2D/3D Pattern Formation Comparison Using Spectral Methods to Solve Nonlinear Partial Differential Equations of Condensed and Soft Matter
Abstract
1. Introduction
2. Materials Mathematical Models and Their Integration Rules
3. Materials and Methods
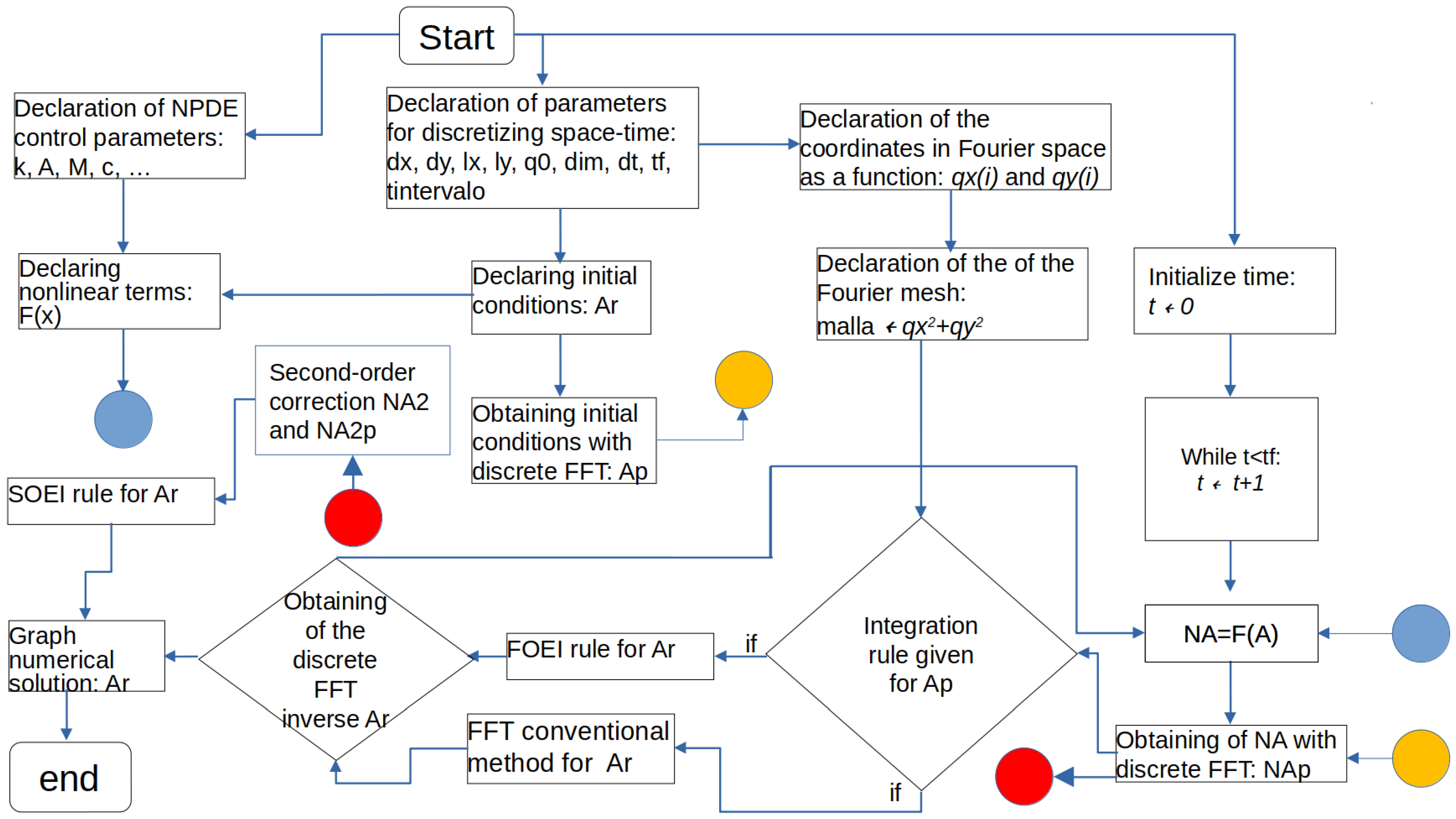
| Algorithm 1: Pseudo-code of spectral methods to solve nonlinear partial differential equation. |
| 1: Declaration of NPDE control parameters: k, A, M, c0, γ, g, λ, ε |
| 2: Declaration of parameters for discretizing space-time: dx, dy, lx, ly, q0, dim = (lx,ly), dt, tf, tintervalo |
| 3: Declaration of the coordinate in Fourier space as a function: qx(i) |
| 4: Declaration of the coordinate in Fourier space as a function: qy(i) |
| 5: Declaration of the Fourier mesh as an array of dimension dim: malla |
| 6: Declaring nonlinear terms as a function: F(X) |
| 7: Declaring initial conditions as an array: Ar |
| 8: Obtaining initial conditions with discrete FFT as an array: Ap |
| 9: Initialize time counter: t ← 0 |
| 10: While t < tf: |
| 11: t ← t + 1 |
| 12: Nonlinear function call: NA = F(Ar) |
| 13: Obtaining of NA with discrete FFT: NAp |
| 14: Integration rule given for Equations (4), (6) and (7) or (6)–(9): Ap |
| 15: Obtaining of the discrete FFT inverse: Ar |
| 16: Nonlinear function call second: NA2 = F(Ar) |
| 17: Obtaining of NA with discrete FFT for the second time: NA2p |
| 18: Second-order correction of the integration rule: Ap |
| 19: Obtaining of the discrete FFT inverse for the second time: Ar |
| 20: if t % tintervalo ← 0: |
| 21: Graph numerical solution: Ar |
| 22: Updates the counter by an order of magnitude: tintervalo |
| 23: end while |
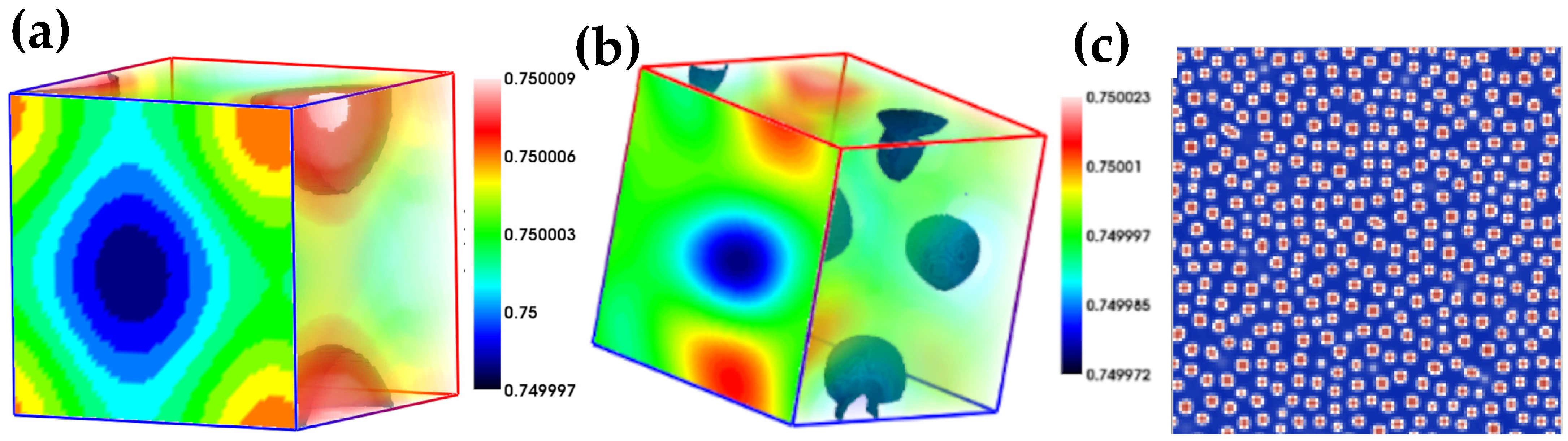
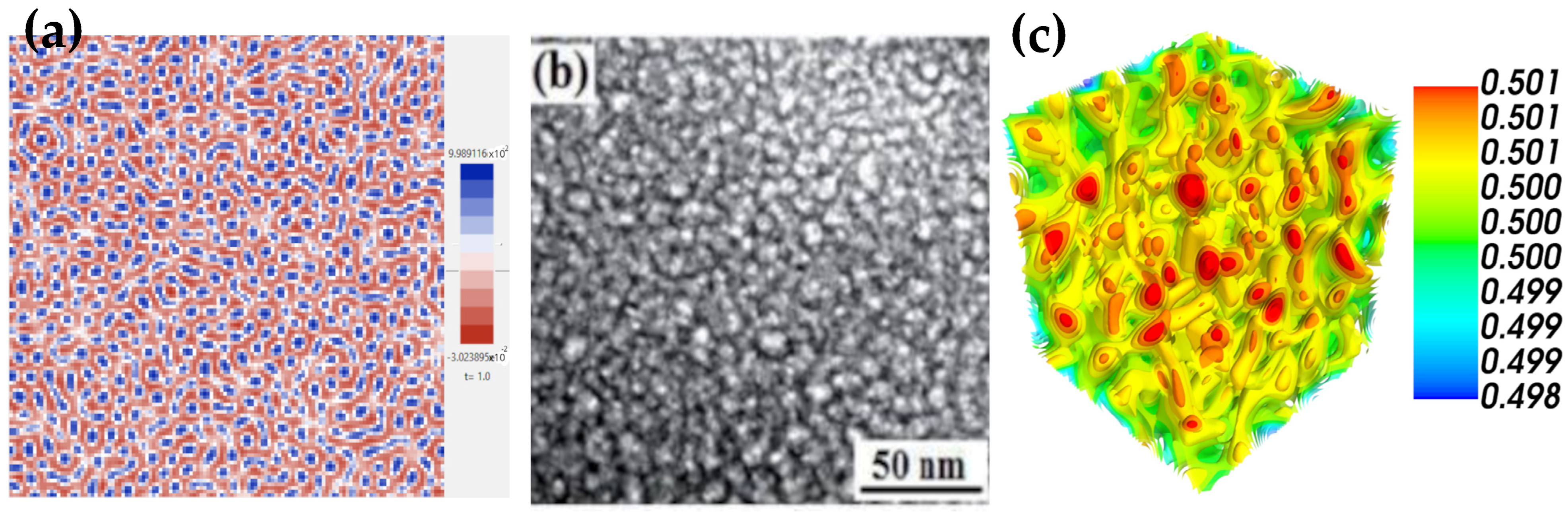
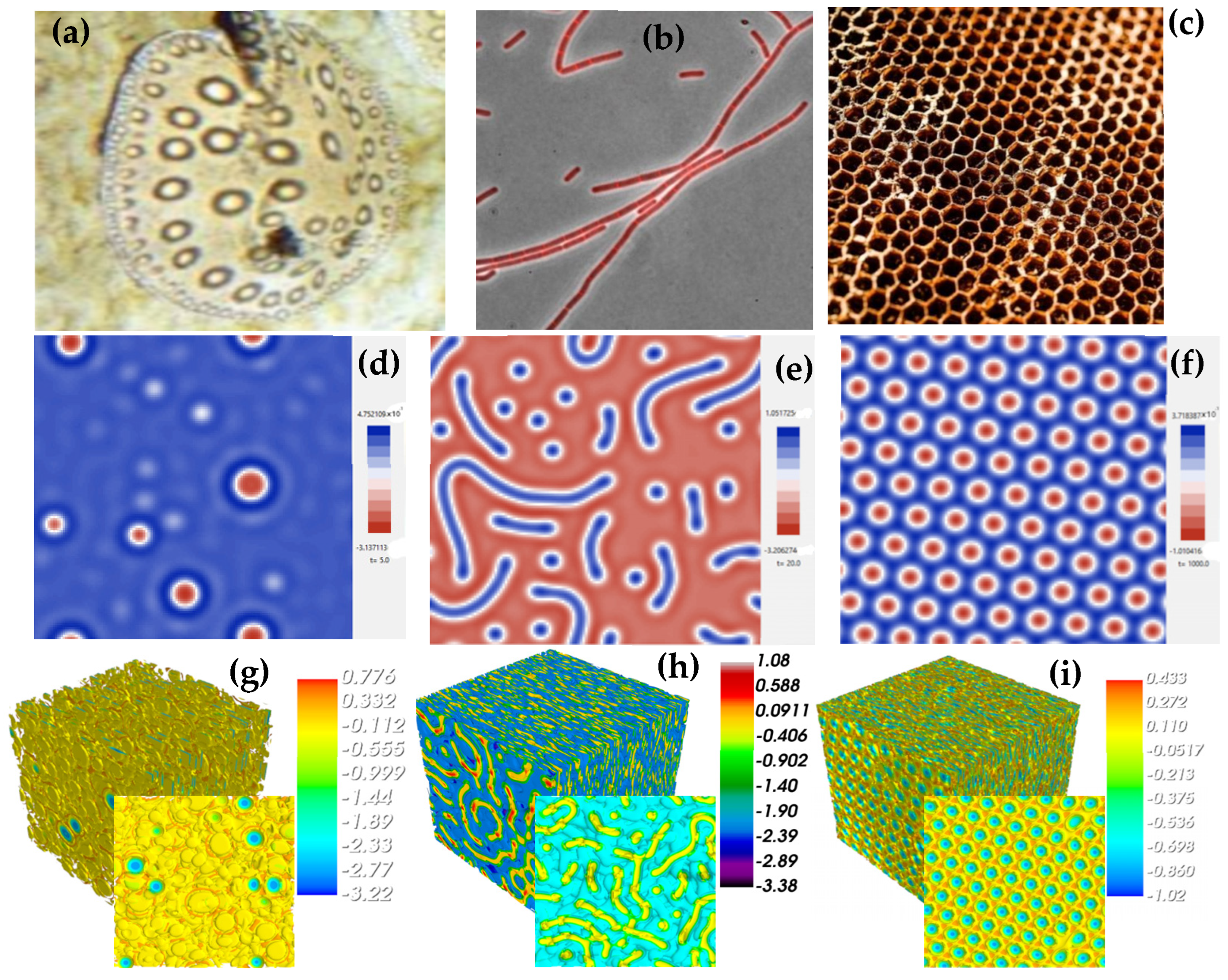
4. Results of the 2D and 3D Simulations
4.1. Python Implementation of the Cahn–Hilliard Equation: Source Code
| 1 | from math import * |
| 2 | from numpy import * |
| 3 | from numpy.fft import * |
| 4 | |
| 5 | k = 1.0 |
| 6 | M = 1.0 #movilidad |
| 7 | A = 1.0 #barrera de energía |
| 8 | c0 = 0.5 |
| 9 | print (“M=“, M, “c0=“, c0) |
| 10 | t0 = 0 |
| 11 | tf = 150,000 |
| 12 | dt = 0.01 (0.001 for 3D pattern) |
| 13 | dx = 0.5 |
| 14 | dy = dx |
| 15 | q0 = 1.0 * dx |
| 16 | q02 = q0 * q0 |
| 17 | lx = 128 |
| 18 | ly = lx |
| 19 | dim = (lx, ly) |
| 20 | tintervalo = 100 |
| 21 | |
| 22 | def qx(i): |
| 23 | if (i < lx/2): |
| 24 | qxi = 2.0 * pi * float(i − 1)/(float(lx) * dx) |
| 25 | else: |
| 26 | qxi = −2.0 * pi * float(lx – i + 1)/(float(lx) * dx) |
| 27 | return qxi |
| 28 | def qy(j): |
| 29 | qyi = ((2.0 * pi) * float(j − 1))/(float(ly) * dy) |
| 30 | return qyi |
| 31 | |
| 32 | malla = array([[qx(i) * qx(i) + qy(j) * qy(j) for j in range(1, int((ly + 2)/2 + 1))] for i in range(1, int(lx + 1))]) |
| 33 | p2 = malla/q02 |
| 34 | print (‘q02’, malla.shape) |
| 35 | |
| 36 | def F(x): |
| 37 | s = 2 * A * x * (1 − x)*(1 − 2 * x) |
| 38 | return s |
| 39 | |
| 40 | noise_str = 0.5 * 10**(−3) |
| 41 | A = uniform((c0 − noise_str), (c0 + noise_str), dim) |
| 42 | Ap = rfft2(A, dim, (−2, −1)) |
| 43 | print (Ap.shape) |
| 44 | import numpy as np |
| 45 | import matplotlib.pyplot as plt |
| 46 | t = t0 |
| 47 | while t < tf: |
| 48 | t = t + 1 |
| 49 | NA = F(A) |
| 50 | Nap = rfft2(NA, dim, (−2, −1)) |
| 51 | Ap = (Ap − dt * (p2 * M) * (NAp))/(1 + dt * (p2 * p2) * (M * k)) |
| 52 | A = irfft2(Ap, dim, (−2, −1)) |
| 53 | if t % tintervalo == 0: |
| 54 | plt.imshow(A, cmap =‘bwr’) |
| 55 | plt.imshow(A, cmap = ‘bwr’) |
| 56 | plt.title(“Cahn–Hilliard equation”) |
| 57 | plt.show() |
| 58 | if t == tintervalo * 10: |
| 59 | tintervalo = 10 * tintervalo |
- 23 def qx(i):
- 24 if (i < lx/2):
- 25 qxi = 2.0 * pi * float(i − 1)/(float(lx) * dx)
- 26 else:
- 27 qxi = −2.0 * pi * float(lx – i + 1)/(float(lx) * dx)
- 28 return qxi
- 29 def qy(j):
- 30 if (j < ly/2):
- 31 qyi = 2.0 * pi * float(j − 1)/(float(ly) * dy)
- 32 else:
- 33 qyi = −2.0*pi*float(ly − j + 1)/(float(ly) * dy)
- 34 return qyi
- 35 def qz(k):
- 36 qzi = ((2.0 * pi) * float(k − 1))/(float(lz) * dz)
- 37 return qzi
- 38 malla = array([[[qx(i) * qx(i) + qy(j) * qy(j) + qz(k) * qz(k) for k in range(1, int((lz + 2)/2 + 1))] for j in range(1, int(ly + 1))] for i in range(1, int(lx + 1))])
- 57 print(A [64, 64, 64], “t=”, t * dt, “n=”, t)
- 58 G.display(A [32, :, :])
- 59 G.time(t * dt)
- 60 archive = open(“test3DFFT”+str(t)+“.pkl”,“wb”)
- 61 pickle.dump (A, archive)
- 62 archive.close()
- 63 if t == tintervalo * 10
- 64 tintervalo = 10 * tintervalo
4.2. Implementation Under Python for Numerically Solving the Swift–Hohenberg Equation
4.3. Python Implementation for Numerically Solving the Cahn–Hilliard–Swift–Hohenberg Equation
4.4. Implementation of the Algorithm ETD2RK for Numerically Solving the Conserved SHE
- 53 b = −q2 * (gamma-lambda(1 − q2)**2) * dt
- 54 Ap = exp(b * dt) − p2 * NAp * (exp(b * dt) − 1)/b
- 55 A = ifft(Ap, dim, (−3, −2, −1))
- 56 NA2 = F(A)
- 57 NAp2 = rfft(NA2, dim, (−3, −2, −1))
- 58 Ap = Ap − p2 * (NAp2 − NAp) * (exp(b * dt) − (1 + b * dt))/(b * dt**2)
- 59 A = ifft(Ap, dim, (−3, −2, −1))
- 60 if t % tintervalo == 0:
- 61 print(A [64, 64, 64], “t=”,t * dt, “n=”, t)
- 62 archive = open(“test3DFFT”+str(t)+ “.pkl”, ”wb”)
- 63 pickle.dump(A, archive)
- 64 archive.close()
- 65 if t == tintervalo * 10:
- 66 tintervalo = 10 * tintervalo
5. Discussion
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MAM | Marco A. Morales |
| DAPM | Diana A. Pérez-Muñoz |
| JAHG | J. Aejandro. Hernández-González |
| MAF | Miguel Alvarado-Flores |
| SRS | Sinuhé Ruiz-Salgado |
Appendix A
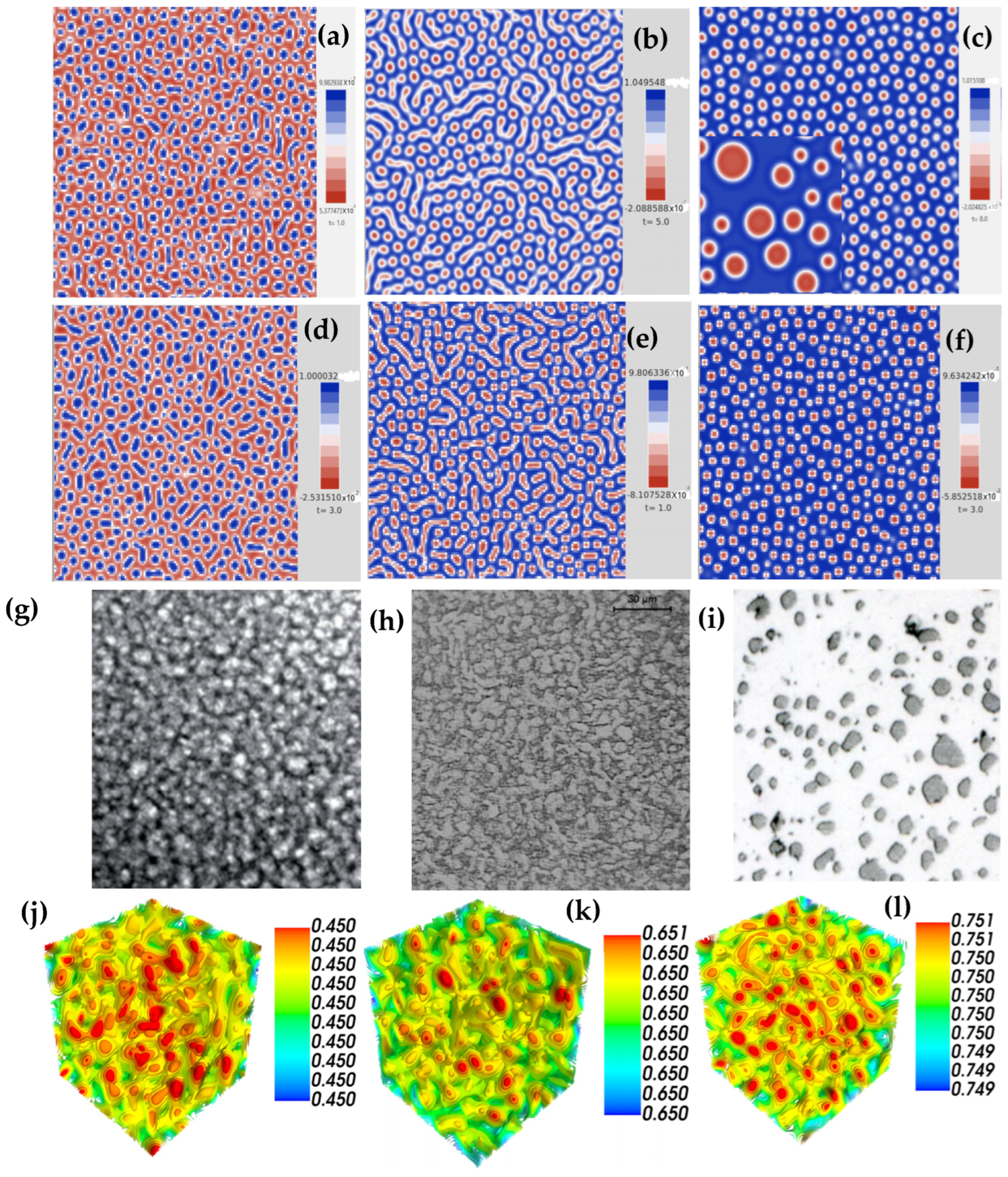
| Parameter | Figure 2a | Figure 2c | Figure 3a | Figure 3b | Figure 3c |
|---|---|---|---|---|---|
| L × L | 1282 | 1283 | 643 | 1283 | 1282 |
| C0 | 0.5 | 0.5 | 0.75 | 0.75 | 0.75 |
| K | 0.01 | 0.001 | 0.001 | 0.001 | 0.001 |
| M | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| A | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| T | 1.0 | 0.1 | 1.0 | 3.0 | 1.0 |
| dx | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
| Dt | 0.001 | 0.001 | 0.0001 | 0.001 | 0.001 |
| Iterations | 1000 | 100 | 10,000 | 3000 | 1000 |
| Parameter | Figure 4d | Figure 4e | Figure 4f | Figure 4g | Figure 4h | Figure 4i |
|---|---|---|---|---|---|---|
| L × L | 1282 | 1282 | 1282 | 1283 | 1283 | 1283 |
| T | 5.0 | 20 | 0.001 | 5.0 | 20 | 0.001 |
| 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | |
| 0.5 | 0.5 | 0.13 | 0.5 | 0.5 | 0.13 | |
| G | −3 | −3 | −1 | −3 | −3 | −1 |
| Dt | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 |
| Iterations | 500 | 2000 | 100,000 | 5500 | 2000 | 100,000 |
| Parameter | Figure 5 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Figure 5a | Figure 5b | Figure 5c | Figure 5d | Figure 5e | Figure 5f | Figure 5j | Figure 5k | Figure 5l | |
| L × L | 2562 | 2562 | 2562 | 1282 | 1282 | 1282 | 1283 | 1283 | 1283 |
| C0 | 0.45 | 0.65 | 0.75 | 0.45 | 0.65 | 0.75 | 0.45 | 0.3 | 0.75 |
| K | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.001 | 0.001 | 0.001 |
| M | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| A | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| T | 6.0 | 5.0 | 8.0 | 1.0 | 1.0 | 1.0 | 0.1 | 20.0 | 3.0 |
| dx | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
| dt | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 |
| Iterations | 8000 | 6000 | 11000 | 4000 | 3000 | 5000 | 1100 | 20000 | 3000 |
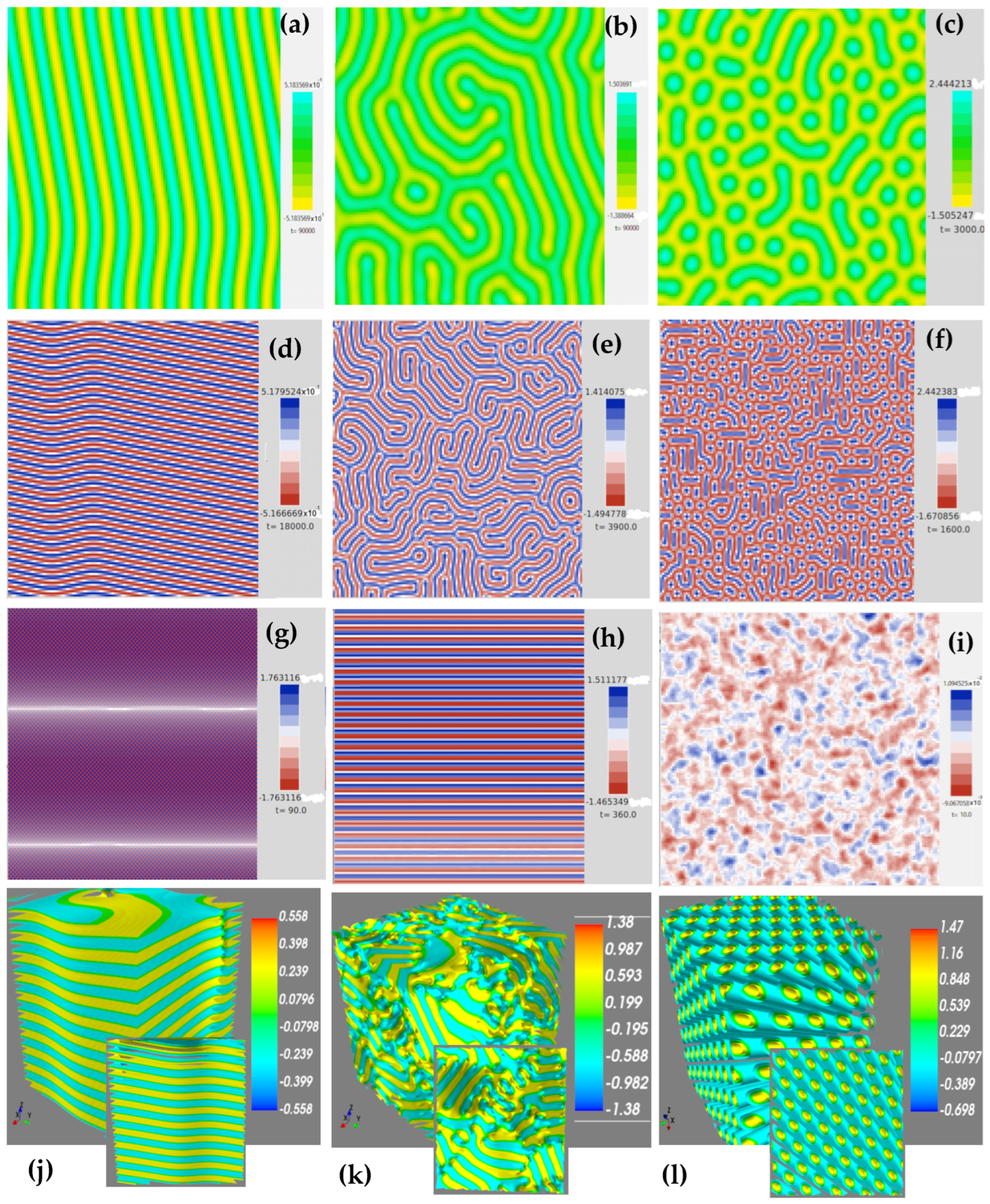
| Parameter | Figure 6 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Figure 6a | Figure 6b | Figure 6c | Figure 6d | Figure 6e | Figure 6f | Figure 6g | Figure 6h | Figure 6i | Figure 6j | Figure 6k | Figure 6l | |
| L × L | 1282 | 1282 | 1282 | 1282 | 1282 | 1282 | 1282 | 1282 | 1282 | 1282 | 1282 | 1282 |
| −0.2 | −1 | −2 | −0.2 | 1 | −2 | −0.2 | 1 | −2 | −0.2 | −1 | −0.4 | |
| 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.8 | |
| G | 0 | 0 | 1.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.0 |
| 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | |
| T | 90,000 | 90,000 | 40,000 | 1.0 | 1.0 | 0.4 | 1.0 | 70.0 | 10.0 | 104 | 104 | 104 |
| Dx | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
| Dt | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| Iterations | 9 × 105 | 9 × 105 | 400,000 | 10 | 10 | 4 | 10 | 700 | 100 | 2 × 105 | 105 | 2 × 105 |
| Algorithm Type | Time (u. a.) | Time Step | Computational Time (s) | Iteration Number | Morphology | |
|---|---|---|---|---|---|---|
| Conventional FFT | 14,000 | 0.01 | 2345.825 | 1400,000 | 5 dx | Lamellar Phase |
| Exponential of 1rt. order | 370 | 0.001 | 728.485 | 360,000 | ||
| Exponential of 2° order | 90,000 | 0.1 | 1804.712 | 900,000 | ||
| Conventional FFT | 2000 | 0.01 | 333.719 | 200,000 | 1.0 | Lamellar Phase |
| Exponential of 1rt. order | 100 | 0.001 | 196.663 | 100,000 | ||
| Exponential of 2° order | 2000 | 0.1 | 43.287 | 20,000 | ||
| Conventional FFT | 150,000 | 0.01 | 265.035 | 15,000 | 5 dx | Labyrinths |
| Exponential of 1rt. order | ---- | 0.001 | ---- | ---- | ||
| Exponential of 2° order | 90,000 | 0.1 | 188.352 | 9000 | ||
| Conventional FFT | 26,000 | 0.01 | 467.210 | 260,000 | 5 dx | Labyrinths |
| Exponential of 1rt. order | ---- | 0.001 | ---- | ---- | ||
| Exponential of 2° order | 8000 | 0.1 | 164.248 | 80,000 | ||
| Conventional FFT | 1600 | 0.01 | 281.799 | 160,000 | 1.0 | Dots and stripes |
| Exponential of 1rt. order | ---- | 0.001 | ---- | ---- | ||
| Exponential of 2° order | 3000 | 0.1 | 60.485 | 30,000 | ||
| Conventional FFT | 10,000 | 0.01 | 190.490 | 100,000 | 5 dx | Dots and stripes |
| Exponential of 1rt. order | ---- | 0.001 | ---- | ---- | ||
| Exponential of 2° order | 3000 | 0.1 | 63.377 | 30,000 |
Appendix B. Source Code for Plot 3D Patterns
- 1 from mayavi.mlab import *
- 2 from numpy import *
- 3 import pickle
- 4 mode = 1
- 5 name = “file_name.pkl”
- 6 out = open(name+“.pkl”, “rb”)
- 7 array_n = load(out, allow_pickle = True)
- 8 out.close()
- 9 if mode == 1:
- 10 patron3d = contour3d(n, colormap = “blue-red”)
- 11 elif mode == 2:
- 12 iso_superficie = contour3d(n, contours=10, transparent = True)
- 13 elif mode == 3:
- 14 for i in range(0, lz):
- 15 patron2D = imshow(n[i], colormap = “blue-red”)
Appendix C. Deduction of the Cahn–Hilliard–Swift–Hohenberg Equation
References
- Yoon, S.; Jeong, D.; Lee, C.; Kim, H.; Kim, S.; Lee, H.G.; Kim, J. Fourier-spectral method for the phase-field equations. Mathematics 2020, 8, 1385. [Google Scholar] [CrossRef]
- Cross, M.C.; Meiron, D.; Tu, Y. Chaotic domains: A numerical investigation. Chaos Interdiscip. J. Nonlinear Sci. 1994, 4, 607–619. [Google Scholar] [CrossRef]
- Boyer, D.; Romeu, D. Modeling grain boundaries in solids using a combined nonlinear and geometrical method. Int. J. Mod. Phys. B 2005, 19, 4047–4056. [Google Scholar] [CrossRef]
- Singh, D.; Mor, K.; Singh, D.; Tiwari, R.S. Phase separation in Ce-based metallic glasses. In Metallic Glasses; IntechOpen: London, UK, 2019; pp. 1–18. [Google Scholar] [CrossRef]
- Oliphant, T.E. Python for scientific computing. Comput. Sci. Eng. 2007, 9, 10–20. [Google Scholar] [CrossRef]
- Johansson, R.; Johansson, R.; John, S. Numerical Python; Apress: Berkeley, CA, USA, 2019; Volume 1. [Google Scholar] [CrossRef]
- Ascher, D.; Dubois, P.F.; Hinsen, K.; Hugunin, J.; Oliphant, T. An Open Source Project: Numerical Python; Lawrence Livermore National Laboratory, The University of California: Livermore, CA, USA, 2001. [Google Scholar]
- Millman, K.J.; Aivazis, M. Python for scientists and engineers. Comput. Sci. Eng. 2011, 13, 9–12. [Google Scholar] [CrossRef]
- Dubois, P.F.; Hinsen, K.; Hugunin, J. Numerical python. Comput. Phys. 1996, 10, 262–267. [Google Scholar] [CrossRef]
- Bulent Biner, S. Programming Phase-Field Modeling; Springer: Cham, Switzerland, 2017. [Google Scholar] [CrossRef]
- Sánchez Pérez-Moreno, S.; Ruiz Chavarría, S.; Ruiz Chavarría, G. Numerical Solution of the Swift–Hohenberg Equation. In Experimental and Computational Fluid Mechanics; Klapp, J., Medina, A., Eds.; Environmental Science and Engineering Series; Springer: Cham, Switzerland, 2014. [Google Scholar] [CrossRef]
- Ramachandran, P.; Varoquaux, G. Mayavi: 3D visualization of scientific data. Comput. Sci. Eng. 2011, 13, 40–51. [Google Scholar] [CrossRef]
- Sullivan, C.; Kaszynski, A. PyVista: 3D plotting and mesh analysis through a streamlined interface for the Visualization Toolkit (VTK). J. Open Source Softw. 2019, 4, 1450. [Google Scholar] [CrossRef]
- Guyer, J.E.; Wheeler, D.; Warren, J.A. FiPy: Partial differential equations with Python. Comput. Sci. Eng. 2009, 11, 6–15. [Google Scholar] [CrossRef]
- Langtangen, H.P.; Logg, A. Solving PDEs in Python: The FEniCS Tutorial I; Springer Nature: Cham, Switzerland, 2017. [Google Scholar] [CrossRef]
- Logg, W.; Wells, G.N.; Bengzon, F. DOLFIN User Manual. 2010. Available online: https://www.csc.kth.se/utbildning/kth/kurser/DN1240/numfcl10/kursmaterial10/dolfin-user-manual-2010-11-04.pdf (accessed on 1 September 2025).
- Kim Doochul, H.; Kim, W.T.; Park, E.S.; Mattern, N.; Eckert, J. Phase separation in metallic glasses. Prog. Mater. Sci. 2013, 58, 1103–1172. [Google Scholar] [CrossRef]
- Morales, M.A.; Fernández-Cervantes, I.; Agustín-Serrano, R.; Anzo, A.; Sampedro, M.P. Patterns formation in ferrofluids and solid dissolutions using stochastic models with dissipative dynamics. Eur. Phys. J. B 2016, 89, 182. [Google Scholar] [CrossRef]
- Nauman, E.B.; He, D.Q. Nonlinear diffusion and phase separation. Chem. Eng. Sci. 2001, 56, 1999–2018. [Google Scholar] [CrossRef]
- Lee, C.; Jeong, D.; Yoon, S.; Kim, J. Porous three-dimensional scaffold generation for 3D printing. Mathematics 2020, 8, 94. [Google Scholar] [CrossRef]
- Permyakova, E.V.; Goldobin, D.S. Exponential Time Differencing for Stiff Systems with Nondiagonal Linear Part. J. Appl. Mech. Tech. Phys. 2020, 61, 1227–1237. [Google Scholar] [CrossRef]
- Martínez-Agustín, F.; Ruiz-Salgado, S.; Zenteno-Mateo, B.; Rubio, E.; Morales, M.A. 3D pattern formation from coupled Cahn-Hilliard and Swift-Hohenberg equations: Morphological phases transitions of polymers, block and diblock copolymers. Comput. Mater. Sci. 2022, 210, 111431. [Google Scholar] [CrossRef]
- Elder, K.R.; Vinals, J.; Grant, M. Ordering dynamics in the two-dimensional stochastic Swift-Hohenberg equation. Phys. Rev. Lett. 1992, 68, 3024. [Google Scholar] [CrossRef]
- Boyer, D.; Viñals, J. Weakly Nonlinear Theory of Grain Boundary Motion in Patterns with Crystalline Symmetry. Phys. Rev. Lett. 2002, 89, 055501. [Google Scholar] [CrossRef]
- Boyer, D.; Vinals, J. Grain-boundary motion in layered phases. Phys. Rev. E 2001, 63, 061704. [Google Scholar] [CrossRef]
- Uwe, T.; Andrew, A.; Mark, R.; Hector, G.; Edgar, K. Localized states in the conserved Swift-Hohenberg equation with cubic nonlinearity. Phys. Rev. E 2013, 87, 042915. [Google Scholar] [CrossRef]
- Morales, M.A.; Rojas, J.F.; Torres, I.; Rubio, E. Modeling ternary mixtures by mean-field theory of polyelectrolytes: Coupled Ginzburg–Landau and Swift–Hohenberg equations. Phys. A Stat. Mech. Appl. 2012, 391, 779–791. [Google Scholar] [CrossRef]
- Boyer, D.; Viñals, J. Grain boundary pinning and glassy dynamics in stripe phases. Phys. Rev. E 2002, 65, 046119. [Google Scholar] [CrossRef] [PubMed][Green Version]
- van Teeffelen, S.; Backofen, R.; Voigt, A.; Löwen, H. Derivation of the phase field crystal model for colloidal solidification. Phys. Rev. E 2009, 79, 051404. [Google Scholar] [CrossRef] [PubMed]
- Morales, M.A.; Ruiz-Salgado, S.; Vázquez-Montiel, R.H.; Agustín-Serrano, R.; Zenteno-Mateo, B. Superconductive phases simulation from a PFC models approach using the non-potential conserved Swift-Hohenberg equation. Commun. Nonlinear Sci. Numer. Simul. 2025, 144, 108667. [Google Scholar] [CrossRef]
- Cox Steven, M.; Paul, C. Matthews. Exponential time differencing for stiff systems. J. Comput. Phys. 2002, 176, 430–455. [Google Scholar] [CrossRef]
- Reis, D.A.P.; de Moura Neto, C.; Couto, A.A.; Silva, C.R.M.; Neto, F.P.; Barboza, M.J.R. A Comparison between CoNiCrAlY Bond Coat and Zirconia Plasma Sprayed Coatings on Creep Tests. Mater. Sci. Forum 2008, 591–593, 30–35. [Google Scholar] [CrossRef]
- García, M.; Martínez, V.; Ortiz, A. Fabricación de Componentes Metálicos Mediante Reoforjado. Supl. Rev. Latinoam. Metal. Mater. 2009, S1, 1297–1308. [Google Scholar] [CrossRef]
- Morales, M.A.; Ruiz-Salgado, S.; Agustín-Serrano, R.; Zenteno-Mateo, B.; Rodríguez-Mora, J.I. Design and mathematical modeling of polymers phases to obtain controlled microporosity materials by 3D printing. Prog. Manuf. Addit. 2021, 8, 1701–7010. [Google Scholar] [CrossRef]
- Morales, M.A.; Rojas, J.F.; Oliveros, J. A new mechanochemical model: Coupled Ginzburg–Landau and Swift–Hohenberg equations in biological patterns of marine animals. J. Theor. Biol. 2015, 368, 37–54. [Google Scholar] [CrossRef]
- Pinna, M.; Zvelindovsky, A.V. Kinetic pathways of gyroid-to-cylinder transitions in diblock copolymers under external fields: Cell dynamics simulation. Soft Matter 2008, 4, 316–327. [Google Scholar] [CrossRef] [PubMed]
- Nikoubashman, A.; Register, R.A.; Panagiotopoulos, A.Z. Simulations of shear-induced morphological transitions in block copolymers. Soft Matter 2013, 9, 9960–9971. [Google Scholar] [CrossRef]
- Nicolaides, D. Mesoscale modeling. Mol. Simul. 2001, 26, 51–72. [Google Scholar] [CrossRef]
- Pinna, M.; Zvelindovsky, A.V.; Todd, S.; Goldbeck-Wood, G. Cubic phases of block copolymers under shear and electric fields by cell dynamics simulation. I. Spherical phase. J. Chem. Phys. 2006, 125, 154905. [Google Scholar] [CrossRef] [PubMed]
| Number Line | Interpretation |
|---|---|
| 1–4 | Libraries used. |
| 5–9 | Simulation-specific material parameters. |
| 7 | Gradient energy coefficient. |
| 10 | Print the value of the control parameters on the screen. |
| 8 | Energy barrier value. |
| 11–13 | Time integration parameters. |
| 14–15 | Parameters of the simulation cell for the mesh. Distance between nodes in “x” and “y” directions. |
| 16 | Fourier mode of mesh. |
| 18 | Number of grid points in the x-direction. |
| 19 | Number of grid points in the y-direction. |
| 20 | Mesh size or total number of simulation grid points. |
| 21 | Iteration interval for plotting a 2D (or 3D) pattern. |
| 23–31 | Discretization of the Fourier space from −2π to 2π and from 0 to 2π. |
| 33 | Definition of the discretized Fourier Space. |
| 34 | Array forming the normalized Fourier space. |
| 35 | Print the dimension of the Fourier space array. |
| 37–39 | Fretché’s functional derivative statement. |
| 41–43 | Statement of random initial conditions from 3D space to the Fourier space. |
| 44 | Print FFT array dimension of initial condition. |
| 45–47 | Graphical part call with PyLab to display the numerical solution. |
| 48–54 | FFT algorithm. |
| 47 | Initialize variable t. |
| 49 | Cycle over time. |
| 50 | Counter. |
| 51 | Call the function of the nonlinear terms. |
| 52 | Transformation of nonlinear terms to Fourier space. |
| 53 | Modified Euler’s rule in Fourier space, Equation (4). |
| 54 | Inverse transforms the solution to real space. |
| 55–57 | Lines of code to display the solution at iteration t or at dimensionless time t dt. |
| 58–59 | Counter update for real-time display for a variable interval. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morales, M.A.; Pérez-Muñoz, D.A.; Hernández-González, J.A.; Alvarado-Flores, M.; Ruiz-Salgado, S. 2D/3D Pattern Formation Comparison Using Spectral Methods to Solve Nonlinear Partial Differential Equations of Condensed and Soft Matter. Algorithms 2025, 18, 585. https://doi.org/10.3390/a18090585
Morales MA, Pérez-Muñoz DA, Hernández-González JA, Alvarado-Flores M, Ruiz-Salgado S. 2D/3D Pattern Formation Comparison Using Spectral Methods to Solve Nonlinear Partial Differential Equations of Condensed and Soft Matter. Algorithms. 2025; 18(9):585. https://doi.org/10.3390/a18090585
Chicago/Turabian StyleMorales, Marco A., Dania A. Pérez-Muñoz, J. Alejandro Hernández-González, Miguel Alvarado-Flores, and Sinuhé Ruiz-Salgado. 2025. "2D/3D Pattern Formation Comparison Using Spectral Methods to Solve Nonlinear Partial Differential Equations of Condensed and Soft Matter" Algorithms 18, no. 9: 585. https://doi.org/10.3390/a18090585
APA StyleMorales, M. A., Pérez-Muñoz, D. A., Hernández-González, J. A., Alvarado-Flores, M., & Ruiz-Salgado, S. (2025). 2D/3D Pattern Formation Comparison Using Spectral Methods to Solve Nonlinear Partial Differential Equations of Condensed and Soft Matter. Algorithms, 18(9), 585. https://doi.org/10.3390/a18090585





