Novel TDP1 Inhibitors: Disubstituted Thiazolidine-2,4-Diones Containing Monoterpene Moieties
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemistry
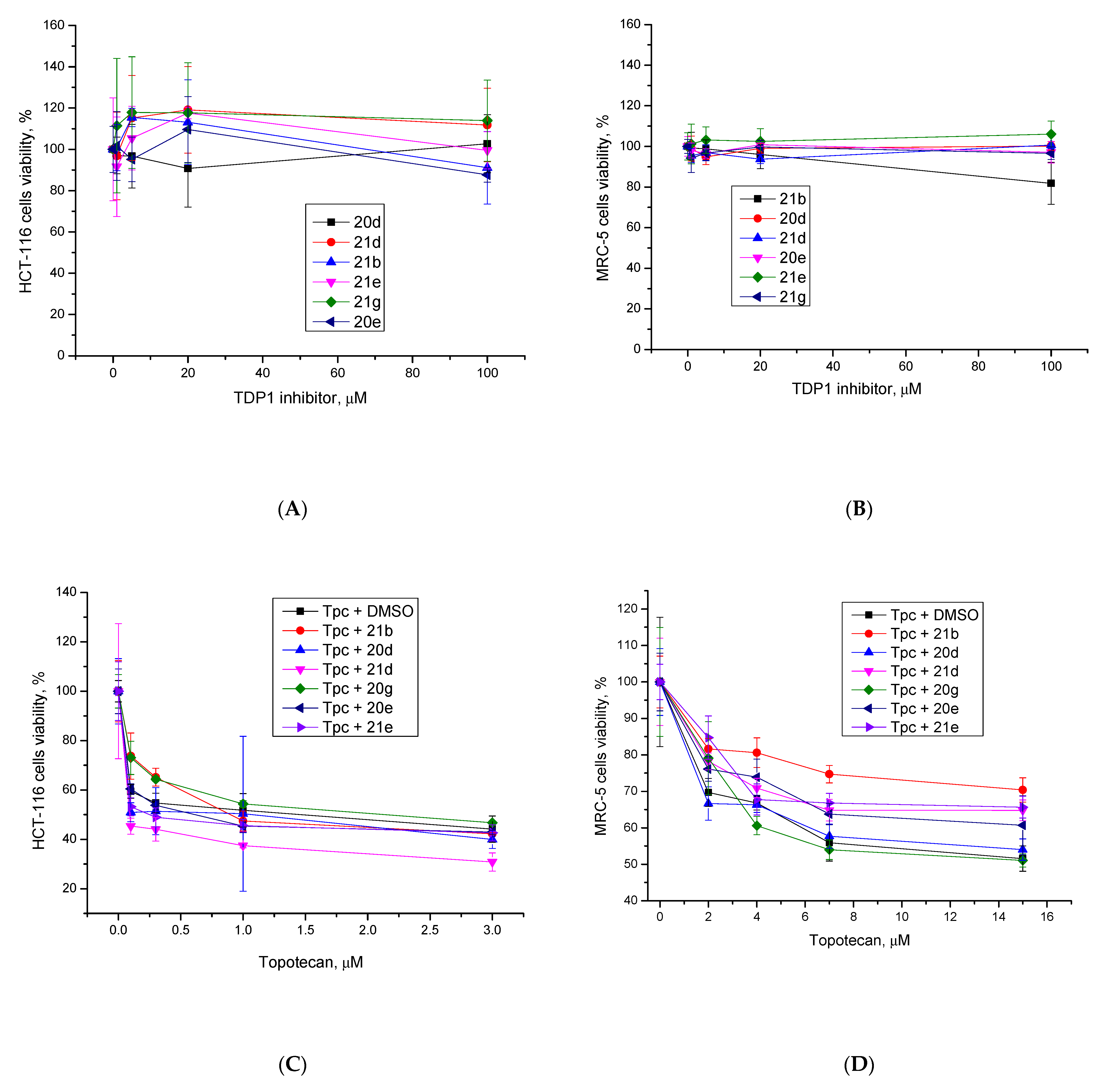
2.2. Biology
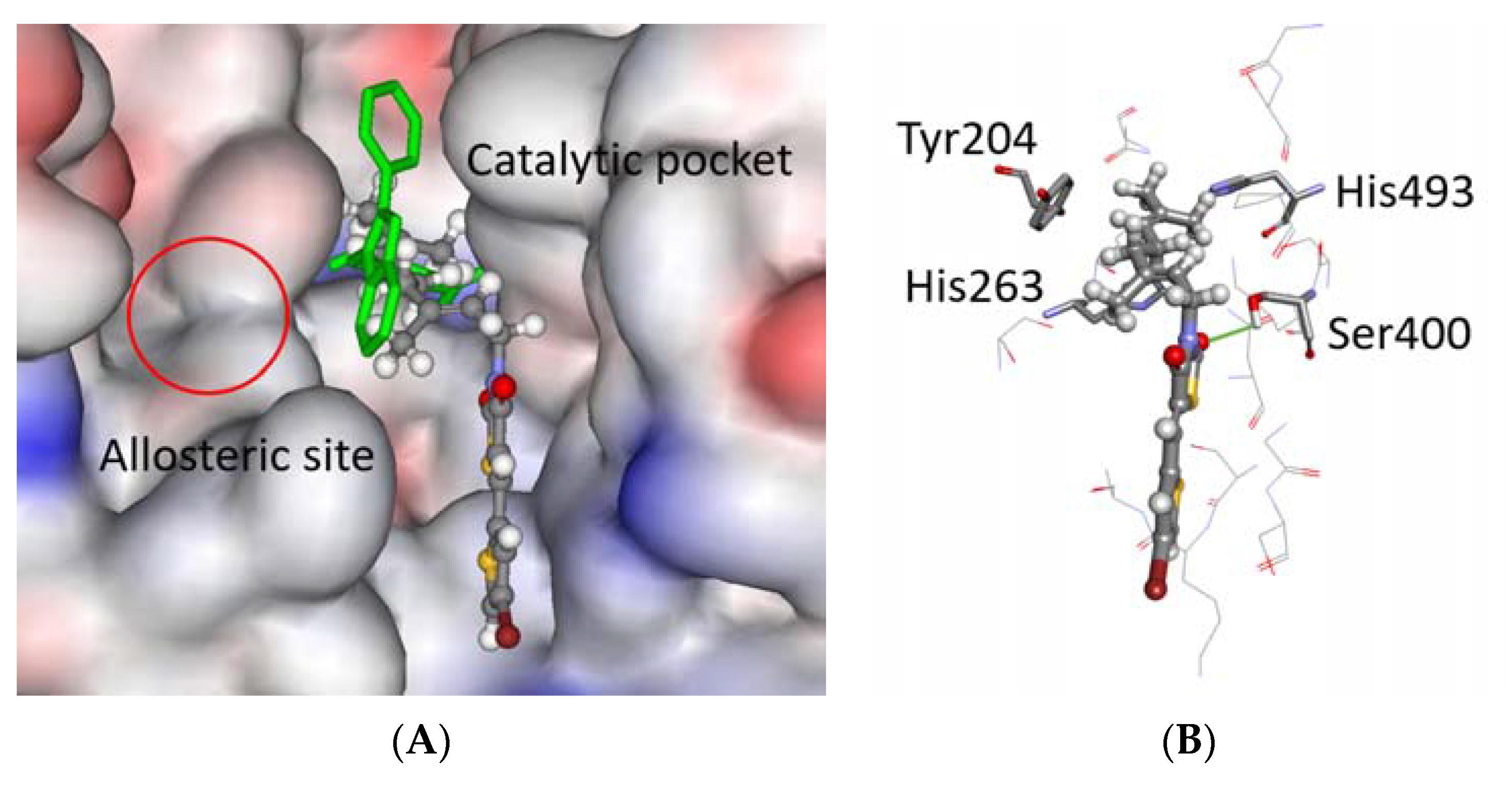
2.3. In Silico
2.3.1. Molecular Modeling
2.3.2. Chemical Space
3. Materials and Methods
3.1. Chemistry Section
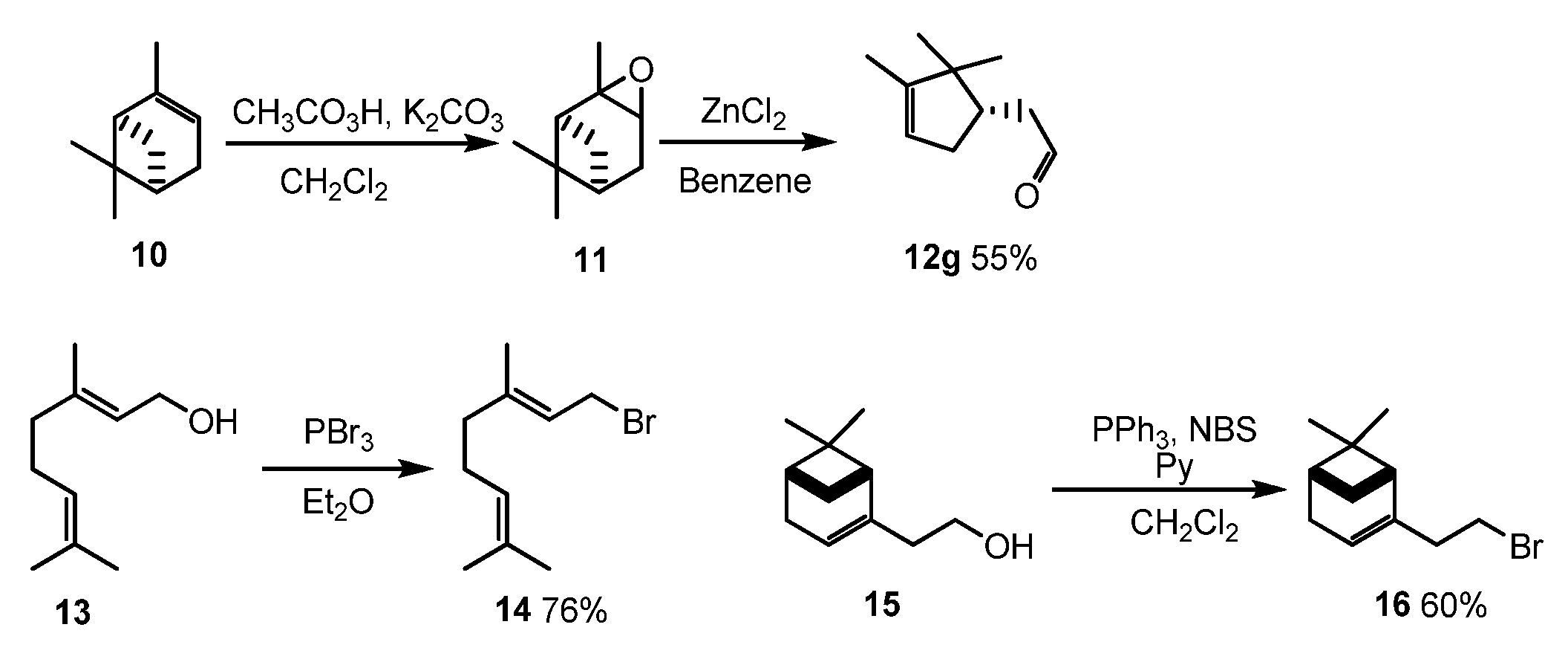
3.1.1. Synthesis of Compounds 12g, 14, and 16
3.1.2. Synthesis of 5-Substituted Thiazolidine-2,4-Diones
3.1.3. Synthesis of 3,5-Disubstituted Thiazolidine-2,4-Diones
3.2. Biology Section
3.2.1. Real-Time Detection of TDP1 Activity
3.2.2. Cytotoxicity Assays
3.3. Modeling Section
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pommier, Y.; Barcelo, J.M.; Rao, V.A.; Sordet, O.; Jobson, A.G.; Thibaut, L.; Miao, Z.H.; Seiler, J.A.; Zhang, H.; Marchand, C.; et al. Repair of Topoisomerase I-Mediated DNA Damage. Prog. Nucleic Acid Res. Mol. Biol. 2006, 81, 179–229. [Google Scholar] [CrossRef]
- Pommier, Y.; Huang, S.; Yin, N.; Gao, R.; Das, B.B.; Murai, J.; Marchand, C. Tyrosyl-DNA-phosphodiesterases (TDP1 and TDP2). DNA Repair (Amst) 2014, 19, 114–129. [Google Scholar] [CrossRef]
- Filimonov, A.S.; Chepanova, A.A.; Luzina, O.A.; Zakharenko, A.L.; Zakharova, O.D.; Ilina, E.S.; Dyrkheeva, N.S.; Kuprushkin, M.S.; Kolotaev, A.V.; Khachatryan, D.S.; et al. New hydrazinothiazole derivatives of usnic acid as potent TDP1 inhibitors. Molecules 2019, 24, 3711. [Google Scholar] [CrossRef]
- Gladkova, E.D.; Nechepurenko, I.V.; Bredikhin, R.A.; Chepanova, A.A.; Zakharenko, A.L.; Luzina, O.A.; Ilina, E.S.; Dyrkheeva, N.S.; Mamontova, E.M.; Anarbaev, R.O.; et al. The first berberine-based inhibitors of tyrosyl-dna phosphodiesterase 1 (TDP1), an important dna repair enzyme. Int. J. Mol. Sci. 2020, 21, 7162. [Google Scholar] [CrossRef]
- Khomenko, T.M.; Zakharenko, A.L.; Chepanova, A.A.; Ilina, E.S.; Zakharova, O.D.; Kaledin, V.I.; Nikolin, V.P.; Popova, N.A.; Korchagina, D.V.; Reynisson, J.; et al. Promising new inhibitors of tyrosyl-DNA phosphodiesterase I (TDP1) combining 4- arylcoumarin and monoterpenoid moieties as components of complex antitumor therapy. Int. J. Mol. Sci. 2020, 21, 126. [Google Scholar] [CrossRef] [PubMed]
- Salomatina, O.V.; Popadyuk, I.I.; Zakharenko, A.L.; Zakharova, O.D.; Chepanova, A.A.; Dyrkheeva, N.S.; Komarova, N.I.; Reynisson, J.; Anarbaev, R.O.; Salakhutdinov, N.F.; et al. Deoxycholic acid as a molecular scaffold for tyrosyl-DNA phosphodiesterase 1 inhibition: A synthesis, structure–activity relationship and molecular modeling study. Steroids 2021, 165, 108771. [Google Scholar] [CrossRef] [PubMed]
- Conda-Sheridan, M.; Reddy, P.V.N.; Morrell, A.; Cobb, B.T.; Marchand, C.; Agama, K.; Chergui, A.; Renaud, A.; Stephen, A.G.; Bindu, L.K.; et al. Synthesis and biological evaluation of indenoisoquinolines that inhibit both tyrosyl-DNA phosphodiesterase i (TDP1) and topoisomerase i (Top1). J. Med. Chem. 2013, 56, 182–200. [Google Scholar] [CrossRef] [PubMed]
- Ivankin, D.I.; Dyrkheeva, N.S.; Zakharenko, A.L.; Ilina, E.S.; Zarkov, T.O.; Reynisson, J.; Luzina, O.A.; Volcho, K.P.; Salakhutdinov, N.F.; Lavrik, O.I. Monoterpene substituted thiazolidin-4-ones as novel TDP1 inhibitors: Synthesis, biological evaluation and docking. Bioorganic Med. Chem. Lett. 2022, 73, 128909. [Google Scholar] [CrossRef]
- Luzina, O.; Filimonov, A.; Zakharenko, A.; Chepanova, A.; Zakharova, O.; Ilina, E.; Dyrkheeva, N.; Likhatskaya, G.; Salakhutdinov, N.; Lavrik, O. Usnic Acid Conjugates with Monoterpenoids as Potent Tyrosyl-DNA Phosphodiesterase 1 Inhibitors. J. Nat. Prod. 2020, 83, 2320–2329. [Google Scholar] [CrossRef]
- Kovaleva, K.; Yarovaya, O.; Ponomarev, K.; Cheresiz, S.; Azimirad, A.; Chernyshova, I.; Zakharenko, A.; Konev, V.; Khlebnikova, T.; Mozhaytsev, E.; et al. Design, synthesis, and molecular docking study of new tyrosyl-dna phosphodiesterase 1 (TDP1) inhibitors combining resin acids and adamantane moieties. Pharmaceuticals 2021, 14, 422. [Google Scholar] [CrossRef]
- Munkuev, A.A.; Mozhaitsev, E.S.; Chepanova, A.A.; Suslov, E.V.; Korchagina, D.V.; Zakharova, O.D.; Ilina, E.S.; Dyrkheeva, N.S.; Zakharenko, A.L.; Reynisson, J.; et al. Novel TDP1 inhibitors based on adamantane connected with monoterpene moieties via heterocyclic fragments. Molecules 2021, 26, 3128. [Google Scholar] [CrossRef]
- Sirivolu, V.R.; Vernekar, S.K.V.; Marchand, C.; Naumova, A.; Chergui, A.; Renaud, A.; Stephen, A.G.; Chen, F.; Sham, Y.Y.; Pommier, Y.; et al. 5-Arylidenethioxothiazolidinones as Inhibitors of Tyrosyl-DNA Phosphodiesterase 1. J. Med. Chem. 2012, 55, 8671–8684. [Google Scholar] [CrossRef]
- Castro, J.M.; Linares-Palomino, P.J.; Salido, S.; Altarejos, J.; Nogueras, M.; Sánchez, A. Enantiospecific synthesis, separation and olfactory evaluation of all diastereomers of a homologue of the sandalwood odorant Polysantol®. Tetrahedron 2005, 61, 11192–11203. [Google Scholar] [CrossRef]
- Lopez, L.; Mele, G.; Fiandanese, V.; Cardellicchio, C.; Nacci, A. Aminium salts catalyzed rearrangement of a=pinene and p-ionone oxides. Tetrahedron Lett. 1994, 50, 9097–9106. [Google Scholar] [CrossRef]
- Yue, L.; Li, J.; Chen, W.; Liu, X.; Jiang, Q.; Xia, W. Geraniol grafted chitosan oligosaccharide as a potential antibacterial agent. Carbohydr. Polym. 2017, 176, 356–364. [Google Scholar] [CrossRef] [PubMed]
- Akgun, B.; Hall, D. Fast and tight boronate formation for click bioorthogonal conjugation. Angew. Chem. 2016, 55, 3909–3913. [Google Scholar] [CrossRef]
- Momose, Y.; Meguro, K.; Ikeda, H.; Hatanaka, C.; Oi, S.; Sohda, T. Studies on Antidiabetic Agents. X. Synthesis and Biological Activities of Pioglitazone and Related Compounds. Chem. Pharm. Bull. 1991, 39, 1440–1445. [Google Scholar] [CrossRef]
- Cortelazzo-Polisini, E.; Boisbrun, M.; Gansmüller, A.H.; Comoy, C. Photoisomerization of Arylidene Heterocycles: Toward the Formation of Fused Heterocyclic Quinolines. J. Org. Chem. 2022, 87, 9699–9713. [Google Scholar] [CrossRef]
- Soccio, R.; Chen, E.; Lazar, M. Thiazolidinediones and the promise of insulin sensitization in type 2 diabetes. Cell Metab. 2014, 20, 573–591. [Google Scholar] [CrossRef]
- Naim, M.; Alam, M.; Ahmad, S.; Nawaz, F.; Shrivastava, N.; Sahu, M.; Alam, O. Therapeutic Journey of 2,4-Thiazolidinediones as a Versatile Scaffold: An Insight into Structure Activity Relationship. Eur. J. Med. Chem. 2017, 129, 218–250. [Google Scholar] [CrossRef] [PubMed]
- Barros, C.; Amato, A.; Bento de Oliveira, T.; Iannini, K.; Lauro da Silva, A.; Gonçalves da Silva, T.; Leite, E.S.; Hernandes, M.Z.; Lima, M. do C.A. de; Galdino, S.; et al. Synthesis and anti-inflammatory activity of new arylidene-thiazolidine-2,4-diones as PPARγ ligands. Bioorganic Med. Chem. 2010, 18, 3805–3811. [Google Scholar] [CrossRef]
- Zhao, X.Z.; Kiselev, E.; Lountos, G.T.; Wang, W.; Tropea, J.E.; Needle, D.; Hilimire, T.A.; Schneekloth, J.S.; Waugh, D.S.; Pommier, Y.; et al. Small molecule microarray identifies inhibitors of tyrosyl-DNA phosphodiesterase 1 that simultaneously access the catalytic pocket and two substrate binding sites. Chem. Sci. 2021, 12, 3876–3884. [Google Scholar] [CrossRef] [PubMed]
- Dyrkheeva, N.S.; Filimonov, A.S.; Luzina, O.A.; Zakharenko, A.L.; Ilina, E.S.; Malakhova, A.A.; Medvedev, S.P.; Reynisson, J.; Volcho, K.P.; Zakian, S.M.; et al. New hybrid compounds combining fragments of usnic acid and monoterpenoids for effective tyrosyl-dna phosphodiesterase 1 inhibition. Biomolecules 2021, 11, 973. [Google Scholar] [CrossRef] [PubMed]
- Gereth, J.; Peter, W.; Glen, R.C.; Leach, R.A.; Taylor, R. Development and Validation of a Genetic Algorithm for Flexible Docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar]
- Eldridge, M.D.; Murray, C.W.; Auton, T.R.; Paolini, G.V.; Mee, R.P. Empirical scoring functions: I. The development of a fast empirical scoring function to estimate the binding affinity of ligands in receptor complexes. J. Comput. Aided. Mol. Des. 1997, 11, 425–445. [Google Scholar] [CrossRef]
- Verdonk, M.L.; Cole, J.C.; Hartshorn, M.J.; Murray, C.W.; Taylor, R.D. Improved Protein–Ligand Docking Using GOLD Marcel. Proteins 2003, 52, 609–623. [Google Scholar] [CrossRef]
- Korb, O.; Stützle, T.; Exner, T.E. Empirical scoring functions for advanced Protein-Ligand docking with Plants. J. Chem. Inf. Model. 2009, 49, 84–96. [Google Scholar] [CrossRef]
- Mooij, W.T.M.; Verdonk, M.L. General and targeted statistical potentials for protein-ligand interactions. Proteins Struct. Funct. Genet. 2005, 61, 272–287. [Google Scholar] [CrossRef]
- Wang, Z.; Sun, H.; Yao, X.; Li, D.; Xu, L.; Li, Y.; Tian, S.; Hou, T. Comprehensive evaluation of ten docking programs on a diverse set of protein-ligand complexes: The prediction accuracy of sampling power and scoring power. Phys. Chem. Chem. Phys. 2016, 18, 12964–12975. [Google Scholar] [CrossRef]
- Bissantz, C.; Folkers, G.; Rognan, D. Protein-based virtual screening of chemical databases. 1. Evaluation of different docking/scoring combinations. J. Med. Chem. 2000, 43, 4759–4767. [Google Scholar] [CrossRef]
- Zakharenko, A.; Luzina, O.; Koval, O.; Nilov, D.; Gushchina, I.; Dyrkheeva, N.; Švedas, V.; Salakhutdinov, N.; Lavrik, O. Tyrosyl-DNA Phosphodiesterase 1 Inhibitors: Usnic Acid Enamines Enhance the Cytotoxic Effect of Camptothecin. J. Nat. Prod. 2016, 79, 2961–2967. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Logan, G.; Reynisson, J. Wine compounds as a source for HTS screening collections. A feasibility study. Mol. Inform. 2012, 31, 847–855. [Google Scholar] [CrossRef] [PubMed]
- Eurtivong, C.; Reynisson, J. The Development of a Weighted Index to Optimise Compound Libraries for High Throughput Screening. Mol. Inform. 2019, 38, 1800068. [Google Scholar] [CrossRef] [PubMed]
- Mahapatra, M.K.; Saini, R.; Kumar, M. Synthesis, anti-hyperglycaemic activity, and in-silico studies of N-substituted 5-(furan-2-ylmethylene)thiazolidine-2,4-dione derivatives. Res. Chem. Intermed. 2016, 42, 8239–8251. [Google Scholar] [CrossRef]
- Muneo, T.; Taketoshi, S.; Kiminori, T. Pyridine Derivatives, Their Production and Use. U.S. Patent 5246948, 21 September 1993. [Google Scholar]
- Akama, T.; Holcomb, R.; Tolman, R.L. Telomerase Inhibitors and Methods of Their Use. U.S. Patent 6452014, 17 September 2002. [Google Scholar]
- Borisova, M.S.; Ivankin, D.I.; Sokolov, D.N.; Luzina, O.A.; Rybalova, T.V.; Tolstikova, T.G.; Salakhutdinov, N.F. Synthesis, antiulcerative, and anti-inflammatory activities of new campholenic derivatives-1,3-thiazolidin-4-ones, 1,3-thiazolidine-2,4-diones, and 1,3-thiazinan-4-ones. Chem. Pap. 2021, 75, 5503–5514. [Google Scholar] [CrossRef]
- Kumar, K.S.; Lakshmana Rao, A.; Rama Sekhara Reddy, D. Design, synthesis, hypoglycemic activity and molecular docking studies of 3-substituted-5-[(Furan-2-yl)-methylene]-thiazolidine-2,4-dione derivatives. Indian J. Pharm. Educ. Res. 2021, 55, 266–275. [Google Scholar] [CrossRef]
- Zakharenko, A.; Khomenko, T.; Zhukova, S.; Koval, O.; Zakharova, O.; Anarbaev, R.; Lebedeva, N.; Korchagina, D.; Komarova, N.; Vasiliev, V.; et al. Synthesis and biological evaluation of novel tyrosyl-DNA phosphodiesterase 1 inhibitors with a benzopentathiepine moiety. Bioorganic Med. Chem. 2015, 23, 2044–2052. [Google Scholar] [CrossRef]
- Dyrkheeva, N.; Anarbaev, R.; Lebedeva, N.; Kuprushkin, M.; Kuznetsova, A.; Kuznetsov, N.; Rechkunova, N.; Lavrik, O. Human Tyrosyl-DNA Phosphodiesterase 1 Possesses Transphosphooligonucleotidation Activity With Primary Alcohols. Front. Cell Dev. Biol. 2020, 8, 604732. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acid Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Berman, H.; Henrick, K.; Nakamura, H. Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 2003, 10, 980. [Google Scholar] [CrossRef]
- Scigress Ultra V. F.J 2.6. (EU 3.1.7); Fujitsu Limited: Minato City, Tokyo, 2016.
- Allinger, N.L.; Yuh, Y.H.; Lii, J.H. Molecular Mechanics. The MM3 Force Field for Hydrocarbons. J. Am. Chem. Soc. 1989, 111, 8551–8566. [Google Scholar] [CrossRef]
- Lii, J.H.; Allinger, N.L. Molecular Mechanics. The MM3 Force Field for Hydrocarbons. 2. Vibrational Frequencies and Thermodynamics. J. Am. Chem. Soc. 1989, 111, 8566–8575. [Google Scholar] [CrossRef]
- Lii, J.H.; Allinger, N.L. Molecular Mechanics. The MM3 Force Field for Hydrocarbons. 3. The van der Waals’ Potentials and Crystal Data for Aliphatic and Aromatic Hydrocarbons. J. Am. Chem. Soc. 1989, 111, 8576–8582. [Google Scholar] [CrossRef]
- Goto, H.; Osawa, E. An efficient algorithm for searching low-energy conformers of cyclic and acyclic molecules. J. Chem. Soc. Perkin Trans. 2 1993, 2, 187–198. [Google Scholar] [CrossRef]
- QikProp, version 6.2; Schrödinger: New York, NY, USA, 2021.
- Ioakimidis, L.; Thoukydidis, L.; Mirza, A.; Naeem, S.; Reynisson, J. Benchmarking the reliability of QikProp. correlation between experimental and predicted values. QSAR Comb. Sci. 2008, 27, 445–456. [Google Scholar] [CrossRef]


| Compounds | 18 | 19 | 20 | 21 | |
|---|---|---|---|---|---|
| H | 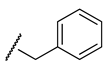 | 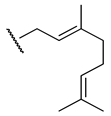 |  | ||
| a | 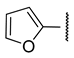 | >100 | >100 | 3.4 ± 0.1 | >100 |
| b | 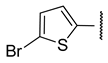 | 1.1 ± 0.2 | 1.9 ± 0.3 | >100 | 2.5 ± 0.4 |
| c | 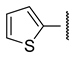 | >100 | >100 | >100 | >100 |
| d |  | >100 | >100 | 0.65 ± 0.07 | 0.55 ± 0.07 |
| e | 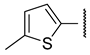 | >100 | >100 | 2.9 ± 0.1 | 2.3 ± 0.1 |
| f |  | >100 | 1.6 ± 0.3 | >100 | >100 |
| g |  | >100 | 2.4 ± 1.1 | 4.1 ± 0.1 | 2.4 ± 0.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ivankin, D.I.; Kornienko, T.E.; Mikhailova, M.A.; Dyrkheeva, N.S.; Zakharenko, A.L.; Achara, C.; Reynisson, J.; Golyshev, V.M.; Luzina, O.A.; Volcho, K.P.; et al. Novel TDP1 Inhibitors: Disubstituted Thiazolidine-2,4-Diones Containing Monoterpene Moieties. Int. J. Mol. Sci. 2023, 24, 3834. https://doi.org/10.3390/ijms24043834
Ivankin DI, Kornienko TE, Mikhailova MA, Dyrkheeva NS, Zakharenko AL, Achara C, Reynisson J, Golyshev VM, Luzina OA, Volcho KP, et al. Novel TDP1 Inhibitors: Disubstituted Thiazolidine-2,4-Diones Containing Monoterpene Moieties. International Journal of Molecular Sciences. 2023; 24(4):3834. https://doi.org/10.3390/ijms24043834
Chicago/Turabian StyleIvankin, Dmitry I., Tatyana E. Kornienko, Marina A. Mikhailova, Nadezhda S. Dyrkheeva, Alexandra L. Zakharenko, Chigozie Achara, Jóhannes Reynisson, Victor M. Golyshev, Olga A. Luzina, Konstantin P. Volcho, and et al. 2023. "Novel TDP1 Inhibitors: Disubstituted Thiazolidine-2,4-Diones Containing Monoterpene Moieties" International Journal of Molecular Sciences 24, no. 4: 3834. https://doi.org/10.3390/ijms24043834
APA StyleIvankin, D. I., Kornienko, T. E., Mikhailova, M. A., Dyrkheeva, N. S., Zakharenko, A. L., Achara, C., Reynisson, J., Golyshev, V. M., Luzina, O. A., Volcho, K. P., Salakhutdinov, N. F., & Lavrik, O. I. (2023). Novel TDP1 Inhibitors: Disubstituted Thiazolidine-2,4-Diones Containing Monoterpene Moieties. International Journal of Molecular Sciences, 24(4), 3834. https://doi.org/10.3390/ijms24043834









