Abstract
Insects offer a promising alternative source of protein to mitigate the environmental consequences of conventional livestock farming. Larvae of the black soldier fly (Hermetia illucens; Linnaeus, 1758) efficiently convert a variety of organic side streams and residues into valuable proteins, lipids, and chitin. Here, we evaluated the suitability of two palm oil industry side streams—empty fruit bunches (EFB) and palm kernel meal (PKM)—as larval feed, and their impact on the larval gut microbiome. Among 69 fungal species we screened, Marasmius palmivorus, Irpex consors, and Bjerkandera adusta achieved the fastest growth and lignin degradation, so these fungi were used for the pretreatment of 7:3 mixtures of EFB and PKM. Larvae reared on the mixture pretreated with B. adusta (BAD) developed significantly more quickly and reached a higher final weight than those reared on the other pretreatments or the non-fermented reference (NFR). Amplicon sequencing of the BAD and NFR groups revealed major differences in the larval gut microbiome. The NFR group was dominated by facultatively anaerobic Enterobacteriaceae (typical of H. illucens larvae) whereas the BAD group favored obligately anaerobic, cellulolytic bacteria (Ruminococcaceae and Lachnospiraceae). We hypothesize that fungal lignin degradation led to an accumulation of mycelia and subsequent cellulolytic breakdown of fiber residues, thus improving substrate digestibility.
1. Introduction
The larvae of the black soldier fly, Hermetia illucens (BSF; Linnaeus, 1758; Diptera: Stratiomyidae), are polyphagous with a remarkably efficient feed conversion ratio, making them suitable for the bioconversion of various animal and vegetable organic substrates into valuable insect proteins and lipids [1,2,3]. BSF larvae can even utilize substrates with a high microbial load, such as livestock manure, reducing the total dry matter and nitrogen content by more than 50% [2,4]. BSF has a short lifecycle of 40–45 d, and can be farmed in space-saving vertically stacked facilities with low technological requirements. BSF larvae therefore offer a promising and sustainable alternative for soy protein in the feed sector, helping to counter the slash and burn clearance of tropical forests. Since 2017, the European Union has also authorized the use of seven insect species including BSF for aquaculture feeding [5].
The versatility of BSF larvae is dependent on the gut microbiome [6,7]. For example, the degradation of complex plant polymers, such as cellulose and lignin, would not be possible without associated microbes. The structure of bacterial and fungal communities in the BSF larval gut is strongly affected by the feed substrate and its properties, but also by extrinsic factors, such as rearing location, scale, or temperature [8,9,10]. In this context, the diet-dependent expression of antimicrobial peptides—defense molecules of the insect innate immune system—plays a key role in the regulation of gut microbial composition [11] although there is also an omnipresent core community [7,8,12,13]. The feeding status (especially periods of starvation) also influence the microbiome and its associated metabolic functions, thus potentially disrupting larval growth and feed conversion efficiency [14]. However, the inoculation of poultry manure with companion bacteria was able to promote larval development and is a promising biotechnological strategy to optimize growth periods [15]. In contrast to the bacterial community, little is known about the mycobiome of BSF larvae. Fungi are important in the digestive capabilities of many insects, including cerambycid beetle larvae, fungus-growing termites, and siricid woodwasps [16]. By secreting a broad enzyme repertoire, fungi mediate many further processes such as the degradation of toxic compounds and the production of bioactive substances, as previously reported in BSF [17]. It is necessary to determine the fungal composition of the larval gut to evaluate their hazard potential as an animal feedstock, given the ability of many fungi to synthesize mycotoxins [18].
The growing worldwide demand for proteins and lipids contrasts with the inefficient use of agro-industrial raw materials and the modern throwaway society. Large amounts of underutilized organic material accumulate as side streams in many parts of the agri-food industry, including palm oil production to meet the increasing demand for vegetable oils [19]. In 2018, 71.5 million tons of crude palm oil was produced on an area of 18.9 million ha [20]. The milling process and subsequent refining and fractionation steps produce side streams and residues such as empty fruit bunches (EFB), palm kernel meal (PKM), as well as sludge and mill effluents (POME), amounting to many millions of tons per year [21]. Although PKM is already used as an inexpensive dietary supplement for ruminants and has been proposed as a sustainable option for BSF rearing [13,22], the utilization of EFB is more challenging due to its high content of lignocellulose. For this reason, it has not been used as a feed, and it is mostly burned so far. The upcycling of palm oil side streams by fermentation and feeding them to insects has not been investigated so far.
The compact structure of lignocelluloses requires special enzyme systems that are exclusively found in wood-degrading fungi. White-rot fungi representing the Basidiomycota are particularly suitable for the depolymerization of lignocelluloses because they produce enzyme cocktails that break down cellulose, hemicellulose, and lignin [23]. This makes them excellent candidates for the pretreatment of lignocellulose-rich EFB by fermentation, although feeding studies using EFB or mixtures of EFB and PKM have not been reported thus far.
Here we investigated the suitability of fermentation as a pretreatment for mixtures of EFB and PKM to improve digestibility, and the corresponding changes in BSF life-history traits as well as diet-specific adaptations in the larval gut microbiome. Extensive screening followed by feeding trials was carried out to find a suitable fungus for the fermentation step. The bacterial and fungal communities in the feed, BSF larval guts, and frass were then characterized by Illumina high-throughput sequencing of the 16S rRNA gene and the fungal internal transcribed spacer (ITS) regions.
2. Materials and Methods
2.1. Fungal Screening and Substrate Preparation
The palm oil side streams were provided by PT Alternative Protein Indonesia (Tebet, Indonesia). EFB was ground to powder (~1 mm particle size) in an SM 2000 cutting mill, autoclaved and set to a moisture content of 60% (Retsch, Haan, Germany). For fungal screening, 50 g of the powder was placed in 250 mL Erlenmeyer flasks before adding 3 mL of homogenized liquid culture representing a particular fungus from the strain collection of the Institute of Food Chemistry and Food Biotechnology (University of Giessen, Germany). Technical duplicates of 69 strains were incubated for 10 d at 30 °C in the dark. Fungal growth and lignocellulose degradation were evaluated visually, with degradation revealed by EFB bleaching [24].
For solid-state fermentation of the feed substrate, submerged cultures of the basidiomycetes Bjerkandera adusta, Irpex consors, and Marasmius palmivorus were processed in a T25 digital Ultra-Turrax homogenizer (IKA, Staufen im Breisgau, Germany) at 10,000 rpm for 30 s. An EFB + PKM mixture in the ratio of 7:3 was chosen because the side streams arise in this ratio. We then inoculated 250 g of the EFB + PKM mixture (7:3 ratio) with 15 mL of the corresponding fungal suspension in 2 L Erlenmeyer flasks (5% w/v) and incubated the cultures for 28 d at 28 °C and 60% relative humidity in the dark. The corresponding fermented feed was described as BAD (B. adusta fermented), ICO (I. consors fermented), or MPA (M. palmivorus fermented), respectively. The unfermented EFB + PKM mixture was used as the control (NFR). The feed was stored at −20 °C and thawed immediately before use. Chicken feed (CF) was used as a positive control, as in previous studies [1,7,10,25,26]. We used GoldDott Eierglück chicken feed (Derby Spezialfutter, Muenster, Germany), which was prepared by grinding in a Mockmill 200 grain mill (Wolfgang Mock, Otzberg, Germany) to a particle size of 0.1–1.5 mm.
2.2. Insect Rearing
BSF were originally obtained from Bio.S Biogas (Grimma, Germany). The larvae were maintained in 19.5 × 16.5 × 9.5 cm (l × w × h) plastic containers within a climate chamber at 27 ± 1 °C, 65 ± 5% relative humidity, in constant darkness [7,27]. Adult flies were kept in 60 × 60 × 90 cm (l × w × h) mesh cages (Bioform, Nuremberg, Germany) in a greenhouse at 25 ± 1 °C, 40 ± 10% relative humidity, and a 12 h photoperiod. Water-soaked paper towels provided drinking water ad libitum. Wooden board stacks held with rubber bands served as an artificial oviposition system, as previously described [27]. We harvested 200 mg eggs per plastic container using a plastic spatula. The containers were sprayed daily with water. Feed was added at 48 h intervals as soon as ≥50% of the eggs had hatched. To prevent contamination, disposable nitrile gloves were worn to handle and administer the feed.
2.3. Developmental Parameters
All developmental parameters were recorded as biological triplicates for each diet group. Data from the CF diet controls were recorded in collaboration with Tegtmeier and colleagues, except for fertility and longevity-related data [7]. We placed 150 mg eggs in three replicate containers per substrate to record the hatching time. Depending on the feed, the larvae reached a manageable size (3–4 mm; instar L3) after 5–10 d, allowing the analysis of growth curves. The mean larval weight was determined (at 48 h intervals until ≥50% of the individuals reached the prepupal stage) by placing random specimens (n = 25) on an AT261 DeltaRange analytical balance (Mettler, Giessen, Germany). The distinction between larval instars was standardized based on the head capsule width and the weight [28]. The survival rates of the larva-prepupa, prepupa-pupa, and pupa-imago stages were determined in 19.5 × 16.5 × 9.5 cm (l × w × h) plastic containers with a stocking density of 100 individuals. The transition between two developmental stages was documented as soon as ≥50% of the population reached the next developmental stage. We recorded the final weight (as above) and final length (using a VHX-2000 digital microscope; Keyence, Osaka, Japan) of 50 L5 larvae, prepupae, pupae and 60 imagines). Adult flies were collected within 24 h of emergence, inactivated by placing on ice and stored at −20 °C before measurement. The sex of adult flies was determined according to external dimorphisms, including antennal and genital structures [29].
Reproductive parameters were evaluated in 20 temporally synchronized pairs of adult flies (within 24 h of emergence) in conical 8.4 × 8.4 × 11.4 cm (l × w × h) polypropylene boxes in the greenhouse. Each pair was placed in its own box, which contained an artificial oviposition system consisting of two stacked 4 × 4 cm wooden boards spaced and held in place by magnetic tape strips. A circular 4.5 cm mesh insert in the lid enabled gas exchange and the daily provision of water by spraying. The adult lifespan and the deposition of eggs were checked and documented daily. Fresh egg clutches were collected, weighed, unclustered carefully using a metal spatula, and counted under an S9i stereomicroscope (Leica Microsystems, Wetzlar, Germany).
2.4. 16S rRNA Gene and ITS Amplicon Sequencing
DNA samples and amplicon sequencing were prepared according to Tegtmeier et al. [7] in order to compare the results to the amplicon data of BSF larvae reared on chicken feed (standard diet for BSF rearing). Briefly, L5 larvae weighing 100–150 mg were collected with spring steel tweezers. Frass samples were taken from three different vertical sites and horizontal levels and pooled to cover a large proportion of the different microbes colonizing the frass. Samples of the NFR and BAD feed were taken at three time points during the feeding period and were also pooled. All samples were stored at −20 °C before processing. The larvae were cleaned with sterile water and then surface sterilized twice with 70% ethanol. To dissect the larval gut, the haemocoel was opened laterally with microscissors and the gut was removed under a stereomicroscope using Dumont (Montignez, Switzerland) No. 5 tweezers. Six single guts per diet as well as three replicates of 100 mg of the corresponding feed and frass samples were disrupted by bead beating in a FastPrep-24 (MP Biomedicals, Solon, OH, USA) for 90 s at 6.5 m∙s−1. DNA was isolated using the NucleoSpin soil kit (Macherey-Nagel, Düren, Germany) according to the manufacturer’s specifications. The yield and purity of the DNA were determined using a Take3 spectrophotometer (BioTek Instruments, Winooski, VT, USA).
Two-step PCR library preparation and amplicon sequencing were carried out by LGC Genomics (Berlin, Germany) on the MiSeq V3 platform (Illumina, San Diego, CA, USA). Primers U341F (5′-CCT AYG GGR BGC ASC AG-3′) and U806R (5′-GGA CTA CNN GGG TAT CTA AT-3′) were used to amplify the 16S rRNA hypervariable V3–V4 region of bacteria and archaea [30], whereas primers fITS7 (5′-GTG ART CAT CGA ATC TTT G-3′) and ITS4 (5′-TCC TCC GCT TAT TGA TAT GC-3′) were used to amplify the fungal ITS2 region [31,32]. We aimed for ~20,000 paired-end reads per sample with a read length of 300 bp. Samples were multiplexed and pooled for sequencing. Demultiplexing and the clipping of adapters and primers was carried out using bcl2fastq 2.17.1.14, followed by analysis with QIIME 2020.6 [33]. Only forward ITS sequencing reads were used, and the read-through adapters and primers were removed using the cutadapt plugin [34]. The DADA2 plugin [35] was used for combined error correction, quality control, filtering chimeric sequences, and the creation of an ASV table showing the number of sequences for each ASV per sample.
Taxonomic classification was achieved using self-trained naïve Bayes classifiers based on SILVA 132 [36] and UNITE 8.2 [37] QIIME-compatible releases with 99% sequence identity for 16S and ITS, respectively. Reference data from SILVA were trimmed before training to include only the 16S rRNA gene region amplified by the primers [38]. The confidence for classification was 0.7 (16S) or 0.94 (ITS) as recommended [39]. Mitochondrial and chloroplast sequences were discarded. Alpha diversity was calculated based on Faith’s phylogenetic diversity [40] and observed ASVs. Rarefied data to equal sequencing depths of 8819 (16S) and 12,846 (ITS) were used to evaluate beta diversity with UniFrac [41] distance metrics.
We used discrete false-discovery rate (DS-FDR) to test for diet-dependent differential abundance in gut samples [42]. We collapsed the classified ASVs at the species level to run the test on log2-transformed data, testing for differences in mean values (p = 0.05, permutations = 1000) with the corresponding QIIME2 plugin. We screened for representatives of the positively and negatively correlated taxa (based on DS-FDR) using the Integrated Microbial Genomes Expert Review (IMG/ER) platform (https://img.jgi.doe.gov/cgi-bin/er/main.cgi, accessed on 28 July 2021) [43], and selected 140 genomes of the 28 most positively correlated taxa (highest p-value) as well as 140 genomes of the 28 most negatively correlated taxa (lowest p-value). Only taxa that were classified to the family level were included. For taxa that were not classified to the genus level, only genomes of representatives with ≥91% sequence identity to the target sequence were included. The genomes were screened for genes involved in cellulose degradation (endoglucanases EC 3.2.1.4, exoglucanases EC 3.2.1.91 and β-glucosidases EC 3.2.1.21) using the KEGG Orthology (KO) database implemented in IMG/ER [44].
2.5. Statistics
Statistical analysis and visualization were carried out using Excel 2016 (Microsoft, Redmond, WA, USA) and R 4.1 [45] with the packages ggpubr, plyr, qiime2R, scales, and tidyverse. Diet-dependent differences in the relative abundance of bacterial or fungal taxa were determined using Student’s (homogeneous variance) or Welch’s (inhomogeneous variance) t-test and an error level of α = 0.05 for statistical significance. The homogeneity of variance was calculated with Levene’s test. Developmental parameters were compared by one-way ANOVA and means were separated using the Bonferroni–Holm test [46]. Sex-specific and total adult longevity were analyzed using the Kaplan–Meier estimation to generate S(t) survival functions, which were compared pairwise by log rank tests at an error level of α = 0.05 for statistical significance in OriginPro 2020b (OriginLab, Northampton, MA, USA). For PERMANOVA [47], we grouped all samples combing feed (NFR, BAD) and source (feed, gut, and frass) separately for 16S and ITS. We used the corresponding unweighted UniFrac distance metric and tested pairwise on the six groups with 999 permutations, with p-value correction for multiple tests based on the Benjamini–Hochberg procedure [48].
3. Results
3.1. Fungal Screening and BSF Growth Performance
In order to identify a suitable fungus for the pretreatment of palm oil side streams, we screened 69 strains, including 61 Basidiomycota and 8 Ascomycota. We found that 29 strains were unable to grow on EFB, and that Mycena pseudocorticola and Omphalotus illudens achieved only weak growth. Members of the family Pleurotaceae showed comparatively fast growth, but the fastest-growing species were the white-rot fungi Marasmius palmivorus, Irpex consors, and Bjerkandera adusta. Bleaching of the dark lignin also indicated these three species and eight others (with Xylaria longipes representing the only ascomycete) as putative candidates for the fermentation of palm oil side streams (Table 1).

Table 1.
Fungal screening for growth 1 and lignin degradation 2 during the solid-state fermentation of empty fruit bunches (EFB).
Based on the initial screen, we used M. palmivorus, I. consors, and B. adusta to ferment the EFB + PKM mixture (7:3 ratio). The corresponding fermented feed was described as MPA, ICO, or BAD, respectively. We carried out comparative feeding studies in BSF larvae using CF as a high-quality control and the untreated EFB + PKM mixture as a low-quality standard diet, described as the non-fermented reference (NFR). The final larval weight differed significantly between the diets, and larvae reared on CF were the heaviest (F4,10 = 58.04; p < 0.002) (Figure 1). Neither MPA nor ICO led to an increase in larval weight compared to NFR, whereas larvae reared on the BAD diet were 25% heavier than their NFR counterparts (F2,6 = 74.84; p = 0.042). All the diets influenced larval development. CF resulted in the shortest developmental period, followed by BAD and ICO. However, MPA extended the larval development phase to 79.0 d, approximately twice the duration of the NFR larvae (F4,10 = 994.61; p < 0.0001). The larvae reared on the BAD diet developed significantly faster than those in the NFR, MPA, and ICO groups (F3,8 = 848.13; p = 0.034).
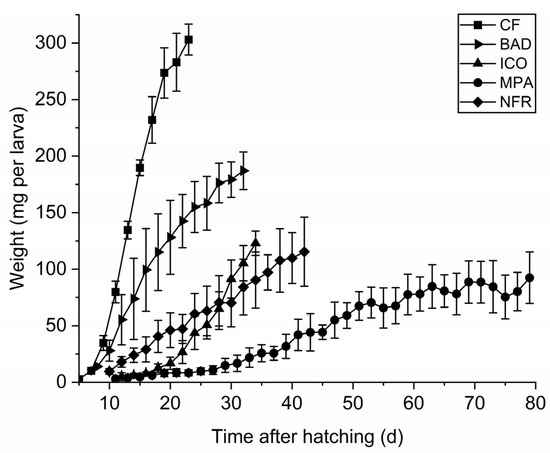
Figure 1.
Growth curves of BSF larvae reared on chicken feed (CF), B. adusta (BAD), I. consors (ICO), or M. palmivorus (MPA) fermented EFB + PKM (7:3) mixtures, as well as a corresponding non-fermented reference (NFR). Data are mean larval weights (±SD) of three replicate boxes per diet (n = 25).
3.2. Life-History Traits
We also compared additional life-history traits between larvae raised on the standard diets (CF, NFR) and the BAD diet. The time until ≥50% of the eggs hatched did not differ between the diets. Larval development differed significantly between the diets, with CF larvae reaching the prepupal stage first (Table 2). Larvae reared on the BAD diet developed 27% faster than their NFR counterparts (F2,6 = 136.50; p = 0.001). Prepupal and pupal development (including metamorphosis) did not differ between the diet groups. However, the total duration of development differed between the groups, being 19% quicker on the BAD diet and 24% quicker on the CF diet compared to the NFR diet (F2,6 = 36.58; p < 0.004). The same pattern was also observed for the total preoviposition period (F2,6 = 39.88; p < 0.002), whereas no significant diet effect was found in the adult preoviposition period. Larvae reared on the CF diet showed the highest larva-to-prepupa developmental success (100%), differing significantly from the BAD diet (F2,6 = 8.10; p = 0.0001). In contrast, the prepupa-to-pupa developmental success was 9% higher for larvae reared on the BAD diet compared to those on the CF diet (F2,6 = 132.58; p = 0.0001). The proportion of adult flies emerging from pupae was 97–100% in all feeding trials. Oviposition lasted 2–10 d. The longevity of adult males (χ2 = 51.67; p < 0.0001), adult females (χ2 = 65.98; p < 0.0001), and all adults (χ2 = 91.31; p < 0.0001) differed significantly between CF and both other diets. In general, male adults lived longer than females in the same diet group (Table 2).

Table 2.
Diet-dependent temporal and physiological parameters of BSF throughout development. BSF larvae were reared on chicken feed (CF), a non-fermented reference (NFR), or a B. adusta fermented diet (BAD). Data are means ± SD. The transition between two developmental stages was defined as when ≥50% of a population reached the next stage. Different letters (a–c) within a row indicate statistically significant differences between diets (p < 0.05; one-way ANOVA; Kaplan–Meier estimator for adult longevity).
The final larval weight (F2,6 = 74.84; p < 0.04) and length (F2,400 = 672.70; p < 0.0001) differed significantly between diets, with the largest and heaviest larvae reared on the CF diet, followed by those reared on the BAD diet. The same profile for weight (F2,6 = 22.22; p < 0.04) and length (F2,400 = 348.58; p < 0.0001) was observed at the prepupal stage. Larvae reared on the CF diet produced pupae that were heavier (F2,6 = 13.26; p = 0.022) and longer (F2,400 = 134.33; p < 0.0001) than NFR counterparts, but there was no statistically significant difference between the CF and BAD diets. Neither the total adult weight nor the sex-specific weight showed significant differences between the diets. The adult males (F2,235 = 96.18; p < 0.0001), adult females (F2,310 = 53.97; p < 0.0001), and all adults (F2,545 = 117.20; p < 0.0001) were shortest in the NFR group, but up to 11% longer in the CF group and up to 6% longer in the BAD group. In general, females were 16–24% heavier and 4–8% longer than the corresponding males, regardless of the diet. All dietary groups had similar sex ratios, with a slight excess of females. The number of egg clutches deposited per female was 5.9-fold higher in the BAD group compared to the NFR group, and even exceeded the performance of the CF group (F2,6 = 55.86; p = 0.0005). Furthermore, the egg clutch size in the BAD group was 2.1-fold higher than the NFR group (F2,27 = 114.63; p < 0.0001), and the egg clutch weight was 2.4-fold higher than the NFR group (F2,27 = 74.36; p < 0.0005), but in neither parameter did the BAD group outperform the CF group. We observed no significant dietary impact on the weight of individual eggs.
3.3. Taxonomic Composition of the Bacterial Gut Microbiome
To identify differences in the bacterial and archaeal gut microbiome between larvae fed on the BAD and NFR diets, we performed 16S rRNA gene sequencing on 24 samples (six guts, three feed samples, and three frass samples per diet). We obtained 527,684 raw read pairs, 406,803 of which remained after quality control and the removal of chimeric sequences (8819–46,354 reads per sample). The mean read length was 421 bp after merging forward and reverse reads. Rarefaction curves suggested a sufficient sequencing depth (Figure 2).
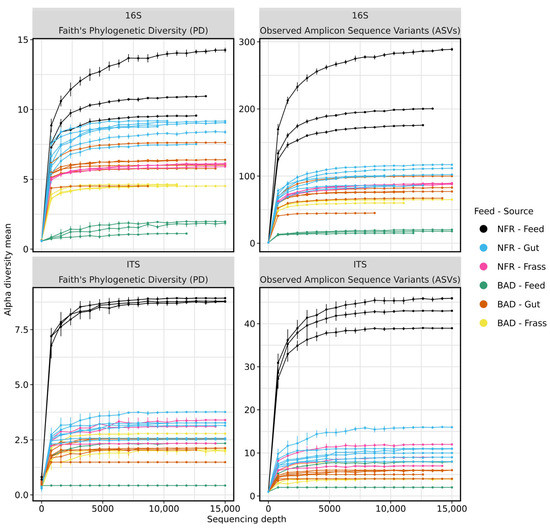
Figure 2.
Rarefaction curves of Faith’s phylogenetic diversity (left) and amplicon sequence variants (right) showing the alpha diversity of 16S rRNA gene and ITS amplicon sequencing. Almost all samples of the non-fermented reference (NFR) and B. adusta fermented diet (BAD) reach a plateau at ~9000 reads. Samples with a lower number of reads also reach the plateau. Greater sequencing depth would therefore not lead to an increase in taxonomic richness. Standard deviations are shown as bars.
Data processing using DADA2, followed by classification using our self-trained naïve Bayes classifier on SILVA 132 and the removal of mitochondrial and chloroplast sequences resolved the dataset to 693 unique amplicon sequence variants (ASVs) across all 24 samples. All the ASVs were assigned to domain Bacteria (i.e., no Archaea were found). We classified 621 of the 693 ASVs down to the genus level. Principal coordinates analysis (PCoA) of weighted UniFrac distance metrics indicated that all frass samples from both diets clustered together with four gut samples from larvae reared on the NFR diet (Figure 3A). In contrast, unweighted distances indicated differences between all sample groups. However, the unweighted distances revealed a higher similarity between gut samples from larvae reared on the BAD diet and the corresponding frass samples (Figure 3B). The BAD and NFR feed samples showed greater differences when comparing unweighted distances and smaller differences when comparing weighted distances. The feed and frass samples of both diets showed little similarity, although the differences in the BAD group were greater (Figure 3A,B).
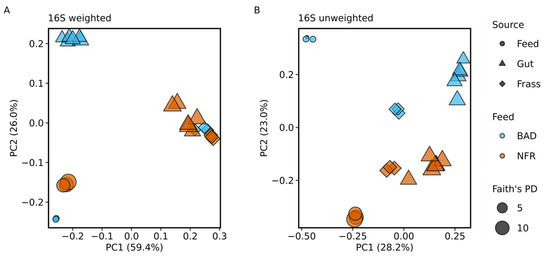
Figure 3.
Principal coordinates analysis (PCoA) when comparing the microbiome between the non-fermented reference (NFR) and B. adusta fermented diet (BAD) groups. (A,B) PCoA comparing the bacterial communities based on weighted and unweighted UniFrac distance metrics. The sample type is represented by symbols (feed, gut, and frass) and the dietary group affiliation by colors. The symbol size represents Faith’s phylogenetic diversity (PD).
Bacteria were detected in all samples. The BAD feed almost exclusively contained Bacillaceae (mostly Bacillus coagulans), with a relative abundance of 99.9%. The NFR feed was more diverse, but was still dominated by Bacillaceae (48–54.3%), followed by Paenibacillaceae (8.8–10.8%), Nocardiopsaceae (4–10.1%), Planococcaceae (5.3–6.1%), and Xanthomonadaceae (3.9–5.7%) (Figure 4). In addition, all NFR feed samples contained seven low-abundance families (0.4–2.3%): Pseudonocardiaceae, Streptomycetaceae, Leuconostocaceae, Staphylococcaceae, Streptosporangiaceae, Micromonosporaceae, and Thermomonosporaceae (Supplementary Table S1). Whereas Bacillaceae were abundant in all diet samples, they were minor components in the gut samples (2.7–4.7% in NFR and just ~0.07% in BAD gut samples). Ruminococcaceae was the predominant family (40.8–50.3%) in all BAD gut samples, although it was not present in the BAD feed. Some members of the Ruminococcaceae were also present at a lower relative abundance in all NFR feed (0.04–0.16%) and NFR gut samples (0.6–8.4%), thus showing remarkable enrichment in the guts of larvae reared on the BAD diet (p < 0.0001). Lachnospiraceae, Enterococcaceae, Rhizobiaceae, and Enterobacteriaceae were core families in all gut samples, regardless of the diet. However, members of the first two families were significantly enriched in BAD gut samples (p < 0.0002 and p = 0.001), whereas family Rhizobiaceae was less abundant in BAD compared to NFR gut samples (p < 0.0002). Enterobacteriaceae was the predominant family (39.7–68.4%) in all replicate NFR gut samples (Figure 4). Most of the taxa identified in the BAD gut samples were not found in the feed or their abundance was low. The family Chitinophagaceae (0.4–1.7%) was exclusively detected in the NFR gut samples, whereas Beutenbergiaceae (2.1–3.9%) and Christensenellaceae (1.3–2.7%) were only found in BAD gut samples (Supplementary Table S1). Interestingly, the family Actinomycetaceae was identified in all replicate gut samples of both diets, but was completely absent in the corresponding feed and frass samples. However, the relative abundance was significantly higher in the BAD gut samples (p < 0.0002).
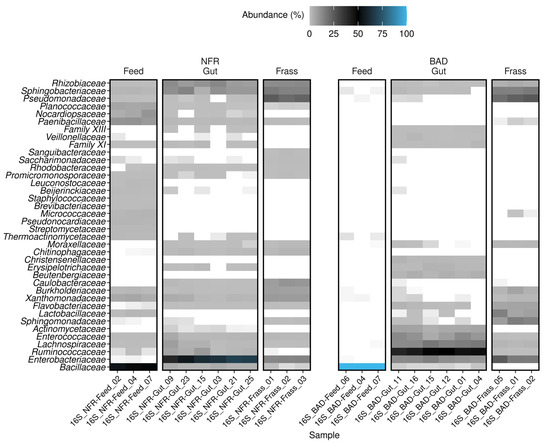
Figure 4.
Heat map showing the family-level composition of the bacterial community characterized by amplicon sequencing of the 16S rRNA gene. Samples (feed, gut, and frass) of the non-fermented reference (NFR) and B. adusta fermented diet (BAD) groups are shown. Only the 38 most abundant classified families are listed. Families without a suitable classification (uncultivated, undefined, and not applicable) are excluded. The relative abundance (%) is shown as a color gradient that runs from light gray through black to blue, the latter representing the highest abundance. White areas indicate that the family was not detected in the sample.
We also found striking differences between the two diet groups at the genus level. Klebsiella was the most prominent genus in all NFR gut samples (31.7–64.8%), but the relative abundance fell to <3% in five of the six replicate BAD gut samples. Instead, Ruminococcaceae UCG-014, Ruminiclostridium 5, and Enterococcus were the dominant genera in all six guts of larvae reared on the BAD diet, with a cumulative abundance of ≥50% (Supplementary Figure S1, Supplementary Table S1). The majority of bacterial families in BSF larval guts could also be found in frass samples, albeit with marked differences in abundance. However, Sanguibacteraceae was only detected in the NFR frass and was completely absent from gut samples. The bacterial community in all BAD frass samples was dominated by the Pseudomonadaceae (20–25.8%), Enterobacteriaceae (16.6–24.9%), Sphingobacteriaceae (14.9–16.8%), Sphingomonadaceae (8.1–16%), Lactobacillaceae (5.8–15.1%), Flavobacteriaceae (5.7–6.4%), Burkholderiaceae (2.2–6.2%), and Rhizobiaceae (almost all Ochrobactrum; 3.3–4.6%), whereas NFR frass was dominated by Pseudomonadaceae (20.1–24.4%), Sphingobacteriaceae (14.1–16.6%), Enterobacteriaceae (13.3–14.1%), Rhizobiaceae (12–12.8%), Caulobacteriaceae (7.8–10%), Xanthomonadaceae (8.1–8.7%), and Burkholderiaceae (6.6–7.5%). The frass samples of both diets showed a common intersection of predominantly represented families, albeit with pronounced differences in relative abundance. The most prominent families in the BAD frass were either absent from the corresponding feed or present at very low abundance in one or two replicate feed samples (0.009–0.038%). In contrast, the most prominent families in the NFR frass were also detected in the feed, with Caulobacteriaceae as the major exception (Supplementary Figure S1, Supplementary Table S1). All NFR feed samples and most gut samples showed higher bacterial alpha diversity than corresponding samples from the BAD group. BAD feed showed the lowest alpha diversity as determined by Faith’s phylogenetic diversity and the observed ASVs (Figure 2 top row, Figure 3A,B). A PERMANOVA revealed significant differences between the diet groups in terms of the beta diversity of the bacterial gut community (Fpseudo = 18.00; padjust = 0.002).
3.4. Taxonomic Composition of the Fungal Gut Microbiome
The same samples discussed above were also used for ITS sequencing to compare the composition of the fungal community in the BAD and NFR groups. The 24 samples yielded 570,372 raw reads, which we reduced to 530,816 after quality control and trimming (12,846–43,666 per sample). The read length was 186–273 bp. Rarefaction curves are shown in Figure 2. We identified 167 unique ASVs across all 24 samples using DADA2. With a minimum confidence of 0.94 for our self-trained naïve Bayes classifier based on UNITE 8.2, we were able to classify 142 of those 167 unique ASVs to the genus level.
As discussed above for the bacterial community, PCoA indicated pronounced differences in the mycobiome between the BAD and NFR feed samples (Figure 5). Weighted UniFrac distances suggested high similarity between gut and frass samples from the NFR group and frass samples from the BAD group, whereas the unweighted PCoA showed greater differences between these groups (Figure 5A,B). Individual gut samples from both diet groups clustered closer together in the weighted UniFrac model than in the unweighted model (Figure 5A). Interestingly, PCoA of unweighted UniFrac distances revealed substantial interindividual differences between BAD feed samples (Figure 5B).
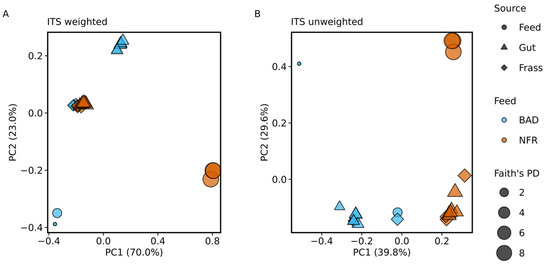
Figure 5.
Principal coordinates analysis (PCoA) when comparing the mycobiome between the non-fermented reference (NFR) and B. adusta fermented diet (BAD) groups. (A,B) PCoA comparing the fungal communities based on weighted and unweighted UniFrac distance metrics. The sample type is represented by symbols (feed, gut, and frass) and the dietary group affiliation by colors. The symbol size represents Faith’s phylogenetic diversity (PD).
Fungi were detected in all samples of both diet groups. The BAD feed almost exclusively featured Phanerochaetaceae (all B. adusta) with a relative abundance of 93.1–100% (100% in two of three replicates). In one BAD feed replicate Trichosporonaceae were detected with a relative abundance of 5.6%. As observed for the bacterial community, the diversity of the fungal community was higher in NFR feed samples. This was anticipated because the feed was inoculated solely with B. adusta after autoclaving. The NFR feed was dominated by ascomycete families: Saccharomycetales family incertae sedis (mostly Candida tropicalis; 53.9–60.8%), Aspergillaceae (mostly Xeromyces; 14.5–21.8%), and Trichocomaceae (mostly Thermomyces; 9.3–11.2%) (Figure 6, Supplementary Figure S2). All NFR feed samples also contained six low-abundance (0.04–1.2%) families: Debaryomycetaceae, Wallemiaceae, Agaricaceae, Saccharomycopsidaceae, Rhizopodaceae, and Cladosporiaceae. The families Chaetomiaceae, Sordariaceae, and Saccharomycetaceae were found in all NFR feed samples (0.1–1.1%) and in at least one frass sample, but were completely absent from the guts of larvae reared on the NFR diet. The members of all predominant families in samples of both feeds were only weakly detected in the corresponding gut samples (0.3–3.1%) and frass samples (0.1–3.1%; Aspergillaceae was identified in two of three replicates) (Supplementary Table S2).
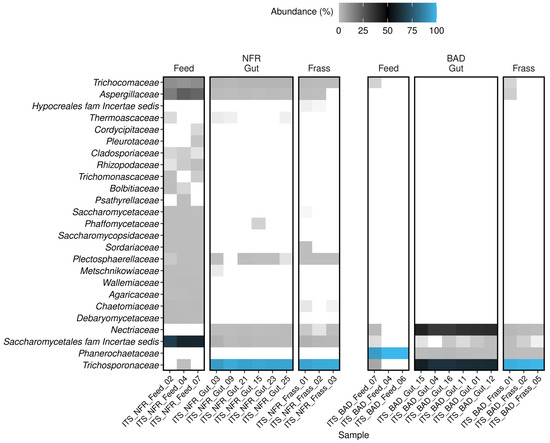
Figure 6.
Heat map showing the family-level composition of the fungal community characterized by ITS amplicon sequencing. Samples (feed, gut, and frass) of the non-fermented reference (NFR) and B. adusta fermented diet (BAD) groups are shown. All 25 classified families are listed. Families without a suitable classification (uncultivated, undefined, and not applicable) are excluded. The relative abundance (%) is shown as a color gradient that runs from light gray through black to blue, the latter representing the highest abundance. White areas indicate that the family was not detected in the sample.
Trichosporonaceae and Nectriaceae were identified in all gut samples, regardless of the diet. A striking enrichment of the first family (entirely Trichosporon asahii) in all guts of larvae reared on the NFR (91.6–95.6%) and BAD (57.9–63.8%) diets was observed, albeit with diet-dependent significant differences in the relative abundance (p < 0.0001). Interestingly, Trichosporonaceae was only weakly detected in a single replicate of NFR feed (0.16%). In contrast to the NFR diet, the Nectriaceae (almost exclusively Fusarium) represented the second most prominent family (34.6–41.2%; p < 0.0001) in the BAD gut samples, although they were only found in one of three feed replicates (~1.1%). However, Nectriaceae was also detected with low abundance in all NFR frass samples but was completely absent in the corresponding feed. The majority of taxa detected in larval guts from the BAD and NFR groups were present in at least one of the feed samples in an inverse relative abundance (Figure 6, Supplementary Table S2). Trichocomaceae (1.5–2.3%), Aspergillaceae (0.4–0.8%), and Plectosphaerellaceae (0.04–0.3% in five replicates) were exclusively detected in the guts of larvae reared on the NFR diet, whereas the family Phanerochaetaceae (0.3–1.6%) was only found in guts of larvae reared on the BAD diet. Most fungal families present in the larval guts were also found in frass samples, albeit with marked differences in abundance. The fungal community in all NFR frass samples was dominated by members of the Trichosporonaceae (94.4–96.6%), followed by Saccharomycetales family incertae sedis (mostly Candida; 1.6–3.1%), Trichocomaceae (all Thermomyces; 0.7–1.2%), Plectosphaerellaceae (all Chordomyces; 0.2–0.3%), and Aspergillaceae (~0.2% in two replicates), whereas BAD frass samples contained Trichosporonaceae (97.8–99.5%), Nectriaceae (0.3–1.5%), Phanerochaetaceae (0.1–0.3%), and Saccharomycetales family incertae sedis (Diutina and Candida; 0.02–0.3%). The frass samples of both diets showed a common intersection of predominantly represented families, albeit with pronounced differences in their relative abundance (p = 0.032). The larval consumption of BAD feed strongly reduced the relative abundance of the fermentation strain B. adusta. The most prominent families in the BAD frass were identified with comparable abundance in one replicate feed sample (except for Phanerochaetaceae and Trichosporonaceae), whereas the most prominent families in the NFR frass, apart from Trichosporonaceae, were detected with a higher abundance in all corresponding feed replicates (Supplementary Figure S2, Supplementary Table S2). When larvae were reared on the NFR diet, almost all gut and frass samples showed higher fungal alpha diversity compared to the BAD diet. Faith’s phylogenetic diversity and the observed ASVs suggested the highest alpha diversity for NFR feed (Figure 2 bottom row, Figure 5A,B). A PERMANOVA revealed significant differences between the diet groups in the beta diversity of the fungal gut community (Fpseudo = 35.17; padjust = 0.001).
4. Discussion
4.1. Rationale for the Pretreatment of Palm Oil Side Streams
BSF larvae do not naturally specialize on lignocellulose degradation in the same manner as termites and other xylophagous species. Lignin in the BSF diet is predominantly metabolized by fungi, which secrete extracellular laccases and peroxidases for this purpose, although there is evidence that some bacteria also facilitate lignocellulose utilization. Nevertheless, the efficiency and capacity of fungi appears higher, as indicated by the symbiosis of several fungus-growing termite (subfamily Macrotermitinae) and ant (tribe Attini) species [49]. Termites that do not cultivate fungi promote the digestion of lignocellulose by deploying mechanical processes, such as initial grinding of plant material with their mandibles and subsequent ball-milling in the gizzard to facilitate chemical and enzymatic digestion [50,51]. Endogenous enzymes such as phenol-oxidizing laccases from the salivary glands, as described for Reticulitermes flavipes (Kollar, 1837; Blattodea: Rhinotermitidae), can modify lignocelluloses and increase accessibility for glycoside hydrolases [52]. The chemical oxidation of lignin by hydroxyl radicals originating from the Fenton reaction is also possible in the foregut. In addition, the alkaline pH of the anterior hindgut compartment promotes autoxidation, which cleaves lignin-carbohydrate complexes [51]. Comparable physiological adaptations are not present in BSF larvae. Therefore, the lignocellulose-rich diet was pretreated by fermentation with a lignin-degrading fungus and we investigated its effects on BSF development and the microbiome of the feed, gut and frass.
4.2. Pretreatment by Fermentation Influences BSF Life-History Traits
This is the first study exploring the growth and development of insects reared on fermented side streams from the palm oil industry. We were able to rear BSF larvae on non-fermented and fermented diets consisting of the palm oil side streams EFB and PKM. Both diets seemed to be sufficient for the full development of flies at the laboratory scale, albeit with significant temporal and physiological differences. Fermentation with the white-rot fungus B. adusta, resulting in the BAD diet, promoted faster larval development and thus achieved a significantly shorter life cycle, which would allow earlier harvesting in an industrial process. In addition, L5 larvae fed on the BAD diet reached a 25% higher final larval weight. Similar weight gain (~30%) and developmental acceleration (up to 10%) were reported when inoculating poultry manure with companion Bacillus subtilis [15]. In addition, feed supplemented with BSF-associated Bacillus licheniformis HI169 improved the growth rate and final larval weight compared to untreated controls [53]. Our observations may reflect both the direct and indirect effects of pretreatment. White-rot fungi are suitable for the degradation of agro-industrial side streams because they are naturally adapted to degrade plant material rich in lignocelluloses by secreting exoenzymes, enabling their saprophytic lifestyle [54,55]. The enrichment of fungal biomass and depolymerized lignocellulose components may have increased the digestibility of the feed and hence nutrient accessibility, explaining the positive effect on the development and reproduction of adult flies. On the other hand, we were able to identify a clear shift in the bacterial and fungal gut community associated with fermentation. Although the relative abundance of B. adusta was almost 100% in the BAD feed, this species was clearly unable to colonize the gut of the BSF larvae. Instead, it was outcompeted by other taxa, as discussed later. However, B. adusta may have broken up rough structural motifs of the lignocellulose in the feed and created a suitable medium for colonization by cellulose-degrading microbes forming the gut microbiome, supporting mutualistic interactions [56,57,58]. In addition to the provision of metabolites or protection against pathogens, symbionts intervene in the feed repertoire of insects to facilitate the utilization of nutritionally unfavorable substrates [59]. Cooperation with bacteria, filamentous fungi, yeast and protozoa contributes to the digestion of plant biomass in phytophagous insects, strongly influencing their development [60,61]. Such cooperation takes many forms, including obligatory intracellular relationships in aphids and other sap-feeding insects, gut-associated communities in beetles and wood bees, and specialized dependent relationships in fungus-cultivating termites, such as Macrotermes annandalei (Silvestri, 1914; Blattodea: Termitidae), with an extra-gastrointestinal mycobiome [59,62].
BSF prepupae do not feed, so the fat body acquired during larval development normally serves as an energy reserve, influencing metamorphosis and adult life-history traits [28,63]. Accordingly, low energy reserves in the NFR larvae may explain the negative effect on reproduction and low fertility, including lower egg clutch size and weight compared to the BAD diet. As an alternative to the fat body, females may also reabsorb oocytes and metabolize them to maintain respiration, which would reduce the egg clutch size [64]. Differences in reproductive success may also reflect microbe–insect interactions. In BSF, oviposition is mediated by bacteria in a conspecific manner [65]. Other dipteran species, including Aedes aegypti (Linnaeus, 1762; Diptera: Culicidae), also recognize specific bacterial compositions, indicating appropriate conditions for egg development and deposition [66]. Bacillus species isolated from BSF eggs reduced the ovipositional response of adult females by more than 50% when added to artificial egg traps [65]. Bacillus was an abundant genus in both diets: 28.5–33% (NFR) and ≥99.8% (BAD). However, there were pronounced differences between the Bacillus species in the BAD and NFR groups. We found that ~68% of the reads in the BAD feed represented B. coagulans, whereas the NFR feed contained a more even distribution of Bacillus species, with B. clausii predominant at ~6% relative abundance (also found in all corresponding gut samples). It is possible that some of these species inhibited oviposition in the NFR group, resulting in fewer egg clutches. Moreover, the microbial community of the NFR feed probably features a higher bacterial load than the BAD feed because only the latter was autoclaved. Gut microbes are known to influence the behavior of several insects. In Drosophila species (Diptera: Drosophilidae), food-induced biases in adult mating behavior were prevented by antibiotic supplements, presumably by inhibiting the growth of sex pheromone-producing bacteria like Lactobacillus plantarum [67]. Such microbially-regulated hormonal changes may also affect BSF mating. It is not clear to what extent fungi play a role in this context. However, fungi can also modify insect host behavior, as shown by various entomopathogenic members of the order Entomophthorales [68].
4.3. Analysis of the Bacterial Communities Associated with BSF
Amplicon sequencing revealed major differences in the microbial communities of the NFR and BAD samples, probably reflecting the pretreatment of the feed. However, we also identified a potential BSF larval core microbiome present in all replicates of both diet groups, including the families Enterobacteriaceae, Enterococcaceae, Actinomycetaceae, Lachnospiraceae, and Ruminococcaceae.
Enterobacteriaceae, Enterococcaceae and Actinomycetaceae have already been recognized as core families in previous studies [7,69] and are prominent members in the guts of BSF larvae reared on chicken feed [7]. Furthermore, representatives of the families Lachnospiraceae and Ruminococcaceae are frequently found in BSF larval guts [9,69], although not in all samples, and the relative abundance of the Ruminococcaceae was low [7]. Our study is the first to demonstrate the significant enrichment of Ruminococcaceae (44–50% relative abundance) in the guts of BSF larvae due to dietary adaptations. Furthermore, Lachnospiraceae and Actinomycetaceae were significantly enriched in larvae reared on the BAD diet, indicating that pretreatment favored these groups. Lachnospiraceae and Ruminococcaceae are obligate anaerobic gut-associated families known for their ability to break down cellulose [70,71]. Therefore, whereas the NFR larval gut was dominated by the facultatively anaerobic Enterobacteriaceae, typically one of the most prominent families in BSF larvae [7,10,69,72], fermentation with B. adusta clearly resulted in a shift towards obligate anaerobic and cellulolytic bacteria.
Only a single draft genome sequence is available for B. adusta (IMG genome ID 2761201615), so we cannot fully evaluate its role in cellulose degradation. However, B. adusta is known to degrade gelatin, chitin, starch, pectin, tributyrin, and carboxymethylcellulose, but not fibrous cellulose, such as filter paper [73]. Therefore, we can assume that lignin, but not fibrous cellulose, was degraded during the pretreatment of EFB-PKM. The degradation of fiber-rich material by bacteria is largely restricted to biomass with a low lignin content [74], so lignin degradation by B. adusta probably facilitated subsequent anaerobic degradation of the remaining fibrous structures by cellulolytic bacteria in the gut. Once lignin is degraded, bacteria can attack cellulose and hemicellulose with multiple carbohydrate-active enzymes (CAZymes). Endoglucanases cleave cellulose chains internally, mainly from the amorphous region, and the released units are further degraded by exoglucanases (cellobiohydrolases) and β-glucosidases. Exoglucanases cleave cellobiose from the end of the polysaccharide chains, and these are subsequently hydrolyzed by β-glucosidases to produce glucose monomers [75]. Endoglucanase and β-glucosidase genes are more abundant in the taxa that were positively correlated with the BAD group, supporting the shift toward a cellulolytic community. Members of the families Ruminococcaceae (e.g., Ruminiclostridium and Ruminococcaceae UCG-014) and Lachnospiraceae (e.g., Anaerocolumna, Anaerosporobacter, and Lachnoclostridium) contained the largest complement of cellulose degradation genes (Figure 7). The genera Actinomyces and Salana as well as the [Eubacterium] fissicatena group were also positively correlated with the larvae reared on the BAD diet and contained a large set of β-glucosidase genes, but none encoding endoglucanases (Supplementary Table S3).
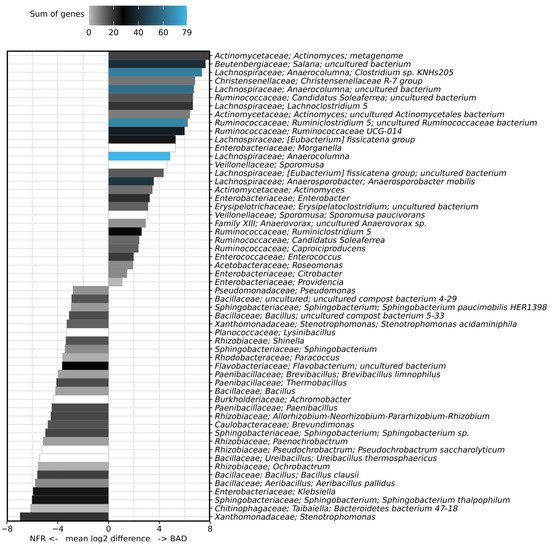
Figure 7.
Sum of genes involved in cellulose degradation (endoglucanases EC 3.2.1.4 and β-glucosidases EC 3.2.1.21) found in the genomes of representatives of the 28 most positively and 28 most negatively correlated taxa (5 genomes per taxon) present in BSF larval guts based on the discrete false-discovery rate (DS-FDR). No genes for exoglucanases (EC: 3.2.1.91) were found. For better illustration, the raw data (Table S3) were transformed by multiplying all values by −1.
Fungal fermentation not only led to lignin degradation and more accessible cellulose in the feed, but probably also lowered the redox potential and created anaerobic conditions. The ingested anaerobic feed substrate may have therefore promoted colonization by obligate anaerobes of the families Lachnospiraceae and Ruminococcaceae, which require negative redox potential for energy metabolism in the gut [76].
The families Rhizobiaceae, Sphingobacteriaceae, Bacillaceae, and Paenibacillaceae were positively correlated with larvae in the NFR group. BSF larvae may acquire members of these families during feeding, given their presence in all replicate NFR feed samples. Although these families primarily feature soil/plant-associated species [77,78,79] the physiochemical conditions in the BSF larval gut nevertheless appear suitable. In contrast, soil/plant-associated microbes were rare in the BAD samples, which was anticipated because the EFB-PKM diet was autoclaved before fermentation. The BAD diet is probably not the major source of bacterial inoculation. Instead, Ruminococcaceae and Lachnospiraceae may be taken up from the eggs or the rearing environment. For example, Lachnospiraceae have been detected in egg samples with a relative abundance of ~11% [72]. The families Lachnospiraceae and Ruminococcaceae feature many spore-forming genera [80,81] and were present in all frass samples in this study and in a previous study [7], suggesting they survive on the pupal surface and are subsequently associated with the flies and their eggs.
Lachnospiraceae and Ruminococcaceae species produce hydrogen during the anaerobic breakdown of cellulose and associated fermentation processes [82]. Such conditions would support colonization by hydrogenotrophic methanogenic archaea, as previously reported in termites and cockroaches [83], but none of the ASVs was assigned to the domain Archaea. In previous studies, methanogenic archaea have been shown to be absent from BSF larval guts [7], present at a low relative abundance [12], or present at a high relative abundance [14]. Other studies have shown that methane emission from BSF farms is generally low and can vary between locations and feeding strategies [84,85,86]. Therefore, the growth of methanogenic archaea in the BSF larval gut and frass should be investigated in more detail when new feed sources are tested. Our data suggest that BSF larvae reared on NFR or BAD diets do not produce methane, which would be climate-friendly in the context of large-scale insect farming.
4.4. Analysis of the Fungal Communities Associated with BSF
Whereas the bacterial community in BSF larvae has been studied intensively over the last decade, the composition and function of the fungal community has been largely overlooked. We found that Trichosporon asahii (Trichosporonaceae) was dominant in all gut and frass samples regardless of the diet, in agreement with earlier reports [7,8], indicating that this yeast is closely associated with BSF larvae and is probably a core species. However, although the fungal microbiome in the NFR group was almost exclusively composed of T. asahii (92–96% relative abundance), the BAD group was additionally colonized with Fusarium species (35–41%), which were not detected in the NFR group nor in BSF larvae reared on chicken feed [7]. As discussed for the bacterial community, cellulose-degrading fungi may be favored by the fermented diet. Although the genus Fusarium includes several entomopathogenic species [87], larval growth performance was clearly not affected by the presence of Fusarium (not assigned to species level). And given that some Fusarium species can break down cellulose [88,89], the presence of these fungi may even boost growth performance by promoting more efficient digestion of the feed.
When new feed substrates are introduced, they must be evaluated for potentially hazardous microorganisms such as mycotoxin-producing fungi. The family Aspergillaceae, which produces aflatoxins and gliotoxins [90], was present at a high relative abundance in the NFR diet, but not in the guts of larvae in the NFR group. BSF does not accumulate aflatoxin B1 [91], but the monitoring of farmed BSF larvae for the accumulation of other mycotoxins would be a wise precaution. The same applies to Fusarium species, which synthesize a wide variety of mycotoxins [92] and were relatively abundant (>34%) in all larvae in the BAD group. Fusarium species are known to produce trichothecenes, such as deoxynivalenol, although species-dependent and strain-specific variations in metabolic profiles have already been described [93].
5. Conclusions
The palm oil industry produces millions of tons of waste biomass annually, which is usually incinerated [94]. Therefore, the bioconversion of these residues into insect protein and lipids can contribute to an environmentally sustainable circular economy with high added value.
We have shown that BSF larvae can be reared on palm oil side streams. The pretreatment of the EFB-PKM mixture by fermentation with B. adusta accelerated development and simultaneously enhanced larval weight compared to the untreated control diet. These observations can be explained by the increase in digestibility through the depolymerization of lignocellulose, the accumulation of fungal mycelia as a potential nutrient source, and the shift to a predominantly anaerobic microbial gut community which subsequently degrades cellulose. Our protocol for the fermentation of EFB and PKM therefore represents a promising approach for the efficient bioconversion of high-fiber side streams by BSF larvae, allowing the production of valuable insect protein that reduces environmental pollution.
6. Patents
Klüber P, Zorn H, Rühl M, Bakonyi D, Pfeiffer J, and Vilcinskas A. Process for the production of an insect substrate, insect substrate and uses thereof. Application number EP22152265.9.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/su14095626/s1, Figure S1: Heat map showing the genus-level composition of the bacterial community characterized by amplicon sequencing of the 16S rRNA gene. Samples (feed, gut, and frass) of the non-fermented reference (NFR) and B. adusta fermented diet (BAD) groups are shown. Only the 38 most abundant classified genera are listed. Genera without a suitable classification (uncultivated, undefined, and not applicable) are excluded. The relative abundance (%) is shown as a color gradient that runs from light gray through black to blue, the latter representing the highest abundance. White areas indicate that the genus was not detected in the sample; Figure S2: Heat map showing the genus-level composition of the fungal community characterized by ITS amplicon sequencing. Samples (feed, gut, and frass) of the non-fermented reference (NFR) and B. adusta fermented diet (BAD) groups are shown. Only the 38 most abundant genera are listed. Genera without a suitable classification (uncultivated, undefined, and not applicable) are excluded. The relative abundance (%) is shown as a color gradient that runs from light gray through black to blue, the latter representing the highest abundance. White areas indicate that the genus was not detected in the sample; Table S1: Interactive spreadsheet showing the detailed composition of the bacterial community in the feed, BSF larval gut, and frass when larvae were reared on a non-fermented reference (NFR) diet or a B. adusta fermented (BAD) diet. Data are based on 16S rRNA gene sequencing and show the relative abundances of ASV counts at different taxonomic levels. The categories on the left allow switching between different taxonomic levels (2 = Phylum; 3 = Class; 4 = Order; 5 = Family; 6 = Genus; and 7 = Species); Table S2: Interactive spreadsheet showing the detailed composition of the fungal community composition in the feed, BSF larval guts, and frass when larvae were reared on a non-fermented reference (NFR) diet or a B. adusta fermented (BAD) diet. Data are based on ITS sequencing and show the relative abundances of ASV counts at different taxonomic levels. The categories on the left allow switching between different taxonomic levels (2 = Phylum; 3 = Class; 4 = Order; 5 = Family; 6 = Genus; and 7 = Species); Table S3: Number of genes involved in cellulose degradation (endoglucanases EC 3.2.1.4 and β-glucosidases EC 3.2.1.21) in the genomes of representatives of the 28 most positively and 28 most negatively correlated taxa found in BSF larval guts based on the discrete false-discovery rate (DS-FDR). Only genomes of representatives which show ≥91% sequence identity to the target sequence are included. Taxa that are not classified up to the family level are excluded.
Author Contributions
Conceptualization, P.K., H.Z., M.R. and D.T.; methodology, P.K., D.T. and J.P.; software, P.K., S.H. and D.T.; validation, P.K., D.T., S.H., H.Z. and M.R.; formal analysis, P.K. and D.T.; investigation, P.K., D.T. and J.P.; resources, H.Z., M.R. and A.V.; data curation, P.K., D.T. and S.H.; writing—original draft preparation, P.K., D.T. and S.H.; writing—review and editing, P.K., D.T., M.R. and H.Z.; visualization, P.K. and S.H.; supervision, P.K.; project administration, P.K.; funding acquisition, H.Z., M.R. and A.V. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Hessian Ministry of Higher Education, Research and the Arts via the LOEWE Center for Insect Biotechnology, and by PT Alternative Protein Indonesia. We acknowledge access to resources financially supported by the BMBF grant FKZ 031A533 within the de.NBI network. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Raw data for 16S rRNA gene and ITS amplicon sequencing are available via NCBI (accession number PRJNA723723).
Acknowledgments
We thank Bio S. Biogas for providing BSF larvae. We also thank Karina Brinkrolf (Justus Liebig University Giessen, Germany) for helpful suggestions on the experimental design, Stefan Janssen (Justus Liebig University Giessen, Germany) for valuable discussions about amplicon data analysis, Christopher Back for milling empty fruit bunches and screening fungal strains, and Richard M. Twyman for manuscript editing.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lalander, C.; Diener, S.; Zurbrügg, C.; Vinneras, B. Effects of feedstock on larval development and process efficiency in waste treatment with black soldier fly (Hermetia illucens). J. Clean. Prod. 2018, 208, 211–219. [Google Scholar] [CrossRef]
- Miranda, C.D.; Cammack, J.A.; Tomberlin, J.K. Life-history traits of the black soldier fly, Hermetia illucens (L.) (Diptera: Stratiomyidae), reared on three manure types. Animals 2019, 9, 281. [Google Scholar] [CrossRef] [PubMed]
- Gold, M.; Cassar, C.M.; Zurbrügg, C.; Kreuzer, M.; Boulos, S.; Diener, S.; Mathys, A. Biowaste treatment with black soldier fly larvae: Increasing performance through the formulation of biowastes based on protein and carbohydrates. Waste Manag. 2019, 102, 319–329. [Google Scholar] [CrossRef]
- Oonincx, D.G.A.B.; Van Huis, A.; Van Loon, J. Nutrient utilisation by black soldier flies fed with chicken, pig, or cow manure. J. Insects Food Feed 2015, 1, 131–139. [Google Scholar] [CrossRef]
- Müller, A.; Wolf, D.; Gutzeit, H.O. The black soldier fly, Hermetia illucens—A promising source for sustainable production of proteins, lipids and bioactive substances. Z. Naturforsch. 2017, 72, 351–363. [Google Scholar] [CrossRef] [PubMed]
- Jeon, H.; Park, S.; Choi, J.; Jeong, G.; Lee, S.-B.; Choi, Y.; Lee, S.-J. The Intestinal Bacterial Community in the Food Waste-Reducing Larvae of Hermetia illucens. Curr. Microbiol. 2011, 62, 1390–1399. [Google Scholar] [CrossRef]
- Tegtmeier, D.; Hurka, S.; Klüber, P.; Brinkrolf, K.; Heise, P.; Vilcinskas, A. Cottonseed press cake as a potential diet for industrially farmed black soldier fly larvae triggers adaptations of their bacterial and fungal gut microbiota. Front. Microbiol. 2021, 12, 634503. [Google Scholar] [CrossRef] [PubMed]
- Varotto Boccazzi, I.; Ottoboni, M.; Martin, E.; Comandatore, F.; Vallone, L.; Spranghers, T.; Eeckhout, M.; Mereghetti, V.; Pinotti, L.; Epis, S. A survey of the mycobiota associated with larvae of the black soldier fly (Hermetia illucens) reared for feed production. PLoS ONE 2017, 12, e0182533. [Google Scholar] [CrossRef]
- Bruno, D.; Bonelli, M.; De Filippis, F.; Di Lelio, I.; Tettamanti, G.; Casartelli, M.; Ercolini, D.; Caccia, S. The Intestinal Microbiota of Hermetia illucens Larvae Is Affected by Diet and Shows a Diverse Composition in the Different Midgut Regions. Appl. Environ. Microbiol. 2019, 85, e01864-18. [Google Scholar] [CrossRef]
- Wynants, E.; Frooninckx, L.; Crauwels, S.; Verreth, C.; De Smet, J.; Sandrock, C.; Wohlfahrt, J.; Van Schelt, J.; Depraetere, S.; Lievens, B.; et al. Assessing the Microbiota of Black Soldier Fly Larvae (Hermetia illucens) Reared on Organic Waste Streams on Four Different Locations at Laboratory and Large Scale. Microb. Ecol. 2018, 77, 913–930. [Google Scholar] [CrossRef]
- Vogel, H.; Müller, A.; Heckel, D.; Gutzeit, H.; Vilcinskas, A. Nutritional immunology: Diversification and diet-dependent expression of antimicrobial peptides in the black soldier fly Hermetia illucens. Dev. Comp. Immunol. 2018, 78, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Klammsteiner, T.; Walter, A.; Bogataj, T.; Heussler, C.D.; Stres, B.; Steiner, F.M.; Schlick-Steiner, B.C.; Arthofer, W.; Insam, H. The Core Gut Microbiome of Black Soldier Fly (Hermetia illucens) Larvae Raised on Low-Bioburden Diets. Front. Microbiol. 2020, 11, 993. [Google Scholar] [CrossRef] [PubMed]
- Klüber, P.; Müller, S.; Schmidt, J.; Zorn, H.; Rühl, M. Isolation of Bacterial and Fungal Microbiota Associated with Hermetia illucens Larvae Reveals Novel Insights into Entomopathogenicity. Microorganisms 2022, 10, 319. [Google Scholar] [CrossRef]
- Yang, F.; Tomberlin, J.K.; Jordan, H.R. Starvation Alters Gut Microbiome in Black Soldier Fly (Diptera: Stratiomyidae) Larvae. Front. Microbiol. 2021, 12, 601253. [Google Scholar] [CrossRef]
- Yu, G.; Cheng, P.; Yang, Z.; Chen, Y.; Li, Y.; Tomberlin, J.K. Inoculating Poultry Manure With Companion Bacteria Influences Growth and Development of Black Soldier Fly (Diptera: Stratiomyidae) Larvae. Environ. Entomol. 2011, 40, 30–35. [Google Scholar] [CrossRef]
- Martin, M.M. The Evolution of Insect-Fungus Associations: From Contact to Stable Symbiosis. Am. Zool. 1992, 32, 593–605. [Google Scholar] [CrossRef][Green Version]
- Correa, Y.; Cabanillas, B.; Jullian, V.; Álvarez, D.; Castillo, D.; Dufloer, C.; Bustamante, B.; Roncal, E.; Neyra, E.; Sheen, P.; et al. Identification and characterization of compounds from Chrysosporium multifidum, a fungus with moderate antimicrobial activity isolated from Hermetia illucens gut microbiota. PLoS ONE 2019, 14, e0218837. [Google Scholar] [CrossRef]
- Schrögel, P.; Wätjen, W. Insects for Food and Feed-Safety Aspects Related to Mycotoxins and Metals. Foods 2019, 8, 288. [Google Scholar] [CrossRef]
- Basiron, Y. Palm oil production through sustainable plantations. Eur. J. Lipid Sci. Technol. 2007, 109, 289–295. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations. FAOSTAT Database. 2018. Available online: http://faostat3.fao.org/home/E (accessed on 15 November 2021).
- Santosa, S.J. Palm Oil Boom in Indonesia: From Plantation to Downstream Products and Biodiesel. CLEAN-Soil Air Water 2008, 36, 453–465. [Google Scholar] [CrossRef]
- Rachmawati, R.; Buchori, D.; Hidayat, P.; Hem, S.; Fahmi, M.R. Perkembangan dan Kandungan Nutrisi Larva Hermetia illucens (Linnaeus) (Diptera: Stratiomyidae) pada Bungkil Kelapa Sawit. Jurnal Entomologi Indonesia 2010, 7, 28. [Google Scholar] [CrossRef]
- Andlar, M.; Rezić, T.; Marđetko, N.; Kracher, D.; Ludwig, R.; Šantek, B. Lignocellulose degradation: An overview of fungi and fungal enzymes involved in lignocellulose degradation. Eng. Life Sci. 2018, 18, 768–778. [Google Scholar] [CrossRef] [PubMed]
- Bajpai, P.; Anand, A.; Bajpai, P.K. Bleaching with lignin-oxidizing enzymes. Biotechnol. Annu. Rev. 2006, 12, 349–378. [Google Scholar] [PubMed]
- Diener, S.; Zurbrügg, C.; Tockner, K. Conversion of organic material by black soldier fly larvae: Establishing optimal feeding rates. Waste Manag. Res. J. Sustain. Circ. Econ. 2009, 27, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Lei, Y.; Rehman, K.U.; Yu, Z.; Zhang, J.; Li, W.; Li, Q.; Tomberlin, J.K.; Zheng, L. Dynamic Effects of Initial pH of Substrate on Biological Growth and Metamorphosis of Black Soldier Fly (Diptera: Stratiomyidae). Environ. Entomol. 2018, 47, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Klüber, P.; Bakonyi, D.; Zorn, H.; Rühl, M. Does light color temperature influence aspects of oviposition by the black soldier fly (Diptera: Stratiomyidae)? J. Econ. Entomol. 2020, 113, 2549–2552. [Google Scholar] [CrossRef]
- Barros, L.M.; Gutjahr, A.L.N.; Keppler, R.L.F.; Martins, R.T. Morphological description of the immature stages of Hermetia illucens (Linnaeus, 1758) (Diptera: Stratiomyidae). Microsc. Res. Tech. 2019, 82, 178–189. [Google Scholar] [CrossRef]
- Rozkosný, R. A Biosystematic Study of the European Stratiomyidae (Diptera): Clitellariinae, Hermediinae, Pachygasterinae and Bibliography; Springer: Dordrecht, The Netherlands, 1983. [Google Scholar]
- Sundberg, C.; Al-Soud, W.A.; Larsson, M.; Alm, E.; Yekta, S.S.; Svensson, B.H.; Sørensen, S.J.; Karlsson, A. 454 pyrosequencing analyses of bacterial and archaeal richness in 21 full-scale biogas digesters. FEMS Microbiol. Ecol. 2013, 85, 612–626. [Google Scholar] [CrossRef]
- Ihrmark, K.; Bödeker, I.T.M.; Cruz-Martinez, K.; Friberg, H.; Kubartova, A.; Schenck, J.; Strid, Y.; Stenlid, J.; Brandström-Durling, M.; Clemmensen, K.E.; et al. New primers to amplify the fungal ITS2 region—Evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol. Ecol. 2012, 82, 666–677. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Caporaso, J.G. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Abarenkov, K.; Zirk, A.; Piirmann, T.; Pöhönen, R.; Ivanov, F.; Nilsson, R.H.; Kõljalg, U. UNITE QIIME Release for Fungi; UNITE Community: London, UK, 2013. [Google Scholar]
- Werner, J.J.; Koren, O.; Hugenholtz, P.; DeSantis, T.Z.; Walters, W.A.; Caporaso, J.G.; Angenent, L.; Knight, R.; Ley, R. Impact of training sets on classification of high-throughput bacterial 16s rRNA gene surveys. ISME J. 2011, 6, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Caporaso, J.G. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Faith, D.P. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 1992, 61, 1–10. [Google Scholar] [CrossRef]
- Lozupone, C.; Knight, R. UniFrac: A New Phylogenetic Method for Comparing Microbial Communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef]
- Jiang, L.; Amir, A.; Morton, J.T.; Heller, R.; Arias-Castro, E.; Knight, R. Discrete False-Discovery Rate Improves Identification of Differentially Abundant Microbes. mSystems 2017, 2, e00092-17. [Google Scholar] [CrossRef]
- Markowitz, V.M.; Mavrommatis, K.; Ivanova, N.; Chen, I.-M.A.; Chu, K.; Kyrpides, N. IMG ER: A system for microbial genome annotation expert review and curation. Bioinformatics 2009, 25, 2271–2278. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Holmes, S. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 1979, 6, 65–70. [Google Scholar]
- Anderson, M.J. Permutational Multivariate Analysis of Variance (PERMANOVA); Balakrishnan, N., Colton, T., Everitt, B., Piegorsch, W., Ruggeri, F., Teugels, J.L., Eds.; Wiley StatsRef: Statistics Reference Online.
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Lee, S.; Kang, M.; Bae, J.-H.; Sohn, J.-H.; Sung, B.H. Bacterial Valorization of Lignin: Strains, Enzymes, Conversion Pathways, Biosensors, and Perspectives. Front. Bioeng. Biotechnol. 2019, 7, 209. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, Y.; Sudo, K.; Shimizu, K. Enzymatic hydrolysis of woods. II. Effect of grinding and alkali treatment on hydrolysis of woods by Trichoderma viride cellulase. J. Jpn. Wood Res. Soc. 1977, 23, 562–570. [Google Scholar]
- Brune, A. Symbiotic digestion of lignocellulose in termite guts. Nat. Rev. Genet. 2014, 12, 168–180. [Google Scholar] [CrossRef]
- Tartar, A.; Wheeler, M.M.; Zhou, X.; Coy, M.R.; Boucias, D.G.; Scharf, M.E. Parallel metatranscriptome analyses of host and symbiont gene expression in the gut of the termite Reticulitermes flavipes. Biotechnol. Biofuels 2009, 2, 25. [Google Scholar] [CrossRef]
- Callegari, M.; Jucker, C.; Fusi, M.; Leonardi, M.G.; Daffonchio, D.; Borin, S.; Savoldelli, S.; Crotti, E. Hydrolytic profile of the culturable gut bacterial community associated with Hermetia illucens. Front. Microbiol. 2020, 11, 1965. [Google Scholar] [CrossRef]
- Martínez, A.T.; Speranza, M.; Ruiz-Dueñas, F.J.; Ferreira, P.; Camarero, S.; Guillén, F.; Martinez, M.J.; Gutiérrez Suárez, A.; del Río Andrade, J.C. Biodegradation of lignocellulosics: Microbial, chemical, and enzymatic aspects of the fungal attack of lignin. Int. Microbiol. 2005, 8, 195–204. [Google Scholar]
- Pérez, J.; Muñoz-Dorado, J.; De La Rubia, T.; Martínez, J. Biodegradation and biological treatments of cellulose, hemicellulose and lignin: An overview. Int. Microbiol. 2002, 5, 53–63. [Google Scholar] [CrossRef]
- Quiroz-Castañeda, R.E.; Balcázar-López, E.; Dantán-González, E.; Martinez, A.; Folch-Mallol, J.; Martínez Anaya, C. Characterization of cellulolytic activities of Bjerkandera adusta and Pycnoporus sanguineus on solid wheat straw medium. Electron. J. Biotechnol. 2009, 12, 5–6. [Google Scholar]
- Quiroz-Castañeda, R.E.; Pérez-Mejía, N.; Martínez-Anaya, C.; Acosta-Urdapilleta, L.; Folch-Mallol, J. Evaluation of different lignocellulosic substrates for the production of cellulases and xylanases by the basidiomycete fungi Bjerkandera adusta and Pycnoporus sanguineus. Biodegradation 2011, 22, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, M.; Pinto, P.; Bezerra, R.; Dias, A.; Guedes, C.; Cardoso, V.; Cone, J.; Ferreira, L.; Colaço, J.; Sequeira, C. Effect of enzyme extracts isolated from white-rot fungi on chemical composition and in vitro digestibility of wheat straw. Anim. Feed Sci. Technol. 2008, 141, 326–338. [Google Scholar] [CrossRef]
- Klepzig, K.D.; Adams, A.S.; Handelsman, J.; Raffa, K.F. Symbioses: A Key Driver of Insect Physiological Processes, Ecological Interactions, Evolutionary Diversification, and Impacts on Humans. Environ. Entomol. 2009, 38, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Shil, R.K.; Mojumder, S.; Sadida, F.F.; Uddin, M.; Sikdar, D. Isolation and Identification of Cellulolytic Bacteria from the Gut of Three Phytophagus Insect Species. Braz. Arch. Biol. Technol. 2014, 57, 927–932. [Google Scholar] [CrossRef]
- Stefanini, I. Yeast-insect associations: It takes guts. Yeast 2018, 35, 315–330. [Google Scholar] [CrossRef]
- Luo, C.; Li, Y.; Chen, Y.; Fu, C.; Long, W.; Xiao, X.; Liao, H.; Yang, Y. Bamboo lignocellulose degradation by gut symbiotic microbiota of the bamboo snout beetle Cyrtotrachelus buqueti. Biotechnol. Biofuels 2019, 12, 70. [Google Scholar] [CrossRef]
- Tomberlin, J.K.; Sheppard, D.C.; Joyce, J.A. Selected Life-History Traits of Black Soldier Flies (Diptera: Stratiomyidae) Reared on Three Artificial Diets. Ann. Entomol. Soc. Am. 2002, 95, 379–386. [Google Scholar] [CrossRef]
- Tomberlin, J.K.; Sheppard, D.C. Factors Influencing Mating and Oviposition of Black Soldier Flies (Diptera: Stratiomyidae) in a Colony. J. Entomol. Sci. 2002, 37, 345–352. [Google Scholar] [CrossRef]
- Zheng, L.; Crippen, T.L.; Holmes, L.; Singh, B.; Pimsler, M.; Benbow, M.E.; Tarone, A.; Dowd, S.; Yu, Z.; VanLaerhoven, S.L.; et al. Bacteria Mediate Oviposition by the Black Soldier Fly, Hermetia illucens (L.), (Diptera: Stratiomyidae). Sci. Rep. 2013, 3, 2563. [Google Scholar] [CrossRef]
- Ponnusamy, L.; Wesson, D.M.; Arellano, C.; Schal, C.; Apperson, C.S. Species Composition of Bacterial Communities Influences Attraction of Mosquitoes to Experimental Plant Infusions. Microb. Ecol. 2010, 59, 158–173. [Google Scholar] [CrossRef]
- Sharon, G.; Segal, D.; Zilber-Rosenberg, I.; Rosenberg, E. Symbiotic bacteria are responsible for diet-induced mating preference in Drosophila melanogaster, providing support for the hologenome concept of evolution. Gut Microbes 2011, 2, 190–192. [Google Scholar] [CrossRef]
- Boyce, G.R.; Gluck-Thaler, E.; Slot, J.C.; Stajich, J.E.; Davis, W.J.; James, T.Y.; Cooley, J.R.; Panaccione, D.G.; Eilenberg, J.; De Fine Licht, H.H.; et al. Psychoactive plant- and mushroom-associated alkaloids from two behavior modifying cicada pathogens. Fungal Ecol. 2019, 41, 147–164. [Google Scholar] [CrossRef] [PubMed]
- Cifuentes, Y.; Glaeser, S.P.; Mvie, J.; Bartz, J.-O.; Müller, A.; Gutzeit, H.O.; Vilcinskas, A.; Kampfer, P. The gut and feed residue microbiota changing during the rearing of Hermetia illucens larvae. Antonie van Leeuwenhoek 2020, 113, 1323–1344. [Google Scholar] [CrossRef] [PubMed]
- Holdemann, L.V.; Cato, E.P.; Moore, W.E.C. (Eds.) Anaerobe Laboratory Manual; Anaerobe Laboratory, Virginia Polytechnic Institute and State University: Blacksburg, VA, USA, 1977. [Google Scholar]
- Biddle, A.; Stewart, L.; Blanchard, J.L.; Leschine, S. Untangling the Genetic Basis of Fibrolytic Specialization by Lachnospiraceae and Ruminococcaceae in Diverse Gut Communities. Diversity 2013, 5, 627–640. [Google Scholar] [CrossRef]
- Zheng, L.; Crippen, T.L.; Singh, B.; Tarone, A.M.; Dowd, S.; Yu, Z.; Wood, T.K.; Tomberlin, J.K. A Survey of Bacterial Diversity From Successive Life Stages of Black Soldier Fly (Diptera: Stratiomyidae) by Using 16S rDNA Pyrosequencing. J. Med. Entomol. 2013, 50, 647–658. [Google Scholar] [CrossRef] [PubMed]
- Korniłłowicz-Kowalska, T.; Rybczyńska-Tkaczyk, K. Growth conditions, physiological properties, and selection of optimal parameters of biodegradation of anticancer drug daunomycin in industrial effluents by Bjerkandera adusta CCBAS930. Int. Microbiol. 2019, 23, 287–301. [Google Scholar] [CrossRef] [PubMed]
- Lynd, L.R.; Weimer, P.J.; van Zyl, W.H.; Pretorius, I.S. Microbial cellulose utilization: Fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 2002, 66, 506–577. [Google Scholar] [CrossRef]
- Béguin, P. Molecular biology of cellulose degradation. Annu. Rev. Microbiol. 1990, 44, 219–248. [Google Scholar] [CrossRef]
- Million, M.; Raoult, D. Linking gut redox to human microbiome. Hum. Microbiome J. 2018, 10, 27–32. [Google Scholar] [CrossRef]
- Alves, L.M.C.; De Souza, J.A.M.; De Mello Varani, A.; De Macedo Lemos, E.G. The family Rhizobiaceae. In The Prokaryotes—Alphaproteobacteria and Betaproteobacteria; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 419–437. [Google Scholar]
- Lambiase, A. The family Sphingobacteriaceae. In The Prokaryotes—Other Major Lineages of Bacteria and Archaea; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 907–914. [Google Scholar]
- Mayilraj, S.; Stackebrandt, E. The family Paenibacillaceae. In The Prokaryotes—Firmicutes and Tenericutes; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 267–280. [Google Scholar]
- Stackebrandt, E. The family Lachnospiraceae. In The Prokaryotes—Firmicutes and Tenericutes; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 197–201. [Google Scholar]
- Rainey, F.A. Family V Lachnospiraceae. In Bergey’s Manual of Systematic Bacteriology; De Vos, P., Garrity, G.M., Jones, D., Krieg, N.R., Ludwig, W., Rainey, F.A., Schleifer, K.H., Whitman, W.B., Eds.; Springer: New York, NY, USA, 2009; p. 921. [Google Scholar]
- Greening, C.; Geier, R.; Wang, C.; Woods, L.C.; Morales, S.E.; McDonald, M.J.; Rushton-Green, R.; Morgan, X.C.; Koike, S.; Leahy, S.C.; et al. Diverse hydrogen production and consumption pathways influence methane production in ruminants. ISME J. 2019, 13, 2617–2632. [Google Scholar] [CrossRef]
- Brune, A. Methanogenesis in the digestive tracts of insects and other arthropods. Biogenesis of hydrocarbons. In Handbook of Hydrocarbon and Lipid Microbiology; Stams, A.J.M., Sousa, D.Z., Eds.; Springer: Cham, Switzerland, 2019; pp. 229–260. [Google Scholar]
- Mertenat, A.; Diener, S.; Zurbrügg, C. Black Soldier Fly biowaste treatment—Assessment of global warming potential. Waste Manag. 2019, 84, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Parodi, A.; De Boer, I.J.M.; Gerrits, W.J.J.; Van Loon, J.J.A.; Heetkamp, M.J.W.; Van Schelt, J.; Bolhuis, J.E.; Van Zanten, H.H.E. Bioconversion efficiencies, greenhouse gas and ammonia emissions during black soldier fly rearing—A mass balance approach. J. Clean Prod. 2020, 271, 122488. [Google Scholar] [CrossRef]
- Ermolaev, E.; Lalander, C.; Vinneras, B. Greenhouse gas emissions from small-scale fly larvae composting with Hermetia illucens. Waste Manag. 2019, 96, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.C.d.S.; Diniz, A.G.; Tiago, P.V.; de Oliveira, N.T. Entomopathogenic Fusarium species: A review of their potential for the biological control of insects, implications and prospects. Fungal Biol. Rev. 2020, 34, 41–57. [Google Scholar] [CrossRef]
- Sutherland, J.B.; Pometto, A.L., III; Crawford, D.L. Lignocellulose degradation by Fusarium species. Can. J. Bot. 1983, 61, 1194–1198. [Google Scholar] [CrossRef]
- Huang, Y.; Busk, P.K.; Lange, L. Cellulose and hemicellulose-degrading enzymes in Fusarium commune transcriptome and functional characterization of three identified xylanases. Enzyme Microb. Technol. 2015, 73, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Bennett, J.W.; Klich, M. Mycotoxins. Clin. Microbiol. Rev. 2003, 16, 497–516. [Google Scholar] [CrossRef]
- Bosch, G.; Van Der Fels-Klerx, H.J.; Rijk, T.C.D.; Oonincx, D.G. Aflatoxin B1 tolerance and accumulation in black soldier fly (Hermetia illucens) and yellow mealworms (Tenebrio molitor). Toxins 2017, 9, 185. [Google Scholar] [CrossRef]
- Langseth, W.; Bernhoft, A.; Rundberget, T.; Kosiak, B.; Gareis, M. Mycotoxin production and cytotoxicity of Fusarium strains isolated from Norwegian cereals. Mycopathologia 1999, 144, 103–113. [Google Scholar] [CrossRef]
- Marasas, W.F.O. Toxigenic fusaria. In Mycotoxins and Animal Foods; Smith, J.E., Henderson, R.S., Eds.; CRC Press: London, UK, 1991. [Google Scholar]
- Dickinson, E.; Harrison, M.; Parker, M.; Dickinson, M.; Donarski, J.; Charlton, A.; Nolan, R.; Rafat, A.; Gschwend, F.; Hallett, J.; et al. From waste to food: Optimizing the breakdown of oil palm waste to provide substrate for insects farmed as animal feed. PLoS ONE 2019, 14, e0224771. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).