wedding, B.B.; Wright, C.; Grauf, S.; Sellin, M.; Fowler, A.; Saunders, T.; Newman, S. Fisheries Research & Development Corporation (Australia); Queensland. Department of Agriculture and Fisheries. In Revolutionising Fish Ageing: Using Near Infrared Spectroscopy to Age Fish; State of Queensland through Department of Agriculture and Fisheries: Queensland, Australia, 2015; ISBN 9780734504494. [Google Scholar]
- DeGabriel, J.L.; Moore, B.; Foley, W.; Johnson, C. The effects of plant defensive chemistry on nutrient availability predict reproductive success in a mammal. Ecology 2009, 90, 711–719. [Google Scholar] [CrossRef] [PubMed]
- McArt, S.H.; Spalinger, D.E.; Collins, W.B.; Schoen, E.R.; Stevenson, T.; Bucho, M. Summer dietary nitrogen availability as a potential bottom-up constraint on moose in south-central Alaska. Ecology 2009, 90, 1400–1411. [Google Scholar] [CrossRef] [PubMed]
- Lyons, R.K.; Stuth, J.W. Fecal NIRS Equations for Predicting Diet Quality of Free-Ranging Cattle. J. Range Manag. 1992, 45, 238. [Google Scholar] [CrossRef]
- Landau, S.; Glasser, T.; Muklada, H.; Dvash, L.; Perevolotsky, A.; Ungar, E.; Walker, J. Fecal NIRS prediction of dietary protein percentage and in vitro dry matter digestibility in diets ingested by goats in Mediterranean scrubland. Small Rumin. Res. 2005, 59, 251–263. [Google Scholar] [CrossRef]
- Tolleson, D.; Osborn, R.G.; Stuth, J.W.; Ginnet, T.F.; Applegath, M.T. Determination of Dietary Tannin Concentration in White-Tailed Deer via Near Infrared Reflectance Spectroscopy of Feces. In Proceedings of the National Conference on Grazinglands, Las Vegas, NV, USA, 5–8 December 2000; pp. 727–733. [Google Scholar]
- Landau, S.; Glasser, T.; Dvash, L.; Perevolotsky, A. Faecal NIRS to Monitor the Diet of Mediterranean Goats. S. Afr. J. Anim. Sci. 2004, 34, 76–80. [Google Scholar]
- Li, H.; Tolleson, D.; Stuth, J.; Bai, K.; Mo, F.; Kronberg, S. Faecal near infrared reflectance spectroscopy to predict diet quality for sheep. Small Rumin. Res. 2007, 68, 263–268. [Google Scholar] [CrossRef]
- Kidane, N.F.; Stuth, J.W.; Tolleson, D.R. Predicting Diet Quality of Donkeys via Fecal-NIRS Calibrations. Rangel. Ecol. Manag. 2008, 61, 232–239. [Google Scholar] [CrossRef]
- Kamler, J.; Homolka, M. Faecal Nitrogen: A Potential Indicator of Red and Roe Deer Diet Quality in Forest Habitats. Folia Zool. 2005, 54, 89–98. [Google Scholar]
- Leslie, D.M.; Bowyer, R.T.; Jenks, J.A. Facts From Feces: Nitrogen Still Measures Up as a Nutritional Index for Mammalian Herbivores. J. Wildl. Manag. 2008, 72, 1420–1433. [Google Scholar] [CrossRef]
- Dhiman, T.; Satter, L. Protein as the First-Limiting Nutrient for Lactating Dairy Cows Fed High Proportions of Good Quality Alfalfa Silage. J. Dairy Sci. 1993, 76, 1960–1971. [Google Scholar] [CrossRef]
- DeGabriel, J.L.; Wallis, I.R.; Moore, B.D.; Foley, W.J. A simple, integrative assay to quantify nutritional quality of browses for herbivores. Oecologia 2008, 156, 107–116. [Google Scholar] [CrossRef]
- Wu, H.; Levin, N.; Seabrook, L.; Moore, B.D.; McAlpine, C. Mapping Foliar Nutrition Using WorldView-3 and WorldView-2 to Assess Koala Habitat Suitability. Remote Sens. 2019, 11, 215. [Google Scholar] [CrossRef]
- Yang, Z.; Han, L.; Zhu, R. Near-Infrared Sensing of Pig Manure Nutrients. In Livestock Environment VII, 18–20 May 2005, Beijing, China; American Society of Agricultural and Biological Engineers: St. Joseph, MI, USA, 2013; Volume 43, pp. 309–315. [Google Scholar] [CrossRef]
- Reeves, J.; Van Kessel, J. Determination of Ammonium-N, Moisture, Total C and Total N in Dairy Manures Using a near Infrared Fibre-Optic Spectrometer. J. Near Infrared Spectrosc. 2000, 8, 151–160. [Google Scholar] [CrossRef]
- Reeves, J.B.; Van Kessel, J.A.S.; Reeves, I.J. Spectroscopic Analysis of Dried Manures. Near- versus Mid-Infrared Diffuse Reflectance Spectroscopy for the Analysis of Dried Dairy Manures. J. Near Infrared Spectrosc. 2002, 10, 93–101. [Google Scholar] [CrossRef]
- Villamuelas, M.; Serrano, E.; Espunyes, J.; Fernández, N.; López-Olvera, J.R.; Garel, M.; e Santos, J.P.V.; Parra-Aguado, M. Ángeles; Ramanzin, M.; Aguilar, X.F.; et al. Predicting herbivore faecal nitrogen using a multispecies near-infrared reflectance spectroscopy calibration. PLoS ONE 2017, 12, e0176635. [Google Scholar] [CrossRef] [PubMed]
- Tolleson, D.R.; Angerer, J.P. The application of near infrared spectroscopy to predict faecal nitrogen and phosphorus in multiple ruminant herbivore species. Rangel. J. 2020, 42, 415–423. [Google Scholar] [CrossRef]
- Leite, E.R.; Stuth, J.W. Influence of Duration of Exposure to Field Conditions on Viability of Fecal Samples for NIRS Analysis. J. Range Manag. 1994, 47, 312. [Google Scholar] [CrossRef]
- Hadinger, U.; Haymerle, A.; Knauer, F.; Schwarzenberger, F.; Walzer, C. Faecal cortisol metabolites to assess stress in wildlife: Evaluation of a field method in free-ranging chamois. Methods Ecol. Evol. 2015, 6, 1349–1357. [Google Scholar] [CrossRef]
- Kaneko, H.; Lawler, I.R. Can Near Infrared Spectroscopy be Used to Improve Assessment of Marine Mammal Diets via Fecal Analysis? Mar. Mammal Sci. 2006, 22, 261–275. [Google Scholar] [CrossRef]
- Corlatti, L. Anonymous fecal sampling and NIRS studies of diet quality: Problem or opportunity? Ecol. Evol. 2020, 10, 6089–6096. [Google Scholar] [CrossRef]
- Fernández-Cabanás, V.M.; Garrido-Varo, A.; Pérez-Marín, D.; Dardenne, P. Evaluation of Pretreatment Strategies for Near-Infrared Spectroscopy Calibration Development of Unground and Ground Compound Feedingstuffs. Appl. Spectrosc. 2006, 60, 17–23. [Google Scholar] [CrossRef]
- Giordanengo, T.; Charpentier, J.-P.; Roger, J.-M.; Roussel, S.; Brancheriau, L.; Chaix, G.; Baillères, H. Correction of moisture effects on near infrared calibration for the analysis of phenol content in eucalyptus wood extracts. Ann. For. Sci. 2008, 65, 803. [Google Scholar] [CrossRef][Green Version]
- Ji, W.; Rossel, R.A.V.; Shi, Z. Accounting for the effects of water and the environment on proximally sensed vis-NIR soil spectra and their calibrations. Eur. J. Soil Sci. 2015, 66, 555–565. [Google Scholar] [CrossRef]
- Bastianelli, D.; Bonnal, L.; Jaguelin-Peyraud, Y.; Noblet, J. Predicting feed digestibility from NIRS analysis of pig faeces. Animal 2015, 9, 781–786. [Google Scholar] [CrossRef]
- McArthur, L.; Greensill, C. Comparison of two NIR systems for quantifying kaolinite in Weipa bauxites. Meas. Sci. Technol. 2007, 18, 3463–3470. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).