Polθ Inhibition: An Anticancer Therapy for HR-Deficient Tumours
Abstract
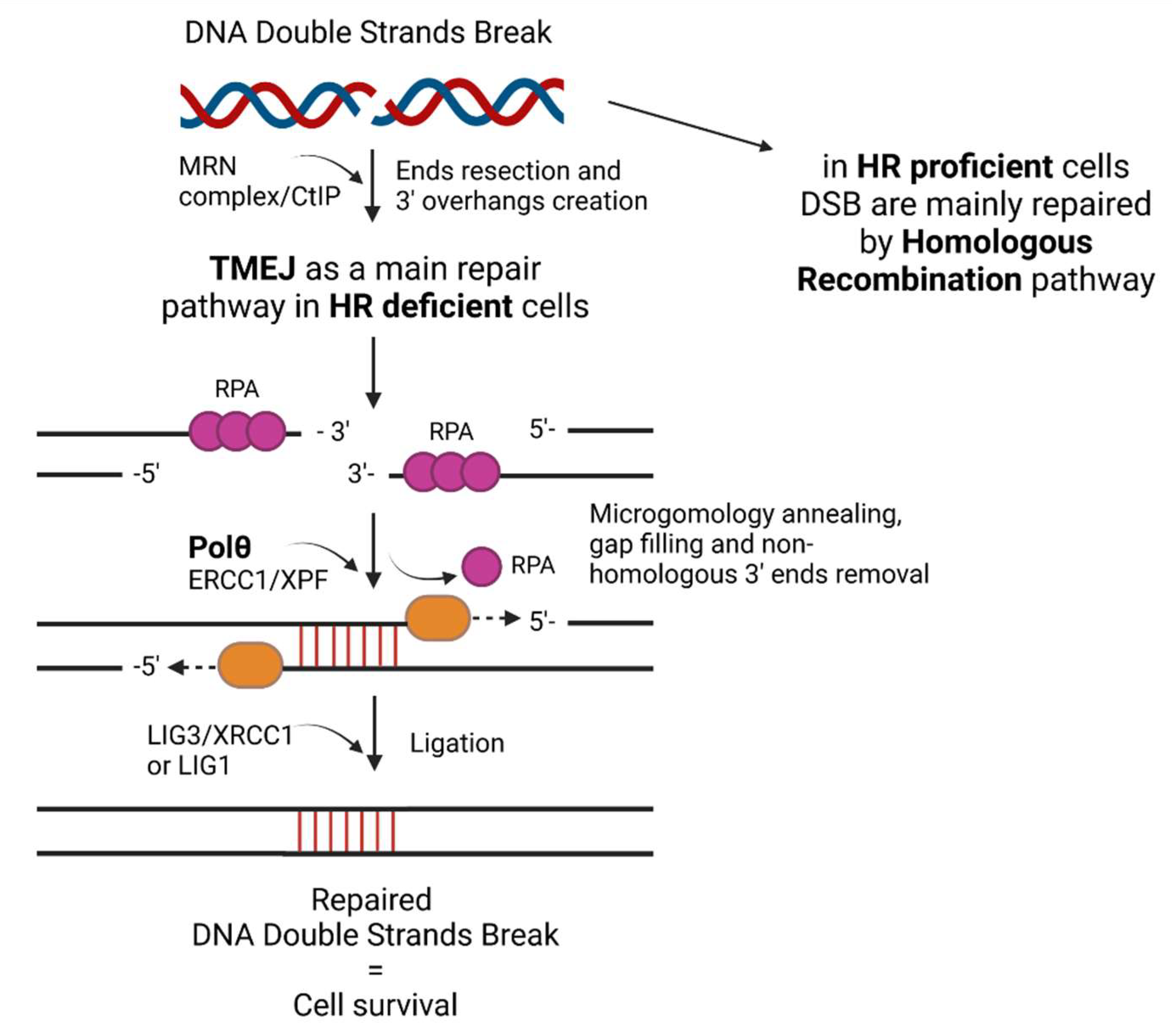
1. Introduction
2. The Role of Polθ—Mediated TMEJ
3. Different Strategies for Polθ Suppression
3.1. RNA Interference Technique—siRNA and shRNA
3.1.1. Description of the Technique
3.1.2. Application in Studies
3.2. CRISPR/Cas9 Technology
3.2.1. Description of the Technique
3.2.2. Application in Studies
4. PolQ Inhibitors
4.1. Novobiocin
4.2. ART558
4.3. RP-6685
5. Conclusions and Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Kaelin, W.G., Jr. The concept of synthetic lethality in the context of anticancer therapy. Nat. Rev. Cancer 2005, 5, 689–698. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Simposon, D.A.; Carvajal-Garcia, J.; Price, B.A.; Kumar, R.J.; Mose, L.E.; Wood, R.D.; Rashid, N.; Purvis, J.E.; Parker, J.S.; et al. Genetic determinants of cellular addiction to DNA polymerase theta. Nat. Commun. 2019, 10, 4286. [Google Scholar] [CrossRef]
- Drzewiecka, M.; Barszczewska-Pietraszek, G.; Czarny, P.; Skorski, T.; Śliwiński, T. Synthetic Lethality Targeting Polθ. Genes 2022, 13, 1101. [Google Scholar] [CrossRef]
- Ashworth, A.; Lord, C.J. Synthetic lethal therapies for cancer: What’s next after PARP inhibitors? Nat. Rev. Clin. Oncol. 2018, 15, 564–576. [Google Scholar] [CrossRef]
- Beagan, K.; Mcvey, M. Linking DNA polymerase theta structure and function in healthand disease. Cell Mol. Life Sci. 2016, 73, 603–615. [Google Scholar] [CrossRef] [PubMed]
- Mateos-gomez, P.A.; Kent, T.; Kashkina, E.; Pomerantz, R.T.; Sfeir, A. The helicase domain of Polθ counteracts RPA to promote alt-NHEJ. Nat. Struct. Mol. Biol. 2017, 24, 1116–1123. [Google Scholar] [CrossRef] [PubMed]
- Ceccaldi, R.; Liu, J.C.; Amunugama, R.; Hajdu, I.; Primack, B.; Petalcorin, M.I.R.; O’Connor, K.W.; Konstantinopoulos, P.A.; Elledge, S.J.; Boulton, S.J.; et al. Homologous recombination-deficient tumors are hyper- dependent on POLQ-mediated repair. Nature 2015, 518, 258–262. [Google Scholar] [CrossRef]
- Pan, Q.; Wang, L.; Liu, Y.; Li, M.; Zhang, Y.; Peng, W.; Deng, T.; Peng, M.-L.; Jiang, J.-Q.; Tang, J.; et al. Knockdown of POLQ interferes the development and progression of hepatocellular carcinoma through regulating cell proliferation, apoptosis and migration. Cancer Cell Int. 2021, 21, 482. [Google Scholar] [CrossRef]
- Lemée, F.; Bergoglio, V.; Fernandez-Vidal, A.; Machado-Silva, A.; Pillaire, M.J.; Bieth, A.; Gentil, C.; Baker, L.; Martin, A.L.; Leduc, C.; et al. DNA polymerase θ up-regulation is associated with poor survival in breast cancer, perturbs DNA replication, and promotes genetic instability. Proc. Natl. Acad. Sci. USA 2010, 107, 13390–13395. [Google Scholar] [CrossRef]
- Zatreanau, D.; Robinson, H.M.R.; Alkhatib, O.; Boursier, M.; Finch, H.; Geo, L.; Grande, D.; Grinkevich, V.; Heald, R.A.; Langdon, S.; et al. Polθ inhibitors elicit BRCA-gene synthetic lethality and target PARP inhibitor resistance. Nat. Commun. 2021, 12, 3636. [Google Scholar] [CrossRef] [PubMed]
- Bubenik, M.; Mader, P.; Mochirian, P.; Vallée, F.; Clark, J.; Truchon, J.F.; Perryman, A.L.; Pau, V.; Kurinov, I.; Zahn, K.E.; et al. Identification of RP-6685, an Orally Bioavailable Compound that Inhibits the DNA Polymerase Activity of Polθ. J. Med. Chem. 2022, 65, 13198–13215. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Gelot, C.; Pantelidou, C.; Li, A.; Yücel, H.; Davis, R.E.; Färkkilä, A.; Kochupurakkal, B.; Syed, A.; Shapiro, G.I.; et al. A first-in-class polymerase theta inhibitor selectively targets homologous-recombination-deficient tumors. Nat. Cancer 2021, 2, 598–610. [Google Scholar] [CrossRef] [PubMed]
- Trenner, A.; Sartori, A.A. Harnessing DNA Double-Strand Break Repair for Cancer Treatment. Front. Oncol. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Kumar, R.J.; Chao, H.X.; Simpson, D.A.; Feng, W.; Cho, M.G.; Roberts, V.R.; Sullivan, A.R.; Shah, S.J.; Wozny, A.S.; Fagan-Solis, K.; et al. Dual inhibition of DNA-PK and DNA polymerase theta overcomes radiation resistance induced by p53 deficiency. NAR Cancer 2020, 2, zcaa038. [Google Scholar] [CrossRef] [PubMed]
- Stoklosa, T.; Poplawski, T.; Koptyra, M.; Nieborowska-Skorska, M.; Basak, G.; Slupianek, A.; Rayevskaya, M.; Seferynska, I.; Herrera, L.; Blasiak, J.; et al. BCR/ABL Inhibits Mismatch Repair to Protect from Apoptosis and Induce Point Mutations. Cancer Res. 2008, 68, 2576–2580. [Google Scholar] [CrossRef]
- Slupianek, A.; Schmutte, C.; Tombline, G.; Nieborowska-Skorska, M.; Hoser, G.; Nowicki, M.O.; Pierce, A.J.; Fishel, R.; Skorski, T. BCR/ABL Regulates Mammalian RecA Homologs, Resulting in Drug Resistance. Mol. Cell 2001, 8, 795–806. [Google Scholar] [CrossRef]
- Schrempf, A.; Slyskova, J.; Loizou, J.I. Targeting the DNA Repair Enzyme Polymerase θ in Cancer Therapy. Trends Cancer 2021, 7, 98–111. [Google Scholar] [CrossRef]
- Brambati, A.; Barry, R.; Sfeir, A. DNA Polymerase theta (Polθ) – an error-prone polymerase necessary for genome stability. Curr. Opin. Genet. Dev. 2020, 60, 119–126. [Google Scholar] [CrossRef]
- Chen, X.S.; Pomerantz, R.T. DNA Polymerase θ: A Cancer Drug Target with Reverse Transcriptase Activity. Genes 2021, 12, 1146. [Google Scholar] [CrossRef]
- Ramsden, D.A.; Carvajal-Garcia, J.; Gupta, G.P. Mechanism, cellular functions and cancer roles of polymerase- mediated DNA end joining. Nat. Rev. 2022, 23, 125–140. [Google Scholar] [CrossRef] [PubMed]
- Hwang, T.; Reh, S.; Dunbayev, Y.; Zhong, Y.; Takata, Y.; Shen, J.; McBride, K.M.; Murnane, J.P.; Bhak, J.; Lee, S.; et al. Defining the mutation signatures of DNA polymerase θ in cancer genomes. NAR Cancer 2020, 2, zcaa017. [Google Scholar] [CrossRef] [PubMed]
- Hanscom, T.; Woodward, N.; Batorsky, R.; Brown, A.J.; Roberts, S.A.; Mcvey, M. Characterization of sequence contexts that favor alternative end joining at Cas9-induced double-strand breaks. Nucleic Acids Res. 2022, 50, 7465–7478. [Google Scholar] [CrossRef] [PubMed]
- Luedeman, M.E.; Stroik, S.; Feng, W.; Luthman, A.J.; Gupta, G.P.; Ramsden, D.A. Poly (ADP) ribose polymerase promotes DNA polymerase theta-mediated end joining by activation of end resection. Nat. Commun. 2022, 13, 4547. [Google Scholar] [CrossRef] [PubMed]
- Schaub, J.M.; Soniat, M.M.; Finkelstein, I.J. Polymerase theta-helicase promotes end joining by stripping single-stranded DNA-binding proteins and bridging DNA ends. Nucleic Acids Res. 2022, 50, 3911–3921. [Google Scholar] [CrossRef] [PubMed]
- Wood, R.D.; Doublié, S. DNA polymerase θ (POLQ), double-strand break repair, and cancer. DNA Repair 2016, 44, 22–32. [Google Scholar] [CrossRef]
- Kawamura, K.; Bahar, R.; Seimiya, M.; Chiyo, M.; Wada, A.; Okada, S.; Hatano, M.; Tokuhisa, T.; Kimura, H.; Watanabe, S.; et al. DNA polymerase θ is preferentially expressed in lymphoid tissues and upregulated in human cancers. Int. J. Cancer 2004, 109, 9–16. [Google Scholar] [CrossRef]
- Schimmel, J.; Kool, H.; van Schendel, R.; Tijsterman, M. Mutational signatures of non-homologous and polymerase theta-mediated end-joining in embryonic stem cells. EMBO J. 2017, 36, 3634–3649. [Google Scholar] [CrossRef]
- Caracciolo, D.; Riillo, C.; Di Martino, M.T.; Tagliaferri, P.; Tassone, P. Alternative Non-Homologous End-Joining: Error-Prone DNA Repair as Cancer’s Achilles’ Heel. Cancers 2021, 13, 1392. [Google Scholar] [CrossRef]
- Carvajal-Garcia, J.; Cho, J.E.; Carvajal-Garcia, P.; Feng, W.; Wood, R.D.; Sekelsky, J.; Gupta, G.P.; Roberts, S.A.; Ramsden, D.A. Mechanistic basis for microhomology identification and genome scarring by polymerase theta. Proc. Natl. Acad. Sci. USA 2020, 117, 8476–8485. [Google Scholar] [CrossRef]
- Liddiard, K.; Aston-Evans, A.N.; Cleal, K.; Hendrickson, E.A.; Baird, D.M. POLQ suppresses genome instability and alterations in DNA repeat tract lengths. NAR Cancer 2022, 4, zcac020. [Google Scholar] [CrossRef] [PubMed]
- Wilson, J.; Loizou, J.I. Exploring the genetic space of the DNA damage response for cancer therapy through CRISPR-based screens. Mol. Oncol. 2022, 16, 3778–3791. [Google Scholar] [CrossRef] [PubMed]
- Ferreira da Silva, J.; Salic, S.; Wiedner, M.; Datlinger, P.; Essletzbichler, P.; Hanzl, A.; Superti-Furga, G.; Bock, C.; Winter, G.; Loizou, J.I. Genome-scale CRISPR screens are efficient in non-homologous end-joining deficient cells. Sci. Rep. 2019, 9, 15751. [Google Scholar] [CrossRef]
- Mara, K.; Charlot, F.; Guyon-Debast, A.; Schaefer, D.G.; Collonnier, C.; Grelon, M.; Nogué, F. POLQ plays a key role in the repair of CRISPR/Cas9-induced double-stranded breaks in the moss Physcomitrella patens. New Phytol. 2019, 222, 1380–1391. [Google Scholar] [CrossRef] [PubMed]
- Dai, C.H.; Chen, P.; Li, J.; Lan, T.; Chen, Y.C.; Qian, H.; Chen, K.; Li, M.Y. Co-inhibition of pol θ and HR genes efficiently synergize with cisplatin to suppress cisplatin-resistant lung cancer cells survival. Oncotarget 2016, 7, 65157–65170. [Google Scholar] [CrossRef] [PubMed]
- Kelso, A.A.; Lopezcolorado, F.W.; Bhargava, R.; Stark, J.M. Distinct roles of RAD52 and POLQ in chromosomal break repair and replication stress response. PLoS Genet. 2019, 15, e1008319. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.A.; Pandupuspitasari, N.S.; Chun-Jie, H.; Ao, Z.; Jamal, M.; Zohaib, A.; Khan, F.A.; Hakim, M.R.; ShuJun, Z. CRISPR/Cas9 therapeutics: A cure for cancer and other genetic diseases. Oncotarget 2016, 7, 52541–52552. [Google Scholar] [CrossRef]
- Ferreira, P.; Choupina, A.B. CRISPR/Cas9 a simple, inexpensive and effective technique for gene editing. Mol. Biol. Rep. 2022, 49, 7079–7086. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, C.; Zhao, W. Recent advances of the biological and biomedical applications of CRISPR/Cas systems. Mol. Biol. Rep. 2022, 49, 7087–7100. [Google Scholar] [CrossRef]
- Pushparaj, P.N.; Aarthi, J.J.; Manikandan, J.; Kumar, S.D. siRNA, miRNA, and shRNA: In vivo Applications. JDR 2008, 87, 992–1003. [Google Scholar] [CrossRef]
- Alshaer, W.; Zureigat, H.; Al Karaki, A.; Al-Kadash, A.; Gharaibeh, L.; Hatmal, M.M.; Aljabali, A.A.A.; Awidi, A. siRNA: Mechanism of action, challenges, and therapeutic approaches. Eur. J. Pharmacol. 2021, 905, 174178. [Google Scholar] [CrossRef] [PubMed]
- Sliva, K.; Schnierle, B.S. Selective gene silencing by viral delivery of short hairpin RNA. Virol. J. 2010, 7, 248. [Google Scholar] [CrossRef] [PubMed]
- McAnuff, M.A.; Rettig, G.R.; Rice, K.G. Potency of siRNA versus shRNA mediated knockdown in vivo. J. Pharm. Sci. 2007, 96, 2922–2930. [Google Scholar] [CrossRef] [PubMed]
- Rao, D.D.; Vorhies, J.S.; Senzer, N.; Nemunaitis, J. siRNA vs. shRNA: Similarities and differences. Adv. Drug Deliv. Rev. 2009, 61, 746–759. [Google Scholar] [CrossRef] [PubMed]
- Dana, H.; Chalbatani, G.M.; Mahmoodzadeh, H.; Karimloo, R.; Rezaiean, O.; Moradzadeh, A.; Mehmandoost, N.; Moazzen, F.; Mazraeh, A.; Marmari, V.; et al. Molecular Mechanisms and Biological Functions of siRNA. Int. J. Biomed. Sci. 2017, 13, 48–57. [Google Scholar]
- Nikam, R.R.; Gore, K.R. Journey of siRNA: Clinical Developments and Targeted Delivery. Nucleic Acid. Ther. 2018, 28, 209–224. [Google Scholar] [CrossRef]
- Lee, W.C.; Berry, R.; Hohenstein, P.; Davies, J. siRNA as a tool for investigating organogenesis: The pitfalls and the promises. Organogenesis 2008, 4, 176–181. [Google Scholar] [CrossRef]
- Moore, C.B.; Guthrie, E.H.; Huang, M.T.; Taxman, D.J. Short Hairpin RNA (shRNA): Design, Delivery, and Assessment of Gene Knockdown. Methods Mol. Biol. 2010, 629, 141–158. [Google Scholar]
- Goullet de Rugy, T.; Bashkurov, M.; Datti, A.; Betous, R.; Guitton-Sert, L.; Cazaux, C.; Durocher, D.; Hoffmann, J.S. Excess Polθ functions in response to replicative stress in homologous recombination-proficient cancer cells. Biol. Open. 2016, 5, 1485–1492. [Google Scholar] [CrossRef]
- Savić, N.; Schwank, G. Advances in therapeutic CRISPR/Cas9 genome editing. Transl. Res. 2016, 168, 15–21. [Google Scholar] [CrossRef]
- Vaghari-Tabari, M.; Hassanpour, P.; Sadeghsoltani, F.; Malakoti, F.; Alemi, F.; Qujeq, D.; Asemi, Z.; Yousefi, B. CRISPR/Cas9 gene editing: A new approach for overcoming drug resistance in cancer. Cell. Mol. Biol. Lett. 2022, 27, 49. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, L.; Huang, X. Genome modification by CRISPR/Cas9. FEBS J. 2014, 281, 5186–5193. [Google Scholar] [CrossRef]
- Lu, Y.; Xue, J.; Deng, T.; Zhou, X.; Yu, K.; Deng, L.; Huang, M.; Yi, X.; Liang, M.; Wang, Y.; et al. Safety and feasibility of CRISPR-edited T cells in patients with refractory non-small-cell lung cancer. Nat. Med. 2020, 26, 732–740. [Google Scholar] [CrossRef] [PubMed]
- Redman, M.; King, A.; Watson, C.; King, D. What is CRISPR/Cas9? Arch. Dis. Child. Educ. Pract. Ed. 2016, 101, 213–215. [Google Scholar] [CrossRef] [PubMed]
- Boettcher, M.; Mcmanus, M.T. Choosing the Right Tool for the Job: RNAi, TALEN or CRISPR. Mol. Cell 2015, 58, 575–585. [Google Scholar] [CrossRef] [PubMed]
- Pugh, K.W.; Zhang, Z.; Wang, J.; Xu, X.; Munthali, V.; Zuo, A.; Blagg, B.S.J. From Bacteria to Cancer: A Benzothiazole-Based DNA Gyrase B Inhibitor Redesigned for Hsp90 C-Terminal Inhibition. ACS Med. Chem. Lett. 2020, 11, 1535–1538. [Google Scholar] [CrossRef]
- Hyun, S.Y.; Le, H.T.; Nguyen, C.T.; Yong, Y.S.; Boo, H.J.; Lee, H.J.; Lee, J.S.; Min, H.Y.; Ann, J.; Chen, J.; et al. Development of a novel Hsp90 inhibitor NCT-50 as a potential anticancer agent for the treatment of non-small cell lung cancer. Sci. Rep. 2018, 8, 13924. [Google Scholar] [CrossRef]
- Forsberg, L.K.; Davis, R.E.; Wimalasena, V.; Blagg BS, J. Exploiting Polarity and Chirality to Probe the Hsp90 C-terminus. Bioorg. Med. Chem. 2019, 26, 3096–3110. [Google Scholar] [CrossRef]
- Zhou, X.; Wen, Y.; Tian, Y.; He, M.; Ke, X.; Huang, Z.; He, Y.; Liu, L.; Scharf, A.; Lu, M.; et al. Heat Shock Protein 90α-Dependent B-Cell-2-Associated Transcription Factor 1 Promotes Hepatocellular Carcinoma Proliferation by Regulating MYC Proto-Oncogene c-MYC mRNA Stability. Hepatology 2019, 69, 1564–1581. [Google Scholar] [CrossRef]
- Garg, G.; Forsberg, L.K.; Zhao, H.; Blagg BS, J. Development of Phenyl Cyclohexylcarboxamides as a Novel Class of Hsp90 C-terminal Inhibitors. Chemistry 2017, 23, 16574–16585. [Google Scholar] [CrossRef]
- Forsberg, L.K.; Liu, W.; Holzbeierlein, J.; Blagg, B.S.J. Modified Biphenyl Hsp90 C-terminal Inhibitors for the Treatment of Cancer. Bioorg. Med. Chem. Lett. 2017, 27, 4514–4519. [Google Scholar] [CrossRef] [PubMed]
- Patterson-Fortin, J.; Bose, A.; Tsai, W.C.; Grochala, C.; Nguyen, H.; Zhou, J.; Parmar, K.; Lazaro, J.B.; Liu, J.; McQueen, K.; et al. Targeting DNA Repair with Combined Inhibition of NHEJ and MMEJ Induces Synthetic Lethality in TP53-Mutant Cancers. Cancer Res. 2022, 88, 3815–3829. [Google Scholar] [CrossRef] [PubMed]
- Higgins, G.S.; Boulton, S.J. Beyond PARP-POLθ as an anticancer target. Science 2018, 359, 1217–1219. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Lu, L.Y.; Yu, X. The role of BRCA1 in DNA damage response. Protein Cell 2010, 1, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Noordermeer, S.M.; Adam, S.; Setiaputra, D.; Barazas, M.; Pettitt, S.J.; Ling, A.K.; Olivieri, M.; Álvarez-Quilón, A.; Moatti, N.; Zimmermann, M.; et al. The shieldin complex mediates 53BP1-dependent DNA repair. Nature 2018, 560, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Dev, H.; Chiang, T.W.; Lescale, C.; de Krijger, I.; Martin, A.G.; Pilger, D.; Coates, J.; Sczaniecka-Clift, M.; Wei, W.; Ostermaier, M.; et al. Shieldin complex promotes DNA end-joining and counters homologous recombination in BRCA1-null cells. Nat Cell Biol. 2018, 20, 954–965. [Google Scholar] [CrossRef] [PubMed]
- Zahn, K.E.; Jensen, R.B. Polymerase θ Coordinates Multiple Intrinsic Enzymatic Activities during DNA Repair. Genes 2021, 12, 1310. [Google Scholar] [CrossRef]
- Eder, J.P.; Wheeler, C.A.; Teicher, B.A.; Schnipper, L.E. A phase I clinical trial of novobiocin, a modulator of alkylating agent cytotoxicity. Cancer Res. 1991, 51, 510–513. [Google Scholar]
- Makridakis, N.M.; Reichardt, J.K.V. Translesion DNA polymerases and cancer. Front. Genet. 2012, 3, 174. [Google Scholar] [CrossRef]
- de Lange, T. Shelterin: The protein complex that shapes and safeguards human telomeres. Genes Dev. 2005, 19, 2100–2110. [Google Scholar] [CrossRef]
- Available online: https://clinicaltrials.gov/ct2/show/NCT04991480 (accessed on 15 November 2022).
- Baxter, J.S.; Zatreanu, D.; Pettitt, S.J.; Lord, C.J. Resistance to DNA repair inhibitors in cancer. Mol. Oncol. 2022, 16, 3811–3827. [Google Scholar] [CrossRef] [PubMed]
- Carvajal-Maldonado, D.; Wood, R.D. Regulating Polθ in Breast Cancer. Cancer Res. 2021, 81, 1441–1442. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barszczewska-Pietraszek, G.; Drzewiecka, M.; Czarny, P.; Skorski, T.; Śliwiński, T. Polθ Inhibition: An Anticancer Therapy for HR-Deficient Tumours. Int. J. Mol. Sci. 2023, 24, 319. https://doi.org/10.3390/ijms24010319
Barszczewska-Pietraszek G, Drzewiecka M, Czarny P, Skorski T, Śliwiński T. Polθ Inhibition: An Anticancer Therapy for HR-Deficient Tumours. International Journal of Molecular Sciences. 2023; 24(1):319. https://doi.org/10.3390/ijms24010319
Chicago/Turabian StyleBarszczewska-Pietraszek, Gabriela, Małgorzata Drzewiecka, Piotr Czarny, Tomasz Skorski, and Tomasz Śliwiński. 2023. "Polθ Inhibition: An Anticancer Therapy for HR-Deficient Tumours" International Journal of Molecular Sciences 24, no. 1: 319. https://doi.org/10.3390/ijms24010319
APA StyleBarszczewska-Pietraszek, G., Drzewiecka, M., Czarny, P., Skorski, T., & Śliwiński, T. (2023). Polθ Inhibition: An Anticancer Therapy for HR-Deficient Tumours. International Journal of Molecular Sciences, 24(1), 319. https://doi.org/10.3390/ijms24010319







