Evolution of the Proto Sex-Chromosome in Solea senegalensis
Abstract
:1. Introduction
2. Results
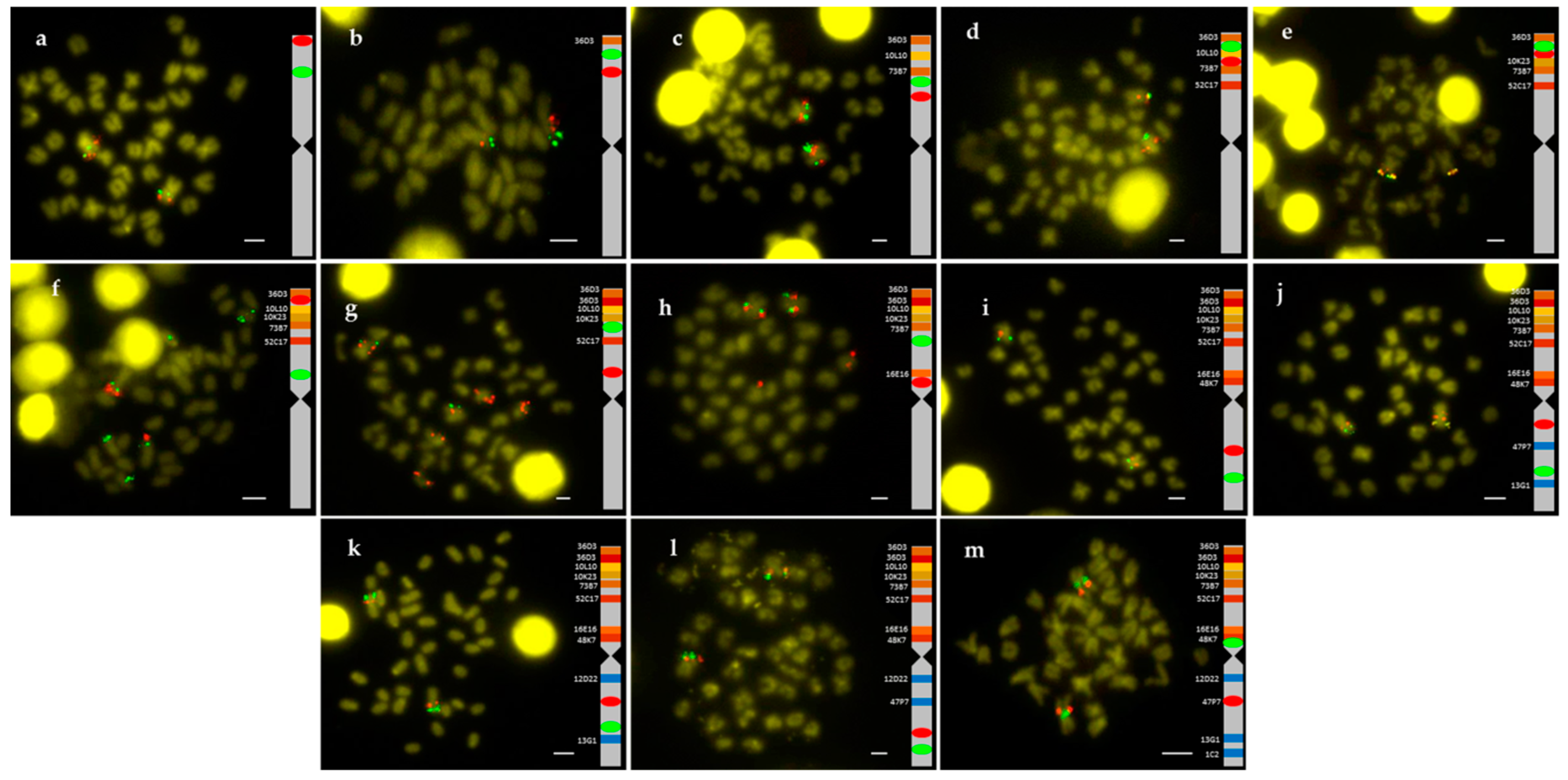
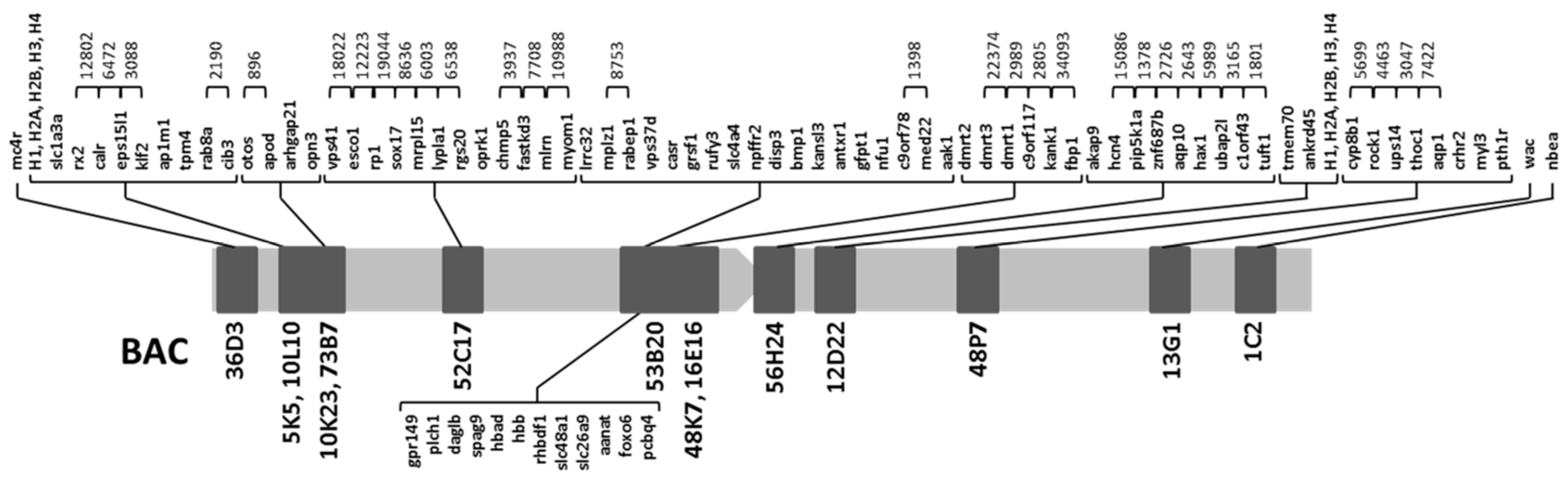
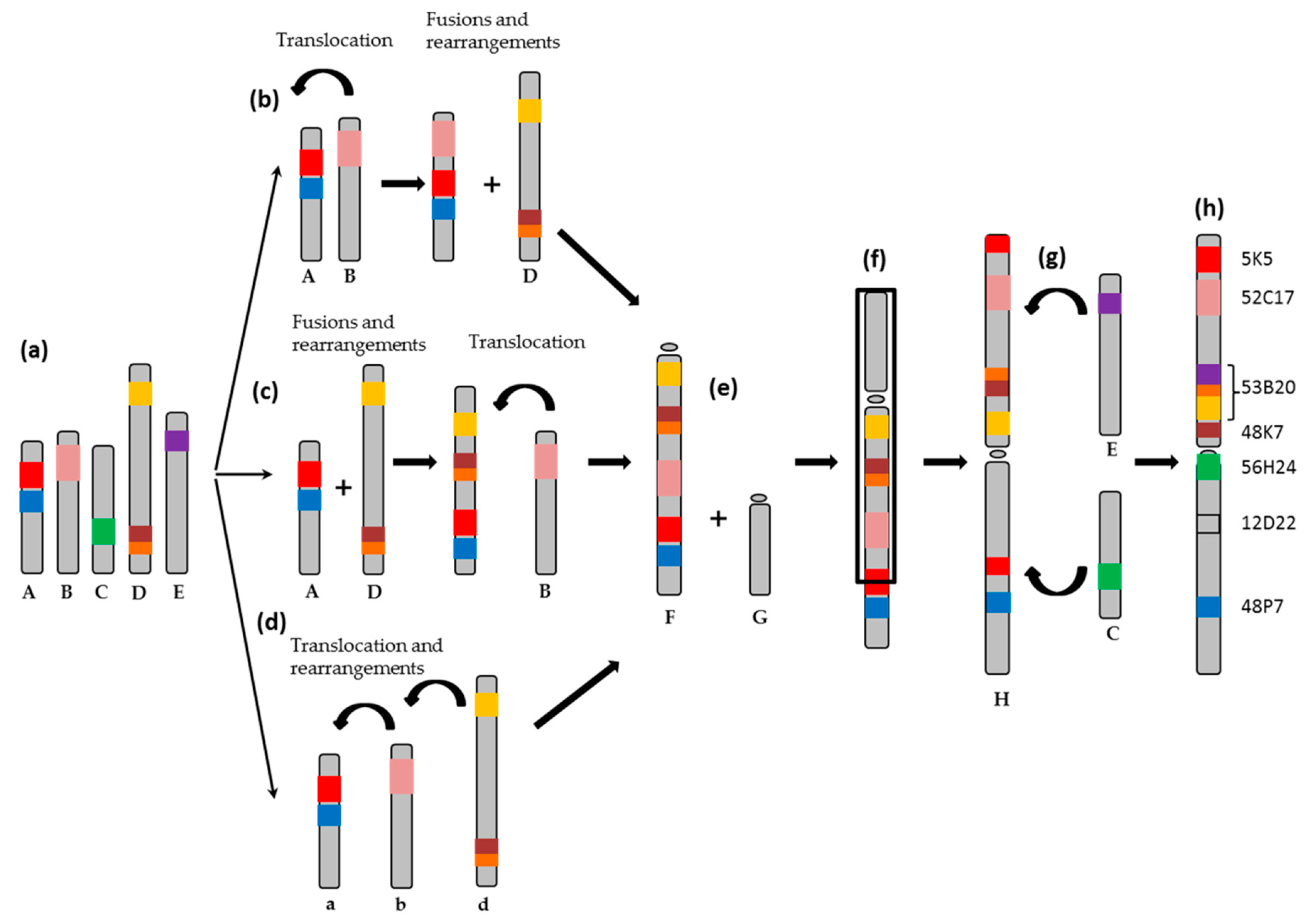
2.1. Obtention of the Integrated Map of Large Metacentric Chromosome 1 of S. senegalensis
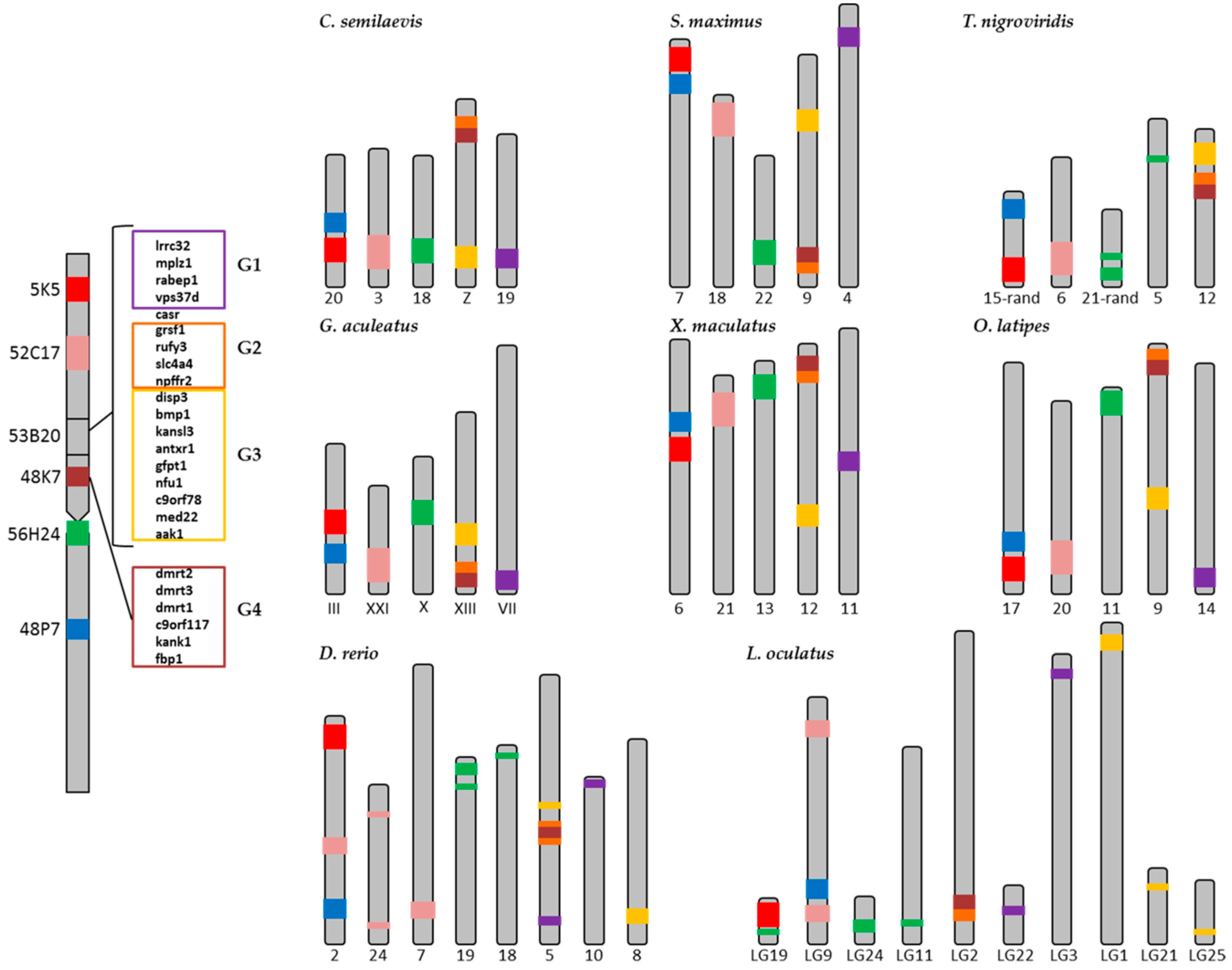
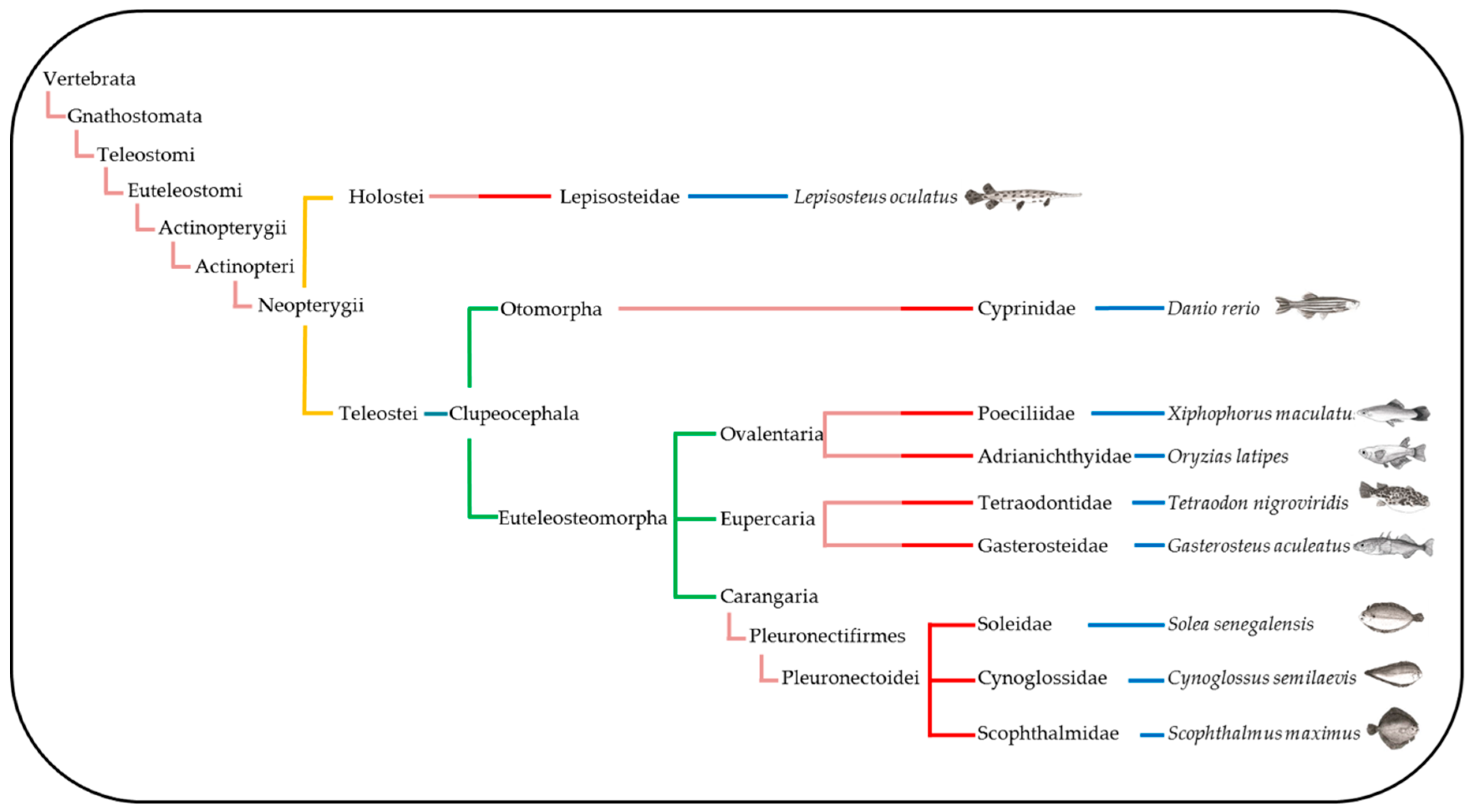
2.2. Synteny Analysis
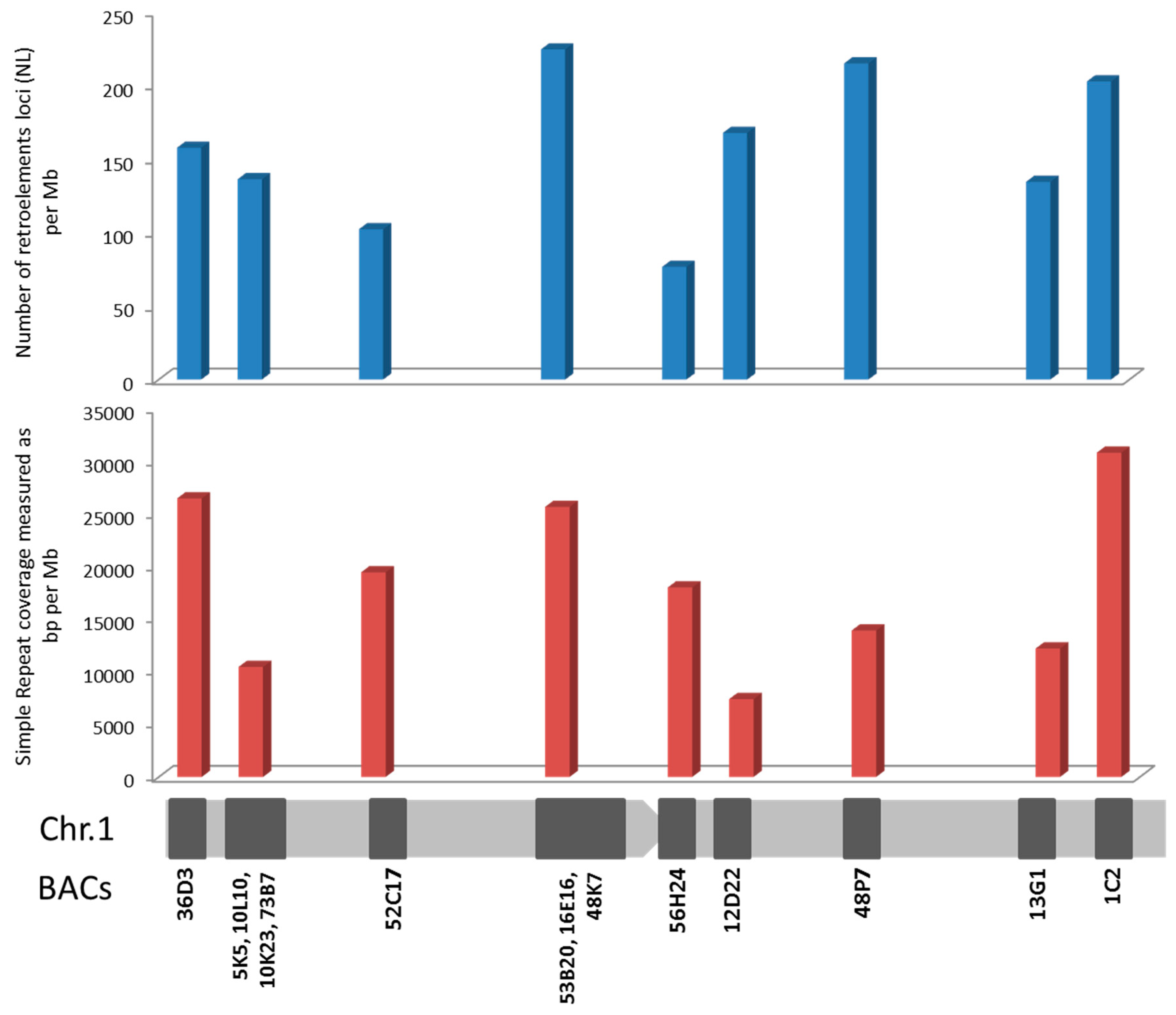
2.3. Repetitive Sequences
3. Discussion
4. Materials and Methods
4.1. BAC Clones
4.2. Chromosome Preparation and Double BAC-FISH
4.3. Sequencing and Bioinformatic Analysis
4.4. Repetitive Elements Analysis
4.5. Synteny Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nelson, J.S. Fishes of the World, 4th ed.; John Willey & Sons, Inc.: Hoboken, NJ, USA, 2006; p. 624. [Google Scholar]
- De Oliveira, E.A.; Sember, A.; Bertollo, L.A.C.; Yano, C.F.; Ezaz, T.; Moreira-Filho, O.; Hatanaka, T.; Trifonov, V.; Liehr, T.; Al-Rikabi, A.B.H.; et al. Tracking the evolutionary pathway of sex chromosomes among fishes: Characterizing the unique XX/XY1Y2 system in Hoplias malabaricus (Teleostei, Characiformes). Chromosoma 2018, 127, 115–128. [Google Scholar] [CrossRef] [PubMed]
- Robles, F.; De La Herrán, R.; Navajas-Pérez, R.; Cano-Roldán, B.; Sola-Campoy, P.J.; García-Zea, J.A.; Ruiz-Rejón, C. Centromeric satellite DNA in flatfish (order Pleuronectiformes) and its relation to speciation processes. J. Hered. 2017, 108, 217–222. [Google Scholar]
- Chen, S.; Zhang, G.; Shao, C.; Huang, Q.; Liu, G.; Zhang, P.; Song, W.; An, N.; Chalopin, D.; Volff, J.N.; et al. Whole-genome sequenced of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat. Genet. 2014, 46, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Molina-Luzón, M.J.; Hermida, M.; Navajas-Pérez, R.; Robles, F.; Navas, J.I.; Ruiz-Rejón, C.; Martínez, P.; De la Herrán, R. First haploid genetic map base don microsatellite markers in Senegalense sole (Solea senegalensis, Kaup, 1858). Mar. Biotecnol. 2015, 17, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Natri, H.M.; Merilä, J.; Shikano, T. The evolution of sex determination associated with a chromosomal inversion. Nat. Commun. 2019, 10, 145. [Google Scholar] [CrossRef]
- Martínez, P.; Viñas, A.M.; Sánchez, L.; Díaz, N.; Ribas, L.; Piferrer, F. Genetic architecture of sex determination in fish: Applications to sex ratio control in aquaculture. Front. Genet. 2014, 5, 340. [Google Scholar] [CrossRef]
- Matsuda, M.; Sakaizumi, M. Evolution of the sex-determining gene in teleostean genus Oryzias. Gen. Comp. Endocrinol. 2016, 239, 80–88. [Google Scholar] [CrossRef]
- Takehana, Y.; Matsuda, M.; Myosho, T.; Suster, M.L.; Kawakami, K.; Shin-I, T.; Kohara, Y.; Kuroki, Y.; Toyoda, A.; Fujiyama, A.; et al. Co-option of sox3 as the male-determining factor on the Y chromosome in fish Oryzias dancena. Nat. Commun. 2014, 5, 4157. [Google Scholar] [CrossRef]
- Hattori, R.S.; Murai, Y.; Oura, M.; Masuda, S.; Majhi, S.K.; Sakamoto, T.; Fernandino, J.I.; Somoza, G.M.; Yokota, M.; Strüssmann, C.A. A Y-linked anti-Müllerian hormone duplication takes over a critical role in sex determination. Proc. Nat. Acad. Sci. USA 2012, 109, 2955–2959. [Google Scholar] [CrossRef]
- Kamiya, T.; Kai, W.; Tasumi, S.; Oka, A.; Matsunaga, T.; Mizuno, N.; Fujita, M.; Suetake, H.; Suzuki, S.; Hosoya, S.; et al. A trans-species missense SNP in Amhr2 is associated with sex determination in the tiger pufferfish, Takifugu rubripes (fugu). PLoS Genet. 2012, 8, e1002798. [Google Scholar] [CrossRef]
- Yano, A.; Nicol, B.; Jouanno, E.; Quillet, E.; Fostier, A.; Guyomard, R.; Guguen, Y. The sexually dimorphic on the Y-chromosome gene (sdY) is a conserved male-specific Y-chromosome sequence in many salmonids. Evol. Appl. 2013, 6, 486–496. [Google Scholar] [CrossRef] [PubMed]
- Pennell, M.W.; Kirkpatrick, M.; Otto, S.P.; Vamosi, J.C.; Peichel, C.L.; Valenzuela, N.; Kitano, J. Y fuse? Sex chromosome fusions in fishes and reptiles. PLoS Genet. 2015, 11, e1005237. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Sember, A.; Zhu, Q.; De Oliveira, E.A.; Liehr, T.; Al-Rikabi, A.B.H.; Xiao, Z.; Song, H.; Cioffi, M.B. Deciphering the origin and evolution of the X1X2Y system in two closely-related Oplegnathus species (Oplegnathidae and Centrarchiformes). Int. J. Mol Sci. 2019, 20, 3517. [Google Scholar] [CrossRef] [PubMed]
- Portela-Bens, S.; Merlo, M.A.; Rodríguez, M.E.; Cross, I.; Manchado, M.; Kosyakova, N.; Liehr, T.; Rebordinos, L. Integrated gene mapping and synteny studies give insights into the evolution of a sex proto-chromosome in Solea senegalensis. Chromosoma 2017, 126, 261–277. [Google Scholar] [CrossRef] [PubMed]
- García-Angulo, A.; Merlo, M.A.; Portela-Bens, S.; Rodríguez, M.E.; García, E.; Al-Rikabi, A.; Liehr, T.; Rebordinos, L. Evidence for a Robertsonian fusion in Solea senegalensis (Kaup, 1858) revealed by zoo-FISH and comparative genome analysis. BMC Genomics 2018, 19, 818. [Google Scholar] [CrossRef]
- Schartl, M.; Schmid, M.; Nanda, I. Dynamics of vertebrate sex chromosome evolution: From equal size to giants and dwarfs. Chromosoma 2016, 125, 553–571. [Google Scholar] [CrossRef]
- Zhou, Q.A. Swimy locus on Y chromosome of the platyfish (Xiphophorus maculatus) is derived from a novel DNA transposon Zisupton. Gene 2012, 503, 254–259. [Google Scholar] [CrossRef]
- Chalopin, D.; Volff, J.N.; Galiana, D.; Anderson, J.L.; Schartl, M. Transposable elements and early evolution of sex chromosome in fish. Chromosome Res. 2015, 23, 545–560. [Google Scholar] [CrossRef]
- Ross, J.A.; Peichel, C.L. Molecular cytogenetic evidence of rearrangements on the Y chromosome of the threespine stickleback fish. Genetics 2008, 179, 2173–2182. [Google Scholar] [CrossRef]
- Matsuda, M.; Nagahama, Y.; Shinomiya, A.; Sato, T.; Matsuda, C.; Kobayashi, T.; Morrey, C.E.; Shibata, N.; Asakawa, S.; Shimizu, N.; et al. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 2002, 417, 559–563. [Google Scholar] [CrossRef]
- Vega, L.; Díaz, E.; Cross, I.; Rebordinos, L. Caracterizaciones citogenética e isoenzimática del lengauado Solea senegalensis Kaup, 1858. Bol. Inst. Esp. Oceanogr. 2002, 18, 245–250. [Google Scholar]
- Merlo, M.A.; Iziga, R.; Portela-Bens, S.; Cross, I.; Kosyakova, N.; Liehr, T.; Manchado, M.; Rebordinos, L. Analysis of the histone cluster in Senegalese sole (Solea senegalensis): Evidence for a divergent evolution of two canonical histone cluster. Genome 2017, 60, 441–453. [Google Scholar] [CrossRef] [PubMed]
- Amores, A.; Catchen, J.; Ferrara, A.; Fontenot, Q.; Postlethwait, J.H. Genome evolution and meiotic maps by massively parallel DNA sequencing: Spotted gar, an outgroup for the teleost genome duplication. Genetics 2011, 188, 799–808. [Google Scholar] [CrossRef] [PubMed]
- Harrington, R.C.; Faircloth, B.C.; Eytan, R.I.; Smith, W.L.; Near, T.J.; Alfaro, M.E.; Friedman, M. Phylogenomic analysis of carangimorph fishes reveals flatfish asymmetry arose in blink of evolutionary eye. BMC Evol. Biol. 2016, 16, 224. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Chen, S.; Kong, X.; Si, L.; Gong, L.; Zhang, Y.; Yu, H. Flatfish monophyly refereed by the relationship of Psettodes in Carangimorphariae. BMC Genom. 2018, 19, 400. [Google Scholar] [CrossRef] [PubMed]
- Braasch, I.; Gehrke, A.R.; Smith, J.J.; Kawasaki, K.; Manousaki, T.; Pasquier, J.; Amores, A.; Desvignes, T.; Batzel, P.; Catchen, L.; et al. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat. Genet. 2016, 48, 427–437. [Google Scholar] [CrossRef] [Green Version]
- Bachtrog, D.; Weiss, S.; Zangerl, B.; Brem, G.; Schlotterer, C. Distribution of dinucleotide microsatellites in the Drosophila melanogaster genome. Mol. Biol. Evol. 1999, 16, 602–610. [Google Scholar] [CrossRef]
- Kubat, Z.; Hobza, R.; Vyskot, B.; Kejnovsky, E. Microsatellite accumulation on the Y chromosome in Silene latifolia. Genome 2008, 51, 350–356. [Google Scholar] [CrossRef]
- Kejnovsky, E.; Hobza, R.; Cermak, T.; Kubat, Z.; Vyskot, B. The role of repetitive DNA in structure and evolution of sex chromosomes in plants. Heredity 2009, 103, 533–541. [Google Scholar] [CrossRef]
- Kejnovský, E.; Michalovova, M.; Steflova, P.; Kejnovska, I.; Manzano, S.; Hobza, R.; Kubat, Z.; Kovarik, J.; Jamilena, M.; Vyskot, B. Expansion of microsatellites on evolutionary young Y chromosome. PLoS ONE 2013, 8, e45519. [Google Scholar] [CrossRef]
- Nadir, E.; Margalit, H.; Gallily, T.; Ben-Sasson, S. Microsatellite spreading in the human genome: Evolutionary mechanisms and structural implications. Proc. Natl. Acad. Sci. USA 1996, 93, 6470–6475. [Google Scholar] [CrossRef] [PubMed]
- Kelkar, Y.D.; Eckert, K.A.; Chiaromonte, F.; Makova, K.D. A matter of life or death: How microsatellites emerge in and vanish from the human genome. Genome Res. 2011, 21, 2038–2048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Temnykh, S.; DeClerk, G.; Lukashova, A.; Lipovich, L.; Cartinhour, S.; McCouh, S. Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): Frequency, length variation, transposon association, and genetic marker potential. Genome Res. 2001, 11, 1441–1452. [Google Scholar] [CrossRef] [PubMed]
- Morgante, M.; Hanafey, M.; Powell, W. Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat. Genet. 2002, 30, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Heule, C.; Salzburger, W.; Böhne, A. Genetics of sexual development: An evolutionary playground for fish. Genetics 2014, 196, 579–591. [Google Scholar] [CrossRef]
- Graves, J.A.M. The rise and fall of SRY. Trends Genet. 2002, 18, 259–264. [Google Scholar] [CrossRef]
- Yamato, K.T.; Ishizaki, K.; Fujisawa, M.; Okada, S.; Nakayama, S.; Fujishita, M.; Bando, H.; Yodoya, K.; Hayashi, K.; Bando, T.; et al. Gene organization of the liverworth Y chromosome reveals distinct sex chromosom evolution in a haploid system. Proc. Natl Acad Sci USA 2007, 104, 6472–6477. [Google Scholar] [CrossRef]
- Charlesworth, B.; Sniegowski, P.; Stephan, W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 1994, 371, 215–220. [Google Scholar] [CrossRef]
- Asakawa, S.; Abe, I.; Kudoh, Y.; Kishi, N.; Wang, Y.; Kubota, R.; Kudoh, J.; Kawasaki, K.; Minoshima, S.; Shimizu, N. Human BAC library: Construction and rapid screening. Gene 1997, 191, 69–79. [Google Scholar] [CrossRef]
- García-Cegarra, A.; Merlo, M.A.; Ponce, M.; Portela-Bens, S.; Cross, I.; Manchado, M.; Rebordinos, L. A preliminary genetic map in Solea senegalensis (Pleuronectiformes, Soleidae) using BAC-FISH and next-generation sequencing. Cytogenet. Genome Res. 2013, 141, 227–240. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Smit, A.F.A.; Hubley, R.; Green, P. RepeatMasker Open-4.0. (2013–2015). Available online: http://www.repeatmasker.org (accessed on 24 May 2018).
| Name of BAC | Gene Annotation | References |
|---|---|---|
| 36D3 | mcr4 | [16] |
| 5K5 | H1, H2a, H2b, H3, H4, slc1a3a, rx2, calr, eps15l1, klf2, ap1m1, tpm4, rab8a, cib3 | [16,23] |
| 10L10 | rx2, calr, eps15l1, klf2 | [16] |
| 10K23 | calr, eps15l1, otos, apod, arhgap21, opn3 | [15] |
| 73B7 | otos, apod, arhgap21 | [16] |
| 52C17 | vps41, esco1, rp1, sox17, mrpl15, lypla1, rgs20, oprk1, chmp5, fastkd3, mlrn, myom1 | [16] |
| 53B20 | lrrc32, mplz1, rabep1, vps37d, casr, gpr149, plch1, grsf1, rufy3, slc4a4, npffr2, daglb, spag9, hbad, hbb, rhbdf1, slc48a1, slc26a9, aanat, foxo6, pcbp4, disp3, bmp1, kansl3, antxr1, gfpt1, nfu1, c9orf78, med22, aak1 | This work |
| 16E16 | dmrt2, dmrt3 | [15,16] |
| 48K7 | dmrt2, dmrt3, dmrt1, c9orf117, kank1, fbp1 | [16] |
| 56H24 | akap9, hcn4, pip5k1a, znf687b, aqp10, hax1, ubap2l, c1orf43, tuft1 | [16] |
| 12D22 | H1, H2a, H2b, H3, H4, tmem70, ankrd45 | [23] |
| 48P7 | cyp8b1, rock1, ups14, thoc1, aqp1, crhr2, myl3, pth1r | [16] |
| 13G1 | wac | [16] |
| 1C2 | nbea | [16] |
| Genes | C. semilaevis | S. maximus | T. nigroviridis | G. aculeatus | X. maculatus | O. latipes | D. rerio | L. oculatus |
|---|---|---|---|---|---|---|---|---|
| gpr149 | 4 | 3 | 16 | I | 18 | 13 | 1 | LG14 |
| plch1 | 4 | 3 | 16 | I | 18 | 13 | 18 | LG14 |
| daglb | 8 | 18 | 2 | V | 10 | 19 | 12 | 1 |
| spag9 | 8 | 18 | 2 | V | 10 | 19 | 12 | LG10 |
| hbad | 8 | 18 | 3 | XI | 10 | 8 | 3 | LG13 |
| hbb | 8 | 18 | 3 | XI | 10 | 19 | 3 | LG13 |
| rhbdf1 | 17 | 18 | 3 | XI | 10 | 8 | 3 | LG13 |
| slc48a1 | 10 | 11 | 11 | XII | 1 | 7 | 23 | LG4 |
| slc26a9 | 1 | 11 | 9 | XII | 1 | 7 | 23 | LG3 |
| aanat | 9 | 8 | 3 | XI | 16 | 8 | 3 | LG13 |
| foxo6 | 13 | 7 | 1 | X | 13 | 11 | 19 | LG6 |
| pcbp4 | 1 | 6 | 9 | XII | 1 | 7 | 22 | LG5 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez, M.E.; Molina, B.; Merlo, M.A.; Arias-Pérez, A.; Portela-Bens, S.; García-Angulo, A.; Cross, I.; Liehr, T.; Rebordinos, L. Evolution of the Proto Sex-Chromosome in Solea senegalensis. Int. J. Mol. Sci. 2019, 20, 5111. https://doi.org/10.3390/ijms20205111
Rodríguez ME, Molina B, Merlo MA, Arias-Pérez A, Portela-Bens S, García-Angulo A, Cross I, Liehr T, Rebordinos L. Evolution of the Proto Sex-Chromosome in Solea senegalensis. International Journal of Molecular Sciences. 2019; 20(20):5111. https://doi.org/10.3390/ijms20205111
Chicago/Turabian StyleRodríguez, María Esther, Belén Molina, Manuel Alejandro Merlo, Alberto Arias-Pérez, Silvia Portela-Bens, Aglaya García-Angulo, Ismael Cross, Thomas Liehr, and Laureana Rebordinos. 2019. "Evolution of the Proto Sex-Chromosome in Solea senegalensis" International Journal of Molecular Sciences 20, no. 20: 5111. https://doi.org/10.3390/ijms20205111






