Microbial Community Response to Various Types of Exogenous Organic Matter Applied to Soil
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemical Properties of EOM
2.2. EOM Effects on Microbiological Soil Parameters
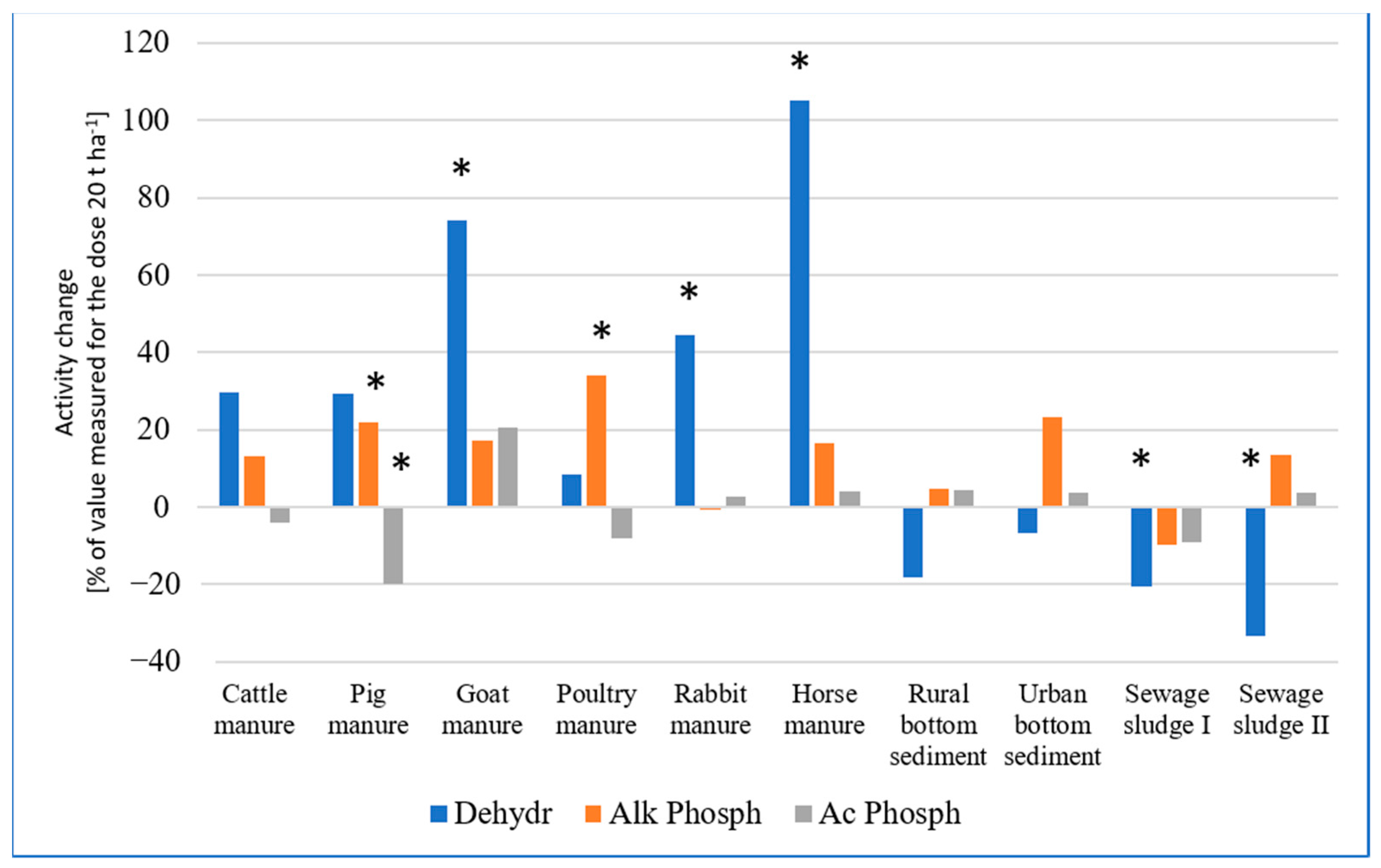
2.2.1. Enzymatic Activity
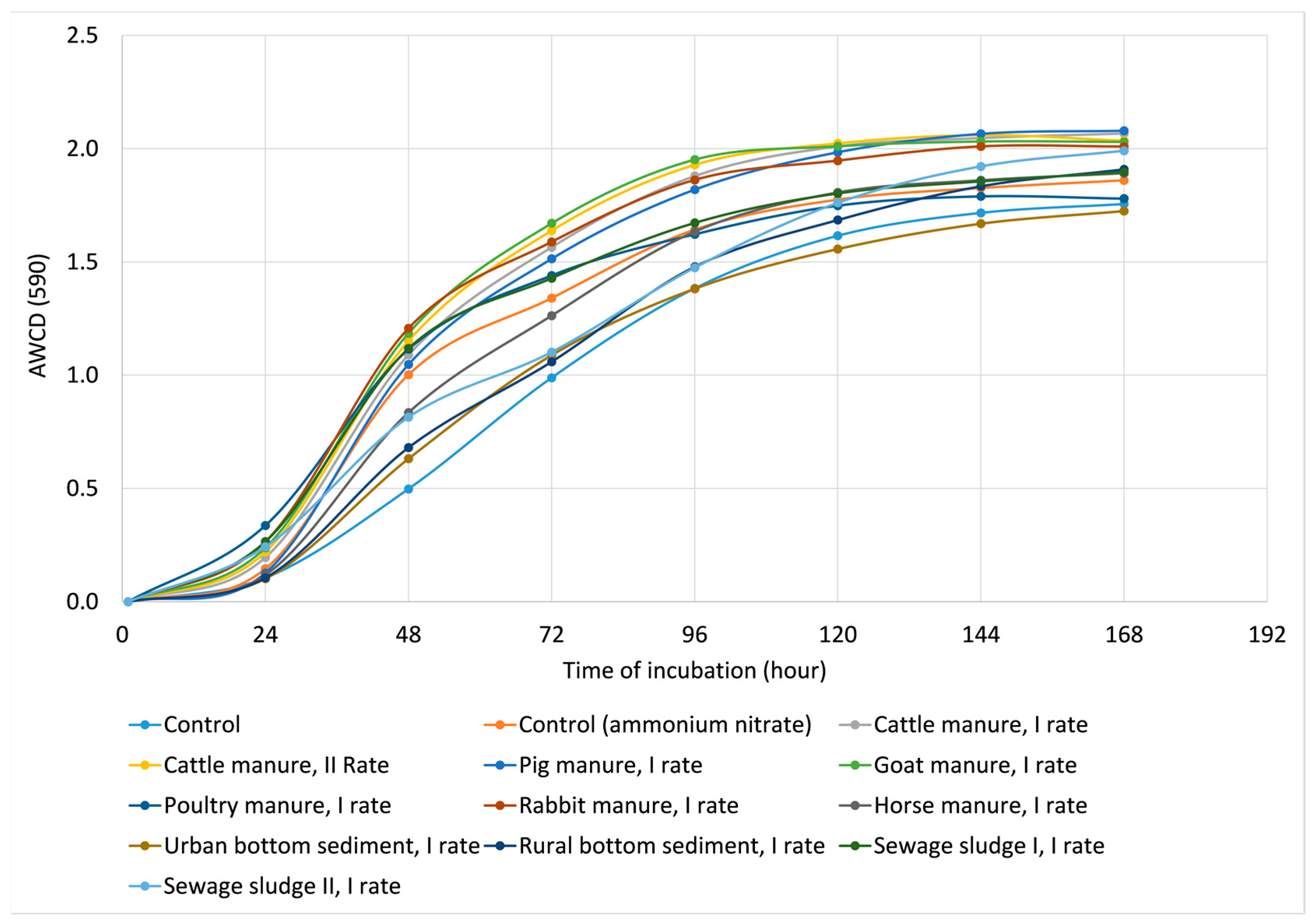
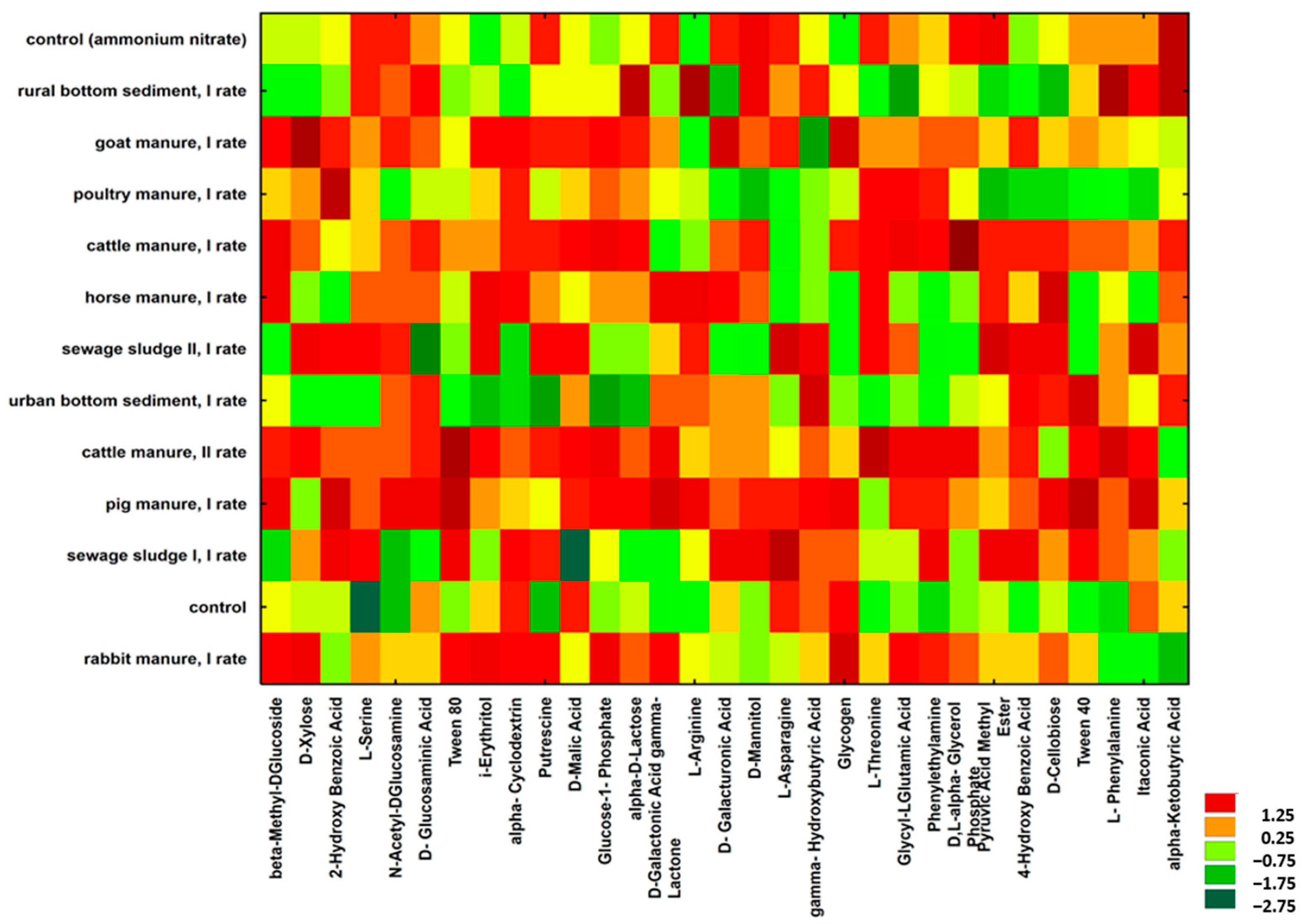
2.2.2. Microbial Functional Diversity
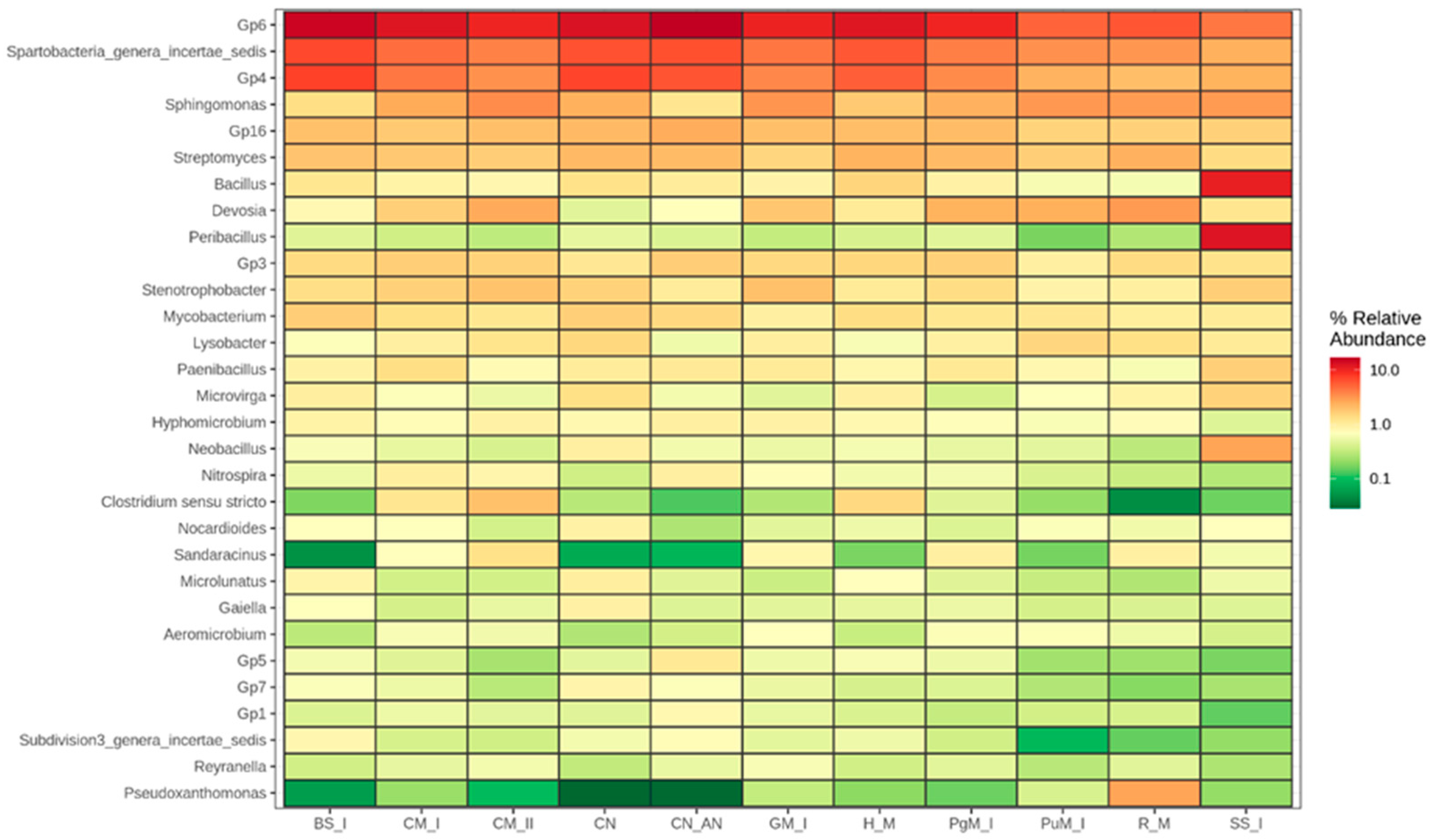
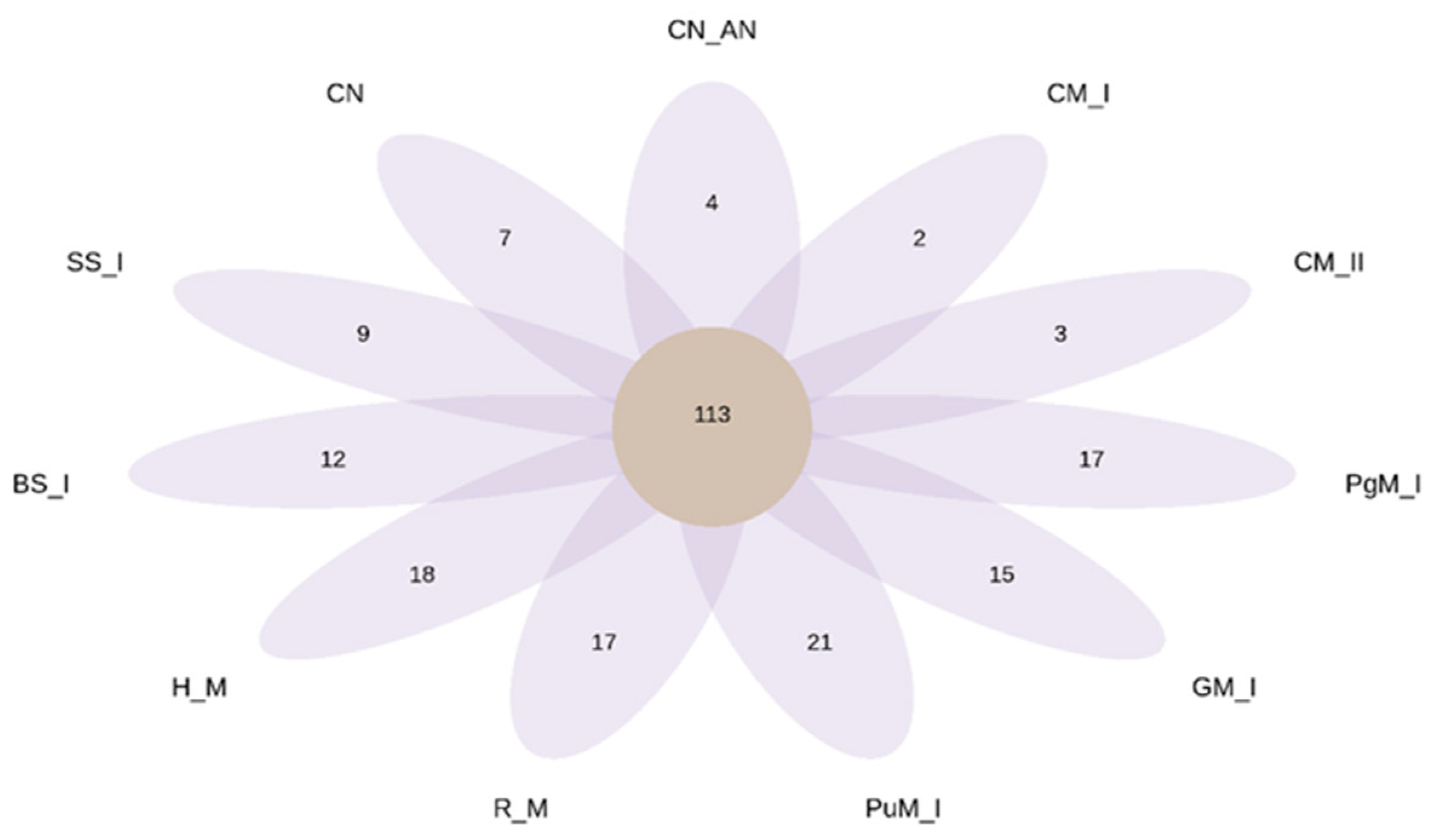
2.2.3. Diversity of Bacteria
2.3. Effect of EOMs on Soil Chemical Properties and Plant Biomass
3. Materials and Methods
3.1. Experimental Scheme
3.2. Chemical Analysis of EOM
3.3. Microbiological Soil Analysis
3.3.1. Enzymatic Activity
3.3.2. Microbial Functional Diversity
3.3.3. NGS—Next Generation Sequencing
3.4. Chemical Soil Analysis
3.5. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lal, R. Soil carbon sequestration impacts on global climate change and food security. Science 2004, 304, 1623–1627. [Google Scholar] [CrossRef]
- Spaccini, R.; Zena, A.; Igwe, C.; Mbagwu, J.; Piccolo, A. Carbohydrates in water-stable aggregates and particle size fractions of forested and cultivated soils in two contrasting tropical ecosystems. Biogeochemistry 2001, 53, 1–22. [Google Scholar] [CrossRef]
- Weil, R.; Magdoff, F. Significance of soil organic matter to soil quality and health. In Soil Organic Matter in Sustainable Agriculture; Magdoff, F., Weil, R., Eds.; CRC Press: Boca Raton, FL, USA, 2004; pp. 1–36. [Google Scholar]
- European Commision; Directorate-General for Research and Innovation; Voglhuber-Slavinsky, A.; Bisoffi, S.; Helming, K.; Giuffre, G.; Ricci, A.; Donitz, E.; Evgrafova, A.; Ratinger, T.; et al. Mission Area: Soil Health and Food: Foresight on Demand Brief in Support of the Horizon Europe Mission Board; Publications Office of the European Union: Luxembourg, 2021; Available online: https://data.europa.eu/doi/10.2777/038626 (accessed on 1 August 2023).
- Mondini, C.; Cayuela, M.L.; Sinicco, T.; Sanchez-Monedero, M.A.; Bertolone, E.; Bardi, L. Soil application of meat and bone meal. Short-term effects on mineralization dynamics and soil biochemical and microbiological properties. Soil Biol. Biochem. 2008, 40, 462–474. [Google Scholar] [CrossRef]
- European Commission. Communication from the Commission: The European Green Deal; COM (2019) 640 Final; European Commission: Brussels, Belgium, 2019. [Google Scholar]
- Darwish, O.H.; Persaud, N.; Martens, D.C. Effect of long-term application of animal manure on physical properties of three soils. Plant Soil 1995, 176, 289–295. [Google Scholar] [CrossRef]
- Stratton, M.L.; Barker, A.V.; Rechcigl, J.E. Compost. In Soil Amendments and Environmental Quality; Rechcigl, J.E., Ed.; CRC Press: Boca Raton, FL, USA, 1995; pp. 249–309. [Google Scholar]
- Raviv, M. Production of high-quality composts for horticultural purposes: A mini-review. Horttechnology 2005, 15, 52–57. [Google Scholar] [CrossRef]
- Leroy, B.L.M.; Herath, S.; Sleutel, S.; De Neve, S. The quality of exogenous organic matter: Short-term effects on soil physical properties and soil organic matter fractions. Soil Use Manag. 2008, 24, 139–147. [Google Scholar] [CrossRef]
- Zhao, H.; Tian, X.; Chen, Y.; Dong, J.; Shi, J. Effect of exogenous substances on soil organic and inorganic carbon sequestration under maize stover addition. Soil Sci. Plant Nutr. 2017, 63, 591–598. [Google Scholar] [CrossRef]
- Chen, Y.; Camps-Arbestain, M.; Shen, Q.; Singh, B.; Cayuela, M. The long-term role of organic amendments in building soil nutrient fertility: A meta-analysis and review. Nutr. Cycl. Agroecosyst. 2018, 111, 103–125. [Google Scholar] [CrossRef]
- Horii, A.; McCue, P.; Shetty, K. Seed vigour studies in corn, soybean and tomato in response to fish protein hydrolysates and consequences on phenolic-linked responses. Bioresour. Technol. 2007, 98, 2170–2177. [Google Scholar] [CrossRef]
- Quilty, J.; Cattle, S. Use and understanding of organic amendments in Australian agriculture: A review. Soil Res. 2011, 49, 1–26. [Google Scholar] [CrossRef]
- Moya, D.; Aldás, C.; López, G.; Kaparaju, P. Municipal solid waste as a valuable renewable energy resource: A worldwide opportunity of energy recovery by using Waste-To-Energy Technologies. Energy Procedia 2017, 134, 286–295. [Google Scholar] [CrossRef]
- Siebielec, G.; Stuczyński, T. Metale śladowe w komunalnych osadach ściekowych wytwarzanych w Polsce. Proc. ECOpole 2008, 2, 479–484. (In Polish) [Google Scholar]
- Siebielec, S.; Siebielec, G.; Smreczak, B. Zanieczyszczenia osadów dennych rzek i zbiorników wodnych. Studia i Raporty IUNG-PIB 2015, 46, 163–183. [Google Scholar]
- Siebielec, S.; Siebielec, G.; Urbaniak, M.; Smreczak, B.; Grzęda, E.; Wyrwicka, A.; Kidd, P. Impact of rhizobacterial inoculants on plant growth and enzyme activities in soil treated with contaminated bottom sediments. Int. J. Phytoremed. 2019, 21, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Siebielec, S.; Siebielec, G.; Klimkowicz-Pawlas, A.; Gałązka, A.; Grządziel, J.; Stuczyński, T. Impact of water stress on microbial community and activity in sandy and loamy soils. Agronomy 2020, 10, 1429. [Google Scholar] [CrossRef]
- Siebielec, S.; Siebielec, G.; Sugier, P.; Woźniak, M.; Grządziel, J.; Gałązka, A.; Stuczynski, T. Activity and diversity of microorganisms in root zone of plant species spontaneously inhabiting smelter waste piles. Molecules 2020, 25, 5638. [Google Scholar] [CrossRef]
- Siebielec, G.; Siebielec, S.; Lipski, D. Long-term impact of sewage sludge, digestate and mineral fertilizers on plant yield and soil biological activity. J. Clean. Prod. 2018, 187, 372–379. [Google Scholar] [CrossRef]
- Doran, J.W.; Parkin, T.B. Defining and assessing soil quality. In Defining Soil Quality for a Sustainable Environment; Doran, J.W., Coleman, D.C., Bezdicek, D.E., Stewart, B.A., Eds.; Soil Science Society of America: Madison, WI, USA, 1994; pp. 3–21. [Google Scholar]
- Doran, J.W.; Sarrantonio, M.; Liebig, M.A. Soil health and sustainability. Adv. Agron. 1996, 56, 1–54. [Google Scholar]
- Marinari, S.; Mancinelli, R.; Campiglia, E.; Grego, S. Chemical and biological indicators of soil quality in organic and conventional farming systems in Italy. Ecol. Indic. 2006, 6, 701–711. [Google Scholar] [CrossRef]
- Smith, J.L.; Paul, E.A. The significance of soil microbial biomass estimations. In Soil Biochemistry; Bollag, J., Stotzky, G., Eds.; Dekker: New York, NY, USA, 1990; pp. 357–396. [Google Scholar]
- Islam, R.; Chauhan, P.S.; Kim, Y.; Kim, M. Community level functional diversity and enzyme activities in paddy soils under different long-term fertilizer management practices. Biol. Fertil. Soils 2011, 47, 599–604. [Google Scholar] [CrossRef]
- Dincă, L.C.; Grenni, P.; Onet, C.; Onet, A. Fertilization and soil microbial community: A review. Appl. Sci. 2022, 12, 1198. [Google Scholar] [CrossRef]
- Albiach, R.; Canet, R.; Pomares, F.; Ingelmo, F. Microbial biomass content and enzymatic activities after the application of organic amendments to a horticultural soil. Bioresour. Technol. 2000, 75, 43–48. [Google Scholar] [CrossRef]
- Debosz, K.; Petersen, S.O.; Kure, L.K.; Ambus, P. Evaluating effects of sewage sludge and household compost on soil physical, chemical and microbiological properties. Appl. Soil Ecol. 2002, 19, 237–248. [Google Scholar] [CrossRef]
- Perucci, P.; Dumontet, S.; Bufo, S.; Mazzatura, A.; Casucci, C. Effects of organic amendment and herbicide treatment on soil microbial biomass. Biol. Fertil. Soils 2000, 32, 17–23. [Google Scholar] [CrossRef]
- Hamm, A.C.; Tenuta, M.; Krause, D.O.; Ominski, K.H.; Tkachuk, V.L.; Flaten, D.N. Bacterial communities of an agricultural soil amended with solid pig and dairy manures, and urea fertilizer. Appl. Soil Ecol. 2016, 103, 61–71. [Google Scholar] [CrossRef]
- Larkin, R.P.; Honeycutt, C.W.; Griffin, T.S. Effect of swine and dairy manure amendments on microbial communities in three soils as influenced by environmental conditions. Biol. Fertil. Soils 2006, 43, 51–61. [Google Scholar] [CrossRef]
- Risberg, K.; Cederlund, H.; Pell, M.; Arthurson, V.; Schnürer, A. Comparative characterization of digestate versus pig slurry and cow manure—Chemical composition and effects on soil microbial activity. Waste Manag. 2017, 61, 529–538. [Google Scholar] [CrossRef]
- Kim, S.Y.; Pramanik, P.; Bodelier, P.L.E.; Kim, P.J. Cattle manure enhances methanogens diversity and methane emissions compared to swine manure under rice paddy. PLoS ONE 2014, 9, e113593. [Google Scholar] [CrossRef]
- Semenov, M.V.; Krasnov, G.S.; Semenov, V.M.; Ksenofontova, N.; Zinyakova, N.B.; van Bruggen, A.H.C. Does fresh farmyard manure introduce surviving microbes into soil or activate soil-borne microbiota? J. Environ. Manag. 2021, 294, 113018. [Google Scholar] [CrossRef]
- Luo, G.; Li, L.; Friman, V.P.; Guo, J.; Guo, S.; Shen, Q.; Ling, N. Organic amendments increase crop yields by improving microbe-mediated soil functioning of agroecosystems: A meta-analysis. Soil Biol. Biochem. 2018, 124, 105–115. [Google Scholar] [CrossRef]
- Ministry of Environment. Rozporządzenie Ministra Środowiska z dnia 6 lutego 2015 r. w sprawie komunalnych osadów ściekowych; Dz.U. 2015 poz.257. Available online: https://isap.sejm.gov.pl/isap.nsf/download.xsp/WDU20150000257/O/D20150257.pdf (accessed on 1 August 2023). (In Polish)
- Ramamoorthy, S.; Ramamoorthy, S. Chlorinated Organic Compounds in the Environment; Lewis Publishers: Boca Raton, FL, USA, 1997. [Google Scholar]
- WHO. Inventory of Evaluations Performed by the Joint Meeting on Pesticide Residues (JMPR) CHLORPYRIFOS-METHYL; World Health Organization: Geneva, Switzerland, 2009; pp. 155–202. [Google Scholar]
- Ding, J.L.; Jiang, X.; Ma, M.C.; Zhou, B.K.; Guan, D.W.; Zhao, B.S.; Zhou, J.; Cao, F.M.; Li, L.; Li, J. Effect of 35 years inorganic fertilizer and manure amendment on structure of bacterial and archaeal communities in black soil of northeast China. Appl. Soil Ecol. 2016, 105, 187–195. [Google Scholar] [CrossRef]
- Jangid, K.; Williams, M.A.; Franzluebbers, A.J.; Sanderlin, J.S.; Reeves, J.H.; Jenkins, M.B.; Endale, D.M.; Coleman, D.C.; Whitman, W.B. Relative impacts of land-use, management intensity and fertilization upon soil microbial community structure in agricultural systems. Soil Biol. Biochem. 2008, 40, 2843–2853. [Google Scholar] [CrossRef]
- Fontaine, S.; Marotti, A.; Abbadie, L. The priming effect of organic matter: A question of microbial competition. Soil Biol. Biochem. 2003, 35, 837–843. [Google Scholar] [CrossRef]
- Nannipieri, P.; Grego, S.; Ceccanti, B. Ecological significance of the biological activity in soil. In Soil Biochemistry; Marcel Dekker: New York, NY, USA, 1990; Volume 6, pp. 293–355. [Google Scholar]
- Moeskops, B.; Buchan, D.; Sleutel, S.; Herawaty, L.; Husen, E.; Saraswati, R.; Setyorini, D.; De Neve, S. Soil microbial communities and activities under intensive organic and conventional vegetable farming in West Java, Indonesia. Appl. Soil Ecol. 2010, 45, 112–120. [Google Scholar] [CrossRef]
- Fernandes, S.A.; Bettiol, W.; Cerri, C.C. Effect of sewage sludge on microbial biomass, basal respiration, metabolic quotient and soil enzymatic activity. Appl. Soil Ecol. 2005, 30, 65–77. [Google Scholar] [CrossRef]
- Pascual, I.; Antolin, M.C.; Garcia, C.; Polo, A.; Sanchez-Diaz, M. Effect of water deficit on microbial characteristics in soil amended with sewage sludge or inorganic fertilizer under laboratory conditions. Bioresour. Technol. 2007, 98, 140–144. [Google Scholar] [CrossRef]
- Stefanowicz, A. The Biolog Plate technique as a tool in ecological studies of microbial communities. Pol. J. Environ. Stud. 2006, 15, 669–676. [Google Scholar]
- Oszust, K.; Gryta, A.; Zieminski, K.; Bilińska-Wielgus, N.; Gałązka, R.; Frąc, M. Characterization of microbial functional and genetic diversity as a novel strategy of biowaste ecotoxicological evaluation. Int. J. Environ. Sci. Techol. 2018, 16, 4261–4274. [Google Scholar] [CrossRef]
- Gryta, A.; Frąc, M.; Oszust, K. Genetic and metabolic diversity of soil microbiome in response to exogenous organic matter amendments. Agronomy 2020, 10, 546. [Google Scholar] [CrossRef]
- Wolińska, A.; Gałązka, A.; Kuźniar, A.; Goraj, W.; Jastrzębska, N.; Grządziel, J.; Stępniewska, Z. Catabolic fingerprinting and diversity of bacteria in mollic gleysol contaminated with petroleum substances. Appl. Sci. 2018, 8, 1970. [Google Scholar] [CrossRef]
- Alvarez, A.; Saez, J.M.; Costa, J.S.D.; Colin, V.L.; Fuentes, M.S.; Cuozzo, C.A.; Benimeli, C.S.; Polti, M.A.; Amoroso, M.J. Actinobacteria: Current research and perspectives for bioremediation of pesticides and heavy metals. Chemosphere 2017, 166, 41–62. [Google Scholar] [CrossRef] [PubMed]
- Larsbrink, J.; McKee, L.S. Bacteroidetes bacteria in the soil: Glycan acquisition, enzyme secretion, and gliding motility. Adv. Appl. Microbiol. 2020, 110, 63–98. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, A.L.; Souza, A.J.; Andrade, P.A.M.; Andreote, F.D.; Coscione, A.R.; Oliveira, F.C.; Regitano, J.B. Sewage sludge microbial structures and relations to their sources, treatments, and chemical attributes. Front. Microbiol. 2018, 9, 1462. [Google Scholar] [CrossRef] [PubMed]
- Veach, A.M.; Zeglin, L.H. Historical drought affects microbial population dynamics and activity during soil drying and re-wet. Microb. Ecol. 2020, 79, 662–674. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Kloepper, J.W.; Huang, P.; McInroy, J.A.; Hu, C.H. Isolation and characterization of N2-fixing bacteria from giant reed and switchgrass for plant growth promotion and nutrient uptake. J. Basic Microbiol. 2018, 58, 459–471. [Google Scholar] [CrossRef]
- Li, H.; Wei, Z.; Song, C.; Chen, X.; Zhang, R.; Liu, Y. Functional keystone drive nitrogen conversion during different animal manures composting. Bioresour. Technol. 2022, 361, 127721. [Google Scholar] [CrossRef]
- Siebielec, S.; Siebielec, G.; Stuczyński, T.; Sugier, P.; Grzęda, E.; Grządziel, J. Long term insight into biodiversity of a smelter wasteland reclaimed with biosolids and by-product lime. Sci. Total Environ. 2018, 636, 1048–1057. [Google Scholar] [CrossRef]
- Boudjabi, S.; Kribaa, M.; Chenchouni, H. Sewage sludge fertilization alleviates drought stress and improves physiological adaptation and yield performances in Durum Wheat (Triticum durum): A double-edged sword. J. King Saud. Univ. Sci. 2017, 31, 336–344. [Google Scholar] [CrossRef]
- Ociepa, E.; Mrowiec, M.; Lach, J. Influence of fertilisation with sewage sludge derived preparation on selected soil properties and prairie cordgrass yield. Environ. Res. 2017, 156, 775–780. [Google Scholar] [CrossRef]
- Ukalska-Jaruga, A.; Smreczak, B.; Siebielec, G. Assessment of pesticide residue content in Polish agricultural soils. Molecules 2020, 25, 587. [Google Scholar] [CrossRef]
- Ukalska-Jaruga, A.; Lewińska, K.; Mammadov, E.; Karczewskam, A.; Smreczakm, B.; Medyńska-Juraszekm, A. Residues of persistent organic pollutants (POPs) in agricultural soils adjacent to historical sources of their storage and distribution. The case study of Azerbaijan. Molecules 2020, 25, 1815. [Google Scholar] [CrossRef] [PubMed]
- Ukalska-Jaruga, A.; Smreczak, B. The impact of organic matter on polycyclic aromatic hydrocarbon (PAH) availability and persistence in soils. Molecules 2020, 25, 2470. [Google Scholar] [CrossRef] [PubMed]
- Tabatabai, M.A.; Bremner, J.M. Use of p-nitrophenylphosphate for assay of soil phosphatase activity. Soil Biol. Biochem. 1969, 1, 301–307. [Google Scholar] [CrossRef]
- Casida, L.E.; Klein, D.A.; Santoro, T. Soil dehydrogenase activity. Soil Sci. 1964, 98, 371–376. [Google Scholar] [CrossRef]
- Pohland, B.; Owen, B. Biolog EcoPlates standard methods. TAS Technol. Bull. Biol. 2009, 1, 1–3. [Google Scholar]
- Garland, J.L. Analytical approaches to the characterization of samples of microbial communities using patterns of potential C source utilization. Soil Biol. Biochem. 1996, 28, 213–221. [Google Scholar] [CrossRef]
- Garland, J.L.; Millis, A.L. Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Appl. Environ. Microbiol. 1991, 57, 2351–2359. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- R Core Team. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Murali, A.; Bhargava, A.; Wright, E.S. IDTAXA: A novel approach for accurate taxonomic classification of microbiome sequences. Microbiome 2018, 6, 140. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S.P. 2013. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Cui, Y.; Li, X.; Yao, M. Microeco: An R package for data mining in microbial community ecology. FEMS Microbiol. Ecol. 2021, 97, fiaa255. [Google Scholar] [CrossRef] [PubMed]
- Kovach, W. MVSP—A Multivariate Statistical Package for Windows, Version 3.1; Kovach Computing Services: Pentraeth, UK, 1999. [Google Scholar]
- White, J.R.; Nagarajan, N.; Pop, M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput. Biol. 2009, 5, e1000352. [Google Scholar] [CrossRef] [PubMed]


| EOM | Carbon | The Content of Nutrients | Trace Metal Content | |||||
|---|---|---|---|---|---|---|---|---|
| Nitrogen | Phosphorus | Potassium | Zinc | Lead | Cadmium | Chromium | ||
| [%] | [mg kg−1] | |||||||
| Cattle manure | 41.8 | 2.06 | 0.62 | 5.37 | 147 | 1.6 | 0.35 | 4.7 |
| Pig manure | 30.1 | 2.26 | 1.62 | 7.18 | 195 | 5.6 | 0.37 | 21.4 |
| Goat manure | 38.5 | 2.35 | 0.99 | 5.75 | 93 | 2.1 | 0.25 | 9.9 |
| Poultry manure | 17.6 | 1.75 | 0.74 | 1.04 | 342 | 86.6 | 0.63 | 63.1 |
| Rabbit manure | 37.1 | 3.25 | 1.36 | 7.62 | 206 | 2.4 | 0.45 | 10.3 |
| Horse manure | 35.9 | 2.43 | 1.29 | 4.18 | 121 | 2.3 | 0.32 | 10.2 |
| Rural bottom sediment | 1.2 | 0.07 | 0.03 | 0.09 | 7 | 4.8 | 0.07 | 5.7 |
| Urban bottom sediment | 1.9 | 0.10 | 0.03 | 0.03 | 65 | 17.7 | 0.22 | 21.2 |
| Sewage sludge I | 27.3 | 3.55 | 2.74 | 0.36 | 344 | 11.1 | 0.53 | 96.4 |
| Sewage sludge II | 32.7 | 5.24 | 1.28 | 0.23 | 1309 | 16.8 | 1.06 | 65.4 |
| Variables | Acid Phosphatase Activity | Alkaline Phosphatase Activity | Dehydrogenases Activity | ||||
|---|---|---|---|---|---|---|---|
| F | p | F | p | F | p | ||
| Main effects | EOM | 60.097 | <0.001 | 20.679 | <0.001 | 23.451 | <0.001 |
| rate | 15.836 | <0.001 | 28.777 | <0.001 | 31.653 | <0.001 | |
| plant | 82.240 | <0.001 | 97.576 | <0.001 | 168.125 | <0.001 | |
| Interaction effects | EOM × rate | 6.317 | <0.001 | 2.051 | 0.044 | 3.903 | <0.001 |
| EOM × plant | 12.153 | <0.001 | 1.614 | 0.125 | 8.248 | <0.001 | |
| rate × plant | 16.352 | <0.001 | 4.683 | 0.033 | 8.329 | 0.005 | |
| EOM × rate × plant | 5.746 | <0.001 | 1.071 | 0.393 | 0.995 | 0.450 | |
| Soil Additive | Rate [t ha−1] | Acid Phosphatase Activity | Alkaline Phosphatase Activity | Dehydrogenase Activity |
|---|---|---|---|---|
| [µg PNP g d.m.−1 h−1] | [µg PNP g d.m.−1 h−1] | [TTC g d.m.−1 h−1] | ||
| Control | 43.8 b B * ± 2.1 | 53.5 ac B ± 5.5 | 9.5 b C ± 1.1 | |
| Control (ammonium nitrate) | 44.7 b B ± 3.2 | 56.0 ac B ± 8.9 | 9.8 b C ± 2.3 | |
| Cattle manure | 20 | 49.9 b ± 4.6 | 80.6 abc ± 25.8 | 28.3 bc ± 9.3 |
| 40 | 47.9 B ± 3.0 | 91.3 AB ± 28.8 | 36.7 ABC ± 6.7 | |
| Pig manure | 20 | 43.9 b ± 0.9 | 60.4 abc ± 5.3 | 23.9 bc ± 10.3 |
| 40 | 35.2 B ± 4.9 | 73.6 AB ± 5.5 | 30.9 BC ± 5.4 | |
| Goat manure | 20 | 40.6 b ± 6.5 | 78.3 abc ± 11.2 | 30.6 abc ± 11.4 |
| 40 | 48.9 B ± 6.6 | 91.8 AB ± 17.8 | 53.3 ABC ± 13.3 | |
| Poultry manure | 20 | 50.6 b ± 8.3 | 94.8 b ± 9.7 | 22.4 bc ± 1.7 |
| 40 | 46.5 B ± 4.3 | 127.1 A ± 13.4 | 24.3 BC ± 2.8 | |
| Rabbit manure | 20 | 41.1 b ± 0.8 | 88.0 ab ± 11.4 | 30.9 abc ± 5.4 |
| 40 | 42.2 B ± 10.8 | 87.3 AB ± 36.4 | 44.7 ABC ± 17.4 | |
| Horse manure | 20 | 50.7 b ± 6.8 | 62.9 abc ± 12.8 | 37.3 ac ± 14.4 |
| 40 | 52.8 B ± 8.5 | 73.4 AB ± 15.1 | 76.5 A ± 17.3 | |
| Rural bottom sediment | 20 | 48.1 b ± 8.5 | 45.5 c ± 2.4 | 18.8 bc ± 1.4 |
| 40 | 50.3 B ± 3.4 | 47.6 B ± 11.7 | 15.4 BC ± 4.2 | |
| Urban bottom sediment | 20 | 49.3 b ± 0.2 | 43.9 c ± 10.4 | 17.5 bc ± 2.1 |
| 40 | 51.2 B ± 2.3 | 54.2 B ± 6.3 | 16.3 BC ± 3.6 | |
| Sewage sludge I | 20 | 48.6 b ± 1.2 | 96.2 b ± 22.5 | 36.4 ac ± 2.6 |
| 40 | 44.2 B ± 6.6 | 86.7 AB ± 33.2 | 28.9 BC ± 2.0 | |
| Sewage sludge II | 20 | 74.8 a ± 11.9 | 58.8 abc ± 5.9 | 50.9 a ± 3.4 |
| 40 | 77.5 B ± 11.5 | 66.7 B ± 11.8 | 34.0 BC ± 14.9 |
| Treatment | H’ | E | R |
|---|---|---|---|
| Control | 3.295 ab * ± 0.035 | 0.982 ab ± 0.005 | 28.667 ab ± 0.577 |
| Control (ammonium nitrate) | 3.338 abc ± 0.027 | 0.985 ab ± 0.007 | 29.667 ab ± 1.155 |
| Cattle manure, I Rate | 3.392 c ± 0.015 | 0.991 ab ± 0.006 | 30.667 b ± 0.577 |
| Cattle manure, II Rate | 3.385 c ± 0.011 | 0.989 ab ± 0.002 | 30.667 b ± 0.577 |
| Pig manure, I rate | 3.374 bc ± 0.008 | 0.986 ab ± 0.006 | 30.667 b ± 0.557 |
| Goat manure, I rate | 3.387 c ± 0.018 | 0.989 ab ± 0.007 | 30.667 b ± 0.557 |
| Poultry manure, I rate | 3.378 bc ± 0.015 | 0.993 ab ± 0.004 | 30.000 ab ± 0.000 |
| Rabbit manure, I rate | 3.379 bc ± 0.013 | 0.987 ab ± 0.002 | 30.667 b ± 0.057 |
| Horse manure, I rate | 3.312 abc ± 0.021 | 0.984 ab ± 0.005 | 29.000 ab ± 1.000 |
| Urban bottom sediment, I rate | 3.258 a ± 0.051 | 0.978 a ± 0.005 | 28.000 ab ± 1.732 |
| Rural bottom sediment, I rate | 3.322 abc ± 0.036 | 0.980 ab ± 0.003 | 29.667 ab ± 1.155 |
| Sewage sludge I, I rate | 3.339 abc ± 0.026 | 0.979 a ± 0.003 | 30.333 ab ± 0.577 |
| Sewage sludge II, I rate | 3.292 ab ± 0.056 | 0.992 ab ± 0.007 | 27.667 a ± 1.528 |
| Treatment | Ammonia-N | Nitrite-N | Nitrate-N | Available P (P2O5) | Available K (K2O) | Soil pH |
|---|---|---|---|---|---|---|
| mg kg−1 | ||||||
| Control | 0.33 abc * ± 0,05 | 0.06 a ± 0.02 | 18.9 a ± 5.33 | 196 a ± 70.9 | 113 b ± 14.49 | 7.23 b ± 0.06 |
| Control—AN | 0.08 a ± 0.02 | 0.04 a ± 0.01 | 60.4 ab ± 19.71 | 212 a ± 4.28 | 168 a ± 9.87 | 6.83 a ± 0.15 |
| Cattle manure | 0.30 abc ± 0.06 | 0.05 a ± 0.01 | 37.7 ab ± 2.03 | 199 a ± 4.62 | 237 a ± 3.21 | 7.17 b ± 0.06 |
| Pig manure | 0.42 bc ± 0.31 | 0.09 a ± 0.02 | 29.4 a ± 1.96 | 268 abc ± 16.38 | 516 d ± 39.36 | 7.37 b ± 0.06 |
| Goat manure | 0.84 d ± 0.13 | 0.07 a ± 0.01 | 28.2 a ± 0.91 | 198 a ± 41.61 | 478 cd ± 24.29 | 7.30 b ± 0.00 |
| Poultry manure | 0.59 cd ± 0.04 | 0.12 a ± 0.03 | 111.5 c ± 4.37 | 281 abc ± 6.43 | 234 a ± 5.19 | 6.70 a ± 0.00 |
| Rabbit manure | 0.34 abc ± 0.14 | 0.15 a ± 0.02 | 94.4 c ± 14.37 | 344 c ± 47.16 | 811 e ± 51.62 | 7.13 b ± 0.12 |
| Horse manure | 0.12 ab ± 0.03 | 0.14 a ± 0.10 | 46.8 ab ± 16.81 | 291 abc ± 34.85 | 390 c ± 100.10 | 7.17 b ± 0.06 |
| Urban bottom sediment | 0.10 ab ± 0.07 | 0.19 a ± 0.16 | 32.7 ab ± 6.96 | 226 ab ± 4.15 | 173 ab ± 36.7 | 7.17 b ± 0.06 |
| Rural bottom sediment | 0.08 a ± 0.05 | 0.06 a ± 0.02 | 38.6 ab ± 6.64 | 234 ab ± 0.07 | 179 ab ± 2.06 | 7.13 b ± 0.06 |
| Sewage sludge I | 0.35 abc ± 0.08 | 0.06 a ± 0.00 | 112.8 c ± 4.19 | 301 bc ± 61.64 | 182 ab ± 5.12 | 7.60 b ± 0.00 |
| Sewage sludge II | 0.35 abc ± 0.08 | 0.09 a ± 0.03 | 233.1 d ± 12.45 | 518 d ± 60.90 | 199 ab ± 28.2 | 6.10 a ± 0.10 |
| Treatment | Ammonia-N | Nitrite-N | Nitrate-N | Available P (P2O5) | Available K (K2O) | Soil pH |
|---|---|---|---|---|---|---|
| mg kg−1 | ||||||
| Control | 0.33 abc * ± 0.05 | 0.06 a ± 0.02 | 18.9 a ± 5.33 | 196 a ± 70.9 | 113 b ± 14.49 | 7.23 b ± 0.06 |
| Control—AN | 0.08 a ± 0.02 | 0.04 a ± 0.01 | 60.4 ab ± 19.71 | 212 a ± 4.28 | 168 a ± 9.87 | 6.83 a ± 0.15 |
| Cattle manure | 0.30 a ± 0,09 | 0.08 a ± 0.01 | 66.1 ab ± 11.57 | 229 ab ± 19.95 | 574 b ± 81.43 | 7.17 b ± 0.06 |
| Pig manure | 0.92 a ± 0.16 | 0.09 a ± 0.00 | 38.5 a ± 6.20 | 403 cd ± 18.99 | 798 d ± 75.13 | 7.50 b ± 0.10 |
| Goat manure | 0.64 a ± 0.11 | 0.10 a ± 0.01 | 47.9 ab ± 12.04 | 303 abc ± 36.90 | 719 cd ± 84.87 | 7.40 b ± 0.10 |
| Poultry manure | 0.65 a ± 0.34 | 0.20 a ± 0.03 | 159.7 cd ± 8.67 | 355 bcd ± 10.28 | 395 e ± 62.16 | 6.43 a ± 0.06 |
| Rabbit manure | 0.36 a ± 0.14 | 0.15 a ± 0.02 | 103.9 bc ± 9.54 | 581 ef ± 79.94 | 1653 f ± 58.94 | 7.57 b ± 0.21 |
| Horse manure | 0.10 a ± 0.00 | 0.09 a ± 0.01 | 73.3 ab ± 11.66 | 388 d ± 19.86 | 602 bc ± 29.54 | 7.07 b ± 0.06 |
| Urban bottom sediment | 0.11 a ± 0.03 | 0.07 a ± 0.02 | 33.0 a ± 6.53 | 236 ab ± 04.78 | 172 a ± 03.97 | 7.23 b ± 0.06 |
| Rural bottom sediment | 0.11 a ± 0.02 | 0.04 a ± 0.00 | 34.5 a ± 1.91 | 300 abc ± 96.76 | 188 a ± 13.30 | 7.13 b ± 0.06 |
| Sewage sludge I | 0.74 a ± 0.18 | 0.09 a ± 0.10 | 170.9 d ± 21.7 | 480 de ± 33.22 | 209 a ± 51.16 | 7.63 b ± 0.06 |
| Sewage sludge II | 11.72 b ± 8.20 | 1.58 a ± 0.71 | 295.2 e ± 58.04 | 706 f ± 58.54 | 202 a ± 10.50 | 5.87 a ± 0.21 |
| Roots | Shoots | Total Biomass | |
|---|---|---|---|
| Control | 0.82 a * ± 0.3 | 4.68 a ± 1.7 | 5.50 a ± 3.0 |
| Control—AN | 0.89 a ± 0.4 | 5.09 a ± 2.6 | 5.98 a ± 3.1 |
| Cattle manure | 0.82 a ± 0.1 | 4.79 a ± 1.1 | 5.61 a ± 1.2 |
| Pig manure | 0.99 a ± 0.1 | 6.15 a ± 1.1 | 7.14 ab ± 1.2 |
| Goat manure | 0.73 a ± 0.3 | 4.64 a ± 1.7 | 5.37 a ± 2.0 |
| Poultry manure | 3.55 c ± 0.6 | 25.12 b ± 0.8 | 28.67 c ± 1.2 |
| Rabbit manure | 1.67 ab ± 0.4 | 13.52 c ± 3.9 | 15.19 b ± 4.3 |
| Horse manure | 1.20 a ± 0.2 | 6.12 a ± 0.9 | 7.32 a ± 1.1 |
| Urban bottom sediment | 0.96 a ± 0.1 | 4.64 a ± 1.0 | 5.59 a ± 1.1 |
| Rural bottom sediment | 1.07 a ± 0.0 | 5.56 a ± 0.9 | 6.64 a ± 0.9 |
| Sewage sludge I | 2.83 bc ± 0.7 | 24.82 b ± 2.7 | 25.72 c ± 4.3 |
| Sewage sludge II | 2.94 bc ± 0.9 | 36.82 d ± 4.0 | 39.77 d ± 4.4 |
| Roots | Shoots | Total Biomass | |
|---|---|---|---|
| Control | 0.82 a * ± 0.3 | 4.68 a ± 1.7 | 5.50 a ± 3.0 |
| Control—AN | 0.89 a ± 0.4 | 5.09 a ± 2.6 | 5.98 a ± 3.1 |
| Cattle manure | 0.95 a ± 0.2 | 6.01 a ± 1.9 | 6.97 a ± 2.1 |
| Pig manure | 0.87 a ± 0.1 | 6.96 a ± 2.3 | 7.83 a ± 2.4 |
| Goat manure | 0.82 a ± 0.3 | 5.60 a ± 1.3 | 6.42 a ± 1.6 |
| Poultry manure | 2.91 b ± 0.1 | 31.01 b ± 3.0 | 33.92 b ± 3.0 |
| Rabbit manure | 3.44 b ± 0.4 | 34.13 b ± 7.2 | 37.57 b ± 6.9 |
| Horse manure | 1.12 a ± 0.1 | 6.67 a ± 1.3 | 7.79 a ± 1.4 |
| Urban bottom sediment | 0.84 a ± 0.3 | 3.85 a ± 0.9 | 4.69 a ± 1.1 |
| Rural bottom sediment | 1.16 a ± 0.1 | 6.70 a ± 1.1 | 7.86 a ± 1.2 |
| Sewage sludge I | 2.87 b ± 0.6 | 27.28 b ± 1.2 | 30.15 b ± 1.7 |
| Sewage sludge II | 3.95 b ± 1.3 | 50.89 c ± 3.7 | 54.83 c ± 4.7 |
| EOM | Per Hectare Equivalent (t) | Per Pot (g pot−1) |
|---|---|---|
| Control without N fertilizer | 0 | 0 |
| Control with ammonium nitrate | 0 | 0 |
| Cattle manure | 20 | 13 |
| 40 | 26 | |
| Pig manure | 20 | 13 |
| 40 | 26 | |
| Goat manure | 20 | 13 |
| 40 | 26 | |
| Poultry manure | 20 | 13 |
| 40 | 26 | |
| Rabbit manure | 20 | 13 |
| 40 | 26 | |
| Horse manure | 20 | 13 |
| 40 | 26 | |
| Bottom sediment rural | 20 | 13 |
| 40 | 26 | |
| Bottom sediment urban | 20 | 13 |
| 40 | 26 | |
| Sewage sludge I | 20 | 13 |
| 40 | 26 | |
| Sewage sludge II | 20 | 13 |
| 40 | 26 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Siebielec, S.; Marzec-Grządziel, A.; Siebielec, G.; Ukalska-Jaruga, A.; Kozieł, M.; Gałązka, A.; Przybyś, M.; Sugier, P.; Urbaniak, M. Microbial Community Response to Various Types of Exogenous Organic Matter Applied to Soil. Int. J. Mol. Sci. 2023, 24, 14559. https://doi.org/10.3390/ijms241914559
Siebielec S, Marzec-Grządziel A, Siebielec G, Ukalska-Jaruga A, Kozieł M, Gałązka A, Przybyś M, Sugier P, Urbaniak M. Microbial Community Response to Various Types of Exogenous Organic Matter Applied to Soil. International Journal of Molecular Sciences. 2023; 24(19):14559. https://doi.org/10.3390/ijms241914559
Chicago/Turabian StyleSiebielec, Sylwia, Anna Marzec-Grządziel, Grzegorz Siebielec, Aleksandra Ukalska-Jaruga, Monika Kozieł, Anna Gałązka, Marcin Przybyś, Piotr Sugier, and Magdalena Urbaniak. 2023. "Microbial Community Response to Various Types of Exogenous Organic Matter Applied to Soil" International Journal of Molecular Sciences 24, no. 19: 14559. https://doi.org/10.3390/ijms241914559
APA StyleSiebielec, S., Marzec-Grządziel, A., Siebielec, G., Ukalska-Jaruga, A., Kozieł, M., Gałązka, A., Przybyś, M., Sugier, P., & Urbaniak, M. (2023). Microbial Community Response to Various Types of Exogenous Organic Matter Applied to Soil. International Journal of Molecular Sciences, 24(19), 14559. https://doi.org/10.3390/ijms241914559











