Discovery of Chalcone-Based Hybrid Structures as High Affinity and Site-Specific Inhibitors against SARS-CoV-2: A Comprehensive Structural Analysis Based on Various Host-Based and Viral Targets
Abstract
1. Introduction
2. Results
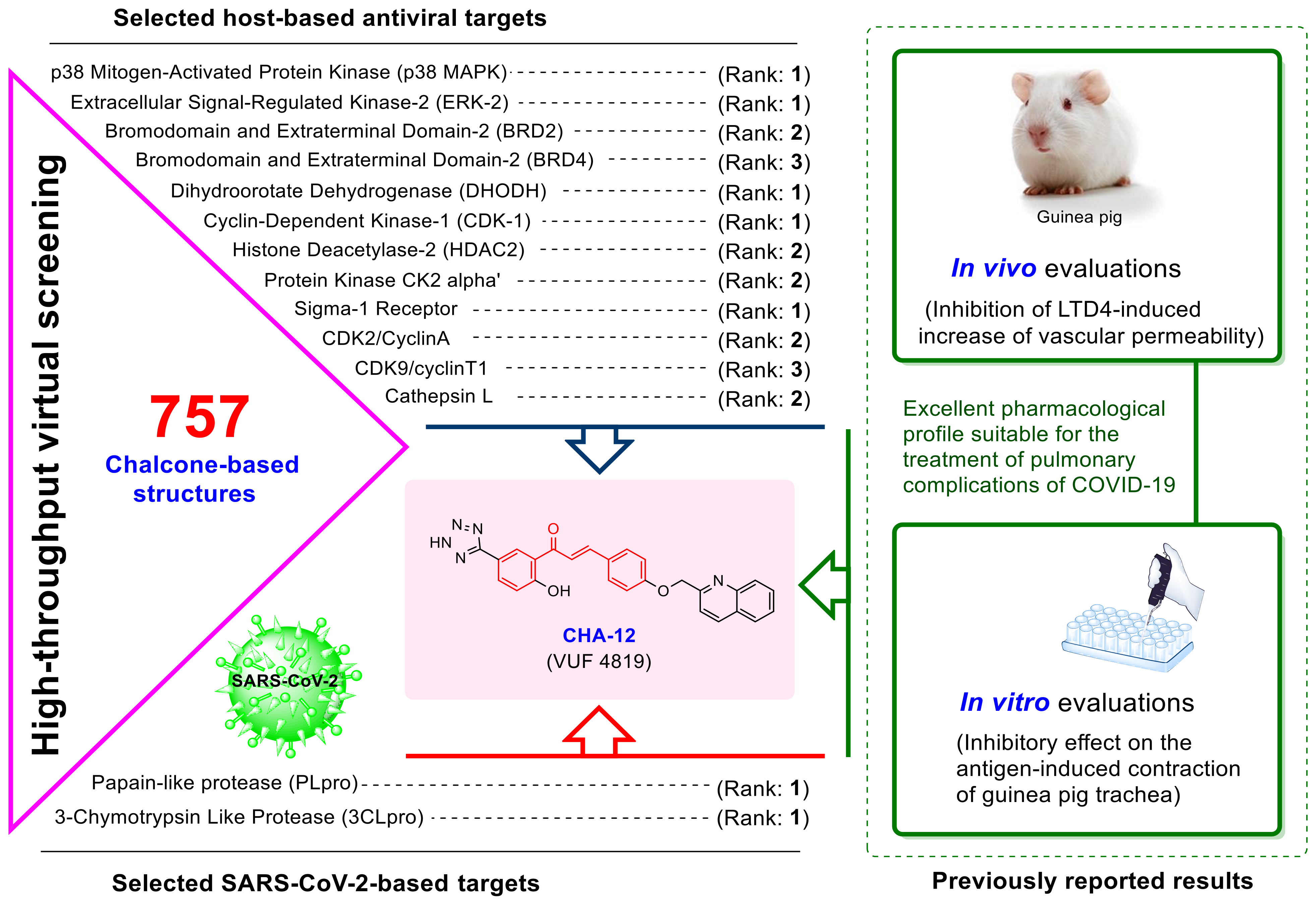
2.1. Virtual Screening
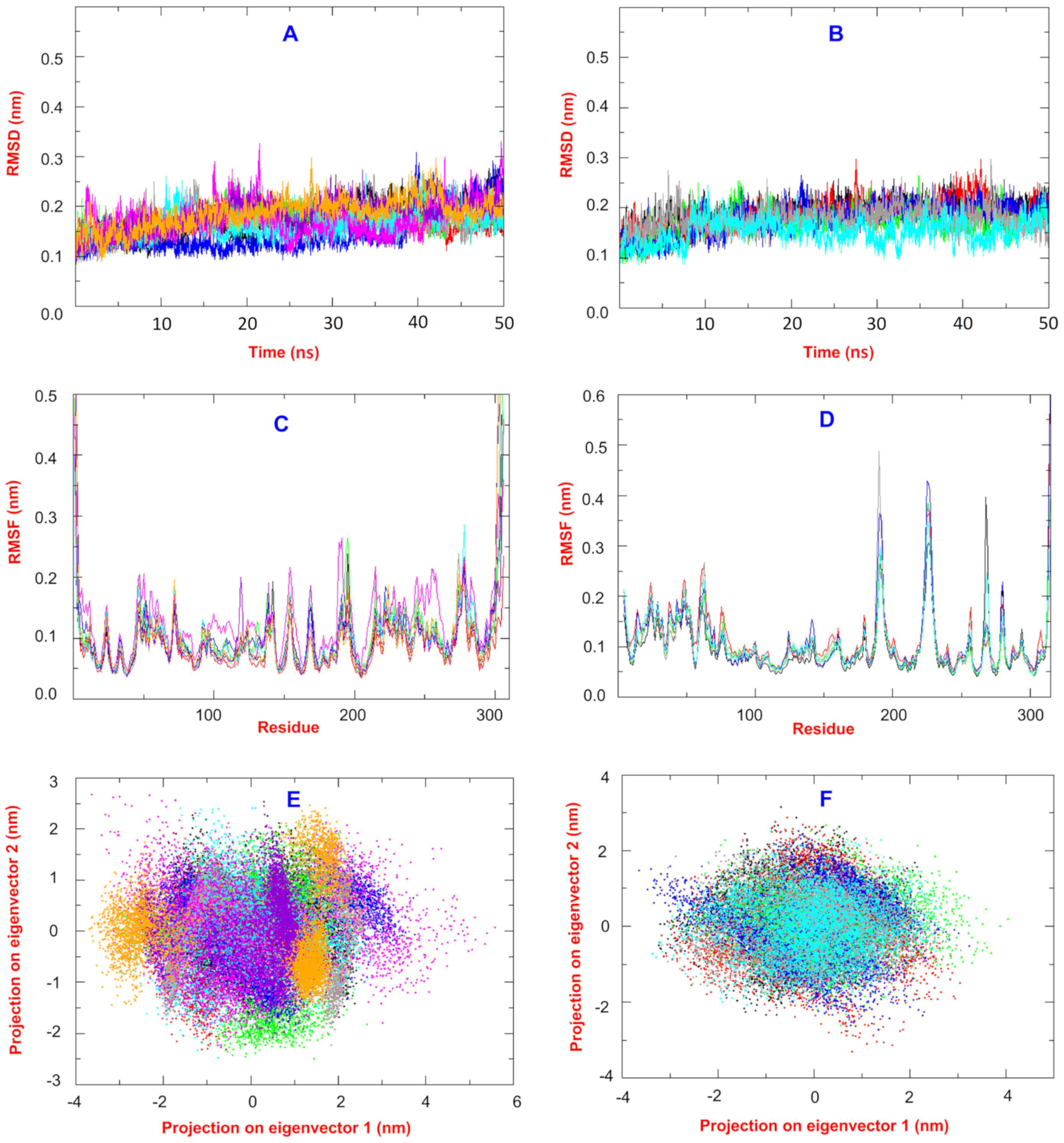
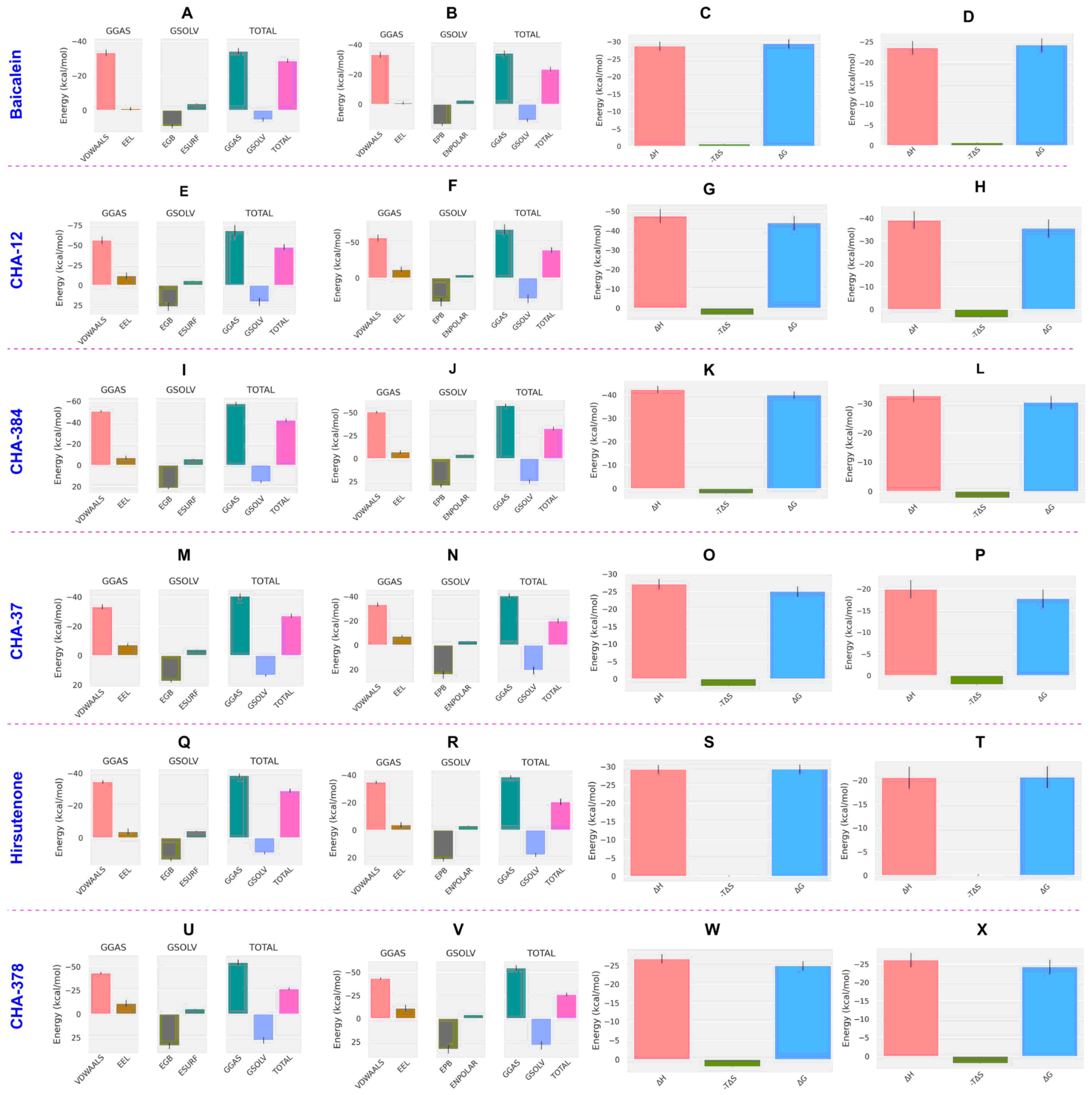
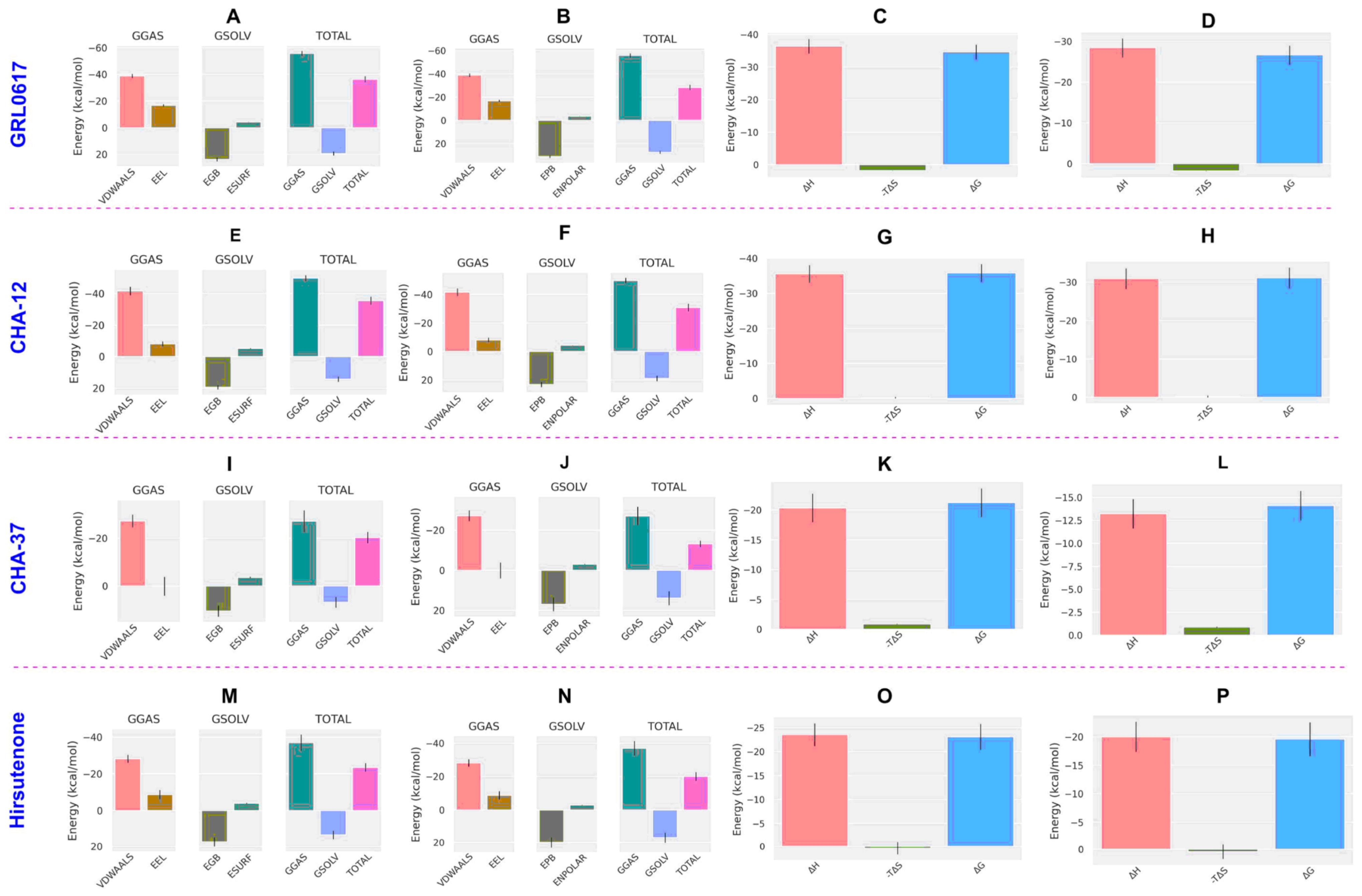
2.2. Molecular Dynamics and MM/PB(GB)SA Analysis
2.3. Druglikeness, ADME, and Toxicity
2.3.1. Oral Bioavailability
2.3.2. Cytochrome P450 Interaction
3. Discussion
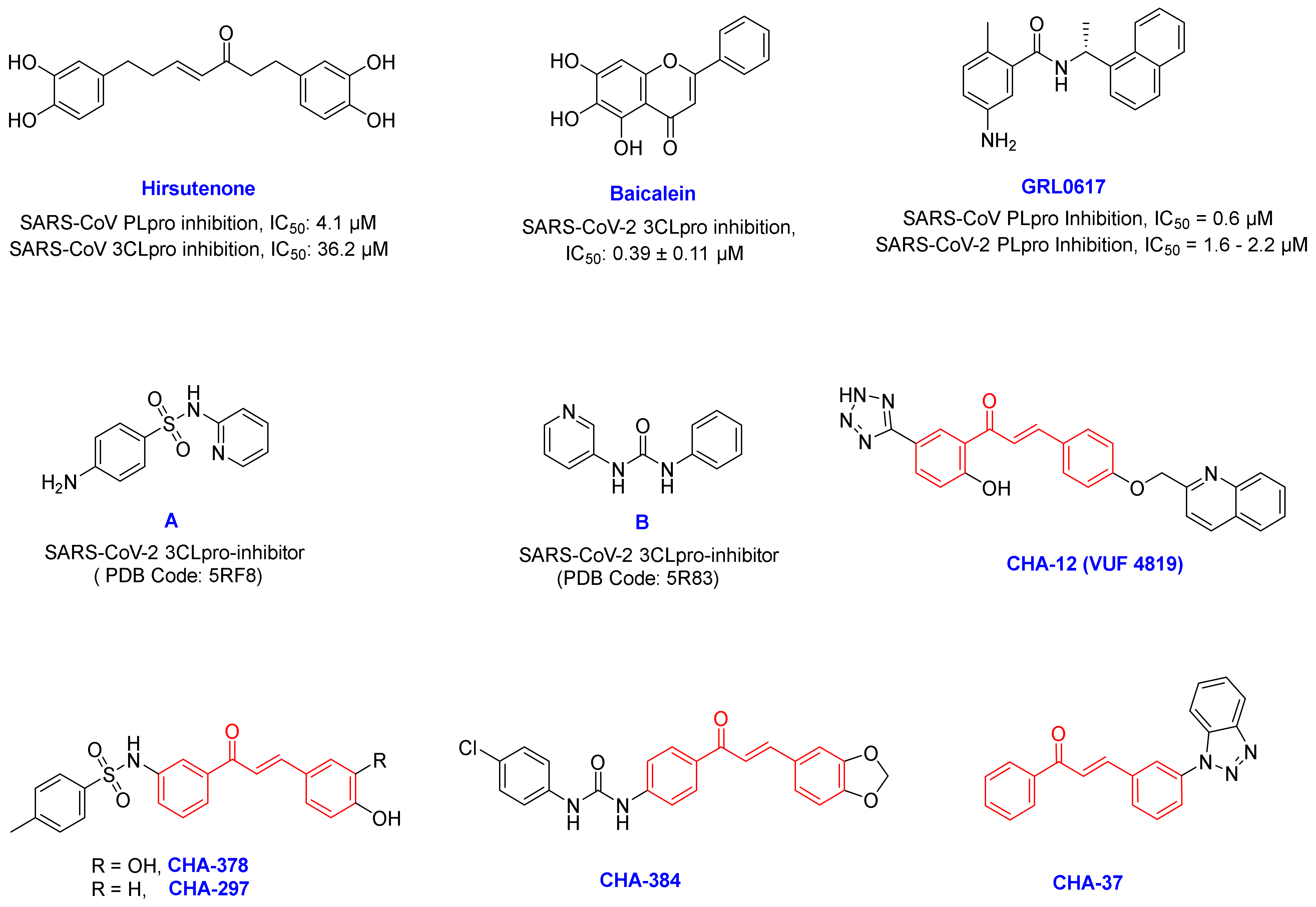
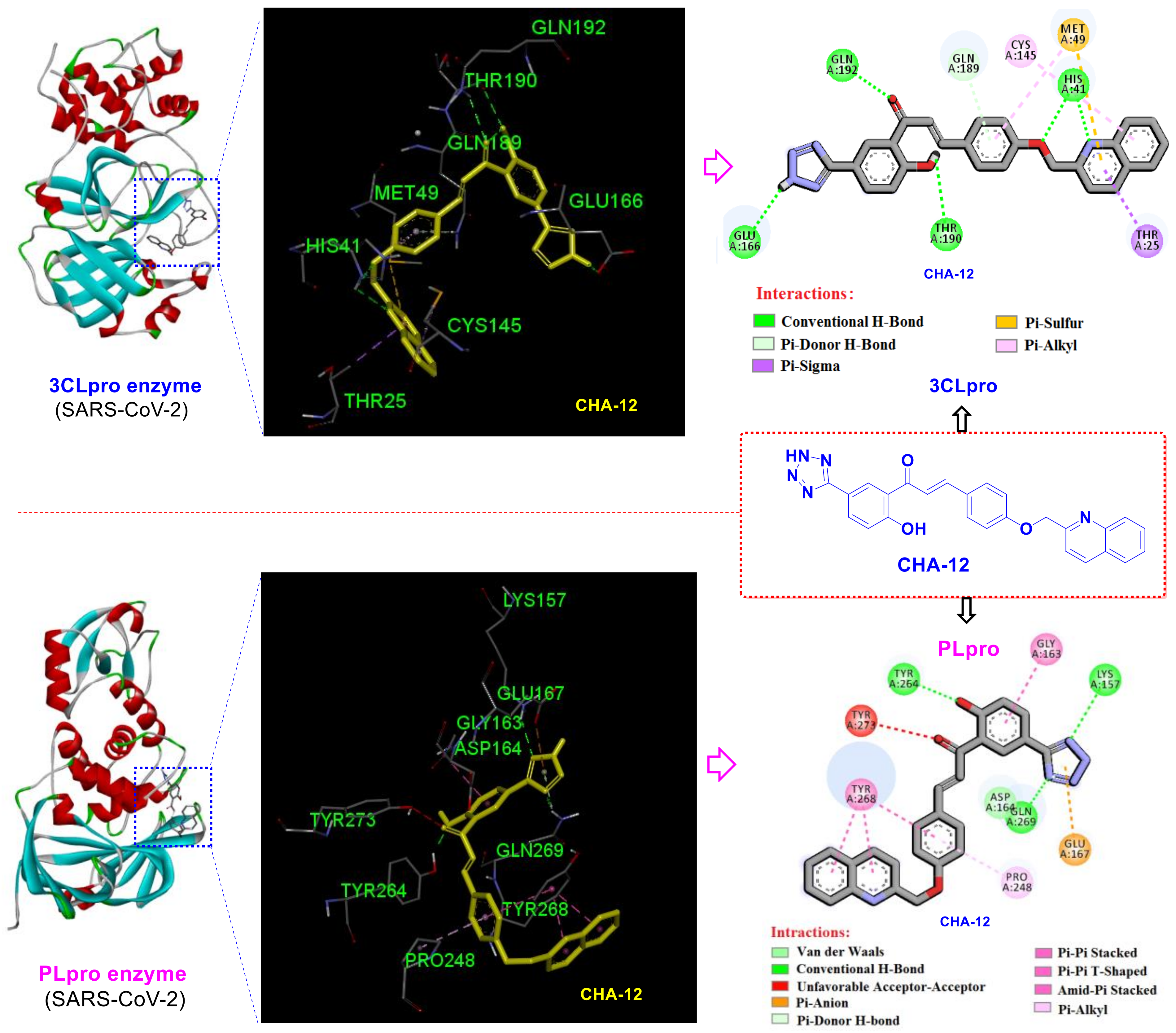
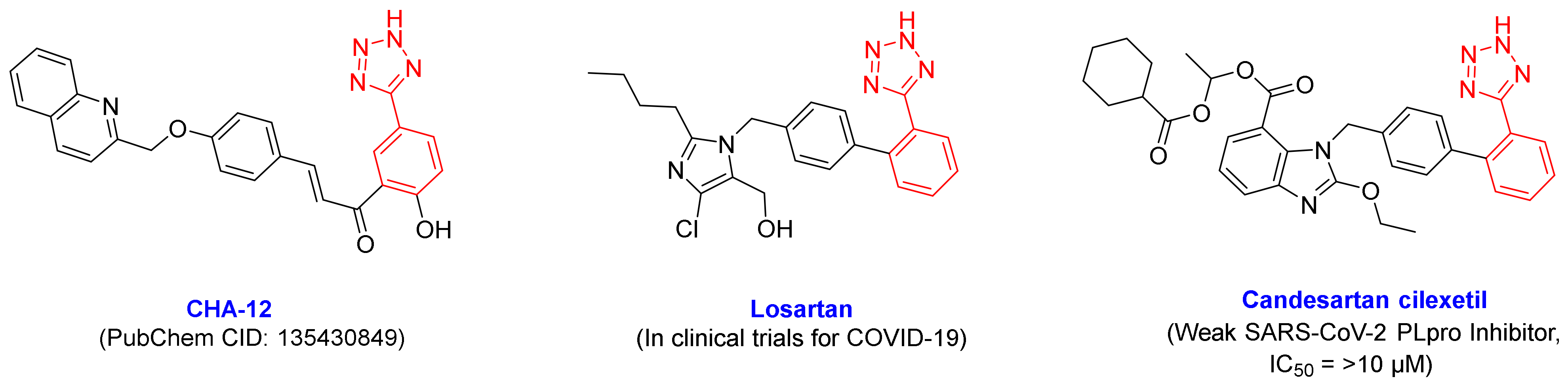
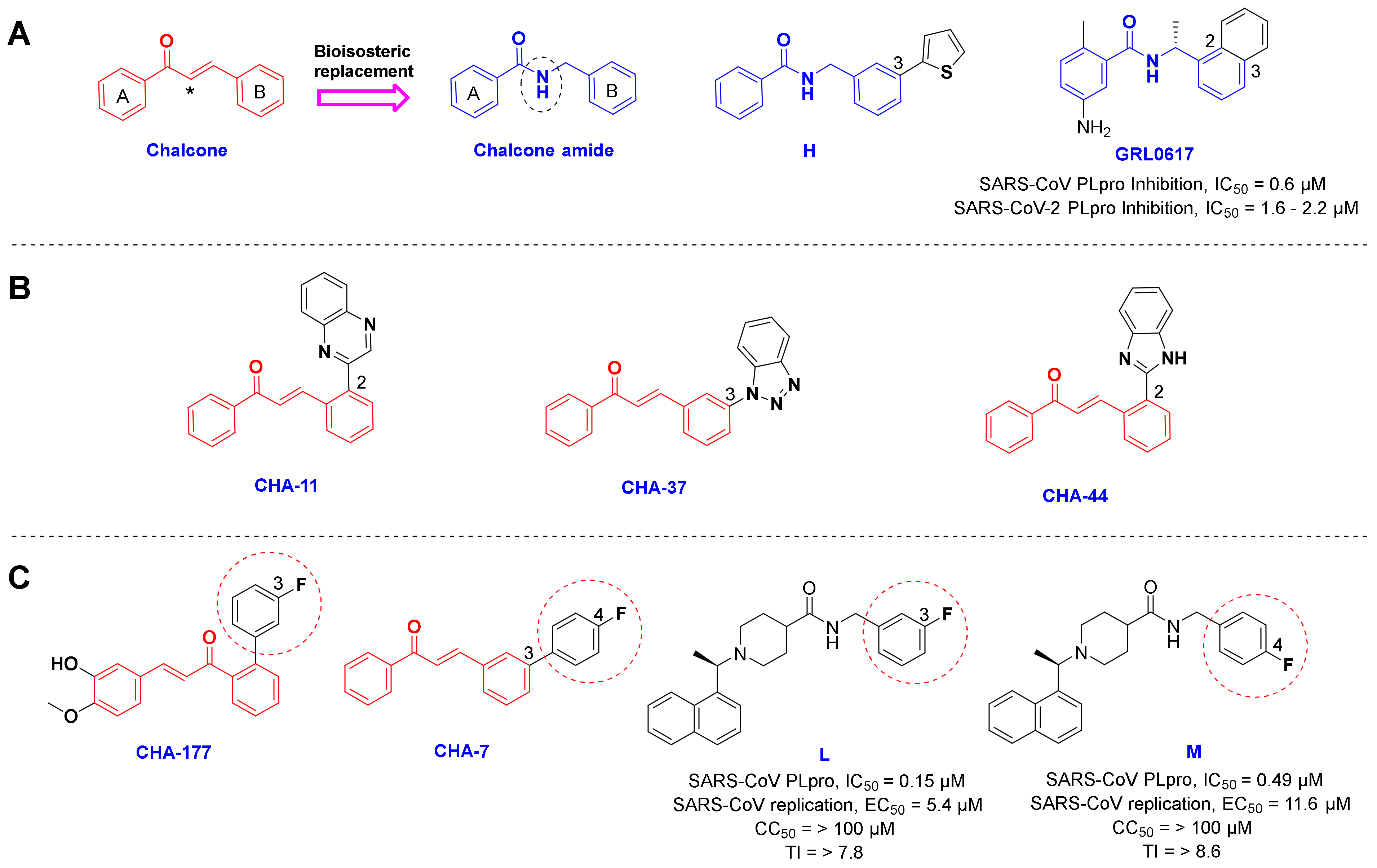
3.1. CHA-12 as a Multi-Target Inhibitor
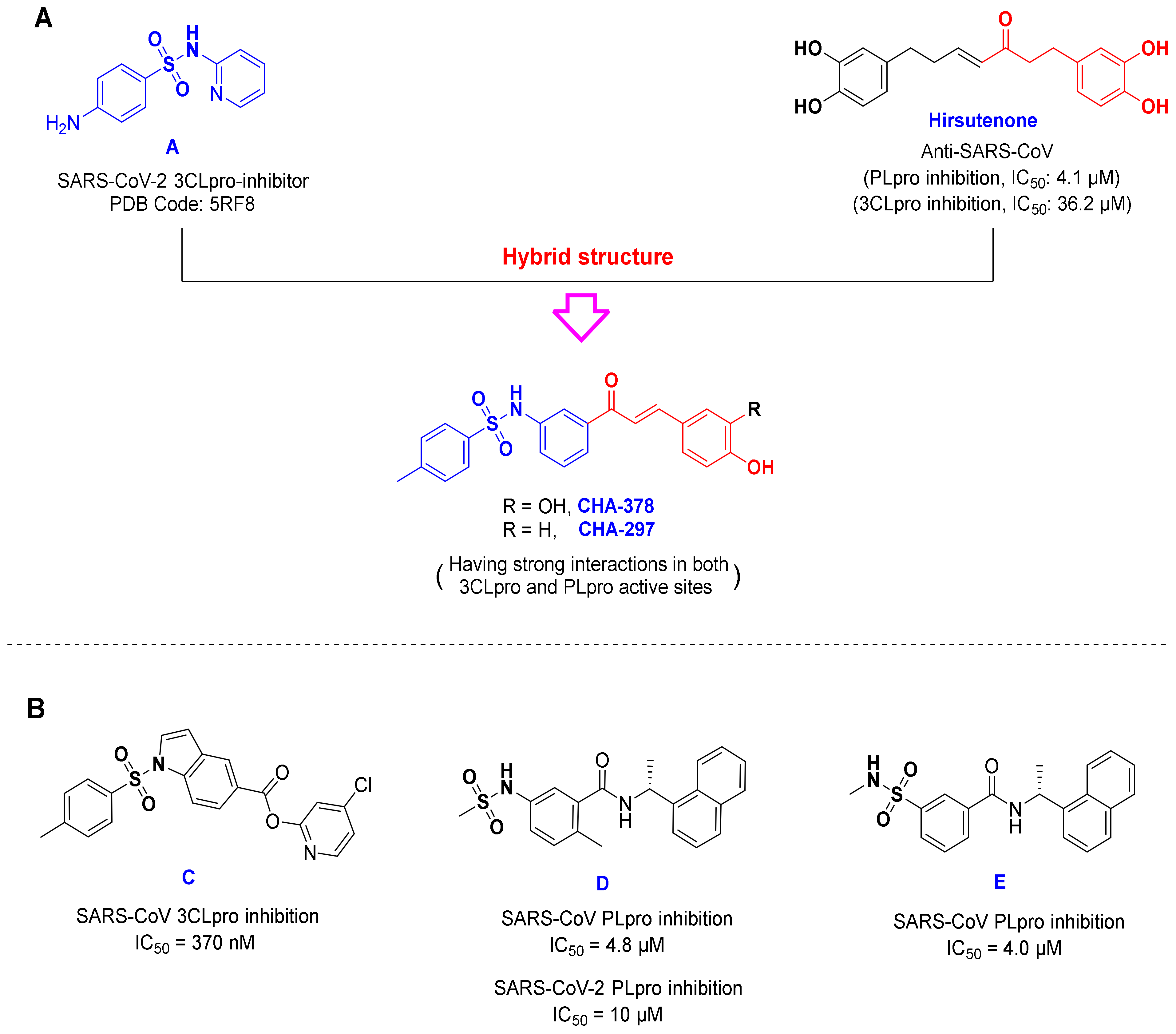
3.2. Chalcones Bearing Sulfonamide Moiety
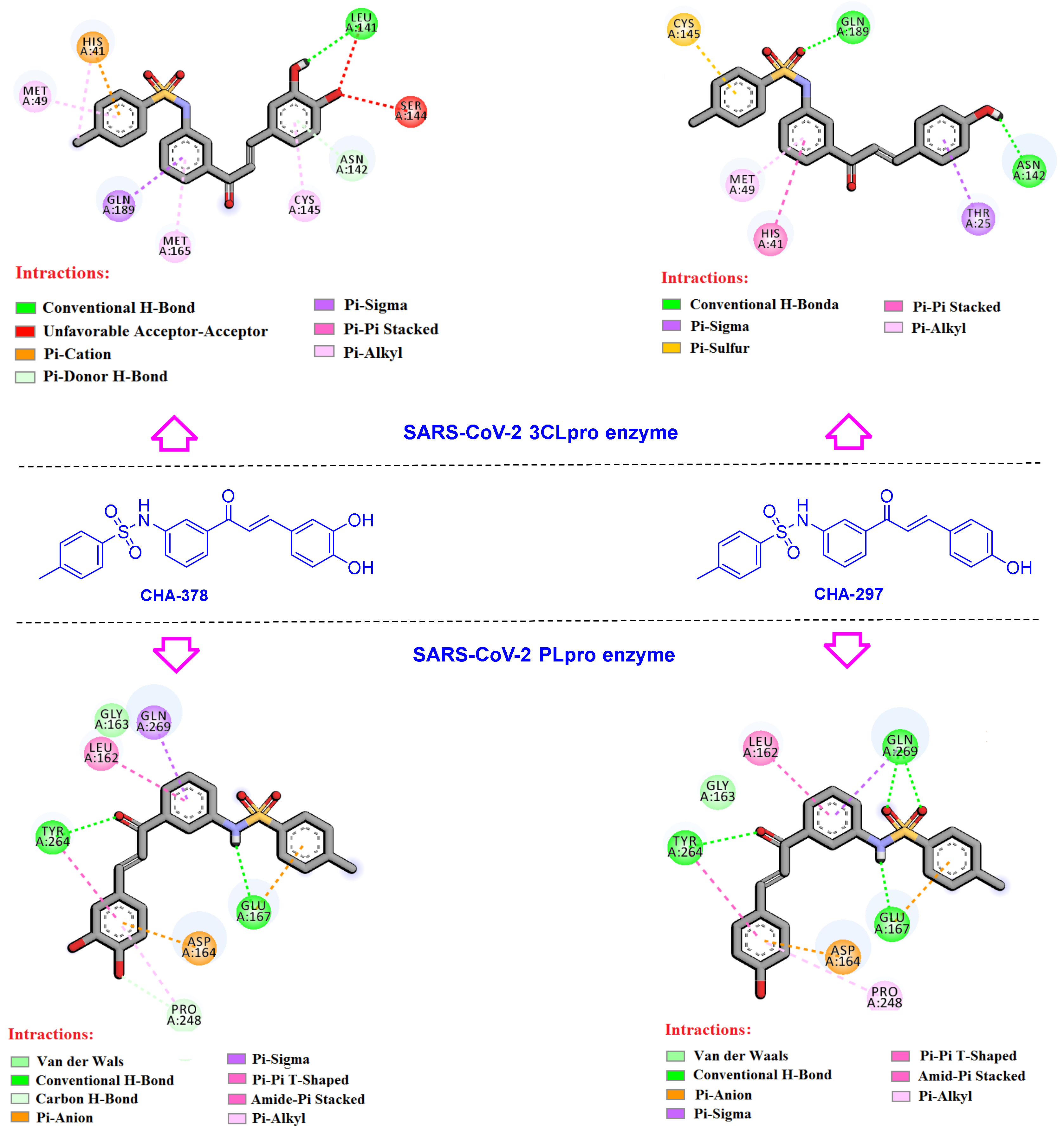
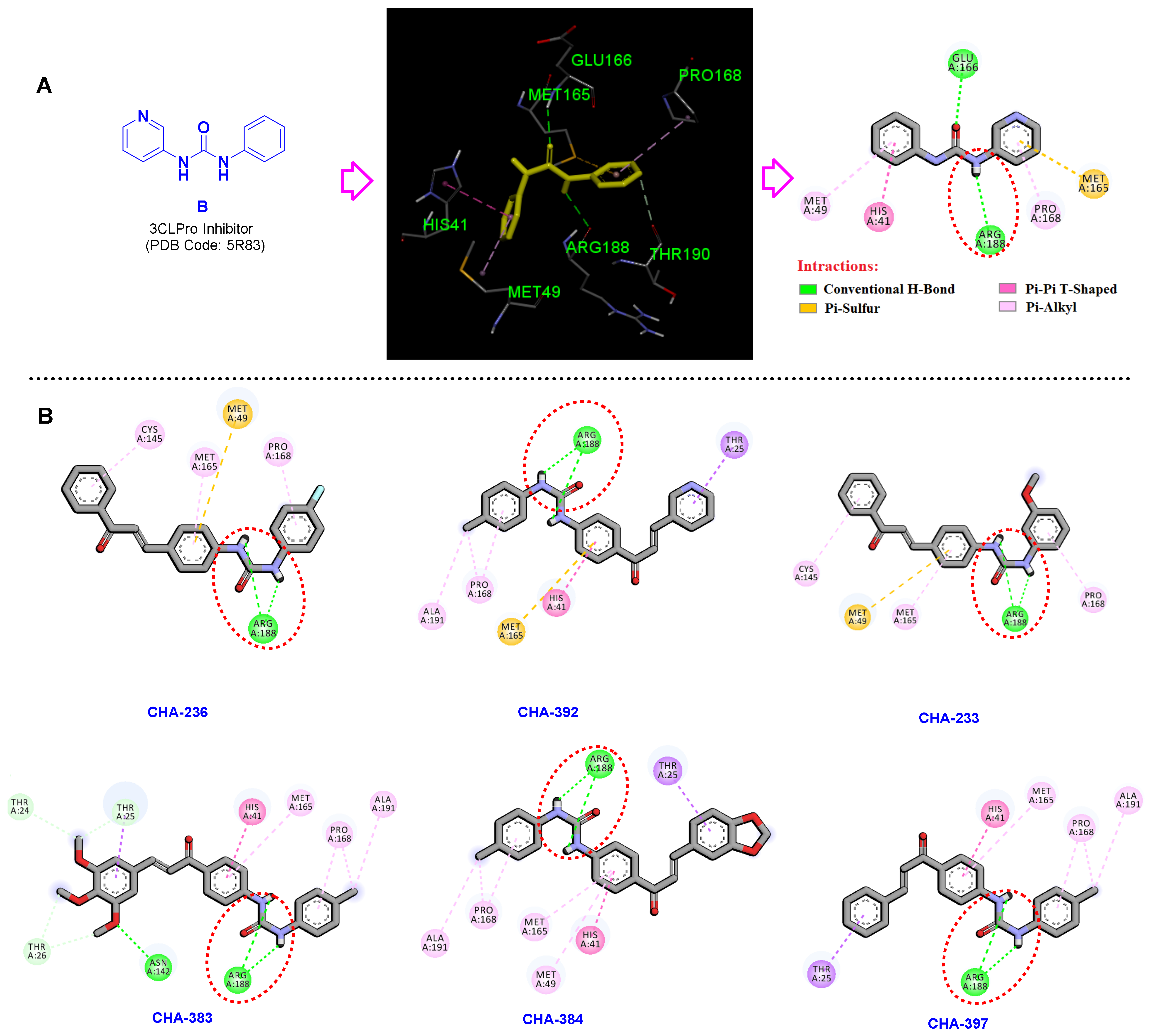
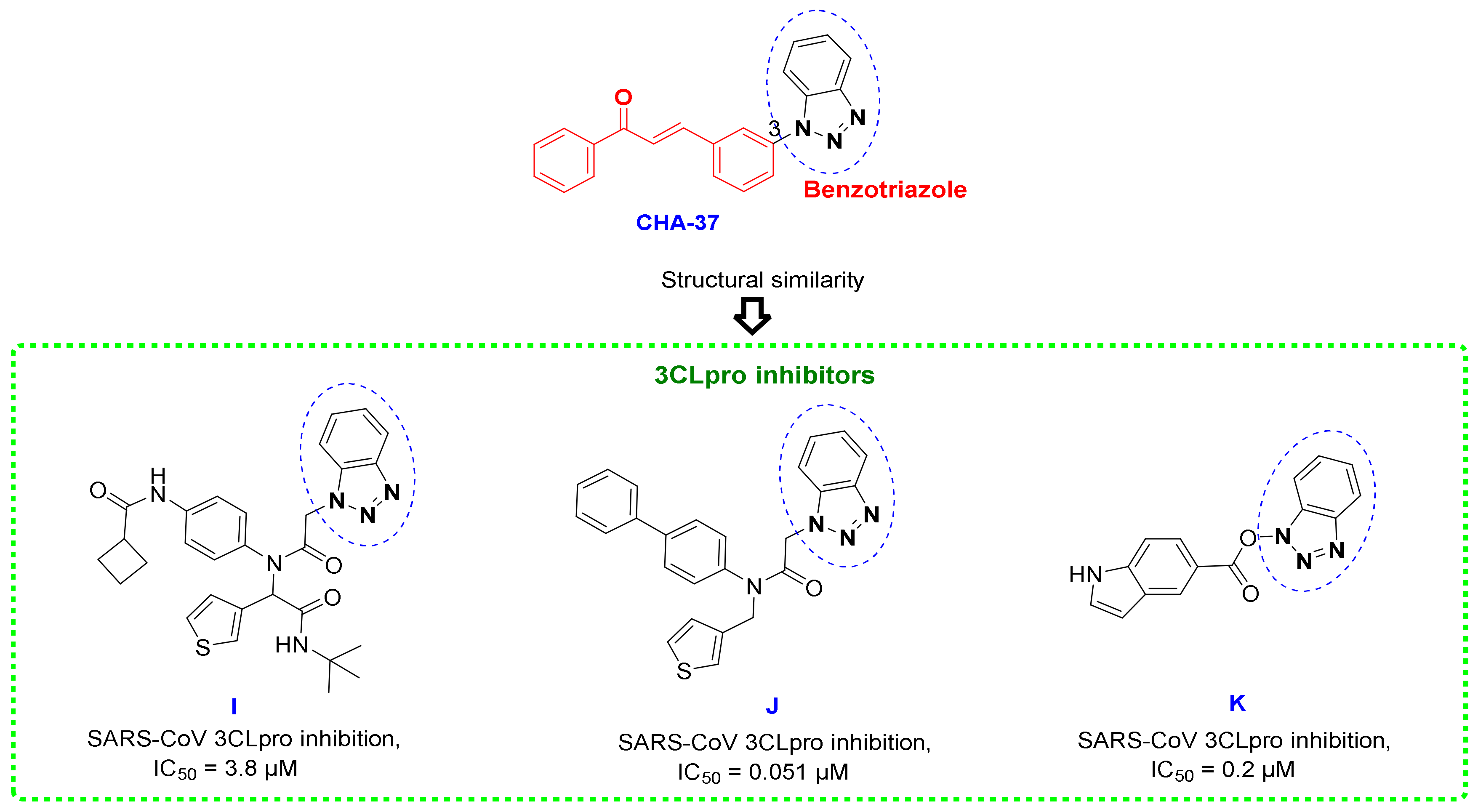
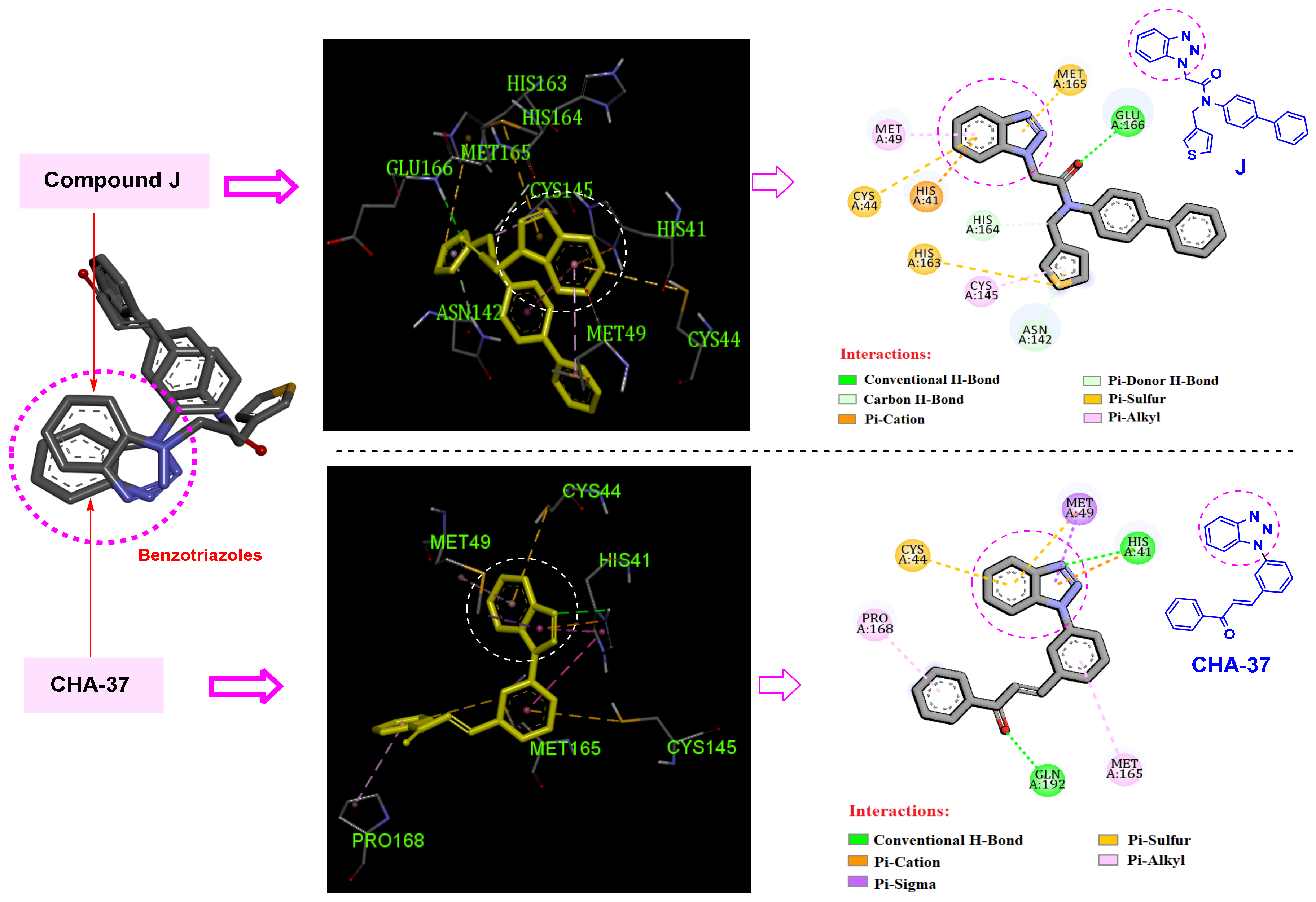
3.3. Active Ligands Selective towards the 3CLpro
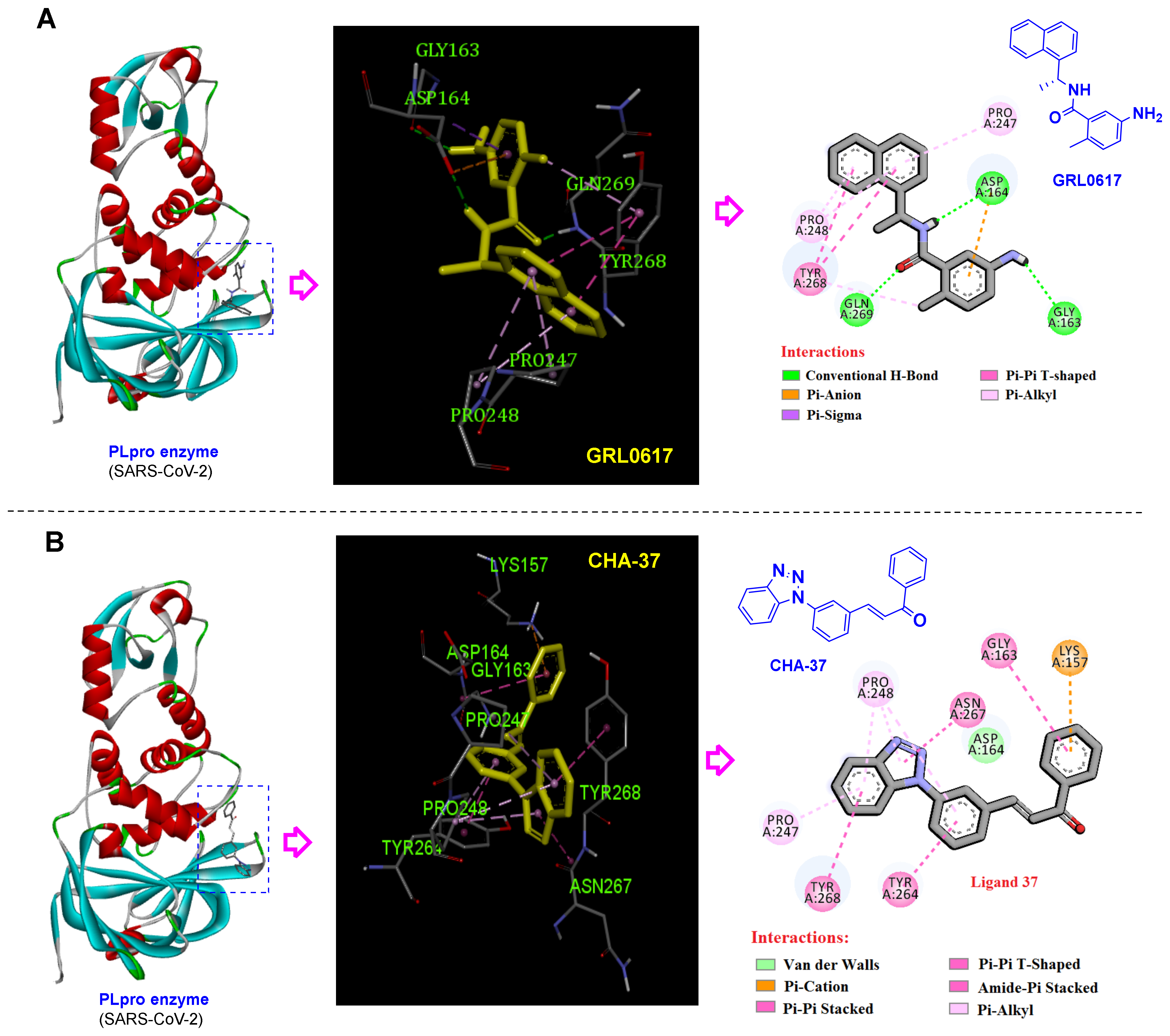
3.4. Selective Ligands towards the SARS-CoV-2 PLpro
3.5. Benzotriazole for Selective 3CLpro Inhibition
4. Materials and Methods
4.1. Compound Library Preparation
4.2. Target Selection and Preparation
4.3. Virtual Screening
4.4. Molecular Dynamics Simulation and MM/PB(GB)SA Calculations
4.5. Druglikeness and Pharmacokinetic Prediction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ahmad, I. The race to treat COVID-19: Potential therapeutic agents for the prevention and treatment of SARS-CoV-2. Eur. J. Med. Chem. 2021, 213, 113157. [Google Scholar]
- Kim, D.; Lee, J.-Y.; Yang, J.-S.; Kim, J.W.; Kim, V.N.; Chang, H. The architecture of SARS-CoV-2 transcriptome. Cell 2020, 181, 914–921.e10. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Cheng, C.; Shi, S.; Wu, Y.; Gao, Y.; Liu, Z.; Liu, M.; Li, Z.; Huo, L.; Pan, X. Identification, optimization, and biological evaluation of 3-O-β-chacotriosyl ursolic acid derivatives as novel SARS-CoV-2 entry inhibitors by targeting the prefusion state of spike protein. Eur. J. Med. Chem. 2022, 238, 114426. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Song, Y.; Chen, Y.; Wu, N.; Xu, J.; Sun, C.; Zhang, J.; Weng, T.; Zhang, Z.; Wu, Z. Molecular architecture of the SARS-CoV-2 virus. Cell 2020, 183, 730–738.e13. [Google Scholar] [CrossRef]
- Kuzikov, M.; Costanzi, E.; Reinshagen, J.; Esposito, F.; Vangeel, L.; Wolf, M.; Ellinger, B.; Claussen, C.; Geisslinger, G.; Corona, A. Identification of inhibitors of SARS-CoV-2 3CL-pro enzymatic activity using a small molecule in vitro repurposing screen. ACS Pharmacol. Transl. Sci. 2021, 4, 1096–1110. [Google Scholar] [CrossRef]
- Xia, Z.; Sacco, M.; Hu, Y.; Ma, C.; Meng, X.; Zhang, F.; Szeto, T.; Xiang, Y.; Chen, Y.; Wang, J. Rational design of hybrid SARS-CoV-2 main protease inhibitors guided by the superimposed cocrystal structures with the peptidomimetic inhibitors GC-376, telaprevir, and boceprevir. ACS Pharmacol. Transl. Sci. 2021, 4, 1408–1421. [Google Scholar] [CrossRef]
- Ma, C.; Wang, J. Validation and invalidation of SARS-CoV-2 papain-like protease inhibitors. ACS Pharmacol. Transl. Sci. 2022, 5, 102–109. [Google Scholar] [CrossRef]
- Glaser, J.; Sedova, A.; Galanie, S.; Kneller, D.W.; Davidson, R.B.; Maradzike, E.; Del Galdo, S.; Labbé, A.; Hsu, D.J.; Agarwal, R. Hit Expansion of a Noncovalent SARS-CoV-2 Main Protease Inhibitor. ACS Pharmacol. Transl. Sci. 2022, 5, 255–265. [Google Scholar] [CrossRef]
- Hammond, J.; Leister-Tebbe, H.; Gardner, A.; Abreu, P.; Bao, W.; Wisemandle, W.; Baniecki, M.; Hendrick, V.M.; Damle, B.; Simón-Campos, A. Oral nirmatrelvir for high-risk, nonhospitalized adults with COVID-19. N. Engl. J. Med. 2022, 386, 1397–1408. [Google Scholar] [CrossRef]
- Douangamath, A.; Fearon, D.; Gehrtz, P.; Krojer, T.; Lukacik, P.; Owen, C.D.; Resnick, E.; Strain-Damerell, C.; Aimon, A.; Ábrányi-Balogh, P. Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease. Nat. Commun. 2020, 11, 5047. [Google Scholar] [CrossRef]
- Mengist, H.M.; Dilnessa, T.; Jin, T. Structural basis of potential inhibitors targeting SARS-CoV-2 main protease. Front. Chem. 2021, 9, 622898. [Google Scholar] [CrossRef]
- Richman, D.D. Antiviral drug resistance. Antivir. Res. 2006, 71, 117–121. [Google Scholar] [CrossRef]
- Strasfeld, L.; Chou, S. Antiviral drug resistance: Mechanisms and clinical implications. Infect. Dis. Clin. 2010, 24, 809–833. [Google Scholar] [CrossRef]
- De Chassey, B.; Meyniel-Schicklin, L.; Vonderscher, J.; André, P.; Lotteau, V. Virus-host interactomics: New insights and opportunities for antiviral drug discovery. Genome Med. 2014, 6, 115. [Google Scholar] [CrossRef]
- Mahajan, S.; Choudhary, S.; Kumar, P.; Tomar, S. Antiviral strategies targeting host factors and mechanisms obliging +ssRNA viral pathogens. Bioorg. Med. Chem. 2021, 46, 116356. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, Y.; Wu, L.; Niu, S.; Song, C.; Zhang, Z.; Lu, G.; Qiao, C.; Hu, Y.; Yuen, K.-Y. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 2020, 181, 894–904.e9. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Dutta, K. Allosteric Site of ACE-2 as a Drug Target for COVID-19. ACS Pharmacol. Transl. Sci. 2022, 5, 179–182. [Google Scholar] [CrossRef]
- Li, X.; Liu, Y.; Song, J.; Zhong, J. Increased plasma ACE2 concentration does not mean increased risk of SARS-CoV-2 infection and increased fatality rate of COVID-19. Acta Pharm. Sin. B 2020, 10, 2010. [Google Scholar] [CrossRef]
- Bian, J.; Li, Z. Angiotensin-converting enzyme 2 (ACE2): SARS-CoV-2 receptor and RAS modulator. Acta Pharm. Sin. B 2021, 11, 1–12. [Google Scholar] [CrossRef]
- Chen, Z.; Du, R.; Achi, J.M.G.; Rong, L.; Cui, Q. SARS-CoV-2 cell entry and targeted antiviral development. Acta Pharm. Sin. B 2021, 11, 3879–3888. [Google Scholar] [CrossRef] [PubMed]
- Sacco, M.D.; Ma, C.; Lagarias, P.; Gao, A.; Townsend, J.A.; Meng, X.; Dube, P.; Zhang, X.; Hu, Y.; Kitamura, N. Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against Mpro and cathepsin L. Sci. Adv. 2020, 6, eabe0751. [Google Scholar] [CrossRef] [PubMed]
- Grimes, J.M.; Grimes, K.V. p38 MAPK inhibition: A promising therapeutic approach for COVID-19. J. Mol. Cell. Cardiol. 2020, 144, 63–65. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Chamorro, L.; Felip, E.; Ezeonwumelu, I.J.; Margelí, M.; Ballana, E. Cyclin-dependent Kinases as Emerging Targets for Developing Novel Antiviral Therapeutics. Trends Microbiol. 2021, 29, 836–848. [Google Scholar] [CrossRef]
- Xu, Y.; Jiang, H. Potential treatment of COVID-19 by inhibitors of human dihydroorotate dehydrogenase. Protein Cell 2020, 11, 699–702. [Google Scholar] [CrossRef]
- Xiang, R.; Yu, Z.; Wang, Y.; Wang, L.; Huo, S.; Li, Y.; Liang, R.; Hao, Q.; Ying, T.; Gao, Y. Recent advances in developing small-molecule inhibitors against SARS-CoV-2. Acta Pharm. Sin. B 2021, 12, 1591–1623. [Google Scholar] [CrossRef]
- Gao, S.; Huang, T.; Song, L.; Xu, S.; Cheng, Y.; Cherukupalli, S.; Kang, D.; Zhao, T.; Sun, L.; Zhang, J. Medicinal chemistry strategies towards the development of effective SARS-CoV-2 inhibitors. Acta Pharm. Sin. B 2021, 12, 581–599. [Google Scholar] [CrossRef]
- Singh, R.; Bhardwaj, V.K.; Purohit, R. Potential of turmeric-derived compounds against RNA-dependent RNA polymerase of SARS-CoV-2: An in-silico approach. Comput. Biol. Med. 2021, 139, 104965. [Google Scholar] [CrossRef]
- Sharma, J.; Bhardwaj, V.K.; Singh, R.; Rajendran, V.; Purohit, R.; Kumar, S. An in-silico evaluation of different bioactive molecules of tea for their inhibition potency against non structural protein-15 of SARS-CoV-2. Food Chem. 2021, 346, 128933. [Google Scholar] [CrossRef]
- Singh, R.; Bhardwaj, V.K.; Sharma, J.; Purohit, R.; Kumar, S. In-silico evaluation of bioactive compounds from tea as potential SARS-CoV-2 nonstructural protein 16 inhibitors. J. Tradit. Complement. Med. 2022, 12, 35–43. [Google Scholar] [CrossRef]
- Singh, R.; Bhardwaj, V.K.; Sharma, J.; Kumar, D.; Purohit, R. Identification of potential plant bioactive as SARS-CoV-2 Spike protein and human ACE2 fusion inhibitors. Comput. Biol. Med. 2021, 136, 104631. [Google Scholar] [CrossRef]
- Singh, R.; Bhardwaj, V.K.; Das, P.; Bhattacherjee, D.; Zyryanov, G.V.; Purohit, R. Benchmarking the ability of novel compounds to inhibit SARS-CoV-2 main protease using steered molecular dynamics simulations. Comput. Biol. Med. 2022, 146, 105572. [Google Scholar] [CrossRef]
- Vijayakumar, B.G.; Ramesh, D.; Joji, A.; Kannan, T. In silico pharmacokinetic and molecular docking studies of natural flavonoids and synthetic indole chalcones against essential proteins of SARS-CoV-2. Eur. J. Pharmacol. 2020, 886, 173448. [Google Scholar] [CrossRef]
- Valipour, M.; Irannejad, H.; Emami, S. Papaverine, a promising therapeutic agent for the treatment of COVID-19 patients with underlying cardiovascular diseases (CVDs). Drug Dev. Res. 2022, 83, 1246–1250. [Google Scholar] [CrossRef]
- Valipour, M. Different Aspects of Emetine’s Capabilities as a Highly Potent SARS-CoV-2 Inhibitor against COVID-19. ACS Pharmacol. Transl. Sci. 2022, 5, 387–399. [Google Scholar] [CrossRef]
- Valipour, M.; Zarghi, A.; Ebrahimzadeh, M.A.; Irannejad, H. Therapeutic potential of chelerythrine as a multi-purpose adjuvant for the treatment of COVID-19. Cell Cycle 2021, 20, 2321–2336. [Google Scholar] [CrossRef]
- Valipour, M.; Irannejad, H.; Emami, S. Application of emetine in SARS-CoV-2 treatment: Regulation of p38 MAPK signaling pathway for preventing emetine-induced cardiac complications. Cell Cycle 2022, 21, 2379–2386. [Google Scholar] [CrossRef]
- Valipour, M. Chalcone-amide, a privileged backbone for the design and development of selective SARS-CoV/SARS-CoV-2 papain-like protease inhibitors. Eur. J. Med. Chem. 2022, 240, 114572. [Google Scholar] [CrossRef]
- Rani, A.; Anand, A.; Kumar, K.; Kumar, V. Recent developments in biological aspects of chalcones: The odyssey continues. Expert Opin. Drug Discov. 2019, 14, 249–288. [Google Scholar] [CrossRef]
- Elkhalifa, D.; Al-Hashimi, I.; Al Moustafa, A.-E.; Khalil, A. A comprehensive review on the antiviral activities of chalcones. J. Drug Target. 2021, 29, 403–419. [Google Scholar] [CrossRef]
- Park, J.-Y.; Ko, J.-A.; Kim, D.W.; Kim, Y.M.; Kwon, H.-J.; Jeong, H.J.; Kim, C.Y.; Park, K.H.; Lee, W.S.; Ryu, Y.B. Chalcones isolated from Angelica keiskei inhibit cysteine proteases of SARS-CoV. J. Enzyme Inhib. Med. Chem. 2016, 31, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Liu, M.; Qin, H.; Lin, H.; An, X.; Shi, Z.; Song, L.; Yang, X.; Fan, H.; Tong, Y. Artemether, artesunate, arteannuin B, echinatin, licochalcone B and andrographolide effectively inhibit SARS-CoV-2 and related viruses in vitro. Front. Cell. Infect. Microbiol. 2021, 11, 680127. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-Y.; Yuk, H.J.; Ryu, H.W.; Lim, S.H.; Kim, K.S.; Park, K.H.; Ryu, Y.B.; Lee, W.S. Evaluation of polyphenols from Broussonetia papyrifera as coronavirus protease inhibitors. J. Enzyme Inhib. Med. Chem. 2017, 32, 504–512. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.W.; Seo, K.H.; Curtis-Long, M.J.; Oh, K.Y.; Oh, J.-W.; Cho, J.K.; Lee, K.H.; Park, K.H. Phenolic phytochemical displaying SARS-CoV papain-like protease inhibition from the seeds of Psoralea corylifolia. J. Enzyme Inhib. Med. Chem. 2014, 29, 59–63. [Google Scholar] [CrossRef]
- Jo, S.; Kim, H.; Kim, S.; Shin, D.H.; Kim, M.S. Characteristics of flavonoids as potent MERS-CoV 3C-like protease inhibitors. Chem. Biol. Drug Des. 2019, 94, 2023–2030. [Google Scholar] [CrossRef]
- Valipour, M. Recruitment of chalcone’s potential in drug discovery of a nti-SARS-CoV-2 agents. Phytother. Res. 2022, 36, 4477–4490. [Google Scholar] [CrossRef]
- Zwaagstra, M.E.; Timmerman, H.; Tamura, M.; Tohma, T.; Wada, Y.; Onogi, K.; Zhang, M.-Q. Synthesis and Structure−Activity Relationships of Carboxylated Chalcones: A Novel Series of CysLT 1 (LTD4) Receptor Antagonists. J. Med. Chem. 1997, 40, 1075–1089. [Google Scholar] [CrossRef]
- Giordo, R.; Zinellu, A.; Eid, A.H.; Pintus, G. Therapeutic potential of resveratrol in COVID-19-associated hemostatic disorders. Molecules 2021, 26, 856. [Google Scholar] [CrossRef]
- Rossi, G.A.; Sacco, O.; Capizzi, A.; Mastromarino, P. Can Resveratrol-Inhaled Formulations Be Considered Potential Adjunct Treatments for COVID-19? Front. Immunol. 2021, 12, 1591. [Google Scholar] [CrossRef]
- Derosa, G.; Maffioli, P.; D’Angelo, A.; Di Pierro, F. A role for quercetin in coronavirus disease 2019 (COVID-19). Phytother. Res. 2021, 35, 1230–1236. [Google Scholar] [CrossRef]
- Yan, Z.; Caldwell, G.W. Metabolism profiling, and cytochrome P450 inhibition & induction in drug discovery. Curr. Top. Med. Chem. 2001, 1, 403–425. [Google Scholar]
- De Groot, M.J. Designing better drugs: Predicting cytochrome P450 metabolism. Drug Discov. Today 2006, 11, 601–606. [Google Scholar] [CrossRef]
- Bengtson, C.D.; Montgomery, R.N.; Nazir, U.; Satterwhite, L.; Kim, M.D.; Bahr, N.C.; Castro, M.; Baumlin, N.; Salathe, M. An open label trial to assess safety of losartan for treating worsening respiratory illness in COVID-19. Front. Med. 2021, 8, 152. [Google Scholar] [CrossRef]
- Nouri-Vaskeh, M.; Kalami, N.; Zand, R.; Soroureddin, Z.; Varshochi, M.; Ansarin, K.; Rezaee, H.; Taghizadieh, A.; Sadeghi, A.; Maleki, M.A. Comparison of losartan and amlodipine effects on the outcomes of patient with COVID-19 and primary hypertension: A randomised clinical trial. Int. J. Clin. Pract. 2021, 75, e14124. [Google Scholar] [CrossRef]
- Puskarich, M.A.; Cummins, N.W.; Ingraham, N.E.; Wacker, D.A.; Reilkoff, R.A.; Driver, B.E.; Biros, M.H.; Bellolio, F.; Chipman, J.G.; Nelson, A.C. A multi-center phase II randomized clinical trial of losartan on symptomatic outpatients with COVID-19. eClinicalMedicine 2021, 37, 100957. [Google Scholar] [CrossRef]
- Geriak, M.; Haddad, F.; Kullar, R.; Greenwood, K.L.; Habib, M.; Habib, C.; Willms, D.; Sakoulas, G. Randomized Prospective Open Label Study Shows No Impact on Clinical Outcome of Adding Losartan to Hospitalized COVID-19 Patients with Mild Hypoxemia. Infect. Dis. Ther. 2021, 10, 1323–1330. [Google Scholar] [CrossRef]
- Zeinalian, M.; Salari-Jazi, A.; Jannesari, A.; Khanahmad, H. A potential protective role of losartan against coronavirus-induced lung damage. Infect. Control Hosp. Epidemiol. 2020, 41, 752–753. [Google Scholar] [CrossRef]
- Shen, Z.; Ratia, K.; Cooper, L.; Kong, D.; Lee, H.; Kwon, Y.; Li, Y.; Alqarni, S.; Huang, F.; Dubrovskyi, O. Potent, Novel SARS-CoV-2 PLpro Inhibitors Block Viral Replication in Monkey and Human Cell Cultures. bioRxiv 2021. [Google Scholar] [CrossRef]
- Park, J.-Y.; Jeong, H.J.; Kim, J.H.; Kim, Y.M.; Park, S.-J.; Kim, D.; Park, K.H.; Lee, W.S.; Ryu, Y.B. Diarylheptanoids from Alnus japonica inhibit papain-like protease of severe acute respiratory syndrome coronavirus. Biol. Pharm. Bull. 2012, 35, 2036–2042. [Google Scholar] [CrossRef]
- Seo, W.D.; Kim, J.H.; Kang, J.E.; Ryu, H.W.; Curtis-Long, M.J.; Lee, H.S.; Yang, M.S.; Park, K.H. Sulfonamide chalcone as a new class of α-glucosidase inhibitors. Bioorg. Med. Chem. Lett. 2005, 15, 5514–5516. [Google Scholar] [CrossRef]
- Williams, S.J.; Goddard-Borger, E.D. α-glucosidase inhibitors as host-directed antiviral agents with potential for the treatment of COVID-19. Biochem. Soc. Trans. 2020, 48, 1287–1295. [Google Scholar] [CrossRef] [PubMed]
- Nichols, C.; Ng, J.; Keshu, A.; Kelly, G.; Conte, M.R.; Marber, M.S.; Fraternali, F.; De Nicola, G.F. Mining the PDB for Tractable Cases Where X-ray Crystallography Combined with Fragment Screens Can Be Used to Systematically Design Protein–Protein Inhibitors: Two Test Cases Illustrated by IL1β-IL1R and p38α–TAB1 Complexes. J. Med. Chem. 2020, 63, 7559–7568. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.K.; Gong, G.; Grum-Tokars, V.; Mulhearn, D.C.; Baker, S.C.; Coughlin, M.; Prabhakar, B.S.; Sleeman, K.; Johnson, M.E.; Mesecar, A.D. Design, synthesis and antiviral efficacy of a series of potent chloropyridyl ester-derived SARS-CoV 3CLpro inhibitors. Bioorg. Med. Chem. Lett. 2008, 18, 5684–5688. [Google Scholar] [CrossRef] [PubMed]
- Welker, A.; Kersten, C.; Müller, C.; Madhugiri, R.; Zimmer, C.; Müller, P.; Zimmermann, R.; Hammerschmidt, S.; Maus, H.; Ziebuhr, J. Structure-Activity Relationships of Benzamides and Isoindolines Designed as SARS-CoV Protease Inhibitors Effective against SARS-CoV-2. ChemMedChem 2021, 16, 340–354. [Google Scholar] [CrossRef] [PubMed]
- Domínguez, J.N.; León, C.; Rodrigues, J.; de Domínguez, N.G.; Gut, J.; Rosenthal, P.J. Synthesis and evaluation of new antimalarial phenylurenyl chalcone derivatives. J. Med. Chem. 2005, 48, 3654–3658. [Google Scholar] [CrossRef]
- Ratia, K.; Pegan, S.; Takayama, J.; Sleeman, K.; Coughlin, M.; Baliji, S.; Chaudhuri, R.; Fu, W.; Prabhakar, B.S.; Johnson, M.E. A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc. Natl. Acad. Sci. USA 2008, 105, 16119–16124. [Google Scholar] [CrossRef]
- Báez-Santos, Y.M.; Barraza, S.J.; Wilson, M.W.; Agius, M.P.; Mielech, A.M.; Davis, N.M.; Baker, S.C.; Larsen, S.D.; Mesecar, A.D. X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human coronavirus papain-like proteases. J. Med. Chem. 2014, 57, 2393–2412. [Google Scholar] [CrossRef]
- Wu, C.-Y.; King, K.-Y.; Kuo, C.-J.; Fang, J.-M.; Wu, Y.-T.; Ho, M.-Y.; Liao, C.-L.; Shie, J.-J.; Liang, P.-H.; Wong, C.-H. Stable benzotriazole esters as mechanism-based inactivators of the severe acute respiratory syndrome 3CL protease. Chem. Biol. 2006, 13, 261–268. [Google Scholar] [CrossRef]
- Turlington, M.; Chun, A.; Tomar, S.; Eggler, A.; Grum-Tokars, V.; Jacobs, J.; Daniels, J.S.; Dawson, E.; Saldanha, A.; Chase, P. Discovery of N-(benzo [1, 2, 3] triazol-1-yl)-N-(benzyl) acetamido) phenyl) carboxamides as severe acute respiratory syndrome coronavirus (SARS-CoV) 3CLpro inhibitors: Identification of ML300 and noncovalent nanomolar inhibitors with an induced-fit binding. Bioorg. Med. Chem. Lett. 2013, 23, 6172–6177. [Google Scholar] [CrossRef]
- Balef, S.S.H.; Piramoon, M.; Hosseinimehr, S.J.; Irannejad, H. In vitro and in silico evaluation of P-glycoprotein inhibition through 99mTc-methoxyisobutylisonitrile uptake. Chem. Biol. Drug Des. 2019, 93, 283–289. [Google Scholar] [CrossRef]
- Goodarzi, A.; Valipour, M.; Irannejad, H. Affinity Prediction of Shikonins towards Sirtuins and the Requisite Structural Motifs for the Selective Inhibition of SIRT2 and SIRT3. Lett. Drug Des. Discov. 2023. [Google Scholar] [CrossRef]
- Valdés-Tresanco, M.S.; Valdés-Tresanco, M.E.; Valiente, P.A.; Moreno, E. gmx_MMPBSA: A New Tool to Perform End-State Free Energy Calculations with GROMACS. J. Chem. Theory Comput. 2021, 17, 6281–6291. [Google Scholar] [CrossRef]

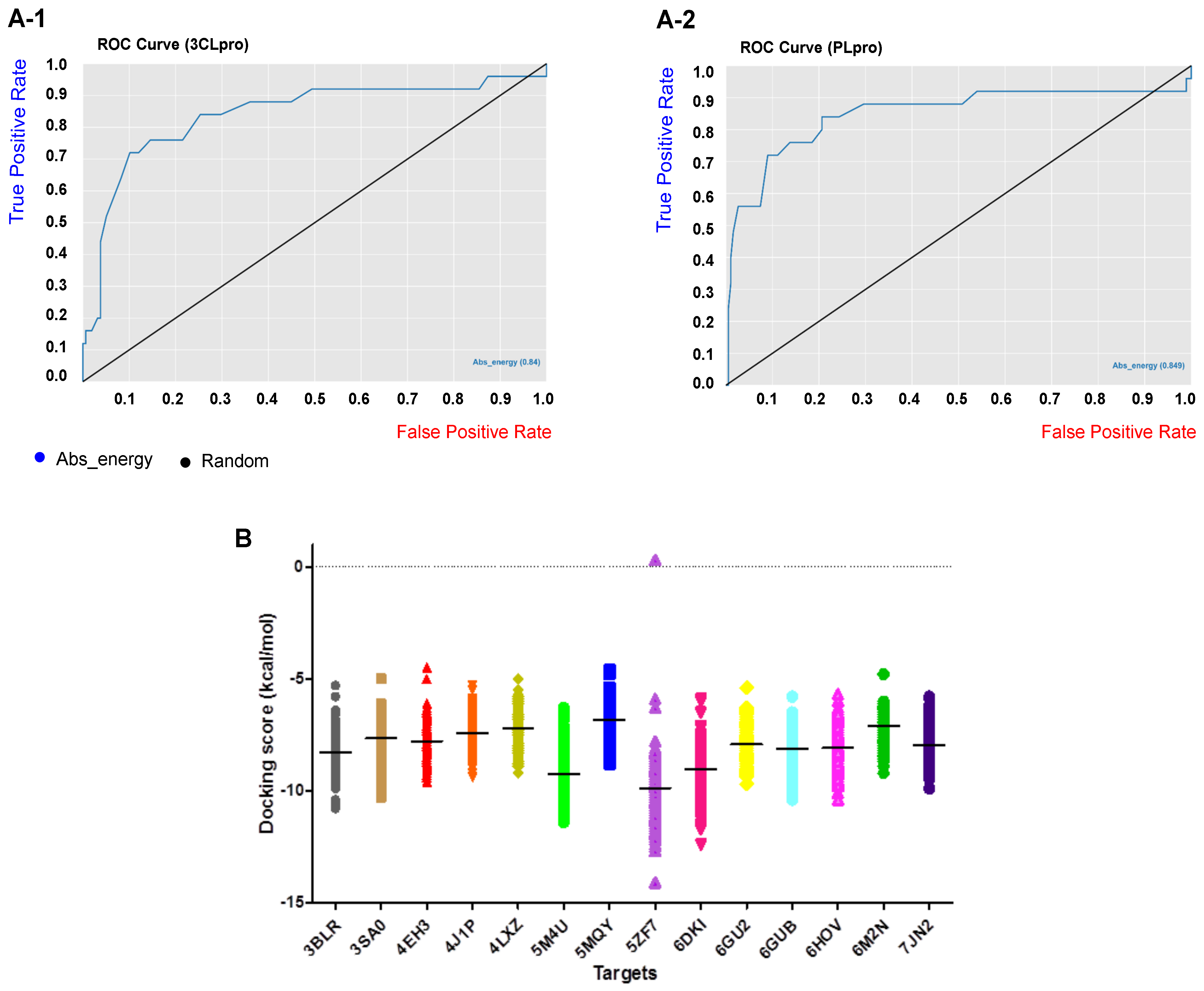



| Target | Correct Classified | Wrong Classified | Accuracy | Error | Cohen’s Kappa (k) |
|---|---|---|---|---|---|
| 3CLpro | 147 | 36 | 80.32% | 19.67% | 0.40% |
| PLpro | 173 | 41 | 80.84% | 19.15% | 0.38% |
| Chalcones | 4EH3 | 5MQY | 6GU2 | 6GUB | 3BLR | 3SA0 | 4LXZ | 5ZF7 | 5M4U | 4J1P | 6HOV | 6DK1 | 6M2N | 7JN2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CHA-7 | 12 | 72 | 5 | 31 | 19 | 17 | 30 | 31 | 6 | 6 | 6 | 4 | 84 | 5 |
| CHA-10 | 86 | 36 | 64 | 33 | 56 | 24 | 1 | 4 | 156 | 85 | 32 | 36 | 27 | 23 |
| CHA-11 | 231 | 5 | 10 | 1 | 2 | 30 | 19 | 25 | 19 | 13 | 2 | 17 | 36 | 6 |
| CHA-12 a | 1 | 2 | 1 | 2 | 3 | 1 | 2 | 1 | 2 | 2 | 3 | 1 | 1 | 1 |
| CHA-15 | 17 | 6 | 6 | 8 | 39 | 24 | 20 | 10 | 11 | 65 | 52 | 8 | 10 | 118 |
| CHA-22 | 153 | 337 | 109 | 263 | 312 | 61 | 566 | 37 | 4 | 191 | 340 | 48 | 161 | 69 |
| CHA-29 | 157 | 220 | 34 | 23 | 25 | 5 | 266 | 145 | 60 | 3 | 43 | 236 | 29 | 99 |
| CHA-30 | 159 | 42 | 53 | 123 | 49 | 33 | 3 | 39 | 209 | 69 | 422 | 52 | 30 | 321 |
| CHA-34 | 4 | 29 | 242 | 96 | 107 | 108 | 14 | 28 | 61 | 50 | 25 | 109 | 52 | 35 |
| CHA-37 | 2 | 4 | 3 | 9 | 7 | 4 | 34 | 9 | 1 | 5 | 1 | 2 | 12 | 3 |
| CHA-44 | 662 | 16 | 8 | 3 | 1 | 28 | 10 | 73 | 118 | 43 | 5 | 53 | 75 | 4 |
| CHA-60 | 5 | 69 | 358 | 65 | 36 | 49 | 448 | 250 | 123 | 631 | 382 | 44 | 187 | 534 |
| CHA-77 | 13 | 206 | 9 | 4 | 37 | 8 | 136 | 653 | 463 | 63 | 14 | 226 | 193 | 298 |
| CHA-84 | 3 | 650 | 29 | 256 | 753 | 431 | 74 | 756 | 755 | 594 | 716 | 757 | 399 | 755 |
| CHA-86 | 190 | 582 | 381 | 307 | 191 | 214 | 310 | 259 | 79 | 84 | 62 | 62 | 114 | 7 |
| CHA-118 | 674 | 684 | 443 | 529 | 503 | 305 | 654 | 315 | 703 | 439 | 672 | 737 | 405 | 8 |
| CHA-134 | 87 | 384 | 65 | 96 | 107 | 53 | 5 | 556 | 685 | 7 | 23 | 228 | 52 | 44 |
| CHA-166 | 410 | 291 | 30 | 19 | 20 | 26 | 78 | 142 | 54 | 4 | 35 | 159 | 90 | 615 |
| CHA-177 | 31 | 39 | 31 | 42 | 6 | 45 | 9 | 48 | 7 | 8 | 9 | 55 | 91 | 9 |
| CHA-178 | 411 | 553 | 623 | 160 | 199 | 223 | 4 | 67 | 107 | 373 | 133 | 128 | 116 | 240 |
| CHA-206 | 18 | 212 | 228 | 21 | 85 | 3 | 79 | 143 | 206 | 9 | 78 | 78 | 159 | 13 |
| CHA-215 | 357 | 7 | 134 | 11 | 58 | 54 | 59 | 3 | 85 | 115 | 16 | 15 | 611 | 567 |
| CHA-233 | 20 | 27 | 39 | 29 | 130 | 17 | 233 | 20 | 43 | 117 | 101 | 12 | 8 | 401 |
| CHA-234 | 197 | 3 | 32 | 22 | 159 | 19 | 146 | 13 | 22 | 37 | 283 | 5 | 40 | 96 |
| CHA-236 | 7 | 1 | 14 | 13 | 251 | 13 | 114 | 7 | 36 | 118 | 79 | 6 | 3 | 249 |
| CHA-282 | 49 | 122 | 309 | 164 | 73 | 71 | 82 | 108 | 58 | 28 | 226 | 107 | 23 | 2 |
| CHA-297 | 158 | 167 | 15 | 6 | 10 | 14 | 63 | 16 | 59 | 10 | 18 | 37 | 11 | 27 |
| CHA-298 | 50 | 56 | 33 | 45 | 15 | 21 | 24 | 21 | 86 | 38 | 10 | 9 | 68 | 34 |
| CHA-375 | 9 | 86 | 41 | 128 | 8 | 15 | 26 | 5 | 62 | 30 | 11 | 18 | 32 | 19 |
| CHA-378 | 10 | 10 | 24 | 5 | 4 | 16 | 15 | 22 | 63 | 1 | 4 | 28 | 4 | 10 |
| CHA-383 | 373 | 344 | 117 | 393 | 660 | 7 | 513 | 55 | 574 | 515 | 430 | 29 | 6 | 663 |
| CHA-384 | 11 | 88 | 7 | 47 | 63 | 2 | 158 | 2 | 25 | 72 | 7 | 3 | 2 | 37 |
| CHA-392 | 74 | 89 | 55 | 58 | 51 | 34 | 579 | 14 | 261 | 73 | 8 | 14 | 5 | 505 |
| CHA-397 | 24 | 90 | 43 | 48 | 43 | 5 | 126 | 12 | 90 | 102 | 12 | 10 | 7 | 260 |
| CHA-485 | 99 | 288 | 4 | 15 | 34 | 142 | 87 | 388 | 119 | 265 | 300 | 206 | 9 | 59 |
| CHA-509 | 175 | 693 | 18 | 61 | 5 | 48 | 39 | 15 | 176 | 16 | 251 | 59 | 79 | 60 |
| CHA-567 | 81 | 12 | 59 | 63 | 77 | 121 | 170 | 51 | 5 | 20 | 26 | 60 | 44 | 64 |
| CHA-581 | 41 | 443 | 99 | 140 | 44 | 49 | 69 | 30 | 3 | 57 | 27 | 7 | 233 | 64 |
| CHA-734 | 43 | 204 | 2 | 40 | 95 | 96 | 56 | 700 | 462 | 12 | 13 | 443 | 111 | 384 |
 Most active ligands selective towards the PLpro target. Lighter colors indicate a lower magnitude of affinity.
Most active ligands selective towards the PLpro target. Lighter colors indicate a lower magnitude of affinity.  Most active ligands towards 3CLpro and PLpro targets. Lighter colors indicate a lower magnitude of affinity.
Most active ligands towards 3CLpro and PLpro targets. Lighter colors indicate a lower magnitude of affinity.  Most active ligands selective towards the 3CLpro target. Lighter colors indicate a lower magnitude of affinity. Lighter colors of each one represent lower significance of the compound.
Most active ligands selective towards the 3CLpro target. Lighter colors indicate a lower magnitude of affinity. Lighter colors of each one represent lower significance of the compound.| Compounds | MM/GBSA | MM/PBSA | |||||||
|---|---|---|---|---|---|---|---|---|---|
| ΔGb | ΔGTotal | ΔGsolv | ΔGgas | ΔGb | ΔGTotal | ΔGsolv | ΔGgas | IE | |
| Baicalein | −29.43 | −28.77 | 5.51 | −34.28 | −24.23 | −23.57 | 10.71 | −34.28 | 0.04 |
| CHA-12 | −44.03 | −47.62 | 20.65 | −68.26 | −35.57 | −39.16 | 29.11 | −68.26 | 0.04 |
| CHA-384 | −40.05 | −42.32 | 15.44 | −57.76 | −30.39 | −32.65 | 25.10 | −57.76 | 0.04 |
| CHA-37 | −25.05 | −27.17 | 13.54 | −40.70 | −17.80 | −19.91 | 20.79 | −40.70 | 0.04 |
| CHA-297 | −14.80 | −21.44 | 26.11 | −47.55 | −14.53 | −21.17 | 26.38 | −47.55 | 0.04 |
| CHA-378 | −24.98 | −26.83 | 27.92 | −54.75 | −24.28 | −26.13 | 28.62 | −54.75 | 0.04 |
| Hirsutenone | −29.37 | −29.28 | 9.42 | −38.70 | −20.66 | −20.58 | 18.12 | −38.70 | 0.1 |
| A | −9.43 | −11.18 | 19.86 | −31.05 | −14.21 | −15.96 | 15.08 | −31.05 | 0.03 |
| B | −19.41 | −20.85 | 8.6 | −29.45 | −13.22 | −14.66 | 14.79 | −29.45 | 1.53 |
| Compounds | MM/GBSA | MM/PBSA | |||||||
|---|---|---|---|---|---|---|---|---|---|
| ΔGb | ΔGT | ΔGsolv | ΔGgas | ΔGb | ΔGT | ΔGsolv | ΔGgas | IE | |
| Co-ligand a | −12.36 | −13.65 | 7.67 | −21.32 | −11.99 | −13.28 | 8.04 | −21.32 | 0.04 |
| GRL0617 | −34.65 | −36.42 | 19.37 | −55.80 | −26.64 | −28.41 | 27.39 | −55.80 | 0.03 |
| CHA−12 | −35.81 | −35.60 | 14.20 | −49.81 | −31.13 | −30.92 | 18.89 | −49.81 | 0.16 |
| CHA-37 | −21.23 | −20.35 | 6.83 | −27.18 | −14.11 | −13.22 | 13.95 | −27.18 | 0.04 |
| CHA-378 | −20.52 | −27.87 | 39.46 | −67.33 | −16.31 | −23.66 | 43.68 | −67.33 | 0.04 |
| Hirsutenone | −23.11 | −23.53 | 13.47 | −37.00 | −19.61 | −20.03 | 16.97 | −37.00 | 0.93 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valipour, M.; Di Giacomo, S.; Di Sotto, A.; Irannejad, H. Discovery of Chalcone-Based Hybrid Structures as High Affinity and Site-Specific Inhibitors against SARS-CoV-2: A Comprehensive Structural Analysis Based on Various Host-Based and Viral Targets. Int. J. Mol. Sci. 2023, 24, 8789. https://doi.org/10.3390/ijms24108789
Valipour M, Di Giacomo S, Di Sotto A, Irannejad H. Discovery of Chalcone-Based Hybrid Structures as High Affinity and Site-Specific Inhibitors against SARS-CoV-2: A Comprehensive Structural Analysis Based on Various Host-Based and Viral Targets. International Journal of Molecular Sciences. 2023; 24(10):8789. https://doi.org/10.3390/ijms24108789
Chicago/Turabian StyleValipour, Mehdi, Silvia Di Giacomo, Antonella Di Sotto, and Hamid Irannejad. 2023. "Discovery of Chalcone-Based Hybrid Structures as High Affinity and Site-Specific Inhibitors against SARS-CoV-2: A Comprehensive Structural Analysis Based on Various Host-Based and Viral Targets" International Journal of Molecular Sciences 24, no. 10: 8789. https://doi.org/10.3390/ijms24108789
APA StyleValipour, M., Di Giacomo, S., Di Sotto, A., & Irannejad, H. (2023). Discovery of Chalcone-Based Hybrid Structures as High Affinity and Site-Specific Inhibitors against SARS-CoV-2: A Comprehensive Structural Analysis Based on Various Host-Based and Viral Targets. International Journal of Molecular Sciences, 24(10), 8789. https://doi.org/10.3390/ijms24108789










