From Single Cell to Plants: Mesophyll Protoplasts as a Versatile System for Investigating Plant Cell Reprogramming
Abstract
1. Introduction
2. Cell Differentiation and De-Differentiation in Planta
3. Experimental Systems for Exploiting Totipotency
4. Using Protoplasts to Study Cell De-Differentiation
4.1. Protoplast Sources
4.1.1. Shoot-Derived Protoplasts
4.1.2. Root Protoplasts
4.1.3. Callus Protoplasts
4.2. Mesophyll Protoplasts to Study Cell De-Differentiation
4.2.1. The Role of Optimal Nutrition in Culture Media for Donor Plant Quality and Protoplast Reprogramming
4.2.2. Competence Window for Leaf Protoplasts
4.2.3. The Protoplast Isolation Step as the Key for Reprogramming
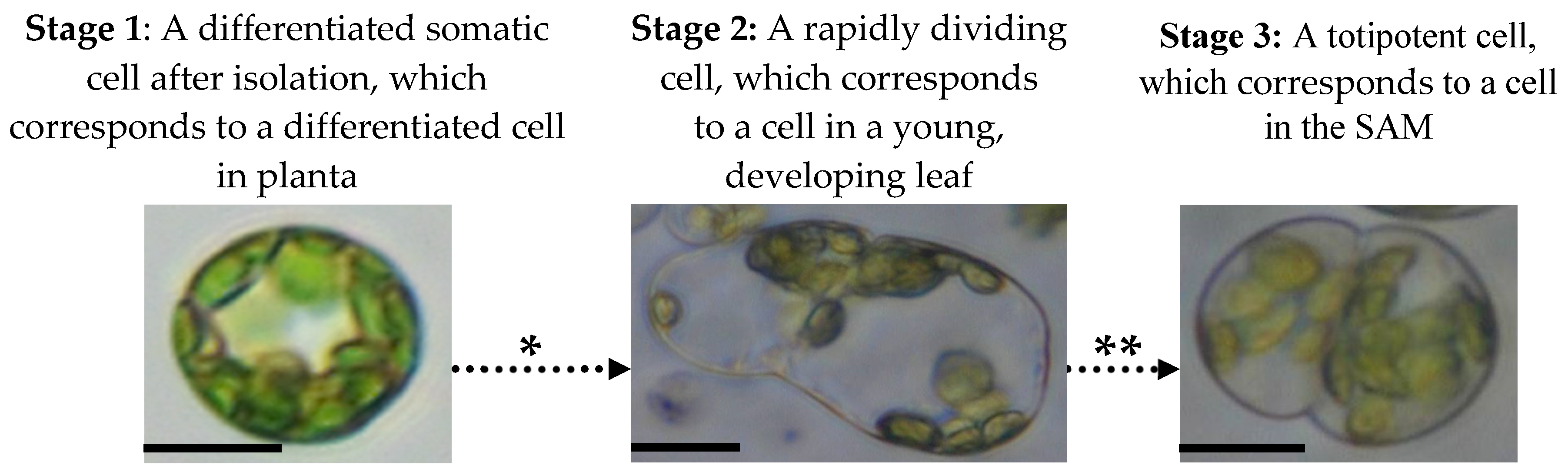
4.2.4. Stages of Mesophyll Protoplast Reprogramming and Accompanying Changes in Their Epigenetic and Physiological Profiles
4.3. Stimuli of Protoplast De-Differentiation: Hormones, Stress and Nutrition
4.4. Types of Cell De-Differentiation
4.5. Induction of Totipotent Stem Cells from Mesophyll Protoplasts
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hofmann, N. A Breakthrough in Monocot Transformation Methods. Plant Cell 2016, 28, 1989. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Zhang, G.; Liu, W.; Shi, J.; Wang, H.; Qi, M.; Li, J.; Qin, P.; Ruan, Y.; Huang, H.; et al. Divergent regeneration-competent cells adopt a common mechanism for callus initiation in angiosperms. Regeneration 2017, 4, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Ikeuchi, M.; Ogawa, Y.; Iwase, A.; Sugimoto, K. Plant regeneration: Cellular origins and molecular mechanisms. Development 2016, 143, 1442–1451. [Google Scholar] [CrossRef] [PubMed]
- Beemster, G.T.; Fiorani, F.; Inzé, D. Cell cycle: The key to plant growth control? Trends Plant Sci. 2003, 8, 154–158. [Google Scholar] [CrossRef]
- Jäger, S.; Maughan, S.; Dewitte, W.; Scofield, S.; Murray, J.A.H. The developmental context of cell-cycle control in plants. Semin. Cell Dev. Boil. 2005, 16, 385–396. [Google Scholar] [CrossRef]
- Velappan, Y.; Signorelli, S.; Considine, M.J. Cell cycle arrest in plants: What distinguishes quiescence, dormancy and differentiated G1? Ann. Bot. 2017, 120, 495–509. [Google Scholar] [CrossRef]
- Pasternak, T.; Dudits, D. Epigenetic Clues to Better Understanding of the Asexual Embryogenesis in planta and in vitro. Front. Plant Sci. 2019, 10, 778. [Google Scholar] [CrossRef]
- Shemer, O.; Landau, U.; Candela, H.; Zemach, A.; Williams, L.E. Competency for shoot regeneration from Arabidopsis root explants is regulated by DNA methylation. Plant Sci. 2015, 238, 251–261. [Google Scholar] [CrossRef]
- Sugimoto, K.; Temman, H.; Kadokura, S.; Matsunaga, S. To regenerate or not to regenerate: Factors that drive plant regeneration. Curr. Opin. Plant Boil. 2019, 47, 138–150. [Google Scholar] [CrossRef]
- Bustillo-Avendaño, E.; Ibáñez, S.; Sanz, O.; Barros, J.A.S.; Gude, I.; Perianez-Rodriguez, J.; Micol, J.L.; del Pozo, J.C.; Moreno-Risueno, M.A.; Perez-Perez, J.M. Regulation of Hormonal Control, Cell Reprogramming, and Patterning during De Novo Root Organogenesis. Plant Physiol. 2017, 176, 1709–1727. [Google Scholar] [CrossRef]
- Ikeuchi, M.; Shibata, M.; Rymen, B.; Iwase, A.; Bagman, A.-M.; Watt, L.; Coleman, D.; Favero, D.S.; Takahashi, T.; Ahnert, S.E.; et al. A Gene Regulatory Network for Cellular Reprogramming in Plant Regeneration. Plant Cell Physiol. 2018, 59, 770–782. [Google Scholar] [CrossRef] [PubMed]
- Huh, J.H.; Bauer, M.J.; Hsieh, T.-F.; Fischer, R.L. Cellular Programming of Plant Gene Imprinting. Cell 2008, 132, 735–744. [Google Scholar] [CrossRef] [PubMed]
- Sussex, I.M. Developmental programming of the shoot meristem. Cell 1989, 56, 225–229. [Google Scholar] [CrossRef]
- Dolzblasz, A.; Gola, E.M.; Sokołowska, K.; Smakowska-Luzan, E.; Twardawska, A.; Janska, H. Impairment of Meristem Proliferation in Plants Lacking the Mitochondrial Protease AtFTSH4. Int. J. Mol. Sci. 2018, 19, 853. [Google Scholar] [CrossRef] [PubMed]
- Veit, B. Stem Cell Signalling Networks in Plants. Plant Mol. Boil. 2006, 60, 793–810. [Google Scholar] [CrossRef]
- Gehring, M. Epigenetic dynamics during flowering plant reproduction: Evidence for reprogramming? New Phytol. 2019, 224, 91–96. [Google Scholar] [CrossRef]
- Sijacic, P.; Bajic, M.; McKinney, E.C.; Meagher, R.B.; Deal, R.B. Changes in chromatin accessibility between Arabidopsis stem cells and mesophyll cells illuminate cell type-specific transcription factor networks. Plant J. 2018, 94, 215–231. [Google Scholar] [CrossRef]
- Morao, A.K.; Bouyer, D.; Roudier, F. Emerging concepts in chromatin-level regulation of plant cell differentiation: Timing, counting, sensing and maintaining. Curr. Opin. Plant Boil. 2016, 34, 27–34. [Google Scholar] [CrossRef]
- Chen, D.-H.; Huang, Y.; Jiang, C.; Si, J.-P. Chromatin-Based Regulation of Plant Root Development. Front. Plant Sci. 2018, 9, 9. [Google Scholar] [CrossRef]
- Mohn, F.; Schübeler, D. Genetics and epigenetics: Stability and plasticity during cellular differentiation. Trends Genet. 2009, 25, 129–136. [Google Scholar] [CrossRef]
- Horsch, R.B.; Fry, J.E.; Hoffman, N.L.; Eichholtz, D.; Rogers, S.G.; Fraley, R.T. A simple and general method for transferring genes into plants. Science 1985, 227, 1229–1231. [Google Scholar]
- Gallois, P.; Marinho, P.; Jones, H. Leaf Disk Transformation Using Agrobacterium tumefaciens-Expression of Heterologous Genes in Tobacco. In Plant Gene Transfer and Expression Protocols; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2003; pp. 39–48. [Google Scholar]
- Ikeuchi, M.; Sugimoto, K.; Iwase, A. Plant callus: Mechanisms of induction and repression. Plant Cell 2013, 25, 3159–3173. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.-Z.; Zuo, E.-H.; Qi, J.-F.; Liu, Y.; Lin, C.-T.; Deng, X. A role of age-dependent DNA methylation reprogramming in regulating the regeneration capacity of Boea hygrometrica leaves. Funct. Integr. Genom. 2019, 20, 133–149. [Google Scholar] [CrossRef] [PubMed]
- Ikeuchi, M.; Favero, D.S.; Sakamoto, Y.; Iwase, A.; Coleman, D.; Rymen, B.; Sugimoto, K. Molecular Mechanisms of Plant Regeneration. Annu. Rev. Plant Boil. 2019, 70, 377–406. [Google Scholar] [CrossRef] [PubMed]
- Rymen, B.; Kawamura, A.; Lambolez, A.; Inagaki, S.; Takebayashi, A.; Iwase, A.; Sakamoto, Y.; Sako, K.; Favero, D.S.; Ikeuchi, M.; et al. Histone acetylation orchestrates wound-induced transcriptional activation and cellular reprogramming in Arabidopsis. Commun. Boil. 2019, 2, 404. [Google Scholar] [CrossRef] [PubMed]
- Atta, R.; Laurens, L.; Guivarc, H.A.; Carnero, E.; Rech, P.; Chriqui, D.; Boucheron-Dubuisson, E.; Giraudat-Pautot, V. Pluripotency of Arabidopsis xylem pericycle underlies shoot regeneration from root and hypocotyl explants grownin vitro. Plant J. 2009, 57, 626–644. [Google Scholar] [CrossRef]
- Kim, J.Y.; Yang, W.; Forner, J.; Lohmann, J.U.; Noh, B.; Noh, Y.S. Epigenetic reprogramming by histone acetyltransferase HAG1/AtGCN5 is required for pluripotency acquisition in Arabidopsis. EMBO J. 2018, 37. [Google Scholar] [CrossRef]
- Betekhtin, A.; Hus, K.; Rojek-Jelonek, M.; Kurczyńska, E.; Nibau, C.; Doonan, J.H.; Hasterok, R. In Vitro Tissue Culture in Brachypodium: Applications and Challenges. Int. J. Mol. Sci. 2020, 21, 1037. [Google Scholar] [CrossRef]
- Pasternak, T.P.; Rudas, V.A.; Lörz, H.; Kumlehn, J. Embryogenic Callus Formation and Plant Regeneration from Leaf Base Segments of Barley (Hordeum vulgare L.). J. Plant Physiol. 1999, 155, 371–375. [Google Scholar] [CrossRef]
- Betekhtin, A.; Rojek, M.; Milewska-Hendel, A.; Gawecki, R.; Karcz, J.; Kurczyńska, E.; Hasterok, R. Spatial Distribution of Selected Chemical Cell Wall Components in the Embryogenic Callus of Brachypodium distachyon. PLoS ONE 2016, 11, e0167426. [Google Scholar] [CrossRef]
- Maddock, S.E.; Lancaster, V.A.; Risiott, R.; Franklin, J. Plant Regeneration from Cultured Immature Embryos and Inflorescences of 25 Cultivars of Wheat (Triticum aestivum). J. Exp. Bot. 1983, 34, 915–926. [Google Scholar] [CrossRef]
- Ozias-Akins, P.; Vasil, I. Plant regeneration from cultured immature embryos and inflorescences of Triticum aestivum L. (wheat): Evidence for somatic embryogenesis. Protoplasma 1982, 110, 95–105. [Google Scholar] [CrossRef]
- Palmer, C.E.; Keller, W.; Kasha, K.J. Haploids in Crop Improvement II; Springer: Berlin/Heidelberg, Germany, 2005; p. 318. [Google Scholar]
- Touraev, A.; Ilham, A.; Vicente, O.; Heberle-Bors, E. Stress-induced microspore embryogenesis in tobacco: An optimized system for molecular studies. Plant Cell Rep. 1996, 15, 561–565. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Sanz, H.; Solís, M.-T.; López, M.-F.; Gómez-Cadenas, A.; Risueño, M.C.; Testillano, P.S. Auxin Biosynthesis, Accumulation, Action and Transport are Involved in Stress-Induced Microspore Embryogenesis Initiation and Progression in Brassica napus. Plant Cell Physiol. 2015, 56, 1401–1417. [Google Scholar] [CrossRef] [PubMed]
- Vladislavovna, I.M. Effect of Growing Conditions of Rice Donor Plants on Anther Culture in Vitro. J. Agric. Sci. Technol. A 2015, 5, 686–694. [Google Scholar] [CrossRef][Green Version]
- Watts, A.; Sankaranarayanan, S.; Raipuria, R.K.; Watts, A. Production and Application of Doubled Haploid in Brassica Improvement. In Brassica Improvement; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2020; pp. 67–84. [Google Scholar]
- Shumilina, D.; Kornyukhin, D.; Domblides, E.A.; Soldatenko, A.; Artemyeva, A. Effects of Genotype and Culture Conditions on Microspore Embryogenesis and Plant Regeneration in Brassica Rapa ssp. Rapa L. Plants 2020, 9, 278. [Google Scholar] [CrossRef] [PubMed]
- Faraco, M.; di Sansebastiano, G.-P.; Spelt, K.; Koes, R.E.; Quattrocchio, F.M. One Protoplast Is Not the Other!1. Plant Physiol. 2011, 156, 474–478. [Google Scholar] [CrossRef]
- Batchelor, S.M.; Elliott, D.C. The Advantages of Isolated Protoplasts for Plant Growth Regulator Studies. In Proceedings of the Protoplasts 1983; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 1983; pp. 178–179. [Google Scholar]
- Yoo, S.-D.; Cho, Y.-H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef]
- Gregory, D.W.; Cocking, E.C. The large-scale isolation of protoplasts from immature tomato fruit. J. Cell Boil. 1965, 24, 143–146. [Google Scholar] [CrossRef]
- Cocking, E.C. Plant Cell Protoplasts-Isolation and Development. Annu. Rev. Plant Physiol. 1972, 23, 29–50. [Google Scholar] [CrossRef]
- Dudits, D.; Kao, K.N.; Constabel, F.; Gamborg, O.L. Embryogenesis and formation of tetraploid and hexaploid plants from carrot protoplasts. Can. J. Bot. 1976, 54, 1063–1067. [Google Scholar] [CrossRef]
- Davey, M.R.; Anthony, P.; Power, J.B.; Lowe, K.C. Plant protoplasts: Status and biotechnological perspectives. Biotechnol. Adv. 2005, 23, 131–171. [Google Scholar] [CrossRef]
- Eeckhaut, T.; Lakshmanan, P.S.; de Ryckere, D.; van Bockstaele, E.; van Huylenbroeck, J. Progress in plant protoplast research. Planta 2013, 238, 991–1003. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto-Shirasu, K.; Roberts, K.J. “Big it up”: Endoreduplication and cell-size control in plants. Curr. Opin. Plant Boil. 2003, 6, 544–553. [Google Scholar] [CrossRef] [PubMed]
- Glimelius, K. High growth rate and regeneration capacity of hypocotyl protoplasts in some Brassicaceae. Physiol. Plant. 1984, 61, 38–44. [Google Scholar] [CrossRef]
- Gendreau, E.; Höfte, H.; Grandjean, O.; Brown, S.; Traas, J. Phytochrome controls the number of endoreduplication cycles in the Arabidopsis thaliana hypocotyl. Plant J. 1998, 13, 221–230. [Google Scholar] [CrossRef]
- Liljebjelke, K.A.; Franceschi, V.R. Differentiation of mesophyll and paraveinal mesophyll in soybean leaf. Bot. Gaz. 1991, 151, 34–41. [Google Scholar] [CrossRef]
- Andriankaja, M.; Dhondt, S.; de Bodt, S.; Vanhaeren, H.; Coppens, F.; de Milde, L.; Mühlenbock, P.; Skirycz, A.; Gonzalez, N.; Beemster, G.T.; et al. Exit from Proliferation during Leaf Development in Arabidopsis thaliana: A Not-So-Gradual Process. Dev. Cell 2012, 22, 64–78. [Google Scholar] [CrossRef]
- Bar, M.; Ori, N. Leaf development and morphogenesis. Development 2014, 141, 4219–4230. [Google Scholar] [CrossRef]
- Wernicke, W.; Milkovits, L. Developmental Gradients in Wheat Leaves—Response of Leaf Segments in Different Genotypes Cultured in vitro. J. Plant Physiol. 1984, 115, 49–58. [Google Scholar] [CrossRef]
- Tallman, G. Guard Cell Protoplasts: Isolation, Culture, and Regeneration of Plants. Plant Cell Cult. Protoc. 2005, 318, 233–252. [Google Scholar] [CrossRef]
- Yao, X.; Zhao, W.; Yang, R.; Wang, J.; Zhao, F.; Wang, S. Preparation and applications of guard cell protoplasts from the leaf epidermis of Solanum lycopersicum. Plant Meth. 2018, 14, 26. [Google Scholar] [CrossRef] [PubMed]
- Jia, N.; Zhu, Y.; Xie, F. An Efficient Protocol for Model Legume Root Protoplast Isolation and Transformation. Front. Plant Sci. 2018, 9, 670. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.-H.; Davey, M.; Cocking, E. Organogenesis from Root Protoplasts of the Forage Legumes Medicago sativa and Trigonella foenum-graecum. Zeitschrift Pflanzenphysiol. 1982, 107, 231–235. [Google Scholar] [CrossRef]
- Xu, Z.-H.; Davey, M.; Cocking, E. Callus formation from root protoplasts of Glycine max (soybean). Plant Sci. Lett. 1982, 24, 111–115. [Google Scholar] [CrossRef]
- Xu, Z.-H.; Davey, M.; Cocking, E. Plant regeneration from root protoplasts of Brassica. Plant Sci. Lett. 1982, 24, 117–121. [Google Scholar] [CrossRef]
- Imanishi, S.; Momose, J.; Hiura, I. Isolation and Culture of Lycopersicon esculentum Root Protoplasts. Plant Tissue Cult. Lett. 1985, 2, 25–26. [Google Scholar] [CrossRef][Green Version]
- Brison, M.; Lamant, A. Callus formation from root protoplasts of Quercus rubra L. (red oak). Plant Cell Rep. 1990, 9, 139–142. [Google Scholar] [CrossRef]
- Faye, M.; David, A. Isolation and culture of gymnosperm root protoplasts (Pinus pinaster). Physiol. Plant. 1983, 59, 359–362. [Google Scholar] [CrossRef]
- Senn, A.; Pilet, P.-E. Isolation and some Morphological Properties of Maize Root Protoplasts. Zeitschrift Pflanzenphysiol. 1980, 100, 299–310. [Google Scholar] [CrossRef]
- Blom-Zandstra, M.; Koot, H.T.M.; van Hattum, J.; Vogelzang, S.A. Isolation of protoplasts for patch-clamp experiments: An improved method requiring minimal amounts of adult leaf or root tissue from monocotyledonous or dicotyledonous plants. Protoplasma 1995, 185, 1–6. [Google Scholar] [CrossRef]
- Kyozuka, J.; Shimamoto, K. Transformation and regeneration of rice protoplasts. In Plant Tissue Culture Manual; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 1991; pp. 243–259. [Google Scholar]
- Rubio, V.; Bustos, R.; Irigoyen, M.L.; Cardona-López, X.; Rojas-Triana, M.; Paz-Ares, J. Plant hormones and nutrient signaling. Plant Mol. Boil. 2008, 69, 361–373. [Google Scholar] [CrossRef]
- Masson, J.; Paszkowski, J. The culture response of Arabidopsis thaliana protoplasts is determined by the growth conditions of donor plants. Plant J. 1992, 2, 829–833. [Google Scholar] [CrossRef]
- Shimada, T.; Yamada, Y. Wheat plants regenerated from embryo cell cultures. Jpn. J. Genet. 1979, 54, 379–385. [Google Scholar] [CrossRef]
- Shimada, T. Plant regeneration from the callus induced from wheat embryo. Jpn. J. Genet. 1978, 53, 371–374. [Google Scholar] [CrossRef]
- Watanabe, M.; Setoguchi, D.; Uehara, K.; Ohtsuka, W.; Watanabe, Y. Apoptosis-like cell death of Brassica napus leaf protoplasts. New Phytol. 2002, 156, 417–426. [Google Scholar] [CrossRef]
- Zhao, J.; Morozova, N.; Williams, L.; Libs, L.; Avivi, Y.; Grafi, G. Two Phases of Chromatin Decondensation during Dedifferentiation of Plant Cells. J. Boil. Chem. 2001, 276, 22772–22778. [Google Scholar] [CrossRef]
- Pasternak, T.; Ötvös, K.; Domoki, M.; Fehér, A. Linked activation of cell division and oxidative stress defense in alfalfa leaf protoplast-derived cells is dependent on exogenous auxin. Plant Growth Regul. 2006, 51, 109–117. [Google Scholar] [CrossRef]
- Butt, A.D.; Bestwick, C.S. Generation of chemiluminescence during enzymatic isolation of protoplasts from leaves of Oryza sativa. J. Plant Physiol. 1997, 150, 729–733. [Google Scholar] [CrossRef]
- Yasuda, K.; Watanabe, Y.; Watanabe, M. Generation of intracellular reactive oxygen species during the isolation of Brassica napus leaf protoplasts. Plant Biotechnol. 2007, 24, 361–366. [Google Scholar] [CrossRef][Green Version]
- Givaty-Rapp, Y.; Yadav, N.S.; Khan, A.; Grafi, G. S1-Type Endonuclease 2 in Dedifferentiating Arabidopsis Protoplasts: Translocation to the Nucleus in Senescing Protoplasts Is Associated with De-Glycosylation. PLoS ONE 2017, 12, e0170067. [Google Scholar] [CrossRef] [PubMed]
- Wiszniewska, A.; Piwowarczyk, B. Activity of selected components of antioxidant system in grass pea and yellow lupine protoplasts after enzymatic isolation. Biotechnology 2015, 4, 285–292. [Google Scholar] [CrossRef]
- Barnes, A.C.; Elowsky, C.; Roston, R.L. An Arabidopsis protoplast isolation method reduces cytosolic acidification and activation of the chloroplast stress sensor Sensitive to Freezing 2. Plant Signal. Behav. 2019, 14, 1629270–1629277. [Google Scholar] [CrossRef] [PubMed]
- Feher, A.; Ötvös, K.; Pasternak, T.; Pettkó-Szandtner, A. The involvement of reactive oxygen species (ROS) in the cell cycle activation (G0-to-G1 transition) of plant cells. Plant Signal. Behav. 2008, 3, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Pasternak, T.; Miskolczi, P.C.; Ayaydin, F.; Mészáros, T.; Dudits, D.; Fehér, A. Exogenous auxin and cytokinin dependent activation of CDKs and cell division in leaf protoplast-derived cells of alfalfa. Plant Growth Regul. 2000, 32, 129–141. [Google Scholar] [CrossRef]
- Pasternak, T.; Asard, H.; Potters, G.; Jansen, M.A.K. The thiol compounds glutathione and homoglutathione differentially affect cell development in alfalfa (Medicago sativa L.). Plant Physiol. Biochem. 2014, 74, 16–23. [Google Scholar] [CrossRef]
- Ondrej, V.; Kitner, M.; Doležalová, I.; Nádvorník, P.; Navrátilová, B.; Lebeda, A. Chromatin structural rearrangement during dedifferentiation of protoplasts of Cucumis sativus L. Mol. Cells 2009, 27, 443–447. [Google Scholar] [CrossRef]
- Feher, A.; Pasternak, T.; Otuus, K.; Dudits, D. Plant protoplasts: Consequences of lost cell walls. In Journey of a Single Cell to a Plant; Murch, S.J., Saxena, P.K., Eds.; CRC Press: Boca Raton, FL, USA, 2005. [Google Scholar]
- Williams, L.; Zhao, J.; Morozova, N.; Li, Y.; Avivi, Y.; Grafi, G. Chromatin reorganization accompanying cellular dedifferentiation is associated with modifications of histone H3, redistribution of HP1, and activation of E2F-target genes. Dev. Dyn. 2003, 228, 113–120. [Google Scholar] [CrossRef]
- Moricová, P.; Ondrej, V.; Navrátilová, B.; Luhová, L. Changes of DNA methylation and hydroxymethylation in plant protoplast cultures. Acta Biochim. Pol. 2013, 60, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Galun, E. Plant Protoplasts as Physiological Tools. Annu. Rev. Plant Physiol. 1981, 32, 237–266. [Google Scholar] [CrossRef]
- Pasternak, T.; Prinsen, E.; Ayaydin, F.; Miskolczi, P.C.; Potters, G.; Asard, H.; van Onckelen, H.A.; Dudits, D.; Fehér, A. The Role of Auxin, pH, and Stress in the Activation of Embryogenic Cell Division in Leaf Protoplast-Derived Cells of Alfalfa1. Plant Physiol. 2002, 129, 1807–1819. [Google Scholar] [CrossRef] [PubMed]
- Sheahan, M.B.; Rose, R.J.; McCurdy, D.W. Actin-filament-dependent remodeling of the vacuole in cultured mesophyll protoplasts. Protoplasma 2007, 230, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Papadakis, A.K. The Generation of Active Oxygen Species Differs in Tobacco and Grapevine Mesophyll Protoplasts. Plant Physiol. 1999, 121, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Papadakis, A.K.; Roubelakis-Angelakis, K.A. Oxidative stress could be responsible for the recalcitrance of plant protoplasts. Plant Physiol. Biochem. 2002, 40, 549–559. [Google Scholar] [CrossRef]
- Papadakis, A.K.; Siminis, C.I.; Roubelakis-Angelakis, K.A. Reduced Activity of Antioxidant Machinery Is Correlated with Suppression of Totipotency in Plant Protoplasts. Plant Physiol. 2001, 126, 434–444. [Google Scholar] [CrossRef][Green Version]
- Schell, J.; Bisseling, T.; Dülz, M.; Franssen, H.; Fritze, K.; John, M.; Kleinow, T.; Leßnick, A.; Miklashevichs, E.; Pawlowski, K.; et al. Re-evaluation of phytohormone-independent division of tobacco protoplast-derived cells. Plant J. 1999, 17, 461–466. [Google Scholar] [CrossRef]
- Winnicki, K. The Winner Takes It All: Auxin—The Main Player during Plant Embryogenesis. Cells 2020, 9, 606. [Google Scholar] [CrossRef]
- Zeng, F.; Zhang, X.; Jin, S.; Cheng, L.; Liang, S.; Hu, L.; Guo, X.; Nie, Y.; Cao, J. Chromatin reorganization and endogenous auxin/cytokinin dynamic activity during somatic embryogenesis of cultured cotton cell. Plant Cell Tissue Organ Cult. 2007, 90, 63–70. [Google Scholar] [CrossRef]
- Wang, S.; Tiwari, S.B.; Hagen, G.; Guilfoyle, T.J. AUXIN RESPONSE FACTOR7 Restores the Expression of Auxin-Responsive Genes in Mutant Arabidopsis Leaf Mesophyll Protoplasts. Plant Cell 2005, 17, 1979–1993. [Google Scholar] [CrossRef]
- Pasternak, T.; Potters, G.; Caubergs, R.; Jansen, M.A.K. Complementary interactions between oxidative stress and auxins control plant growth responses at plant, organ, and cellular level. J. Exp. Bot. 2005, 56, 1991–2001. [Google Scholar] [CrossRef]
- Potters, G.; Jansen, M.A.K.; Horemans, N.; Guisez, Y.; Pasternak, T. Dehydroascorbate and glutathione regulate the cellular development of Nicotiana tabacum L. SR-1 protoplasts. Vitr. Cell. Dev. Boil. Anim. 2010, 46, 289–297. [Google Scholar] [CrossRef]
- Ötvös, K.; Pasternak, T.; Miskolczi, P.C.; Domoki, M.; Dorjgotov, D.; Szucs, A.; Bottka, S.; Dudits, D.; Fehér, A. Nitric oxide is required for, and promotes auxin-mediated activation of, cell division and embryogenic cell formation but does not influence cell cycle progression in alfalfa cell cultures. Plant J. 2005, 43, 849–860. [Google Scholar] [CrossRef]
- Ageeva-Kieferle, A.; Rudolf, E.E.; Lindermayr, C. Redox-Dependent Chromatin Remodeling: A New Function of Nitric Oxide as Architect of Chromatin Structure in Plants. Front. Plant Sci. 2019, 10, 625. [Google Scholar] [CrossRef]
- Lavrekha, V.V.; Pasternak, T.; Ivanov, V.; Palme, K.; Mironova, V. 3D analysis of mitosis distribution highlights the longitudinal zonation and diarch symmetry in proliferation activity of the Arabidopsis thaliana root meristem. Plant J. 2017, 92, 834–845. [Google Scholar] [CrossRef]
- Murray, J.A.H.; Jones, A.; Godin, C.; Traas, J. Systems Analysis of Shoot Apical Meristem Growth and Development: Integrating Hormonal and Mechanical Signaling. Plant Cell 2012, 24, 3907–3919. [Google Scholar] [CrossRef]
- Xiao, L.; Zhang, L.; Yang, G.; Zhu, H.; He, Y. Transcriptome of Protoplasts Reprogrammed into Stem Cells in Physcomitrella patens. PLoS ONE 2012, 7, e35961. [Google Scholar] [CrossRef]
- Sakakibara, K.; Reisewitz, P.; Aoyama, T.; Friedrich, T.; Ando, S.; Sato, Y.; Tamada, Y.; Nishiyama, T.; Hiwatashi, Y.; Kurata, T.; et al. WOX13-like genes are required for reprogramming of leaf and protoplast cells into stem cells in the moss Physcomitrella patens. Development 2014, 141, 1660–1670. [Google Scholar] [CrossRef]
- Domoki, M.; Györgyey, J.; Biro, J.; Pasternak, T.; Zvara, A.; Bottka, S.; Puskás, L.; Dudits, D.; Fehér, A. Identification and characterization of genes associated with the induction of embryogenic competence in leaf-protoplast-derived alfalfa cells. Biochim. Biophys. Acta Gene Struct. Expr. 2006, 1759, 543–551. [Google Scholar] [CrossRef]
| Leaf | Hypocotyl/Cotyledon | Root | Callus | |
|---|---|---|---|---|
| Homogeneity | yes | yes | no | yes |
| Reprogramming from differentiated to proliferating cells | yes | yes | no | no |
| Potential for totipotency | high for dicots, limited for monocots | high for young explants | high for dicots, limited for monocots | high for dicots and monocots |
| Species | Approach | Process | References |
|---|---|---|---|
| Nicotiana tabacum | fluorescence-activated cell sorter (FACS); gel electrophoresis of DNA after micrococcal nuclease (MNase) digestion | chromatin condensation/decondensation | [72] |
| Cucumis sativus | FACS; fluorescence in situ hybridisation | chromocentre and repeat reassembly | [82] |
| Medicago sativa | flow cytometry; nucleus morphology | chromatin relaxation; DNA stainability | [51,80,83] |
| Nicotiana tabacum | nucleus morphology; gene expression | histone H3 modifications; redistribution of HP1; activation of the E2F transcription factor genes | [84] |
| Brassica oleracea; Cucumis sativus | quantification of methylated and hydroxymethylated DNA | temporal changes in the amount of 5-mC and 5-hmC | [85] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pasternak, T.; Lystvan, K.; Betekhtin, A.; Hasterok, R. From Single Cell to Plants: Mesophyll Protoplasts as a Versatile System for Investigating Plant Cell Reprogramming. Int. J. Mol. Sci. 2020, 21, 4195. https://doi.org/10.3390/ijms21124195
Pasternak T, Lystvan K, Betekhtin A, Hasterok R. From Single Cell to Plants: Mesophyll Protoplasts as a Versatile System for Investigating Plant Cell Reprogramming. International Journal of Molecular Sciences. 2020; 21(12):4195. https://doi.org/10.3390/ijms21124195
Chicago/Turabian StylePasternak, Taras, Kateryna Lystvan, Alexander Betekhtin, and Robert Hasterok. 2020. "From Single Cell to Plants: Mesophyll Protoplasts as a Versatile System for Investigating Plant Cell Reprogramming" International Journal of Molecular Sciences 21, no. 12: 4195. https://doi.org/10.3390/ijms21124195
APA StylePasternak, T., Lystvan, K., Betekhtin, A., & Hasterok, R. (2020). From Single Cell to Plants: Mesophyll Protoplasts as a Versatile System for Investigating Plant Cell Reprogramming. International Journal of Molecular Sciences, 21(12), 4195. https://doi.org/10.3390/ijms21124195







