Adaptive Radiation from a Chromosomal Perspective: Evidence of Chromosome Set Stability in Cichlid Fishes (Cichlidae: Teleostei) from the Barombi Mbo Lake, Cameroon
Abstract
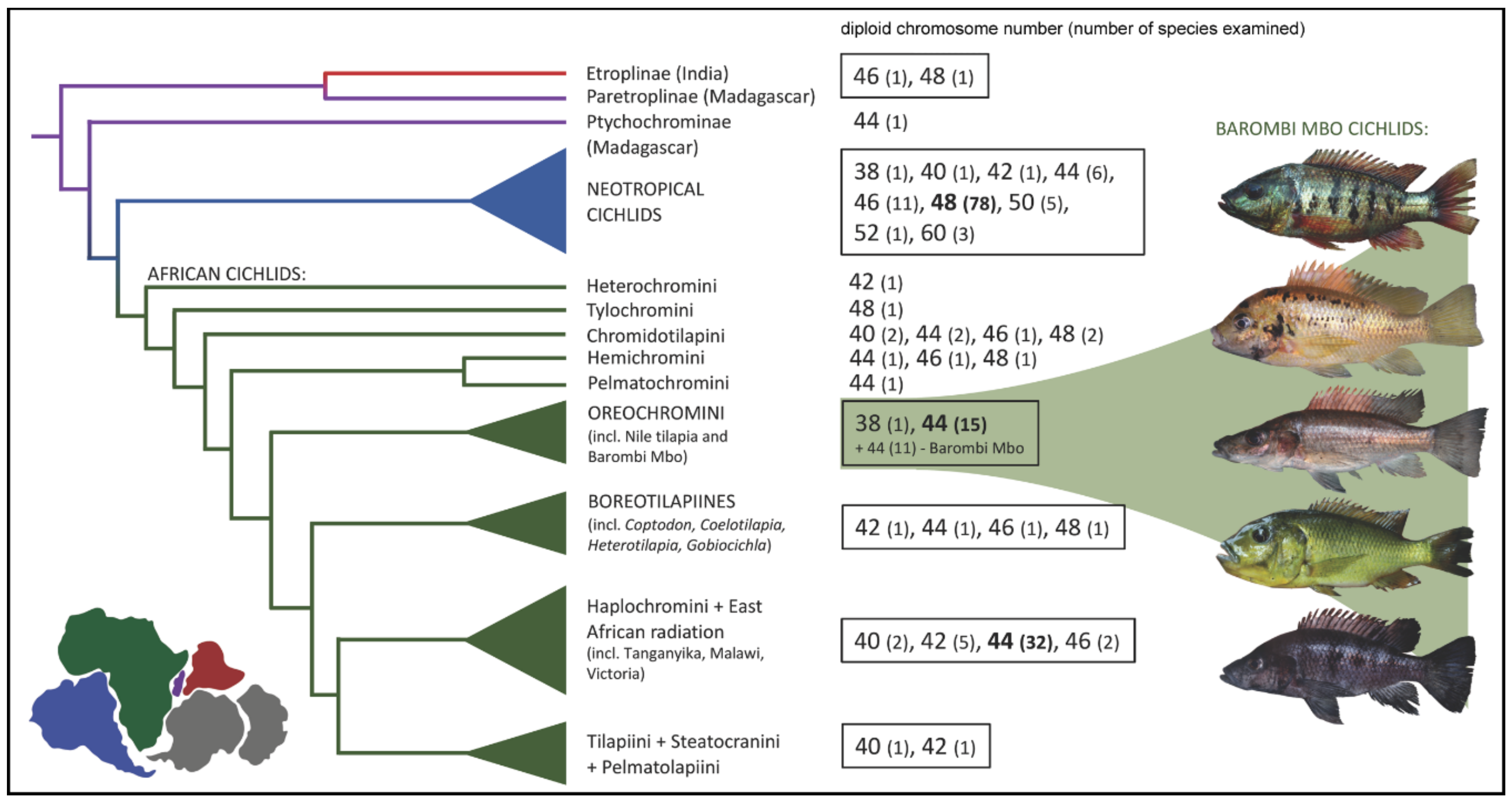
:1. Introduction
2. Results
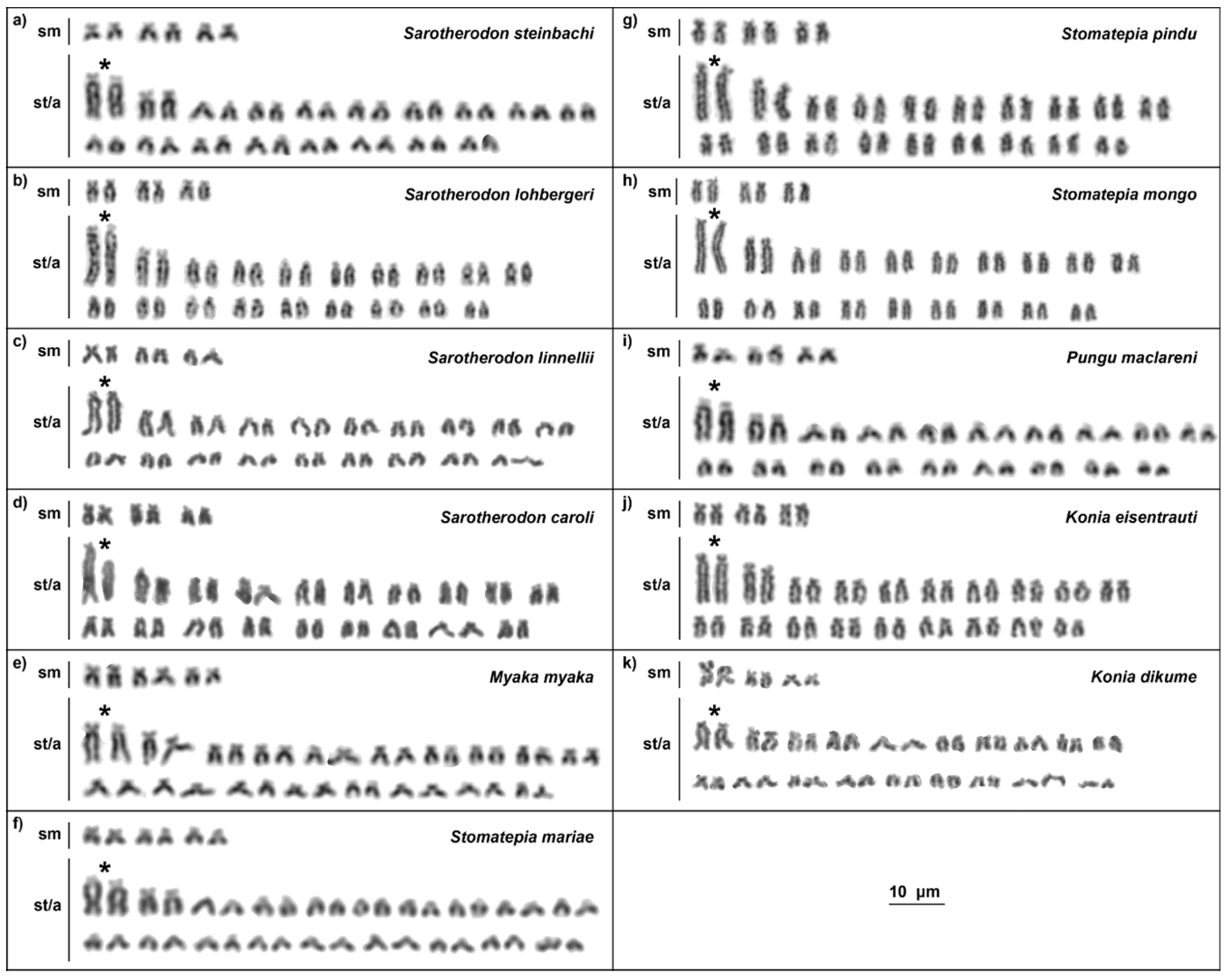
2.1. Karyotypes
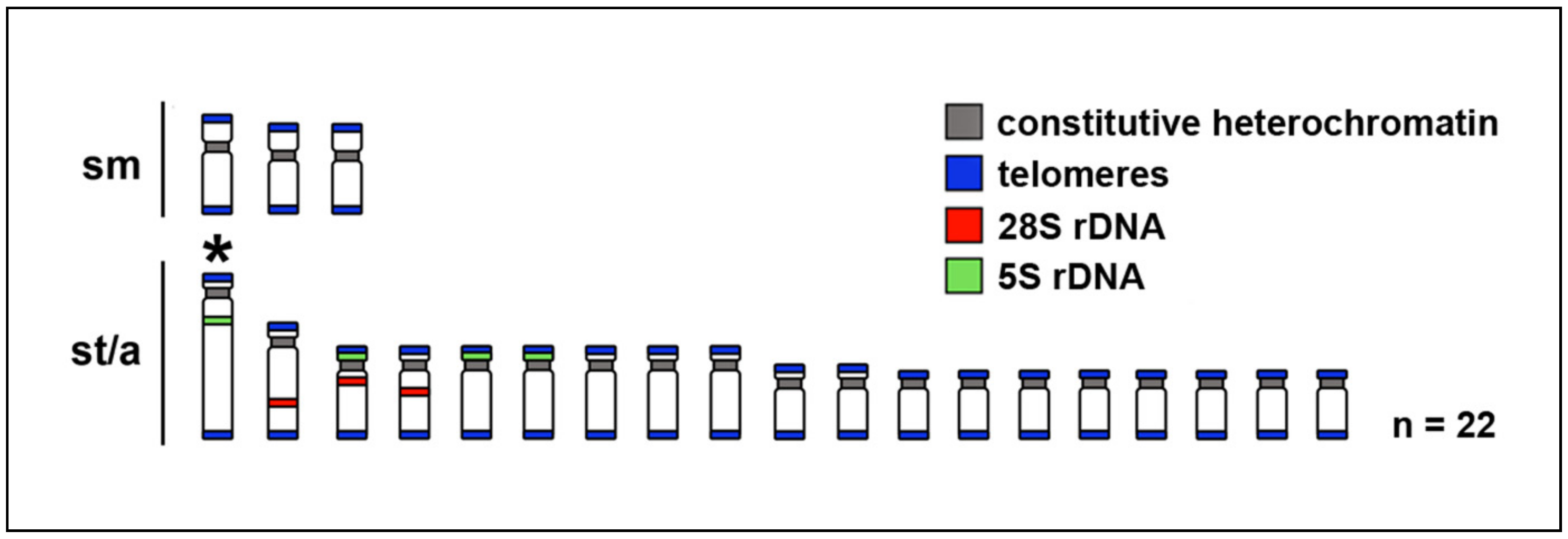
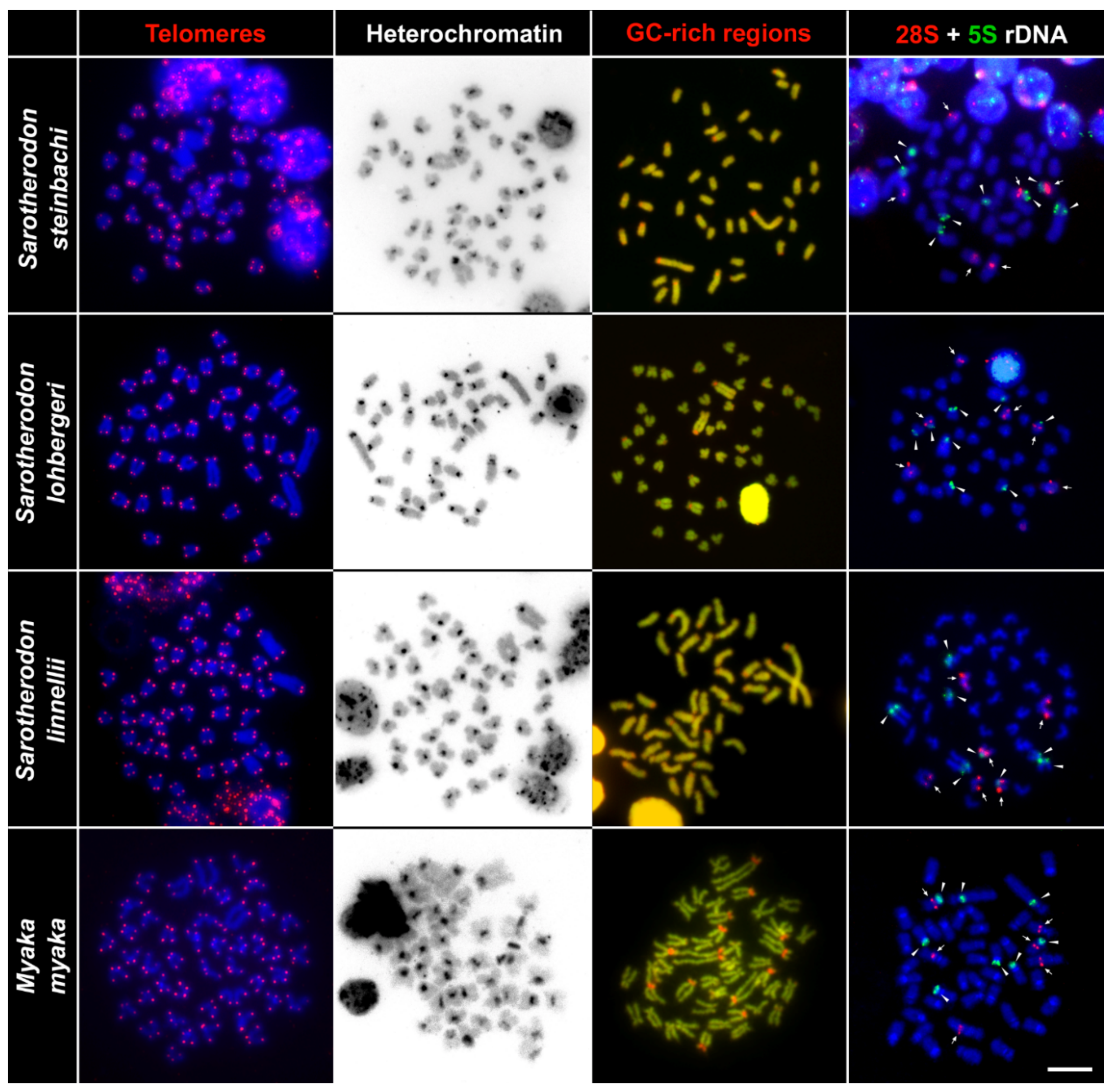
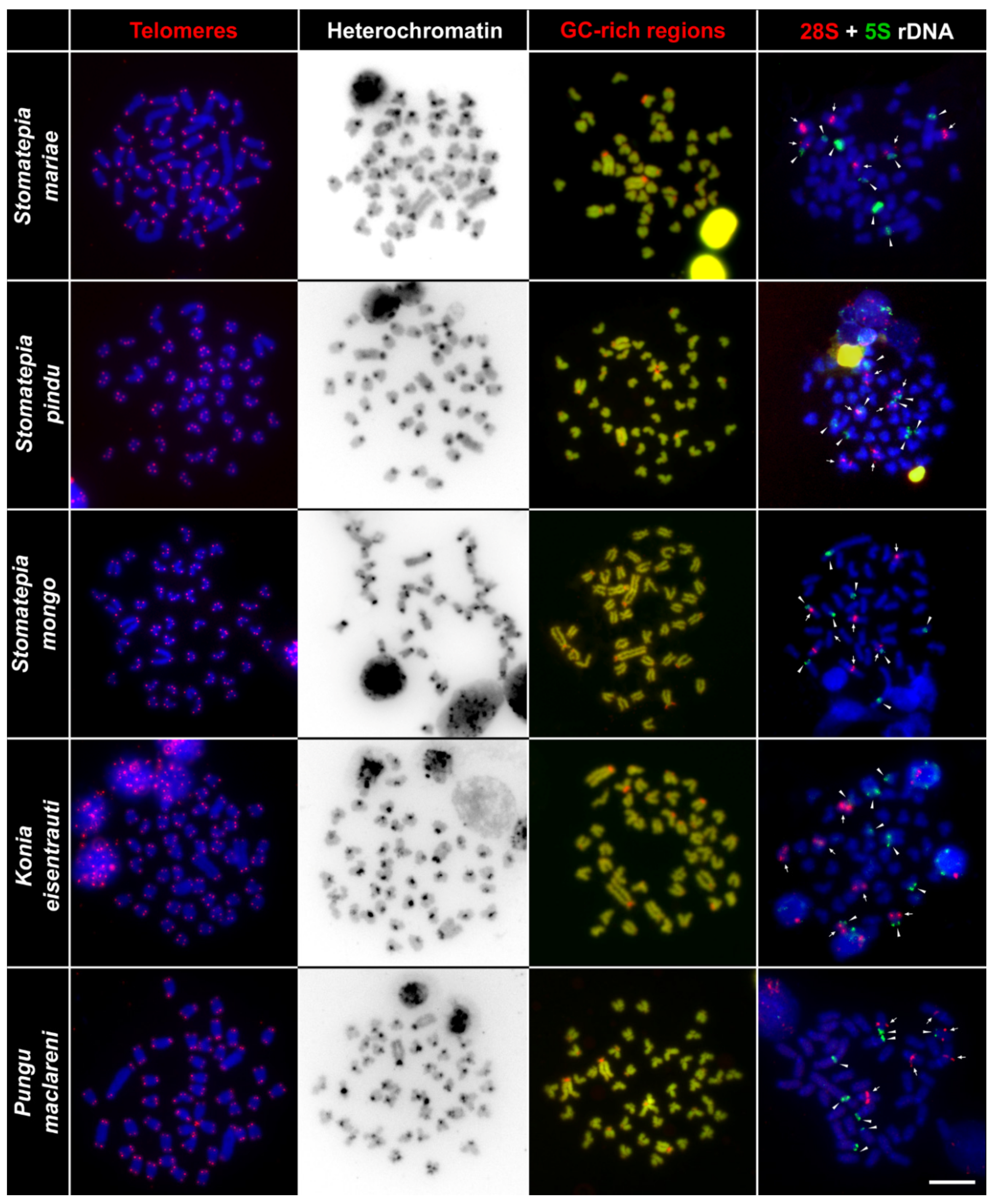
2.2. Telomere Mapping
2.3. C-Banding
2.4. CMA3/DAPI Staining
2.5. Fluorescence In Situ Hybridization with rDNA Genes
3. Discussion
4. Materials and Methods
4.1. Specimens
4.2. Chromosome Preparation and Giemsa Staining, CMA3 Staining, and C-Banding
4.3. Fluorescence in situ Hybridization with Telomeric Probe and rDNA Genes
4.4. Microscopy and Image Processing
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Seehausen, O. Process and pattern in cichlid radiations—Inferences for understanding unusually high rates of evolutionary diversification. New Phytol. 2015, 207, 304–312. [Google Scholar] [CrossRef] [PubMed]
- Salzburger, W. Understanding explosive diversification through cichlid fish genomics. Nat. Rev. Genet. 2018, 19, 705–717. [Google Scholar] [CrossRef] [PubMed]
- Fryer, G. Comparative aspects of adaptive radiation and speciation in Lake Baikal and the great rift lakes of Africa. Hydrobiologia 1991, 211, 137–146. [Google Scholar] [CrossRef]
- Goto, A.; Yokoyama, R.; Sideleva, V.G. Evolutionary diversification in freshwater sculpins (Cottoidea): A review of two major adaptive radiations. Environ. Biol. Fishes 2014, 98, 307–335. [Google Scholar] [CrossRef]
- Kornfield, I.; Carpenter, K. The Cyprinids of Lake Lanao, Philippines: Taxonomic Validity, Evolutionary Rates and Speciation Scenarios. In Evolution of Fish Species Flocks; Echelle, A.A., Kornfield, I., Eds.; University of Maine Press: Orono, MN, USA, 1984; pp. 69–84. ISBN 089-1-0105-80. [Google Scholar]
- De Graaf, M.; Dejen, E.; Osse, J.W.M.; Sibbing, F.A. Adaptive radiation of Lake Tana’s (Ethiopia) Labeobarbus species flock (Pisces, Cyprinidae). Mar. Freshw. Res. 2008, 59, 391–407. [Google Scholar] [CrossRef]
- Præbel, K.; Knudsen, R.; Siwertsson, A.; Karhunen, M.; Kahilainen, K.K.; Ovaskainen, O.; Østbye, K.; Peruzzi, S.; Fevolden, S.-E.; Amundsen, P.-A. Ecological speciation in postglacial European whitefish: Rapid adaptive radiations into the littoral, pelagic, and profundal lake habitats. Ecol. Evol. 2013, 3, 4970–4986. [Google Scholar] [CrossRef]
- Hudson, A.G.; Lundsgaard-Hansen, B.; Lucek, K.; Vonlanthen, P.; Seehausen, O. Managing cryptic biodiversity: Fine-scale intralacustrine speciation along a benthic gradient in Alpine whitefish (Coregonus spp.). Evol. Appl. 2017, 10, 251–266. [Google Scholar] [CrossRef]
- Feulner, P.G.D.; Seehausen, O. Genomic insights into the vulnerability of sympatric whitefish species flocks. Mol. Ecol. 2019, 28, 615–629. [Google Scholar] [CrossRef]
- Brooks, J.L. Speciation in ancient lakes (concluded). Q. Rev. Biol. 1950, 25, 131–176. [Google Scholar] [CrossRef]
- Martens, K. Speciation in ancient lakes. Trends Ecol. Evol. Amst. 1997, 12, 177–182. [Google Scholar] [CrossRef]
- Cristescu, M.E.; Adamowicz, S.J.; Vaillant, J.J.; Haffner, D.G. Ancient lakes revisited: From the ecology to the genetics of speciation. Mol. Ecol. 2010, 19, 4837–4851. [Google Scholar] [CrossRef] [PubMed]
- Von Rintelen, T.; von Rintelen, K.; Glaubrecht, M.; Schubart, C.; Herder, F. Aquatic Biodiversity Hotspots in Wallacea: The Species Flocks in the Ancient Lakes of Sulawesi, Indonesia. In Biotic Evolution and Environmental Change in Southeast Asia; Gower, D., Johnson, K., Ridchardson, J., Rosen, B., Rüber, L., Williams, S., Eds.; Cambridge University Press: Cambridge, UK, 2012; pp. 290–315. ISBN 978-1-1070-0130-5. [Google Scholar]
- Kottelat, M.; Xin-Luo, C. Revision of Yunnanilus with descriptions of a miniature species flock and six new species from China (Cypriniformes: Homalopteridae). Environ. Biol. Fish. 1988, 23, 65–94. [Google Scholar] [CrossRef]
- Parenti, L.R. Biogeography of the Andean Killifish Genus Orestias with Comments on the Species Flock Concept. In Evolution of Fish Species Flocks; Echelle, A.A., Kornfield, I., Eds.; University of Maine Press: Orono, MN, USA, 1984; pp. 85–92. ISBN 089-1-0105-80. [Google Scholar]
- Vila, I.; Scott, S.; Lam, N.; Méndez, A.M. Karyological and Morphological Analysis of Divergence among Species of the Killifish Genus Orestias (Teleostei: Cyprinodontidae) from the Southern Altiplano. In Origin and Phylogenetic Interrelationships of Teleosts; Nelson, J.S., Schultze, H.-P., Wilson, M.V.H., Eds.; Verlag Dr. Friedrich Pfeil: New York, NY, USA, 2010; pp. 471–480. ISBN 3-89937-107-0. [Google Scholar]
- Schliewen, U.K.; Tautz, D.; Pääbo, S. Sympatric speciation suggested by monophyly of crater lake cichlids. Nature 1994, 368, 629. [Google Scholar] [CrossRef] [PubMed]
- Schliewen, U.K.; Klee, B. Reticulate sympatric speciation in Cameroonian crater lake cichlids. Front. Zool. 2004, 1, 5. [Google Scholar] [CrossRef] [PubMed]
- Symonová, R.; Majtánová, Z.; Sember, A.; Staaks, G.B.O.; Bohlen, J.; Freyhof, J.; Rábová, M.; Ráb, P. Genome differentiation in a species pair of coregonine fishes: An extremely rapid speciation driven by stress-activated retrotransposons mediating extensive ribosomal DNA multiplications. BMC Evol. Biol. 2013, 13, 42. [Google Scholar] [CrossRef] [PubMed]
- Dion-Côté, A.-M.; Symonová, R.; Ráb, P.; Bernatchez, L. Reproductive isolation in a nascent species pair is associated with aneuploidy in hybrid offspring. Proc. R. Soc. B-Biol. Sci. 2015, 282, 20142862. [Google Scholar] [CrossRef] [PubMed]
- Dion-Côté, A.-M.; Symonová, R.; Lamaze, F.C.; Pelikánová, Š.; Ráb, P.; Bernatchez, L. Standing chromosomal variation in Lake Whitefish species pairs: The role of historical contingency and relevance for speciation. Mol. Ecol. 2017, 26, 178–192. [Google Scholar] [CrossRef] [PubMed]
- Ozouf-Costaz, C.; Coutanceau, J.P.; Bonillo, C.; Mercot, H.; Fermon, Y.; Guidi-Rontani, C. New insights into the chromosomal differentiation patterns among cichlids from Africa and Madagascar. Cybium 2017, 41, 35–43. [Google Scholar]
- Poletto, A.B.; Ferreira, I.A.; Cabral-de-Mello, D.C.; Nakajima, R.T.; Mazzuchelli, J.; Ribeiro, H.B.; Venere, P.C.; Nirchio, M.; Kocher, T.D.; Martins, C. Chromosome differentiation patterns during cichlid fish evolution. BMC Genet. 2010, 11, 50. [Google Scholar] [CrossRef]
- Nelson, J.S.; Grande, T.C.; Wilson, M.V.H. Fishes of the World, 5th ed.; Wiley: Hoboken, NJ, USA, 2016; ISBN 978-1-118-34233-6. [Google Scholar]
- Kocher, T.D. Adaptive evolution and explosive speciation: The cichlid fish model. Nat. Rev. Genet. 2004, 5, 288. [Google Scholar] [CrossRef]
- Genner, M.J.; Seehausen, O.; Lunt, D.H.; Joyce, D.A.; Shaw, P.W.; Carvalho, G.R.; Turner, G.F. Age of cichlids: New dates for ancient lake fish radiations. Mol. Biol. Evol. 2007, 24, 1269–1282. [Google Scholar] [CrossRef] [PubMed]
- Smith, W.L.; Chakrabarty, P.; Sparks, J.S. Phylogeny, taxonomy, and evolution of Neotropical cichlids (Teleostei: Cichlidae: Cichlinae). Cladistics 2008, 24, 625–641. [Google Scholar] [CrossRef]
- Schedel, F.D.B.; Musilová, Z.; Schliewen, U.K. East African cichlid lineages (Teleostei: Cichlidae) might be older than their ancient host lakes: New divergence estimates for the east African cichlid radiation. BMC Evol. Biol. 2019, 19, 94. [Google Scholar] [CrossRef] [PubMed]
- Matschiner, M.; Musilová, Z.; Barth, J.M.I.; Starostová, Z.; Salzburger, W.; Steel, M.; Bouckaert, R. Bayesian phylogenetic estimation of clade ages supports trans-Atlantic dispersal of cichlid fishes. Syst. Biol. 2017, 66, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Wagner, C.E.; Harmon, L.J.; Seehausen, O. Ecological opportunity and sexual selection together predict adaptive radiation. Nature 2012, 487, 366–369. [Google Scholar] [CrossRef] [PubMed]
- Musilová, Z.; Indermaur, A.; Nyom, A.R.B.; Tropek, R.; Martin, C.; Schliewen, U.K. Persistence of Stomatepia mongo, an endemic cichlid fish of the Barombi Mbo crater lake, southwestern Cameroon, with notes on its life history and behavior. Copeia 2014, 2014, 556–560. [Google Scholar] [CrossRef]
- Trewavas, E.; Green, J.; Corbet, S.A. Ecological studies on crater lakes in West Cameroon Fishes of Barombi Mbo. J. Fish Zool. 1972, 167, 41–95. [Google Scholar] [CrossRef]
- Baldo, L.; Pretus, J.L.; Riera, J.L.; Musilová, Z.; Bitja, A.N.; Salzburger, W. Convergence of gut microbiotas in the adaptive radiations of African cichlid fishes. ISME J. 2017, 11, 1975–1987. [Google Scholar] [CrossRef] [Green Version]
- Musilová, Z.; Indermaur, A.; Bitja-Nyom, A.R.; Omelchenko, D.; Kłodawska, M.; Albergati, L.; Remišová, K.; Salzburger, W. Evolution of visual sensory system in cichlid fishes from crater lake Barombi Mbo in Cameroon. Mol. Ecol. 2019. [Google Scholar] [CrossRef]
- Friedman, M.; Keck, B.P.; Dornburg, A.; Eytan, R.I.; Martin, C.H.; Hulsey, C.D.; Wainwright, P.C.; Near, T.J. Molecular and fossil evidence place the origin of cichlid fishes long after Gondwanan rifting. Proc. Biol. Sci. 2013, 280, 20131733. [Google Scholar] [CrossRef]
- Cornen, G.; Bande, Y.; Giresse, P.; Maley, J. The nature and chronostratigraphy of Quaternary pyroclastic accumulations from Lake Barombi Mbo (West-Cameroon). J. Volcanol. Geoth. Res. 1992, 51, 357–374. [Google Scholar] [CrossRef]
- Tchamabé, B.; Youmen, D.; Owona, S.; Issa; Ohba, T.; Németh, K.; Ngapna, M.; Asaah, A.; Aka, F.; Tanyileke, G.; et al. Eruptive history of the Barombi Mbo Maar, Cameroon Volcanic Line, Central Africa: Constraints from volcanic facies analysis. Open Geosci. 2014, 5, 480–496. [Google Scholar]
- Dunz, A.R.; Schliewen, U.K. Molecular phylogeny and revised classification of the haplotilapiine cichlid fishes formerly referred to as “Tilapia”. Mol. Phylogenet. Evol. 2013, 68, 64–80. [Google Scholar] [CrossRef] [PubMed]
- Dominey, W.J.; Snyder, A.M. Kleptoparasitism of freshwater crabs by cichlid fishes endemic to Lake Barombi Mbo, Cameroon, West Africa. Environ. Biol. Fish. 1988, 22, 155. [Google Scholar] [CrossRef]
- Richards, E.J.; Poelstra, J.W.; Martin, C.H. Don’t throw out the sympatric speciation with the crater lake water: Fine-scale investigation of introgression provides equivocal support for causal role of secondary gene flow in one of the clearest examples of sympatric speciation. Evol. Lett. 2018, 2, 524–540. [Google Scholar] [CrossRef] [PubMed]
- Coyne, J.A.; Orr, H.A. Speciation; Oxford University Press: Oxford, UK; New York, NY, USA, 2004; ISBN 978-0-87893-089-0. [Google Scholar]
- Dobzhansky, T. Genetics and the Origin of Species; Columbia University Press: New York, NY, USA, 1937; ISBN 978-0-231-05475-1. [Google Scholar]
- Rieseberg, L.H. Chromosomal rearrangements and speciation. Trends Ecol. Evol. 2001, 16, 351–358. [Google Scholar] [CrossRef]
- Navarro, A.; Barton, N.H. Chromosomal speciation and molecular divergence-accelerated evolution in rearranged chromosomes. Science 2003, 300, 321–324. [Google Scholar] [CrossRef]
- Searle, J.B.; Polly, P.D.; Zima, J. Shrews, Chromosomes and Speciation; Cambridge University Press: Cambridge, UK, 2019; Volume 6, ISBN 978-0-511-89553-1. [Google Scholar]
- Potter, S.; Bragg, J.G.; Blom, M.P.K.; Deakin, J.E.; Kirkpatrick, M.; Eldridge, M.D.B.; Moritz, C. Chromosomal speciation in the genomics era: Disentangling phylogenetic evolution of rock-wallabies. Front. Genet. 2017, 8, 10. [Google Scholar] [CrossRef]
- Livingstone, K.; Rieseberg, L. Chromosomal evolution and speciation: A recombination-based approach. New Phytol. 2004, 161, 107–112. [Google Scholar] [CrossRef]
- Kandul, N.P.; Lukhtanov, V.A.; Pierce, N.E. Karyotypic diversity and speciation in Agrodiaetus butterflies. Evolution 2007, 61, 546–559. [Google Scholar] [CrossRef]
- Supiwong, W.; Pinthong, K.; Seetapan, K.; Saenjundaeng, P.; Bertollo, L.A.C.; de Oliveira, E.A.; Yano, C.F.; Liehr, T.; Phimphan, S.; Tanomtong, A.; et al. Karyotype diversity and evolutionary trends in the Asian swamp eel Monopterus albus (Synbranchiformes, Synbranchidae): A case of chromosomal speciation? BMC Evol. Biol. 2019, 19, 73. [Google Scholar] [CrossRef] [PubMed]
- Bertollo, L.A.C. Chromosome Evolution in the Neotropical Erythrinidae Fish Family: An Overview. In Fish Cytogenetics; Pisano, E., Ozouf-Costaz, C., Foresti, F., Kapoor, B.G., Eds.; Science Publishers: Enfield, NH, USA, 2007; pp. 195–211. ISBN 978-1-5780-8330-5. [Google Scholar]
- Conte, M.A.; Joshi, R.; Moore, E.C.; Nandamuri, S.P.; Gammerdinger, W.J.; Roberts, R.B.; Carleton, K.L.; Lien, S.; Kocher, T.D. Chromosome-scale assemblies reveal the structural evolution of African cichlid genomes. Gigascience 2019, 8, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Arai, R. Fish Karyotypes: A Check List, 2011 ed.; Springer: Tokyo, Japan; New York, NY, USA, 2011; ISBN 978-4-431-53876-9. [Google Scholar]
- Schneider, C.H.; Gross, M.C.; Terencio, M.L.; Artoni, R.F.; Vicari, M.R.; Martins, C.; Feldberg, E. Chromosomal evolution of neotropical cichlids: The role of repetitive DNA sequences in the organization and structure of karyotype. Rev. Fish Biol. Fisher. 2013, 23, 201–214. [Google Scholar] [CrossRef]
- Marescalchi, O. Karyotype and mitochondrial 16S gene characterizations in seven South American Cichlasomatini species (Perciformes, Cichlidae). J. Zool. Syst. Evol. Res. 2005, 43, 22–28. [Google Scholar] [CrossRef]
- Hodaňová, L.; Kalous, L.; Musilová, Z. Comparative cytogenetics of Neotropical cichlid fishes (Nannacara, Ivanacara and Cleithracara) indicates evolutionary reduction of diploid chromosome numbers. Comp. Cytogenet. 2014, 8, 169–183. [Google Scholar]
- Krajáková, L.; Musilová, Z.; Kalous, L. Karyotype Characterizations in Two South American Cichlasomatini Species (Perciformes, Cichlidae). In Proceedings of the Workshop on Animal Biodiversity, Jevany, Czech Republic, 7 July 2010; pp. 78–80. [Google Scholar]
- Valente, G.T.; de Andrade Vitorino, C.; Cabral-de-Mello, D.C.; Oliveira, C.; Souza, I.L.; Martins, C.; Venere, P.C. Comparative cytogenetics of ten species of cichlid fishes (Teleostei, Cichlidae) from the Araguaia River system, Brazil, by conventional cytogenetic methods. Comp. Cytogenet. 2012, 6, 163–181. [Google Scholar]
- Oliveira, C.; Wright, J.M. Molecular cytogenetic analysis of heterochromatin in the chromosomes of tilapia, Oreochromis niloticus (Teleostei: Cichlidae). Chromosome Res. 1998, 6, 205–211. [Google Scholar] [CrossRef]
- Kornfield, I.; Smith, P.F. African cichlid fishes: Model systems for evolutionary biology. Annu. Rev. Ecol. Syst. 2000, 31, 163–196. [Google Scholar] [CrossRef]
- Green, J.; Corbet, S.A.; Betney, E. Ecological studies on crater lakes in West Cameroon the blood of endemic cichlids in Barombi Mbo in relation to stratification and their feeding habits. J. Zool. 1973, 170, 299–308. [Google Scholar] [CrossRef]
- Thieme, M.L.; Abell, R.; Stiassny, M.L.J.; Skelton, P. Freshwater Ecoregions of Africa and Madagascar: A Conservation Assessment; Island Press: Washington, DC, USA, 2005; ISBN 978-1-55963-365-9. [Google Scholar]
- Malinsky, M.; Challis, R.J.; Tyers, A.M.; Schiffels, S.; Terai, Y.; Ngatunga, B.P.; Miska, E.A.; Durbin, R.; Genner, M.J.; Turner, G.F. Genomic islands of speciation separate cichlid ecomorphs in an East African crater lake. Science 2015, 350, 1493–1498. [Google Scholar] [CrossRef] [Green Version]
- Schluter, D.; McPhail, J.D. Ecological character displacement and speciation in sticklebacks. Am. Nat. 1992, 140, 85–108. [Google Scholar] [CrossRef] [PubMed]
- Danley, P.D.; Kocher, T.D. Speciation in rapidly diverging systems: Lessons from Lake Malawi. Mol. Ecol. 2001, 10, 1075–1086. [Google Scholar] [CrossRef] [PubMed]
- Skúlason, S.; Snorrason, S.S.; Noakes, D.L.G.; Ferguson, M.M.; Malmquist, H.J. Segregation in spawning and early life history among polymorphic Arctic charr, Salvelinus alpinus, in Thingvallavatn, Iceland. J. Fish Biol. 1989, 35, 225–232. [Google Scholar] [CrossRef]
- Volff, J.N. Genome evolution and biodiversity in teleost fish. Heredity 2005, 94, 280–294. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, I.A.; Poletto, A.B.; Kocher, T.D.; Mota-Velasco, J.C.; Penman, D.J.; Martins, C. Chromosome evolution in African cichlid fish: Contributions from the physical mapping of repeated DNAs. Cytogenet. Genome Res. 2010, 129, 314–322. [Google Scholar] [CrossRef] [PubMed]
- Valente, G.T.; Mazzuchelli, J.; Ferreira, I.A.; Poletto, A.B.; Fantinatti, B.E.A.; Martins, C. Cytogenetic mapping of the retroelements Rex1, Rex3 and Rex6 among cichlid fish: New insights on the chromosomal distribution of transposable elements. Cytogenet. Genome Res. 2011, 133, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Mazzuchelli, J.; Kocher, T.D.; Yang, F.; Martins, C. Integrating cytogenetics and genomics in comparative evolutionary studies of cichlid fish. BMC Genom. 2012, 13, 463. [Google Scholar] [CrossRef]
- Nakajima, R.T.; Cabral-de-Mello, D.C.; Valente, G.T.; Venere, P.C.; Martins, C. Evolutionary dynamics of rRNA gene clusters in cichlid fish. BMC Evol. Biol. 2012, 12, 198. [Google Scholar] [CrossRef]
- Clark, F.E.; Conte, M.A.; Ferreira-Bravo, I.A.; Poletto, A.B.; Martins, C.; Kocher, T.D. Dynamic sequence evolution of a sex-associated B chromosome in Lake Malawi cichlid fish. J. Hered. 2017, 108, 53–62. [Google Scholar] [CrossRef]
- Clark, F.E.; Conte, M.A.; Kocher, T.D. Genomic Characterization of a B chromosome in Lake Malawi cichlid fishes. Genes 2018, 9, 610. [Google Scholar] [CrossRef]
- Feldberg, E.; Ivan, J.; Porto, R.; Antonio, L.; Bertollo, C. Chromosomal Changes and Adaptation of Cichlid Fishes during Evolution. In Fish Adaptation; Val, A.L., Kapoor, B.G., Eds.; Science Publishers: Enfield, NH, USA, 2003; pp. 285–308. ISBN 978-1-5780-8249-0. [Google Scholar]
- Sochorová, J.; Garcia, S.; Gálvez, F.; Symonová, R.; Kovařík, A. Evolutionary trends in animal ribosomal DNA loci: Introduction to a new online database. Chromosoma 2018, 127, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Gornung, E. Twenty years of physical mapping of major ribosomal RNA genes across the Teleosts: A review of research. Cytogenet. Genome Res. 2013, 141, 90–102. [Google Scholar] [CrossRef] [PubMed]
- Cioffi, M.B.; Bertollo, L.A.C. Chromosomal distribution and evolution of repetitive DNAs in fish. Genome Dyn. 2012, 7, 197–221. [Google Scholar] [PubMed]
- Martins, C.; Wasko, A.P.; Oliveira, C.; Porto-Foresti, F.; Parise-Maltempi, P.P.; Wright, J.M.; Foresti, F. Dynamics of 5S rDNA in the tilapia (Oreochromis niloticus) genome: Repeat units, inverted sequences, pseudogenes and chromosome loci. Cytogenet. Genome Res. 2002, 98, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Symonová, R.; Majtánová, Z.; Arias-Rodriguez, L.; Mořkovský, L.; Kořínková, T.; Cavin, L.; Pokorná, M.J.; Doležálková, M.; Flajšhans, M.; Normandeau, E.; et al. Genome compositional organization in gars shows more similarities to mammals than to oher ray-finned fish. J. Exp. Zool. B Mol. Dev. Evol. 2017, 328, 607–619. [Google Scholar] [CrossRef] [PubMed]
- Escobar, J.S.; Glémin, S.; Galtier, N. GC-biased gene conversion impacts ribosomal DNA evolution in vertebrates, angiosperms, and other eukaryotes. Mol. Biol. Evol. 2011, 28, 2561–2575. [Google Scholar] [CrossRef] [PubMed]
- Bertollo, L.A.C.; Cioffi, M.B.; Moreira-Filho, O. Direct Chromosome Preparation from Freshwater Teleost Fishes. In Fish Cytogenetic Techniques; Ozouf-Costaz, C., Pisano, E., Foresti, F., Foresti de Almeida-Toledo, L., Eds.; CRC Press, Inc.: Enfield, NH, USA, 2015; pp. 21–26. ISBN 978-0-3673-7755-7. [Google Scholar]
- Völker, M.; Ráb, P. Direct Chromosome Preparation from Regenerating Fin Tissue. In Fish Cytogenetic Techniques; Ozouf-Costaz, C., Pisano, E., Foresti, F., Foresti de Almeida-Toledo, L., Eds.; CRC Press, Inc.: Enfield, NH, USA, 2015; pp. 37–41. ISBN 978-0-3673-7755-7. [Google Scholar]
- Sumner, A.T. A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 1972, 75, 304–306. [Google Scholar] [CrossRef]
- Pokorná, M.; Rens, W.; Rovatsos, M.; Kratochvíl, L. A ZZ/ZW sex chromosome system in the thick-tailed gecko (Underwoodisaurus milii; Squamata: Gekkota: Carphodactylidae), a member of the ancient gecko lineage. Cytogenet. Genome Res. 2014, 142, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Sola, L.; Rossi, A.R.; Iaselli, V.; Rasch, E.M.; Monaco, P.J. Cytogenetics of bisexual/unisexual species of Poecilia. II. Analysis of heterochromatin and nucleolar organizer regions in Poecilia mexicana mexicana by C-banding and DAPI, quinacrine, chromomycin A3, and silver staining. Cytogenet. Cell Genet. 1992, 60, 229–235. [Google Scholar] [CrossRef]
- Zhang, Q.; Cooper, R.K.; Tiersch, T.R. Chromosomal location of the 28S ribosomal RNA gene of channel catfish by in situ polymerase chain reaction. J. Fish Biol. 2000, 56, 388–397. [Google Scholar] [CrossRef]
- Komiya, H.; Takemura, S. Nucleotide sequence of 5S ribosomal RNA from rainbow trout (Salmo gairdnerii) liver. J. Biochem. 1979, 86, 1067–1080. [Google Scholar] [CrossRef] [PubMed]
- Majtánová, Z.; Moy, K.G.; Unmack, P.J.; Ráb, P.; Ezaz, T. Characterization of the karyotype and accumulation of repetitive sequences in Australian Darling hardyhead Craterocephalus amniculus (Atheriniformes, Teleostei). PeerJ 2019, 7, e7347. [Google Scholar] [CrossRef] [PubMed]
- Symonová, R.; Sember, A.; Majtánová, Z.; Ráb, P. Characterization of Fish Genomes by GISH and CGH. In Fish Cytogenetic Techniques; Ozouf-Costaz, C., Pisano, E., Foresti, F., de Almeida, L., Eds.; CRC Press: Boca Raton, FL, USA, 2015; pp. 118–131. ISBN 978-1-4822-1198-6. [Google Scholar]
- Levan, A.; Fredga, K.; Sandberg, A.A. Nomenclature for centromeric position on chromosomes. Hereditas 1964, 52, 201–220. [Google Scholar] [CrossRef]
| Species | No. of Individuals | No. of Cells Examined | ||||
|---|---|---|---|---|---|---|
| Giemsa | C-Banding | CMA3 | Telomeres | FISH | ||
| Konia eisentrauti | 1♂, 2♀ | 35 | 30 | 30 | 30 | 15 |
| Konia dikume | 1 | 8 | n/a | n/a | n/a | n/a |
| Myaka myaka | 4♂ | 35 | 30 | 30 | 30 | 16 |
| Pungu maclareni | 1♀ | 35 | 30 | 30 | 30 | 15 |
| Sarotherodon steinbachi | 3 juveniles | 60 | 30 | 30 | 30 | 12 |
| Sarotherodon lohbergeri | 2 juveniles | 35 | 30 | 30 | 30 | 15 |
| Sarotherodon linnellii | 2♀, 1 juveniles | 45 | 30 | 30 | 30 | 18 |
| Sarotherodon caroli | 2 | 10 | n/a | n/a | n/a | n/a |
| Stomatepia mariae | 4 juveniles | 35 | 30 | 30 | 30 | 10 |
| Stomatepia pindu | 3 juveniles | 40 | 30 | 30 | 30 | 13 |
| Stomatepia mongo | 3 juveniles | 30 | 30 | 30 | 30 | 12 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Majtánová, Z.; Indermaur, A.; Bitja Nyom, A.R.; Ráb, P.; Musilova, Z. Adaptive Radiation from a Chromosomal Perspective: Evidence of Chromosome Set Stability in Cichlid Fishes (Cichlidae: Teleostei) from the Barombi Mbo Lake, Cameroon. Int. J. Mol. Sci. 2019, 20, 4994. https://doi.org/10.3390/ijms20204994
Majtánová Z, Indermaur A, Bitja Nyom AR, Ráb P, Musilova Z. Adaptive Radiation from a Chromosomal Perspective: Evidence of Chromosome Set Stability in Cichlid Fishes (Cichlidae: Teleostei) from the Barombi Mbo Lake, Cameroon. International Journal of Molecular Sciences. 2019; 20(20):4994. https://doi.org/10.3390/ijms20204994
Chicago/Turabian StyleMajtánová, Zuzana, Adrian Indermaur, Arnold Roger Bitja Nyom, Petr Ráb, and Zuzana Musilova. 2019. "Adaptive Radiation from a Chromosomal Perspective: Evidence of Chromosome Set Stability in Cichlid Fishes (Cichlidae: Teleostei) from the Barombi Mbo Lake, Cameroon" International Journal of Molecular Sciences 20, no. 20: 4994. https://doi.org/10.3390/ijms20204994
APA StyleMajtánová, Z., Indermaur, A., Bitja Nyom, A. R., Ráb, P., & Musilova, Z. (2019). Adaptive Radiation from a Chromosomal Perspective: Evidence of Chromosome Set Stability in Cichlid Fishes (Cichlidae: Teleostei) from the Barombi Mbo Lake, Cameroon. International Journal of Molecular Sciences, 20(20), 4994. https://doi.org/10.3390/ijms20204994





