Molecular Characterization and Mutational Analysis of Clarithromycin- and Levofloxacin-Resistance Genes in Helicobacter pylori from Gastric Biopsies in Southern Croatia
Abstract
1. Introduction
2. Results
2.1. Genetic Mutation Study
2.1.1. Nucleotide Sequence Analysis of the V Domain of the 23S rRNA Gene of H. pylori Isolates and Molecular Docking
2.1.2. Nucleotide Sequence Analysis of the DNA Gyrase, Subunit A (gyrA) Gene from H. pylori Isolates and Molecular Docking
2.1.3. Nucleotide Sequence Analysis of the DNA Gyrase, Subunit B (gyrB) Gene from H. pylori Isolates and Molecular Docking
3. Discussion
- Identification of primary and secondary resistance: Distinguishing between primary and secondary resistance is critical for making informed treatment decisions. Comprehensive patient data could provide insight into whether resistance arose from previous antibiotic exposure or from intrinsic genetic factors.
- Population genotyping and resistance correlation: MLST analysis would elucidate the genetic diversity of local H. pylori strains. Linking this diversity to resistance patterns could provide insight into population dynamics that influence the evolution of antibiotic resistance.
- Treatment optimization: A more nuanced understanding of patient demographics, clinical symptoms, and resistance mechanisms would enable the development of personalized treatment regimens and an increase in treatment success rates.
- Epidemiologic insights: Comprehensive research would contribute valuable data to the global understanding of H. pylori antibiotic resistance, supporting the formulation of effective public health strategies.
4. Materials and Methods
4.1. Isolation of H. pylori
4.2. Isolation of Genomic DNA from H. pylori
4.3. Amplification of the 23S rRNA, gyrA, and gyrB Genes
4.4. Sequencing and Identification of the Mutation
4.5. Molecular Docking Analysis
4.5.1. GyrA and GyrB with LVX
4.5.2. 23S rRNA and CAM
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Murray, P.R.; Rosenthal, K.; Pfaller, M.A. Medical Microbiology, 9th ed.; Elsevier: Houston, TX, USA, 2021; p. 872. [Google Scholar]
- Baj, J.; Forma, A.; Sitarz, M.; Portincasa, P.; Garruti, G.; Krasowska, D.; Maciejewski, R. Helicobacter pylori Virulence Factors-Mechanisms of Bacterial Pathogenicity in the Gastric Microenvironment. Cells 2021, 10, 27. [Google Scholar] [CrossRef] [PubMed]
- Öztekin, M.; Yılmaz, B.; Ağagündüz, D.; Capasso, R. Overview of Helicobacter pylori Infection: Clinical Features, Treatment, and Nutritional Aspects. Diseases 2021, 9, 66. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Chey, W.D.; Leontiadis, G.I.; Howden, C.W.; Moss, S.F. ACG Clinical Guideline: Treatment of Helicobacter pylori Infection. Am. J. Gastroenterol. 2017, 112, 212–239. [Google Scholar] [CrossRef]
- Fock, K.M.; Graham, D.Y.; Malfertheiner, P. Helicobacter pylori research: Historical insights and future directions. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 495–500. [Google Scholar] [CrossRef]
- Malfertheiner, P.; Megraud, F.; O’Morain, C.A.; Gisbert, J.P.; Kuipers, E.J.; Axon, A.T.; Bazzoli, F.; Gasbarrini, A.; Atherton, J.; Graham, D.Y.; et al. Management of Helicobacter pylori infection-the Maastricht V/Florence Consensus Report. Gut 2017, 66, 6–30. [Google Scholar] [CrossRef]
- Plummer, M.; Franceschi, S.; Vignat, J.; Forman, D.; de Martel, C. Global burden of gastric cancer attributable to Helicobacter pylori. Int. J. Cancer 2015, 136, 487–490. [Google Scholar] [CrossRef]
- Mommersteeg, M.C.; Yu, J.; Peppelenbosch, M.P.; Fuhler, G.M. Genetic host factors in Helicobacter pylori-induced carcinogenesis: Emerging new paradigms. Biochim. Biophys. Acta Rev. Cancer 2018, 1869, 42–52. [Google Scholar] [CrossRef]
- Sugano, K.; Tack, J.; Kuipers, E.J.; Graham, D.Y.; El-Omar, E.M.; Miura, S.; Haruma, K.; Asaka, M.; Uemura, N.; Malfertheiner, P.; et al. Kyoto global consensus report on Helicobacter pylori gastritis. Gut 2015, 64, 1353–1367. [Google Scholar] [CrossRef]
- Bennett, J.E.; Dolin, R.; Blaser, M.J. Principles and Practice of Infectious Diseases, 8th ed.; John, E., Bennett, R.D., Blaser, M.J., Eds.; Elsevier/Saunders: Philadelphia, PA, USA, 2015; Volume 1, p. 3577. [Google Scholar]
- Malfertheiner, P.; Megraud, F.; O’Morain, C.; Hungin, A.; Jones, R.; Axon, A.; Graham, D.; Tytgat, G.; Asaka, M.; Bazzoli, F.; et al. Current concepts in the management of Helicobacter pylori infection—The Maastricht 2–2000 Consensus Report. Aliment. Pharmacol. Ther. 2002, 16, 167–180. [Google Scholar] [CrossRef]
- Savoldi, A.; Carrara, E.; Graham, D.Y.; Conti, M.; Tacconelli, E. Prevalence of Antibiotic Resistance in Helicobacter pylori: A Systematic Review and Meta-analysis in World Health Organization Regions. Gastroenterology 2018, 155, 1372–1382.e17. [Google Scholar] [CrossRef] [PubMed]
- Thung, I.; Aramin, H.; Vavinskaya, V.; Gupta, S.; Park, J.Y.; Crowe, S.E.; Valasek, M.A. Review article: The global emergence of Helicobacter pylori antibiotic resistance. Aliment. Pharm. 2016, 43, 514–533. [Google Scholar] [CrossRef] [PubMed]
- Morehead, M.S.; Scarbrough, C. Emergence of Global Antibiotic Resistance. Prim. Care 2018, 45, 467–484. [Google Scholar] [CrossRef] [PubMed]
- Megraud, F.; Bruyndonckx, R.; Coenen, S.; Wittkop, L.; Huang, T.D.; Hoebeke, M.; Bénéjat, L.; Lehours, P.; Goossens, H.; Glupczynski, Y.; et al. resistance to antibiotics in Europe in 2018 and its relationship to antibiotic consumption in the community. Gut 2021, 70, 1815–1822. [Google Scholar] [CrossRef]
- Mukherjee, S.; Saha, N. Correlation of Recommendations of Treatment Guidelines and Frequently Prescribed Antibiotics: Evaluation of Their Pharmaceutical Pack Size. Basic Clin. Pharmacol. Toxicol. 2018, 122, 317–321. [Google Scholar] [CrossRef]
- Tshibangu-Kabamba, E.; Yamaoka, Y. Helicobacter pylori infection and antibiotic resistance—From biology to clinical implications. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 613–629. [Google Scholar] [CrossRef]
- Versalovic, J.; Shortridge, D.; Kibler, K.; Griffy, M.; Beyer, J.; Flamm, R.; Tanaka, S.; Graham, D.; Go, M. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 1996, 40, 477–480. [Google Scholar] [CrossRef]
- Garcia, M.; Raymond, J.; Garnier, M.; Cremniter, J.; Burucoa, C. Distribution of Spontaneous gyrA Mutations in 97 Fluoroquinolone-Resistant Helicobacter pylori Isolates Collected in France. Antimicrob. Agents Chemother. 2012, 56, 550–551. [Google Scholar] [CrossRef]
- Perkovic, N.; Mestrovic, A.; Bozic, J.; Ivelja, M.P.; Vukovic, J.; Kardum, G.; Sundov, Z.; Tonkic, M.; Puljiz, Z.; Vukojevic, K.; et al. Randomized Clinical Trial Comparing Concomitant and Tailored Therapy for Eradication of Helicobacter pylori infection. J. Pers. Med. 2021, 11, 534. [Google Scholar] [CrossRef]
- Skinner, R.; Cundliffe, E.; Schmidt, F.J. Site of action of a ribosomal RNA methylase responsible for resistance to erythromycin and other antibiotics. J. Biol. Chem. 1983, 258, 12702–12706. [Google Scholar] [CrossRef]
- Albasha, A.M.; Elnosh, M.M.; Osman, E.H.; Zeinalabdin, D.M.; Fadl, A.A.M.; Ali, M.A.; Altayb, H.N. Helicobacter pylori 23S rRNA gene A2142G, A2143G, T2182C, and C2195T mutations associated with clarithromycin resistance detected in Sudanese patients. BMC Microbiol. 2021, 21, 38. [Google Scholar] [CrossRef] [PubMed]
- Marques, A.T.; Vítor, J.M.B.; Santos, A.; Oleastro, M.; Vale, F.F. Trends in Helicobacter pylori resistance to clarithromycin: From phenotypic to genomic approaches. Microb. Genom. 2020, 6. [Google Scholar] [CrossRef] [PubMed]
- Khan, R.; Nahar, S.; Sultana, J.; Ahmad, M.M.; Rahman, M. T2182C mutation in 23S rRNA is associated with clarithromycin resistance in Helicobacter pylori isolates obtained in Bangladesh. Antimicrob. Agents Chemother. 2004, 48, 3567–3569. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.S.; Kang, J.O.; Eun, C.S.; Han, D.S.; Choi, T.Y. Mutations in the 23S rRNA gene of Helicobacter pylori associated with clarithromycin resistance. J. Korean Med. Sci. 2002, 17, 599–603. [Google Scholar] [CrossRef]
- Bińkowska, A.; Biernat, M.M.; Łaczmański, Ł.; Gościniak, G. Molecular Patterns of Resistance among Helicobacter pylori Strains in South-Western Poland. Front. Microbiol. 2018, 9, 3154. [Google Scholar] [CrossRef]
- Chu, A.; Wang, D.; Guo, Q.; Lv, Z.; Yuan, Y.; Gong, Y. Molecular detection of H. pylori antibiotic-resistant genes and molecular docking analysis. FASEB J. 2020, 34, 610–618. [Google Scholar] [CrossRef]
- Phan, T.N.; Santona, A.; Tran, V.H.; Tran, T.N.; Le, V.A.; Cappuccinelli, P.; Rubino, S.; Paglietti, B. High rate of levofloxacin resistance in a background of clarithromycin- and metronidazole-resistant Helicobacter pylori in Vietnam. Int. J. Antimicrob. Agents 2015, 45, 244–248. [Google Scholar] [CrossRef]
- Zhang, W.; Li, Z.; Sun, Y.; Cui, P.; Liang, J.; Xing, Q.; Wu, J.; Xu, Y.; Zhang, Y.; He, L.; et al. Cryo-EM structure of Mycobacterium tuberculosis 50S ribosomal subunit bound with clarithromycin reveals dynamic and specific interactions with macrolides. Emerg. Microbes Infect. 2022, 11, 293–305. [Google Scholar] [CrossRef]
- Kannan, K.; Vázquez-Laslop, N.; Mankin, A.S. Selective protein synthesis by ribosomes with a drug-obstructed exit tunnel. Cell 2012, 151, 508–520. [Google Scholar] [CrossRef]
- Correia, S.; Poeta, P.; Hébraud, M.; Capelo, J.L.; Igrejas, G. Mechanisms of quinolone action and resistance: Where do we stand? J. Med. Microbiol. 2017, 66, 551–559. [Google Scholar] [CrossRef]
- Matsuzaki, J.; Suzuki, H.; Tsugawa, H.; Nishizawa, T.; Hibi, T. Homology model of the DNA gyrase enzyme of Helicobacter pylori, a target of quinolone-based eradication therapy. J. Gastroenterol. Hepatol. 2010, 25, S7–S10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wen, Y.; Xiao, Q.; Zheng, W.; Long, G.; Chen, B.; Shu, X.; Jiang, M. Mutations in the Antibiotic Target Genes Related to Clarithromycin, Metronidazole and Levofloxacin Resistance in Helicobacter pylori Strains from Children in China. Infect. Drug Resist. 2020, 13, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Lok, C.H.; Zhu, D.; Wang, J.; Ren, Y.T.; Jiang, X.; Li, S.J.; Zhao, X.Y. Phenotype and Molecular Detection of Clarithromycin and Levofloxacin Resistance in Helicobacter pylori Clinical Isolates in Beijing. Infect. Drug Resist. 2020, 13, 2145–2153. [Google Scholar] [CrossRef] [PubMed]
- Miyachi, H.; Miki, I.; Aoyama, N.; Shirasaka, D.; Matsumoto, Y.; Toyoda, M.; Mitani, T.; Morita, Y.; Tamura, T.; Kinoshita, S.; et al. Primary levofloxacin resistance and gyrA/B mutations among Helicobacter pylori in Japan. Helicobacter 2006, 11, 243–249. [Google Scholar] [CrossRef]
- Domanovich-Asor, T.; Craddock, H.; Motro, Y.; Khalfin, B.; Peretz, A.; Moran-Gilad, J. Unraveling antimicrobial resistance in Helicobacter pylori: Global resistome meets global phylogeny. Helicobacter 2021, 26, e12782. [Google Scholar] [CrossRef]
- Jaka, H.; Rüttgerodt, N.; Bohne, W.; Mueller, A.; Gross, U.; Kasang, C.; Mshana, S.E. Mutations Conferring Resistance to Fluoroquinolones and Clarithromycin among Dyspeptic Patients Attending a Tertiary Hospital, Tanzania. Can. J. Gastroenterol. Hepatol. 2019, 2019, 8481375. [Google Scholar] [CrossRef]
- Tanih, N.F.; Ndip, R.N. Molecular Detection of Antibiotic Resistance in South African Isolates of Helicobacter pylori. Gastroenterol. Res. Pract. 2013, 2013, 259457. [Google Scholar] [CrossRef]
- Losurdo, G.; Giorgio, F.; Pricci, M.; Girardi, B.; Russo, F.; Riezzo, G.; Martulli, M.; Piazzolla, M.; Cocomazzi, F.; Abbruzzi, F.; et al. Primary and Secondary Genotypic Resistance to Clarithromycin and Levofloxacin Detection in Stools: A 4-Year Scenario in Southern Italy. Antibiotics 2020, 9, 723. [Google Scholar] [CrossRef]
- Tamayo, E.; Montes, M.; Fernández-Reyes, M.; Lizasoain, J.; Ibarra, B.; Mendarte, U.; Zapata, E.; Mendiola, J.; Pérez-Trallero, E. Clarithromycin resistance in Helicobacter pylori and its molecular determinants in Northern Spain, 2013–2015. J. Glob. Antimicrob. Resist. 2017, 9, 43–46. [Google Scholar] [CrossRef]
- Lauener, F.N.; Imkamp, F.; Lehours, P.; Buissonnière, A.; Benejat, L.; Zbinden, R.; Keller, P.M.; Wagner, K. Genetic Determinants and Prediction of Antibiotic Resistance Phenotypes in Helicobacter pylori. J. Clin. Med. 2019, 8, 53. [Google Scholar] [CrossRef]
- Sanches, B.S.; Martins, G.M.; Lima, K.; Cota, B.; Moretzsohn, L.D.; Ribeiro, L.T.; Breyer, H.P.; Maguilnik, I.; Maia, A.B.; Rezende-Filho, J.; et al. Detection of Helicobacter pylori resistance to clarithromycin and fluoroquinolones in Brazil: A national survey. World J. Gastroenterol. 2016, 22, 7587–7594. [Google Scholar] [CrossRef] [PubMed]
- Zerbetto De Palma, G.; Mendiondo, N.; Wonaga, A.; Viola, L.; Ibarra, D.; Campitelli, E.; Salim, N.; Corti, R.; Goldman, C.; Catalano, M. Occurrence of Mutations in the Antimicrobial Target Genes Related to Levofloxacin, Clarithromycin, and Amoxicillin Resistance in Helicobacter pylori Isolates from Buenos Aires City. Microb. Drug. Resist. 2017, 23, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Camorlinga-Ponce, M.; Gómez-Delgado, A.; Aguilar-Zamora, E.; Torres, R.C.; Giono-Cerezo, S.; Escobar-Ogaz, A.; Torres, J. Phenotypic and Genotypic Antibiotic Resistance Patterns in Helicobacter pylori Strains From Ethnically Diverse Population in México. Front. Cell Infect. Microbiol. 2020, 10, 539115. [Google Scholar] [CrossRef] [PubMed]
- Tshibangu-Kabamba, E.; Ngoma-Kisoko, P.J.; Tuan, V.P.; Matsumoto, T.; Akada, J.; Kido, Y.; Tshimpi-Wola, A.; Tshiamala-Kashala, P.; Ahuka-Mundeke, S.; Ngoy, D.M.; et al. Next-Generation Sequencing of the Whole Bacterial Genome for Tracking Molecular Insight into the Broad-Spectrum Antimicrobial Resistance of Helicobacter pylori Clinical Isolates from the Democratic Republic of Congo. Microorganisms 2020, 8, 887. [Google Scholar] [CrossRef]
- López-Gasca, M.; Peña, J.; García-Amado, M.A.; Michelangeli, F.; Contreras, M. Point Mutations at gyrA and gyrB Genes of Levofloxacin-Resistant Helicobacter pylori Isolates in the Esophageal Mucosa from a Venezuelan Population. Am. J. Trop. Med. Hyg. 2018, 98, 1051–1055. [Google Scholar] [CrossRef]
- Saranathan, R.; Levi, M.H.; Wattam, A.R.; Malek, A.; Asare, E.; Behin, D.S.; Pan, D.H.; Jacobs, W.R.; Szymczak, W.A. Helicobacter pylori Infections in the Bronx, New York: Surveying Antibiotic Susceptibility and Strain Lineage by Whole-Genome Sequencing. J. Clin. Microbiol. 2020, 58, 10–1128. [Google Scholar] [CrossRef]
- Moore, R.A.; Beckthold, B.; Wong, S.; Kureishi, A.; Bryan, L.E. Nucleotide sequence of the gyrA gene and characterization of ciprofloxacin-resistant mutants of Helicobacter pylori. Antimicrob. Agents Chemother. 1995, 39, 107–111. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhong, Z.; Hu, S.; Wang, J.; Deng, Y.; Li, X.; Chen, X.; Li, X.; Tang, Y.; Li, X.; et al. A Survey of Helicobacter pylori Antibiotic-Resistant Genotypes and Strain Lineages by Whole-Genome Sequencing in China. Antimicrob. Agents Chemother. 2022, 66, e02188-21. [Google Scholar] [CrossRef]
- Puah, S.; Goh, K.; Ng, H.; Chua, K. Current status of Helicobacter pylori resistance to Clarithromycin and Levofloxacin in Malaysia-findings from a molecular based study. PeerJ 2021, 9, e11518. [Google Scholar] [CrossRef]
- Farzi, N.; Yadegar, A.; Sadeghi, A.; Asadzadeh Aghdaei, H.; Marian Smith, S.; Raymond, J.; Suzuki, H.; Zali, M.R. High Prevalence of Antibiotic Resistance in Iranian. J. Clin. Med. 2019, 8, 2004. [Google Scholar] [CrossRef]
- Miftahussurur, M.; Aftab, H.; Shrestha, P.K.; Sharma, R.P.; Subsomwong, P.; Waskito, L.A.; Doohan, D.; Fauzia, K.A.; Yamaoka, Y. Effective therapeutic regimens in two South Asian countries with high resistance to major Helicobacter pylori antibiotics. Antimicrob. Resist. Infect. Control 2019, 8, 40. [Google Scholar] [CrossRef] [PubMed]
- Versalovic, J.; Osato, M.S.; Spakovsky, K.; Dore, M.P.; Reddy, R.; Stone, G.G.; Shortridge, D.; Flamm, R.K.; Tanaka, S.K.; Graham, D.Y. Point mutations in the 23S rRNA gene of Helicobacter pylori associated with different levels of clarithromycin resistance. J. Antimicrob. Chemother. 1997, 40, 283–286. [Google Scholar] [CrossRef] [PubMed][Green Version]
- The European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 9.0, 2019. Available online: http://www.eucast.org (accessed on 15 January 2019).
- Rimbara, E.; Sasatsu, M.; Graham, D.Y. PCR detection of Helicobacter pylori in clinical samples. Methods Mol. Biol. 2013, 943, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, N.; Rimbara, E.; Kato, A.; Tanaka, A.; Tokunaga, K.; Kawai, T.; Takahashi, S.; Sasatsu, M. Detection of mixed clarithromycin-resistant and -susceptible Helicobacter pylori using nested PCR and direct sequencing of DNA extracted from faeces. J. Med. Microbiol. 2007, 56, 1174–1180. [Google Scholar] [CrossRef]
- Wani, F.A.; Bashir, G.; Khan, M.A.; Zargar, S.A.; Rasool, Z.; Qadri, Q. Antibiotic resistance in Helicobacter pylori: A Mutational Analysis from a Tertiary Care Hospital in Kashmir, India. Indian J. Med. Microbiol. 2018, 36, 265–272. [Google Scholar] [CrossRef]
- Hall, T. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Larkin, M.; Blackshields, G.; Brown, N.; Chenna, R.; McGettigan, P.; McWilliam, H.; Valentin, F.; Wallace, I.; Wilm, A.; Lopez, R.; et al. Clustal W and clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A. Software News and Update AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Honorato, R.V.; Koukos, P.I.; Jiménez-García, B.; Tsaregorodtsev, A.; Verlato, M.; Giachetti, A.; Rosato, A.; Bonvin, A.M.J.J. Structural Biology in the Clouds: The WeNMR-EOSC Ecosystem. Front. Mol. Biosci. 2021, 8, 729513. [Google Scholar] [CrossRef]
- van Zundert, G.C.P.; Rodrigues, J.P.G.L.; Trellet, M.; Schmitz, C.; Kastritis, P.L.; Karaca, E.; Melquiond, A.S.J.; van Dijk, M.; de Vries, S.J.; Bonvin, A.M.J.J. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J. Mol. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2023 update. Nucleic Acids Res. 2023, 51, D1373–D1380. [Google Scholar] [CrossRef] [PubMed]
- Jubb, H.C.; Higueruelo, A.P.; Ochoa-Montaño, B.; Pitt, W.R.; Ascher, D.B.; Blundell, T.L. Arpeggio: A Web Server for Calculating and Visualising Interatomic Interactions in Protein Structures. J. Mol. Biol. 2017, 429, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Szulc, N.A.; Mackiewicz, Z.; Bujnicki, J.M.; Stefaniak, F. fingeRNAt—A novel tool for high-throughput analysis of nucleic acid-ligand interactions. PLoS Comput. Biol. 2022, 18, e1009783. [Google Scholar] [CrossRef]

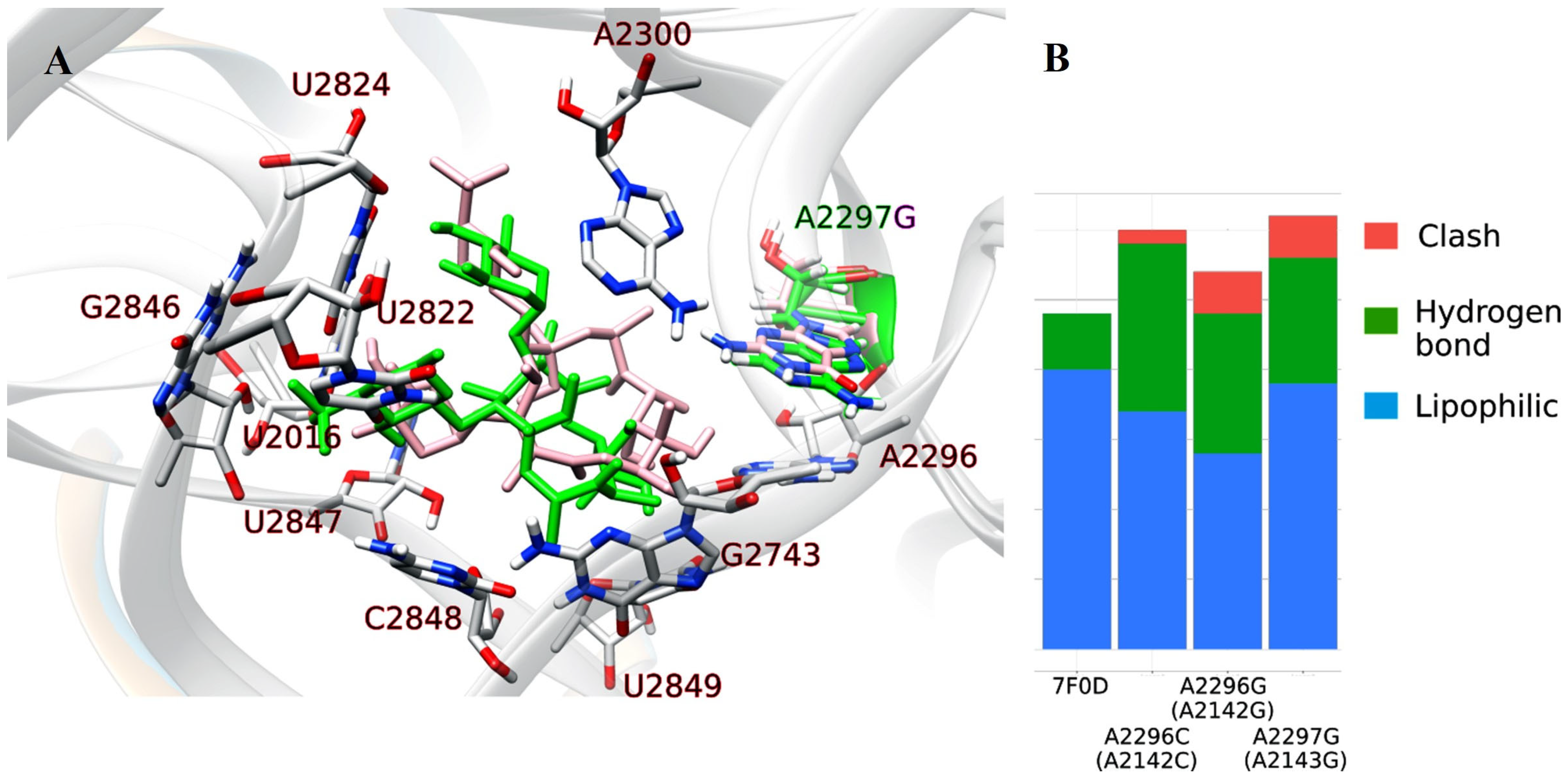
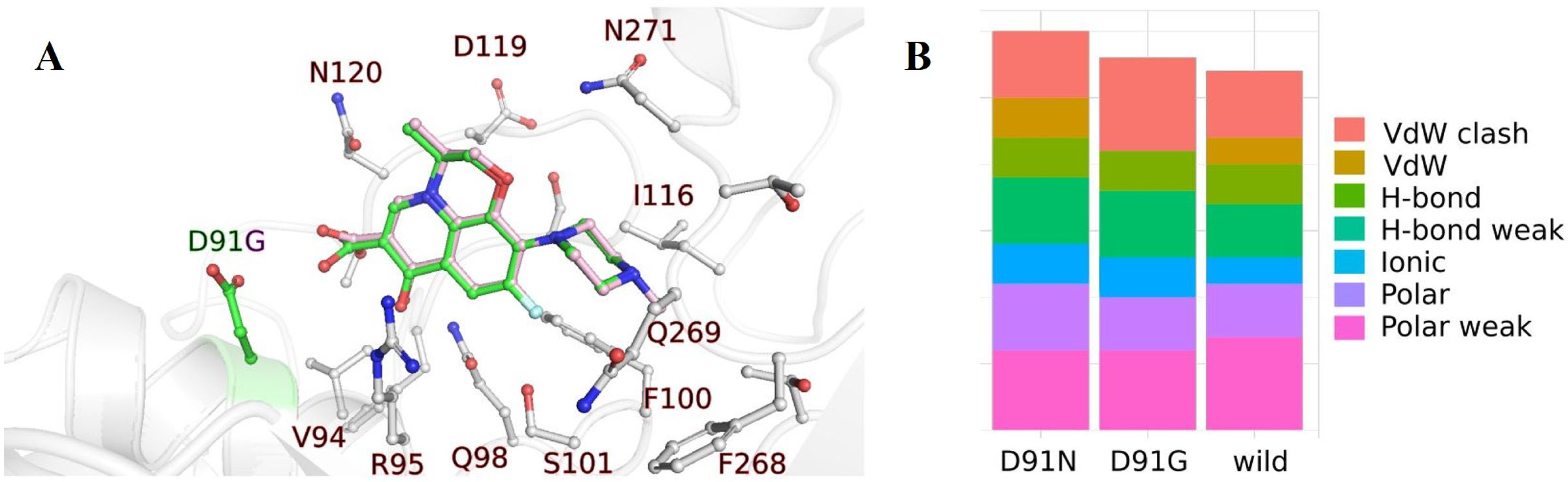
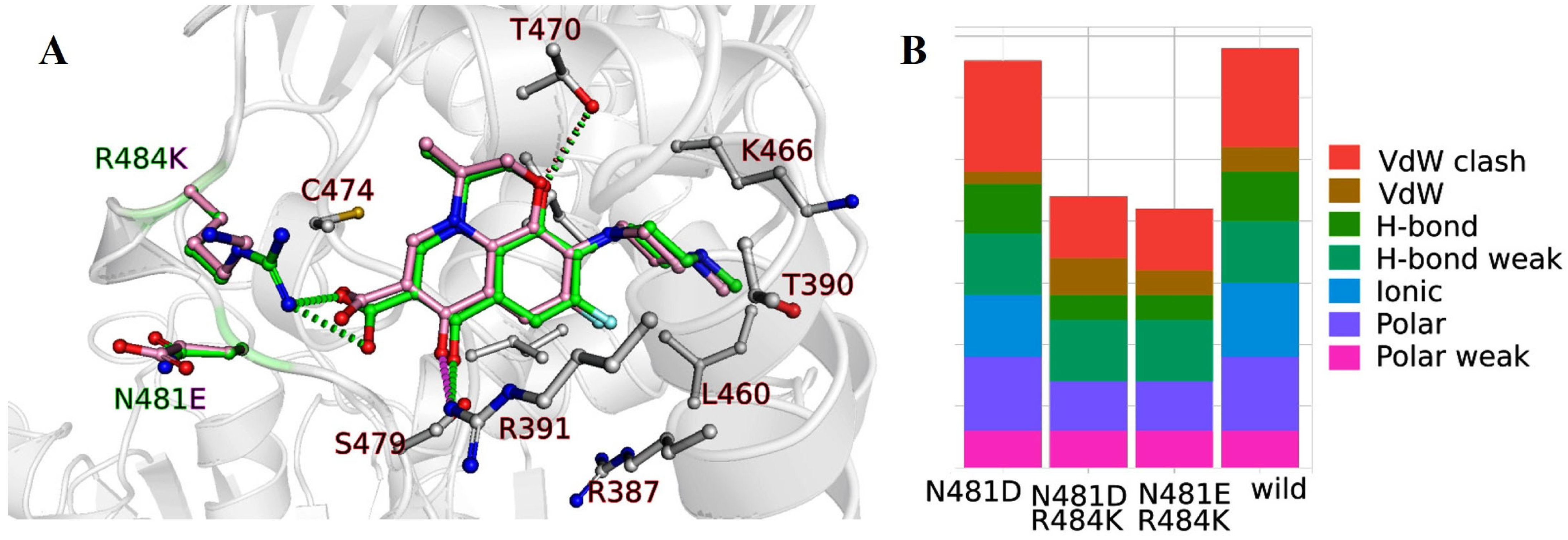
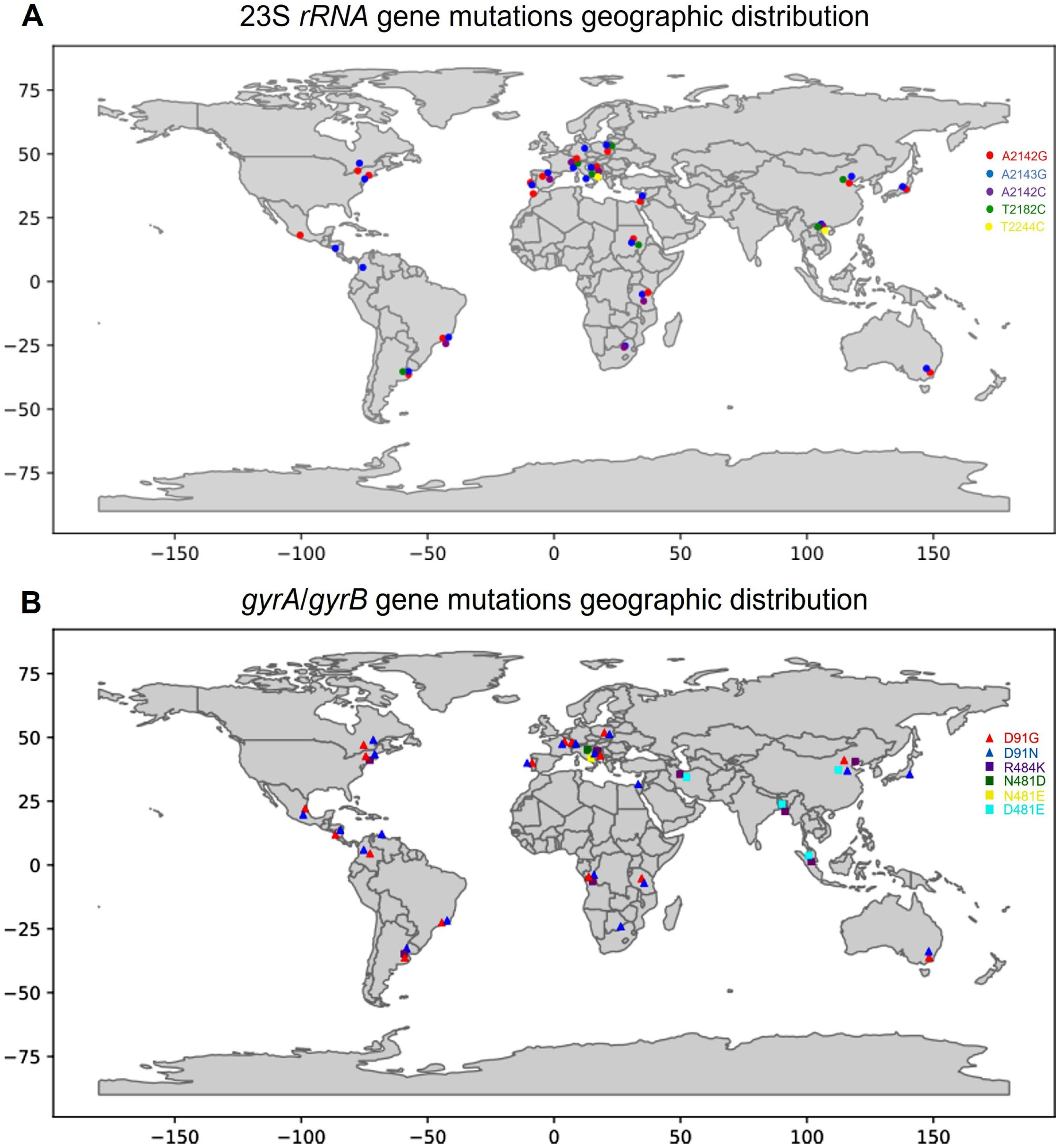
| Isolate | Sensitivity to CAM | Mutation Site |
|---|---|---|
| HP2 | resistant | A2143G, T2182C, T2244C |
| HP5 | resistant | A2143G, T2244C |
| HP8 | resistant | A2142G, T2182C, T2244C |
| HP9 | sensitive | T2182C, T2244C |
| HP10 | resistant | A2142G, T2182C, T2244C |
| HP11 | resistant | A2143G, T2182C, T2244C |
| HP14 | sensitive | T2244C |
| HP19 | resistant | A2142G, T2182C, T2244C |
| HP20 | resistant | A2142C, T2244C |
| Isolate | Base Mutation | Amino Acid Change | Point Mutation |
|---|---|---|---|
| HP5, HP11 | AAC → AAT | - | - |
| HP5, HP8 | GAT → AAT | D → N | D91N |
| HP2 | GAT → GGT | D → G | D91G |
| Isolate | Base Mutation | Amino Acid Change | Point Mutation |
|---|---|---|---|
| HP2, HP5, HP8, HP9, | AAT → GAG | N → E | N481E |
| HP10, HP11, HP14 | AAT → GAT | N → D | N481D |
| HP2, HP5, HP8, HP9, HP11 | AGA → AAA | R → K | R484K |
| Isolate | Susceptible to LVX | Point Mutation in Subunit A of DNA Gyrase | Point Mutation in Subunit B of DNA Gyrase |
|---|---|---|---|
| HP2 | resistant | D91G | N481E, R484K |
| HP5 | resistant | D91N | N481E, R484K |
| HP8 | resistant | D91N | N481E, R484K |
| HP9 | sensitive | no mutation | N481E, R484K |
| HP10 | sensitive | no mutation | N481D |
| HP11 | sensitive | no mutation | N481D, R484K |
| HP14 | no data | no mutation | N481D |
| Isolate | Susceptibility to CAM | Mutation Site in 23s rRNA Gene | Susceptibility to LVX | Point Mutation in GyrA | Point Mutation in GyrB |
|---|---|---|---|---|---|
| HP2 | resistant | A2143G, T2182C, T2244C | resistant | D91G | N481E, R484K |
| HP5 | resistant | A2143G, T2244C | resistant | D91N | N481E, R484K |
| HP8 | resistant | A2142G, T2182C, T2244C | resistant | D91N | N481E, R484K |
| HP9 | sensitive | T2182C, T2244C | sensitive | no mutation | N481E, R484K |
| HP10 | resistant | A2142G, T2182C, T2244C | sensitive | no mutation | N481D |
| HP11 | resistant | A2143G, T2182C, T2244C | sensitive | no mutation | N481D, R484K |
| HP14 | sensitive | T2244C | no data | no mutation | N481D |
| HP19 | resistant | A2142G, T2182C, T2244C | resistant | no mutation | N481E, R484K |
| HP20 | resistant | A2142C, T2244C | sensitive | no mutation | no mutation |
| Gene | Primer | Primer Sequence (5′→3′) | PCR Conditions | Reaction Mixture (30 µL) per Sample | Amplicon Size (bp) | Reference |
|---|---|---|---|---|---|---|
| 23S rRNA | Hp23S 1942F Hp23S 2308R | AGGATGCGTCAGTCGCAAGAT CCTGTGGATAACACAGGCCAGT | initial denaturation at 95 °C for 2 min, followed by 5 cycles: 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s; then 30 cycles: 94 °C for 15 s, 60 °C for 15 s, and 72 °C for 20 s, with a final extension at 72 °C for 7 min | 3 µL PCR buffer (10×), 0.3 µL bovine serum albumin (10 μg/μL), 3 µL dNTPs (2 mM), 1.5 µL each of forward (10 µM) and reverse primers (10 µM), 25 ng/μL of DNA template, and 0.3 µL Taq polymerase (5 U/µL) * | 367 | [56,57] |
| gyrA | gyr APF gyr APR | AGCTTATTCCATGAGCGTGA TCAGGCCCTTTGACAAATTC | initial denaturation 95 °C, 5 min; 35 cycles of amplification: 94 °C, 1 min; 53 °C, 1 min; 72 °C, 1 min; final extension 72 °C, 10 min | 3 µL PCR buffer (10×), 0.3 µL bovine serum albumin (10 μg/μL), 3 µL dNTPs (2 mM), 1.5 µL each of gyrA and gyrB forward (10 µM) and reverse primers (10 µM), 25 ng/μL of DNA template, and 0.3 µL Taq polymerase (5 U/µL) * | 582 | [58] |
| gyrB | gyr BPF gyr BPR | CCCTAACGAAGCCAAAATCA GGGCGCAAATAACGATAGAA | 465 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Šamanić, I.; Dadić, B.; Sanader Maršić, Ž.; Dželalija, M.; Maravić, A.; Kalinić, H.; Vrebalov Cindro, P.; Šundov, Ž.; Tonkić, M.; Tonkić, A.; et al. Molecular Characterization and Mutational Analysis of Clarithromycin- and Levofloxacin-Resistance Genes in Helicobacter pylori from Gastric Biopsies in Southern Croatia. Int. J. Mol. Sci. 2023, 24, 14560. https://doi.org/10.3390/ijms241914560
Šamanić I, Dadić B, Sanader Maršić Ž, Dželalija M, Maravić A, Kalinić H, Vrebalov Cindro P, Šundov Ž, Tonkić M, Tonkić A, et al. Molecular Characterization and Mutational Analysis of Clarithromycin- and Levofloxacin-Resistance Genes in Helicobacter pylori from Gastric Biopsies in Southern Croatia. International Journal of Molecular Sciences. 2023; 24(19):14560. https://doi.org/10.3390/ijms241914560
Chicago/Turabian StyleŠamanić, Ivica, Blanka Dadić, Željka Sanader Maršić, Mia Dželalija, Ana Maravić, Hrvoje Kalinić, Pavle Vrebalov Cindro, Željko Šundov, Marija Tonkić, Ante Tonkić, and et al. 2023. "Molecular Characterization and Mutational Analysis of Clarithromycin- and Levofloxacin-Resistance Genes in Helicobacter pylori from Gastric Biopsies in Southern Croatia" International Journal of Molecular Sciences 24, no. 19: 14560. https://doi.org/10.3390/ijms241914560
APA StyleŠamanić, I., Dadić, B., Sanader Maršić, Ž., Dželalija, M., Maravić, A., Kalinić, H., Vrebalov Cindro, P., Šundov, Ž., Tonkić, M., Tonkić, A., & Vuković, J. (2023). Molecular Characterization and Mutational Analysis of Clarithromycin- and Levofloxacin-Resistance Genes in Helicobacter pylori from Gastric Biopsies in Southern Croatia. International Journal of Molecular Sciences, 24(19), 14560. https://doi.org/10.3390/ijms241914560








