Subcellular Localization Signals of bHLH-PAS Proteins: Their Significance, Current State of Knowledge and Future Perspectives
Abstract
1. Introduction
2. Regulation of the Subcellular Localization of Class I bHLH-PAS Transcription Factors
2.1. AHR Localization Regulation
2.2. HIF-1-3α Localization Regulation
2.3. SIM1-2 Localization Regulation
2.4. CLOCK Localization Regulation
2.5. NPAS1-4 Localization Regulation
3. Regulation of the Subcellular Localization of Class II of bHLH-PAS Transcription Factors: ARNT1-4
4. Regulation of Subcellular Localization of Drosophila melanogaster bHLH-PAS Transcription Factors
5. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 20E | 20-hydroxyecdysone |
| bHLH-PAS | basic helix-loop-helix/ Period-ARNT-Single minded |
| AHRHIF | Aryl hydrocarbon ReceptorHypoxia Inducible Factor |
| ARNT | Aryl Hydrocarbon Nuclear Translocator |
| ARNTL | Aryl hydrocarbon receptor nuclear translocator-like protein |
| cNLS | classical NLS |
| CRM1 | Chromosome region maintenance 1 protein homolog |
| CRY | Cryptochrome |
| ER | Endoplasmatic reticulum |
| ERK | Extracellular signal-regulated kinase |
| GCE | germ cell-expressed protein |
| HRE | Hypoxia Response Element |
| HSCs | hematopoietic stem cells |
| JH | juvenile hormone |
| LMB | Leptomycin B |
| MET | Methoprene tolerant protein |
| NES | nuclear export signal |
| NLS | nuclear localization signal |
| NPAS | Neuronal PAS domain-containing protein |
| PAC | C-terminal to PAS domain |
| PER | Period protein |
| pVHL | von Hippel–Lindau tumor suppressor |
| RPD | repression domain |
| SIM | Single-minded homolog protein |
| SIMA | similar |
| TAD | transcription activation domain |
| TF | transcription factor |
| TRH | Trachealess protein |
References
- Crews, S.T. Control of cell lineage-specific development and transcription by bHLH-PAS proteins. Genes Dev. 1998, 12, 607–620. [Google Scholar] [CrossRef] [PubMed]
- Kewley, R.J.; Whitelaw, M.L.; Chapman-Smith, A. The mammalian basic helix–loop–helix/PAS family of transcriptional regulators. Int. J. Biochem. Cell Biol. 2004, 36, 189–204. [Google Scholar] [CrossRef]
- Furness, S.G.B.; Lees, M.J.; Whitelaw, M.L. The dioxin (aryl hydrocarbon) receptor as a model for adaptive responses of bHLH/PAS transcription factors. FEBS Lett. 2007, 581, 3616–3625. [Google Scholar] [CrossRef] [PubMed]
- Nebert, D.W. Aryl hydrocarbon receptor (AHR): “pioneer member” of the basic-helix/loop/helix per-Arnt-sim (bHLH/PAS) family of “sensors” of foreign and endogenous signals. Prog. Lipid Res. 2017, 67, 38–57. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Duan, X.; Jiang, H.; Sun, Y.; Tang, Y.; Yuan, Z.; Guo, J.; Liang, W.; Chen, L.; Yin, J.; et al. Genome-Wide Analysis of Basic/Helix-Loop-Helix Transcription Factor Family in Rice and Arabidopsis. PLANT Physiol. 2006, 141, 1167–1184. [Google Scholar] [CrossRef]
- Ponting, C.P.; Aravind, L. PAS: A multifunctional domain family comes to light. Curr. Biol. 1997, 7, R674–R677. [Google Scholar] [CrossRef]
- Henry, J.T.; Crosson, S. Ligand-binding PAS domains in a genomic, cellular, and structural context. Annu. Rev. Microbiol. 2011, 65, 261–286. [Google Scholar] [CrossRef]
- Hefti, M.H.; Françoijs, K.-J.; De Vries, S.C.; Dixon, R.; Vervoort, J. The PAS fold. Eur. J. Biochem. 2004, 271, 1198–1208. [Google Scholar] [CrossRef]
- Wu, D.; Rastinejad, F. Structural characterization of mammalian bHLH-PAS transcription factors. Curr. Opin. Struct. Biol. 2017, 43, 1–9. [Google Scholar] [CrossRef]
- Partch, C.L.; Gardner, K.H. Coactivator recruitment: A new role for PAS domains in transcriptional regulation by the bHLH-PAS family. J. Cell Physiol. 2010, 223, 553–557. [Google Scholar] [CrossRef]
- Fribourgh, J.L.; Partch, C.L. Assembly and function of bHLH-PAS complexes. Proc. Natl. Acad. Sci. USA 2017, 114, 5330–5332. [Google Scholar] [CrossRef] [PubMed]
- Kallio, P.J.; Okamoto, K.; Brien, S.O.; Carrero, P.; Makino, Y.; Tanaka, H.; Poellinger, L. Signal transduction in hypoxic cells: Inducible nuclear translocation and recruitment of the CBP/p300 coactivator by the hypoxia-inducible factor-1 α. EMBO J. 1998, 17, 6573–6586. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-H.; Hannink, M. Molecular mechanisms that regulate transcription factor localization suggest new targets for drug development. Adv. Drug Deliv. Rev. 2003, 55, 717–731. [Google Scholar] [CrossRef]
- Kumar, S.; Saradhi, M.; Chaturvedi, N.K.; Tyagi, R.K. Intracellular localization and nucleocytoplasmic trafficking of steroid receptors: An overview. Mol. Cell Endocrinol. 2006, 246, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Sorokin, A.V.; Kim, E.R.; Ovchinnikov, L.P. Nucleocytoplasmic transport of proteins. Biochemistry (Mosc) 2007, 72, 1439–1457. [Google Scholar] [CrossRef] [PubMed]
- Lange, A.; Mills, R.E.; Lange, C.J.; Stewart, M.; Devine, S.E.; Corbett, A.H. Classical Nuclear Localization Signals: Definition, Function, and Interaction with Importin α. J. Biol. Chem. 2007, 282, 5101–5105. [Google Scholar] [CrossRef] [PubMed]
- Gerace, L. Nuclear Export Signals and the Fast Track to the Cytoplasm. Cell 1995, 82, 341–344. [Google Scholar] [CrossRef][Green Version]
- Kudo, N.; Matsumori, N.; Taoka, H.; Fujiwara, D.; Schreiner, E.P.; Wolff, B.; Yoshida, M.; Horinouchi, S. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. USA 1999, 96, 9112–9117. [Google Scholar] [CrossRef]
- Greb-Markiewicz, B.; Orlowski, M.; Dobrucki, J.; Ozyhar, A. Sequences that direct subcellular traffic of the Drosophila methoprene-tolerant protein (MET) are located predominantly in the PAS domains. Mol. Cell Endocrinol. 2011, 345, 16–26. [Google Scholar] [CrossRef]
- Greb-Markiewicz, B.; Sadowska, D.; Surgut, N.; Godlewski, J.; Zarębski, M.; Ożyhar, A. Mapping of the Sequences Directing Localization of the Drosophila Germ Cell-Expressed Protein (GCE). PLoS ONE 2015, 10, e0133307. [Google Scholar] [CrossRef][Green Version]
- Greb-Markiewicz, B.; Zarębski, M.; Ożyhar, A. Multiple sequences orchestrate subcellular trafficking of neuronal PAS domain-containing protein 4 (NPAS4). J. Biol. Chem. 2018, 293, 11255–11270. [Google Scholar] [CrossRef] [PubMed]
- Kosugi, S.; Hasebe, M.; Tomita, M.; Yanagawa, H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. USA 2009, 106, 10171–10176. [Google Scholar] [CrossRef] [PubMed]
- Nguyen Ba, A.N.; Pogoutse, A.; Provart, N.; Moses, A.M. NLStradamus: A simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinform. 2009, 10, 202. [Google Scholar] [CrossRef] [PubMed]
- Brameier, M.; Krings, A.; MacCallum, R.M. NucPred--predicting nuclear localization of proteins. Bioinformatics 2007, 23, 1159–1160. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.-R.; Mondal, A.; Liu, R.; Hu, J. Minimalist ensemble algorithms for genome-wide protein localization prediction. BMC Bioinform. 2012, 13, 157. [Google Scholar] [CrossRef]
- La Cour, T.; Kiemer, L.; Mølgaard, A.; Gupta, R.; Skriver, K.; Brunak, S. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 2004, 17, 527–536. [Google Scholar] [CrossRef]
- Brar, S.S.; Watson, M.; Diaz, M. Activation-induced cytosine deaminase (AID) is actively exported out of the nucleus but retained by the induction of DNA breaks. J. Biol. Chem. 2004, 279, 26395–26401. [Google Scholar] [CrossRef]
- Xu, D.; Marquis, K.; Pei, J.; Fu, S.-C.; Cağatay, T.; Grishin, N.V.; Chook, Y.M. LocNES: A computational tool for locating classical NESs in CRM1 cargo proteins. Bioinformatics 2015, 31, 1357–1365. [Google Scholar] [CrossRef]
- Dinkel, H.; Michael, S.; Weatheritt, R.J.; Davey, N.E.; Van Roey, K.; Altenberg, B.; Toedt, G.; Uyar, B.; Seiler, M.; Budd, A.; et al. ELM--the database of eukaryotic linear motifs. Nucleic Acids Res. 2012, 40, 242–251. [Google Scholar] [CrossRef]
- Dinkel, H.; Van Roey, K.; Michael, S.; Davey, N.E.; Weatheritt, R.J.; Born, D.; Speck, T.; Krüger, D.; Grebnev, G.; Kuban, M.; et al. The eukaryotic linear motif resource ELM: 10 years and counting. Nucleic Acids Res. 2014, 42, D259–D266. [Google Scholar] [CrossRef]
- Gouw, M.; Michael, S.; Sámano-Sánchez, H.; Kumar, M.; Zeke, A.; Lang, B.; Bely, B.; Chemes, L.B.; Davey, N.E.; Deng, Z.; et al. The eukaryotic linear motif resource—2018 update. Nucleic Acids Res. 2018, 46, D428–D434. [Google Scholar] [CrossRef] [PubMed]
- Petrulis, J.R.; Kusnadi, A.; Ramadoss, P.; Hollingshead, B.; Perdew, G.H. The hsp90 Co-chaperone XAP2 Alters Importin β Recognition of the Bipartite Nuclear Localization Signal of the Ah Receptor and Represses Transcriptional Activity. J. Biol. Chem. 2003, 278, 2677–2685. [Google Scholar] [CrossRef] [PubMed]
- Perdew, G.H. Association of the Ah receptor with the 90-kDa heat shock protein. J. Biol. Chem. 1988, 263, 13802–13805. [Google Scholar] [PubMed]
- Pongratz, I.; Mason, G.G.; Poellinger, L. Dual roles of the 90-kDa heat shock protein hsp90 in modulating functional activities of the dioxin receptor. Evidence that the dioxin receptor functionally belongs to a subclass of nuclear receptors which require hsp90 both for ligand binding activity and repression of intrinsic DNA binding activity. J. Biol. Chem. 1992, 267, 13728–13734. [Google Scholar] [PubMed]
- Ikuta, T.; Eguchi, H.; Tachibana, T.; Yoneda, Y.; Kawajiri, K. Nuclear Localization and Export Signals of the Human Aryl Hydrocarbon Receptor. J. Biol. Chem. 1998, 273, 2895–2904. [Google Scholar] [CrossRef]
- Shimizu, Y.; Nakatsuru, Y.; Ichinose, M.; Takahashi, Y.; Kume, H.; Mimura, J.; Fujii-Kuriyama, Y.; Ishikawa, T. Benzo[a]pyrene carcinogenicity is lost in mice lacking the aryl hydrocarbon receptor. Proc. Natl. Acad. Sci. USA 2000, 97, 779–782. [Google Scholar] [CrossRef] [PubMed]
- Gasiewicz, T.A.; Singh, K.P.; Casado, F.L. The aryl hydrocarbon receptor has an important role in the regulation of hematopoiesis: Implications for benzene-induced hematopoietic toxicity. Chem. Biol. Interact. 2010, 184, 246–251. [Google Scholar] [CrossRef]
- Mulero-Navarro, S.; Fernandez-Salguero, P.M. New Trends in Aryl Hydrocarbon Receptor Biology. Front. Cell Dev. Biol. 2016, 4, 45. [Google Scholar] [CrossRef]
- Xue, P.; Fu, J.; Zhou, Y. The Aryl Hydrocarbon Receptor and Tumor Immunity. Front. Immunol. 2018, 9, 286. [Google Scholar] [CrossRef]
- Tkachenko, A.; Henkler, F.; Brinkmann, J.; Sowada, J.; Genkinger, D.; Kern, C.; Tralau, T.; Luch, A. The Q-rich/PST domain of the AHR regulates both ligand-induced nuclear transport and nucleocytoplasmic shuttling. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef]
- Pollenz, R.S.; Barbour, E.R. Analysis of the Complex Relationship between Nuclear Export and Aryl Hydrocarbon Receptor-Mediated Gene Regulation. Mol. Cell Biol. 2000, 20, 6095–6104. [Google Scholar] [CrossRef] [PubMed]
- Berg, P.; Pongratz, I. Differential usage of nuclear export sequences regulates intracellular localization of the dioxin (aryl hydrocarbon) receptor. J. Biol. Chem. 2001, 276, 43231–43238. [Google Scholar] [CrossRef] [PubMed]
- Kietzmann, T.; Mennerich, D.; Dimova, E.Y. Hypoxia-Inducible Factors (HIFs) and Phosphorylation: Impact on Stability, Localization, and Transactivity. Front. Cell Dev. Biol. 2016, 4, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-W.; Bae, S.-H.; Jeong, J.-W.; Kim, S.-H.; Kim, K.-W. Hypoxia-inducible factor (HIF-1)α: Its protein stability and biological functions. Exp. Mol. Med. 2004, 36, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Berchner-Pfannschmidt, U.; Frede, S.; Wotzlaw, C.; Fandrey, J. Imaging of the hypoxia-inducible factor pathway: Insights into oxygen sensing. Eur. Respir. J. 2008, 32, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Cockman, M.E.; Masson, N.; Mole, D.R.; Jaakkola, P.; Chang, G.-W.; Clifford, S.C.; Maher, E.R.; Pugh, C.W.; Ratcliffe, P.J.; Maxwell, P.H. Hypoxia Inducible Factor-α Binding and Ubiquitylation by the von Hippel-Lindau Tumor Suppressor Protein. J. Biol. Chem. 2000, 275, 25733–25741. [Google Scholar] [CrossRef]
- Ema, M.; Hirota, K.; Mimura, J.; Abe, H.; Yodoi, J.; Sogawa, K.; Poellinger, L.; Fujii-Kuriyama, Y. Molecular mechanisms of transcription activation by HLF and HIF1alpha in response to hypoxia: Their stabilization and redox signal-induced interaction with CBP/p300. EMBO J. 1999, 18, 1905–1914. [Google Scholar] [CrossRef] [PubMed]
- Mylonis, I.; Chachami, G.; Paraskeva, E.; Simos, G. Atypical CRM1-dependent nuclear export signal mediates regulation of hypoxia-inducible factor-1alpha by MAPK. J. Biol. Chem. 2008, 283, 27620–27627. [Google Scholar] [CrossRef]
- Mylonis, I.; Kourti, M.; Samiotaki, M.; Panayotou, G.; Simos, G. Mortalin-mediated and ERK-controlled targeting of HIF-1α to mitochondria confers resistance to apoptosis under hypoxia. J. Cell Sci. 2017, 130, 466–479. [Google Scholar] [CrossRef]
- Depping, R.; Jelkmann, W.; Kosyna, F.K. Nuclear-cytoplasmatic shuttling of proteins in control of cellular oxygen sensing. J. Mol. Med. 2015, 93, 599–608. [Google Scholar] [CrossRef]
- Dekanty, A.; Lavista-Llanos, S.; Irisarri, M.; Oldham, S.; Wappner, P. The insulin-PI3K/TOR pathway induces a HIF-dependent transcriptional response in Drosophila by promoting nuclear localization of HIF-α/Sima. J. Cell Sci. 2005, 118, 5413–5441. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.C.; Shibuya, M. A variant of nuclear localization signal of bipartite-type is required for the nuclear translocation of hypoxia inducible factors (1alpha, 2alpha and 3alpha). Oncogene 2001, 20, 1435–1444. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Ruas, J.L.; Cao, R.; Salomons, F.A.; Cao, Y.; Poellinger, L.; Pereira, T. Cell-type-specific regulation of degradation of hypoxia-inducible factor 1 alpha: Role of subcellular compartmentalization. Mol. Cell. Biol. 2006, 26, 4628–4641. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chun, Y.; Choi, E.; Kim, T.; Kim, M.; Park, J. A dominant-negative isoform lacking exons 11 and 12 of the human hypoxia-inducible factor-1α gene. Biochem. J. 2002, 79, 71–79. [Google Scholar] [CrossRef]
- Romero, N.M.; Irisarri, M.; Roth, P.; Cauerhff, A.; Samakovlis, C.; Wappner, P. Regulation of the Drosophila hypoxia-inducible factor alpha Sima by CRM1-dependent nuclear export. Mol. Cell Biol. 2008, 28, 3410–3423. [Google Scholar] [CrossRef] [PubMed]
- Gkotinakou, I.-M.; Befani, C.; Simos, G.; Liakos, P. ERK1/2 phosphorylates HIF-2α and regulates its activity by controlling its CRM1-dependent nuclear shuttling. J. Cell Sci. 2019, 132, jcs225698. [Google Scholar] [CrossRef] [PubMed]
- Woods, S.L.; Whitelaw, M.L. Differential Activities of Murine Single Minded 1 (SIM1) and SIM2 on a Hypoxic Response Element. J. Biol. Chem. 2002, 277, 10236–10243. [Google Scholar] [CrossRef]
- Yamaki, A.; Kudoh, J.; Shimizu, N.; Shimizu, Y. A novel nuclear localization signal in the human single-minded proteins SIM1 and SIM2. Biochem. Biophys. Res. Commun. 2004, 313, 482–488. [Google Scholar] [CrossRef]
- Edgar, R.S.; Green, E.W.; Zhao, Y.; van Ooijen, G.; Olmedo, M.; Qin, X.; Xu, Y.; Pan, M.; Valekunja, U.K.; Feeney, K.A.; et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature 2012, 485, 459–464. [Google Scholar] [CrossRef]
- Asher, G.; Sassone-Corsi, P. Time for Food: The Intimate Interplay between Nutrition, Metabolism, and the Circadian Clock. Cell 2015, 161, 84–92. [Google Scholar] [CrossRef]
- Mauvoisin, D. Circadian rhythms and proteomics: It’s all about posttranslational modifications! Wiley Interdiscip. Rev. Syst. Biol. Med. 2019, 11, e1450. [Google Scholar] [CrossRef] [PubMed]
- Gekakis, N.; Staknis, D.; Nguyen, H.B.; Davis, F.C.; Wilsbacher, L.D.; King, D.P.; Takahashi, J.S.; Weitz, C.J. Role of the CLOCK protein in the mammalian circadian mechanism. Science 1998, 280, 1564–1569. [Google Scholar] [CrossRef] [PubMed]
- Honma, S.; Ikeda, M.; Abe, H.; Tanahashi, Y.; Namihira, M.; Honma, K.; Nomura, M. Circadian oscillation of BMAL1, a partner of a mammalian clock gene Clock, in rat suprachiasmatic nucleus. Biochem. Biophys. Res. Commun. 1998, 250, 83–87. [Google Scholar] [CrossRef] [PubMed]
- Tamanini, F.; Chaves, I.; Bajek, M.I.; van der Horst, G.T.J. Structure Function Analysis of Mammalian Cryptochromes. Cold Spring Harb. Symp. Quant. Biol. 2007, 72, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Sancar, A. Regulation of the mammalian circadian clock by cryptochrome. J. Biol. Chem. 2004, 279, 34079–34082. [Google Scholar] [CrossRef] [PubMed]
- Kondratov, R.V.; Chernov, M.V.; Kondratova, A.A.; Gorbacheva, V.Y.; Gudkov, A.V.; Antoch, M.P. BMAL1-dependent circadian oscillation of nuclear CLOCK: Posttranslational events induced by dimerization of transcriptional activators of the mammalian clock system. Genes Dev. 2003, 17, 1921–1932. [Google Scholar] [CrossRef] [PubMed]
- Yoshitane, H.; Takao, T.; Satomi, Y.; Du, N.-H.; Okano, T.; Fukada, Y. Roles of CLOCK Phosphorylation in Suppression of E-Box-Dependent Transcription. Mol. Cell Biol. 2009, 29, 3675–3686. [Google Scholar] [CrossRef] [PubMed]
- Ward, S.M.; Fernando, S.J.; Hou, T.Y.; Duffield, G.E. The transcriptional repressor ID2 can interact with the canonical clock components CLOCK and BMAL1 and mediate inhibitory effects on mPer1 expression. J. Biol. Chem. 2010, 285, 38987–39000. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.-D.; Barnard, M.; Tian, H.; Li, X.; Ring, H.Z.; Francke, U.; Shelton, J.; Richardson, J.; Russell, D.W.; McKnight, S.L. Molecular characterization of two mammalian bHLH-PAS domain proteins selectively expressed in the central nervous system. Proc. Natl. Acad. Sci. USA 1997, 94, 713–718. [Google Scholar] [CrossRef] [PubMed]
- Erbel-Sieler, C.; Dudley, C.; Zhou, Y.; Wu, X.; Estill, S.J.; Han, T.; Diaz-Arrastia, R.; Brunskill, E.W.; Potter, S.S.; McKnight, S.L. Behavioral and regulatory abnormalities in mice deficient in the NPAS1 and NPAS3 transcription factors deficient in the NPAS1 and NPAS3 transcription factors. Proc. Natl. Acad. Sci. USA 2004, 101, 13648–13653. [Google Scholar] [CrossRef]
- Teh, C.H.L.; Lam, K.K.Y.; Loh, C.C.; Loo, J.M.; Yan, T.; Lim, T.M. Neuronal PAS domain protein 1 is a transcriptional repressor and requires arylhydrocarbon nuclear translocator for its nuclear localization. J. Biol. Chem. 2006, 281, 34617–34629. [Google Scholar] [CrossRef] [PubMed]
- Ohsawa, S.; Hamada, S.; Kakinuma, Y.; Yagi, T.; Miura, M. Novel function of neuronal PAS domain protein 1 in erythropoietin expression in neuronal cells. J. Neurosci. Res. 2005, 79, 451–458. [Google Scholar] [CrossRef] [PubMed]
- Dioum, E.M.; Rutter, J.; Tuckerman, J.R.; Gonzalez, G.; Gilles-Gonzalez, M.-A.; McKnight, S.L. NPAS2: A Gas-Responsive Transcription Factor. Science 2002, 298, 2385–2387. [Google Scholar] [CrossRef] [PubMed]
- Kondratov, R.V.; Kondratova, A.A.; Lee, C.; Gorbacheva, V.Y.; Chernov, M.V.; Antoch, M.P. Post-translational regulation of circadian transcriptional CLOCK(NPAS2)/BMAL1 complex by CRYPTOCHROMES. Cell Cycle 2006, 5, 890–895. [Google Scholar] [CrossRef] [PubMed]
- Brunskill, E.W.; Witte, D.P.; Shreiner, A.B.; Potter, S.S. Characterization of Npas3, a novel basic helix-loop-helix PAS gene expressed in the developing mouse nervous system. Mech. Dev. 1999, 88, 237–241. [Google Scholar] [CrossRef]
- Kamnasaran, D. Disruption of the neuronal PAS3 gene in a family affected with schizophrenia. J. Med. Genet. 2003, 40, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Luoma, L.M.; Berry, F.B. Molecular analysis of NPAS3 functional domains and variants. BMC Mol. Biol. 2018, 19, 1–19. [Google Scholar] [CrossRef]
- Ooe, N.; Saito, K.; Mikami, N.; Nakatuka, I.; Kaneko, H. Identification of a Novel Basic Helix-Loop-Helix-PAS Factor, NXF, Reveals a Sim2 Competitive, Positive Regulatory Role in Dendritic-Cytoskeleton Modulator Drebrin Gene Expression. Mol. Cell Biol. 2003, 24, 608–616. [Google Scholar] [CrossRef]
- Esser, J.S.; Charlet, A.; Schmidt, M.; Heck, S.; Allen, A.; Lother, A.; Epting, D.; Patterson, C.; Bode, C.; Moser, M. The neuronal transcription factor NPAS4 is a strong inducer of sprouting angiogenesis and tip cell formation. Cardiovasc. Res. 2017, 113, 222–223. [Google Scholar] [CrossRef]
- Sabatini, P.V.; Krentz, N.A.J.; Zarrouki, B.; Westwell-Roper, C.Y.; Nian, C.; Uy, R.A.; Shapiro, A.M.J.; Poitout, V.; Lynn, F.C. Npas4 is a novel activity-regulated cytoprotective factor in pancreatic β-cells. Diabetes 2013, 62, 2808–2820. [Google Scholar] [CrossRef]
- Shamloo, M.; Soriano, L.; Von Schack, D.; Rickhag, M.; Chin, D.J.; Gonzalez-Zulueta, M.; Gido, G.; Urfer, R.; Wieloch, T.; Nikolich, K. Npas4, a novel helix-loop-helix PAS domain protein, is regulated in response to cerebral ischemia. Eur. J. Neurosci. 2006, 24, 2705–2720. [Google Scholar] [CrossRef]
- Ooe, N.; Saito, K.; Kaneko, H. Characterization of functional heterodimer partners in brain for a bHLH-PAS factor NXF. Biochim. Biophys. Acta 2009, 1789, 192–197. [Google Scholar] [CrossRef]
- Moser, M.; Knoth, R.; Bode, C.; Patterson, C. LE-PAS, a novel Arnt-dependent HLH-PAS protein, is expressed in limbic tissues and transactivates the CNS midline enhancer element. Mol. Brain Res. 2004, 128, 141–149. [Google Scholar] [CrossRef]
- Sullivan, A.E.; Peet, D.J.; Whitelaw, M.L. MAGED1 is a novel regulator of a select subset of bHLH PAS transcription factors. FEBS J. 2016, 283, 3488–3502. [Google Scholar] [CrossRef]
- Fan, W.; Long, Y.; Lai, Y.; Wang, X.; Chen, G.; Zhu, B. NPAS4 Facilitates the Autophagic Clearance of Endogenous Tau in Rat Cortical Neurons. J. Mol. Neurosci. 2016, 58, 401–410. [Google Scholar] [CrossRef]
- Dougherty, E.P.R. Comprehensive Toxicology. 2.13—ARNT: A Key bHLH/PAS Regulatory Protein Across Multiple Pathways; Elsevier Limited: Auburn, AL, USA, 2019; ISBN 9780080468686. [Google Scholar]
- Hirose, K.; Morita, M.; Ema, M.; Mimura, J.; Hamada, H.; Fujii, H.; Saijo, Y.; Gotoh, O.; Sogawa, K.; Fujii-Kuriyama, Y. cDNA cloning and tissue-specific expression of a novel basic helix-loop-helix/PAS factor (Arnt2) with close sequence similarity to the aryl hydrocarbon receptor nuclear translocator (Arnt). Mol. Cell Biol. 1996, 16, 1706–1713. [Google Scholar] [CrossRef]
- Dougherty, E.J. Analysis of the Role of bHLH/PAS Proteins in Aryl Hydrocarbon Receptor Signaling Scholar Commons Citation. Graduate Theses and Dissertations. 2008. Available online: https://scholarcommons.usf.edu/etd/218 (accessed on 23 September 2019).
- Ikeda, M.; Yu, W.; Hirai, M.; Ebisawa, T.; Honma, S.; Yoshimura, K.; Honma, K.I.; Nomura, M. cDNA cloning of a novel bHLH-PAS transcription factor superfamily gene, BMAL2: Its mRNA expression, subcellular distribution, and chromosomal localization. Biochem. Biophys. Res. Commun. 2000, 275, 493–502. [Google Scholar] [CrossRef]
- Dang, F.; Sun, X.; Ma, X.; Wu, R.; Zhang, D.; Chen, Y.; Xu, Q.; Wu, Y.; Liu, Y. Insulin post-transcriptionally modulates Bmal1 protein to affect the hepatic circadian clock. Nat. Commun. 2016, 7, 1–12. [Google Scholar] [CrossRef]
- Holmes, J.L.; Pollenz, R.S. Determination of aryl hydrocarbon receptor nuclear translocator protein concentration and subcellular localization in hepatic and nonhepatic cell culture lines: Development of quantitative Western blotting protocols for calculation of aryl hydrocarbon rec. Mol. Pharmacol. 1997, 52, 202–211. [Google Scholar] [CrossRef]
- Eguchi, H.; Ikuta, T.; Tachibana, T.; Yoneda, Y.; Kawajiri, K. A nuclear localization signal of human aryl hydrocarbon receptor nuclear translocator/hypoxia-inducible factor 1beta is a novel bipartite type recognized by the two components of nuclear pore-targeting complex. J. Biol. Chem. 1997, 272, 17640–17647. [Google Scholar] [CrossRef]
- Bersten, D.C.; Wright, J.A.; McCarthy, P.J.; Whitelaw, M.L. Regulation of the neuronal transcription factor NPAS4 by REST and microRNAs. Biochim. Biophys. Acta Gene Regul. Mech. 2014, 1839, 13–24. [Google Scholar] [CrossRef]
- Kwon, I.; Lee, J.; Chang, S.H.; Jung, N.C.; Lee, B.J.; Son, G.H.; Kim, K.; Lee, K.H. BMAL1 shuttling controls transactivation and degradation of the CLOCK/BMAL1 heterodimer. Mol. Cell. Biol. 2006, 26, 7318–7330. [Google Scholar] [CrossRef]
- Tamaru, T.; Hirayama, J.; Isojima, Y.; Nagai, K.; Norioka, S.; Takamatsu, K.; Sassone-Corsi, P. CK2α phosphorylates BMAL1 to regulate the mammalian clock. Nat. Struct. Mol. Biol. 2009, 16, 446–448. [Google Scholar] [CrossRef]
- Truman, J.W.; Riddiford, L.M. The origins of insect metamorphosis. Nature 1999, 401, 447–452. [Google Scholar] [CrossRef]
- Dubrovsky, E.B. Hormonal cross talk in insect development. Trends Endocrinol. Metab. 2005, 16, 6–11. [Google Scholar] [CrossRef]
- Charles, J.-P.; Iwema, T.; Epa, V.C.; Takaki, K.; Rynes, J.; Jindra, M. Ligand-binding properties of a juvenile hormone receptor, Methoprene-tolerant. Proc. Natl. Acad. Sci. USA 2011, 108, 21128–21133. [Google Scholar] [CrossRef]
- Parthasarathy, R.; Tan, A.; Palli, S.R. bHLH-PAS family transcription factor methoprene-tolerant plays a key role in JH action in preventing the premature development of adult structures during larval-pupal metamorphosis. Mech. Dev. 2008, 125, 601–616. [Google Scholar] [CrossRef]
- Baumann, A.; Barry, J.; Wang, S.; Fujiwara, Y.; Wilson, T.G. Paralogous genes involved in juvenile hormone action in Drosophila melanogaster. Genetics 2010, 185, 1327–1336. [Google Scholar] [CrossRef]
- Abdou, M.; Peng, C.; Huang, J.; Zyaan, O.; Wang, S.; Li, S.; Wang, J. Wnt Signaling Cross-Talks with JH Signaling by Suppressing Met and gce Expression. PLoS ONE 2011, 6, e26772. [Google Scholar] [CrossRef]
- Godlewski, J.; Wang, S.; Wilson, T.G. Interaction of bHLH-PAS proteins involved in juvenile hormone reception in Drosophila. Biochem. Biophys. Res. Commun. 2006, 342, 1305–1311. [Google Scholar] [CrossRef]
- Shemshedini, L.; Wilson, T.G. Resistance to juvenile hormone and an insect growth regulator in Drosophila is associated with an altered cytosolic juvenile hormone-binding protein. Proc. Natl. Acad. Sci. USA 1990, 87, 2072–2076. [Google Scholar] [CrossRef]
- Miura, K.; Oda, M.; Makita, S.; Chinzei, Y. Characterization of the Drosophila Methoprene -tolerant gene product. Juvenile hormone binding and ligand-dependent gene regulation. FEBS J. 2005, 272, 1169–1178. [Google Scholar] [CrossRef]
- He, Q.; Wen, D.; Jia, Q.; Cui, C.; Wang, J.; Palli, S.R.; Li, S. Heat shock protein 83 (Hsp83) facilitates methoprene-tolerant (Met) nuclear import to modulate juvenile hormone signaling. J. Biol. Chem. 2014, 289, 27874–27885. [Google Scholar] [CrossRef]
- Baumann, A.; Fujiwara, Y.; Wilson, T.G. Evolutionary divergence of the paralogs Methoprene tolerant (Met) and germ cell expressed (gce) within the genus Drosophila. J. Insect Physiol. 2010, 56, 1445–1455. [Google Scholar] [CrossRef]
- Magdalena Romero, N.; Dekanty, A.; Wappner, P. Cellular and Developmental Adaptations to Hypoxia: A Drosophila Perspective. Methods Enzymol. 2007, 435, 123–144. [Google Scholar]
- Lavista-llanos, S.; Irisarri, M.; Russo, D.M.; Gleadle, J.M.; Bocca, S.N.; Muzzopappa, M.; Ratcliffe, P.J.; Wappner, P. Control of the Hypoxic Response in Drosophila melanogaster by the Basic Helix-Loop-Helix PAS Protein Similar. Mol. Cell Biol. 2002, 22, 6842–6853. [Google Scholar] [CrossRef]
- Ward, M.P.; Mosher, J.T.; Crews, S.T. Regulation of bHLH-PAS protein subcellular localization during Drosophila embryogenesis. Development 1998, 1608, 1599–1608. [Google Scholar]
- Uniacke, J.; Holterman, C.E.; Lachance, G.; Franovic, A.; Jacob, M.D.; Fabian, M.R.; Payette, J.; Holcik, M.; Pause, A.; Lee, S. An oxygen-regulated switch in the protein synthesis machinery. Nature 2012, 486, 126–129. [Google Scholar] [CrossRef]
- Lipton, J.O.; Yuan, E.D.; Boyle, L.M.; Ebrahimi-Fakhari, D.; Kwiatkowski, E.; Nathan, A.; Güttler, T.; Davis, F.; Asara, J.M.; Sahin, M. The Circadian Protein BMAL1 Regulates Translation in Response to S6K1-Mediated Phosphorylation. Cell 2015, 161, 1138–1151. [Google Scholar] [CrossRef]
- Michael, A.K.; Asimgil, H.; Partch, C.L. Cytosolic BMAL1 moonlights as a translation factor. Trends Biochem. Sci. 2015, 40, 489–490. [Google Scholar] [CrossRef]

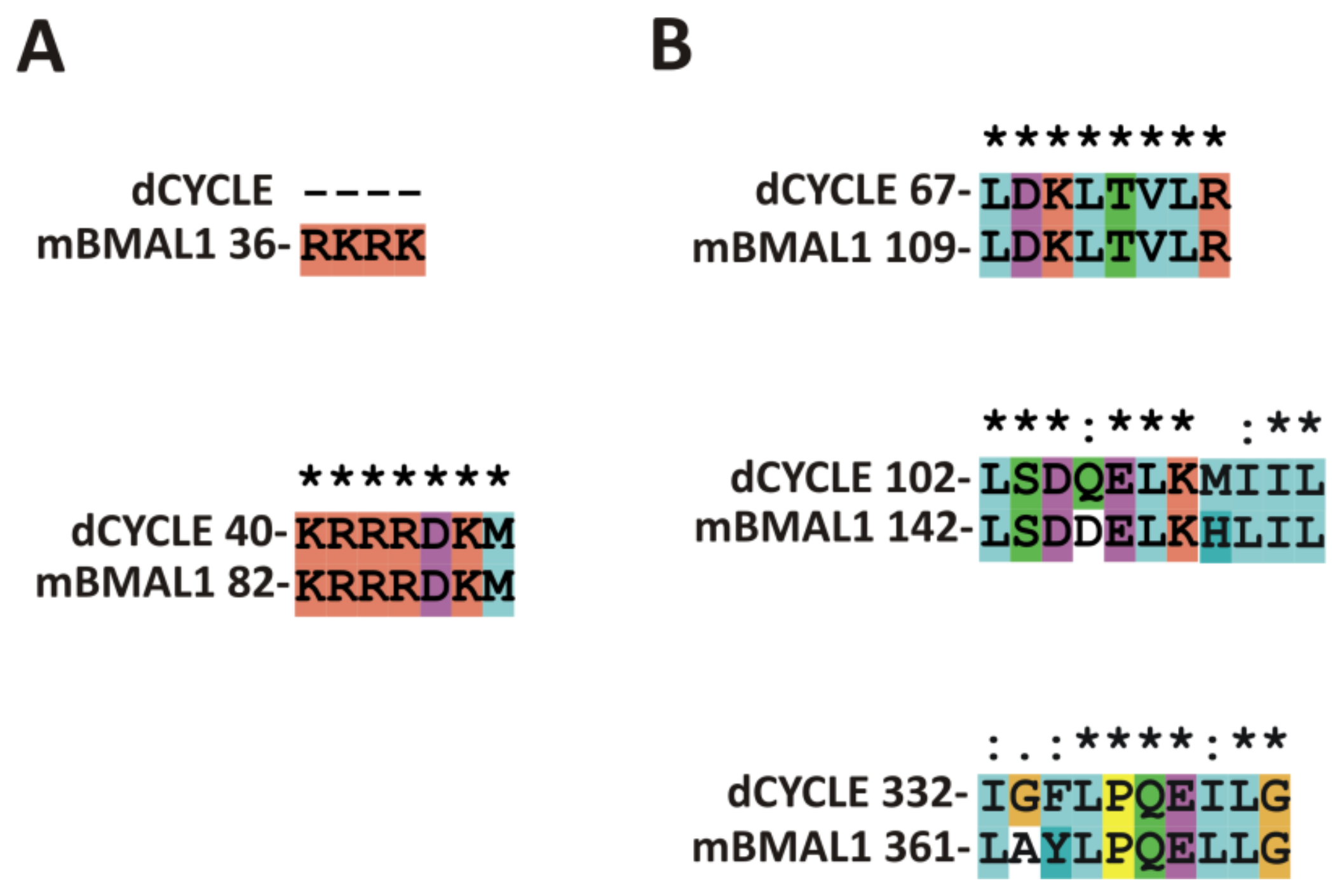
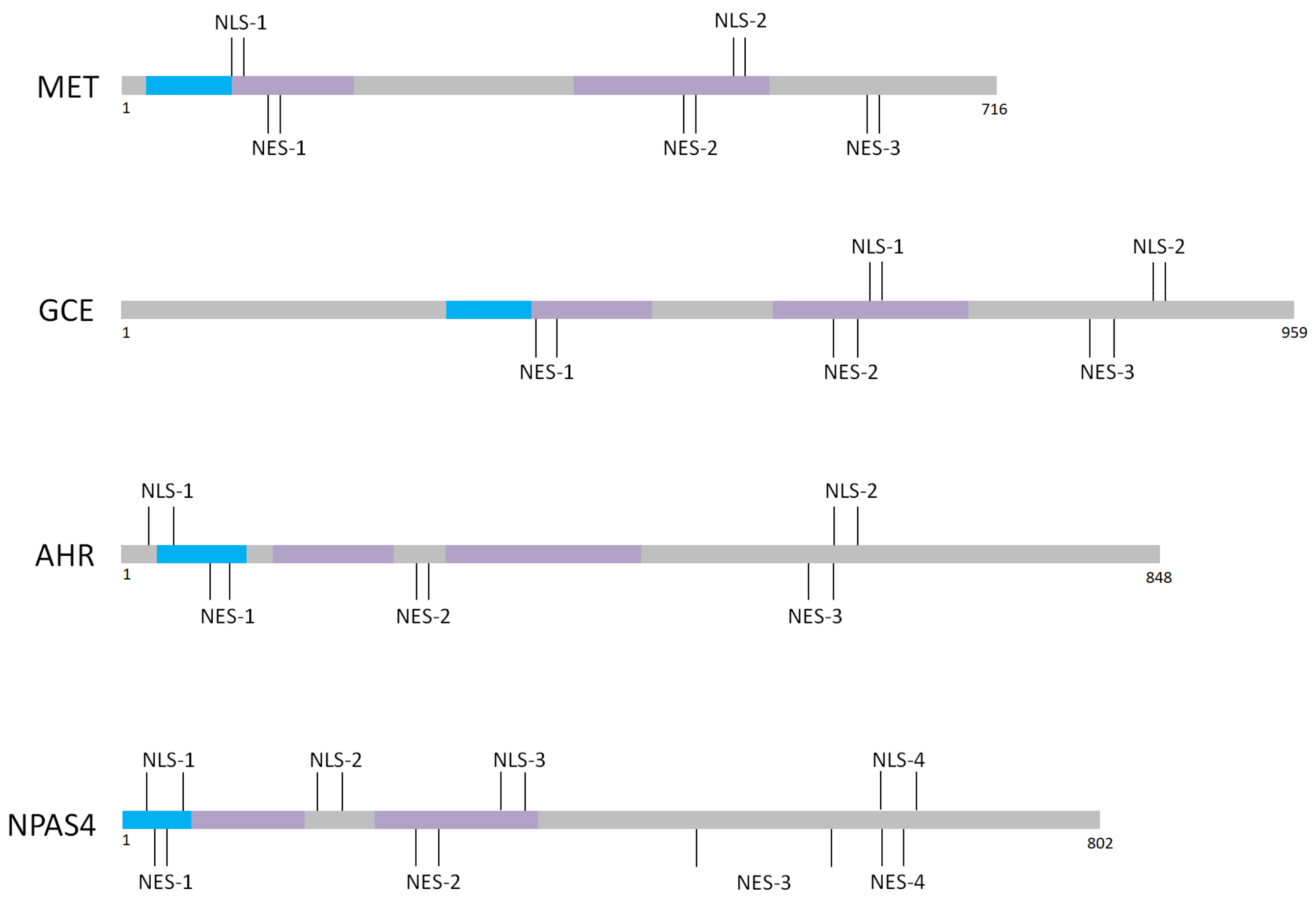
| Protein Name | Domains According to UniProt | Predicted NLS aa Area | Predicted NES aa Area | EXPERIMENTAL NLS | EXPERIMENTALNES |
|---|---|---|---|---|---|
| hAHR UniProtKB - P35869 | 27-80 bHLH 111-181 PAS-1 275-342 PAS-2 348-386 PAC | 11-43abcdef 78-106a 247-280abf 368-398a 640-668a 746-776a | 47-56g 59-77ghi 258-272gi 417-431gi 538-552fg 735-746gi | 13-39 [35] 648-671 [40] | 55-75 [42] 214-222 [42] NES around V647 [40] |
| hHIF-1α UniProtKB - Q16665 | 17-70 bHLH 85-158 PAS-1 228-298PAS-2 302-345 PAC | 3-41abde 58-60e 68-97a 155-160b 245-278a 365-394a 709-746abcdf | 37-46gh? 52-69gi 82-97fg 390-399h 558-572ghi? 627-647i L737h? | 17-33 [86] 718-721 [12] 717-757 [52] | 616-658 [48] |
| hHIF-2α (EPAS1) UniProtKB - Q99814 | 14-67 bHLH 84-154 PAS-1 230-300 PAS-2 304-347 PAC | 3-33abcde 144-177ab 680-735ab 737-765ab 793-840a | 33-47gi 49-64hi 82-102ghi? 497-511ghi? 525-539ghi? 698-712gi 772-786gi | 14-30 705-742 [52] | 705-738 [55] |
| hHIF-3α UniProtKB - Q9Y2N7 | 14-67 bHLH 82-154 PAS-1 227-297 PAS-2 | 7-53abe 153-182a 196-226a 361-392a 438-465a 512-574ab 563-598abc 590-620a | 34-43g 51-70ghi 114-128hi L221h? 477-497ghi 603-617gi 629-643gi | 77-100 Uniprot 561-591 [52] | 230-274 Uniprot 561-586 [55] |
| hSIM1 UniProtKB - P81133 | 1-53 bHLH 77-147 PAS-1 218-288 PAS-2 292-335 PAC | 2-33ab 74-104a 163-197a 235-267a 347-386ab 600-628a 703-734a | 19-33gi 42-56i 231-249gi? 340-347h | 368-388 [58] | 295-333? [58] |
| hSIM2 UniProtKB - Q14190 | 1-53 bHLH 77-149 PAS-1 218-288 PAS-2 292-335 PAC | 2-35abe 74—105a 163-195a 351-386ab 556-586ab 637-665a | 8-17g 19-33i 35-49i 42-56i 65-74g 102-116i? 275-289gi? 325-344hi? | 367-386 [58] | - |
| hCLOCK UniProtKB - O15516 | 34-84 bHLH 107-177 PAS-1 262-332 PAS-2 336-379 PAC | 27-85abcde 256-290a 411-443a 451-481a 526-553a | 111-127hi 507-516g 546-561fgi? 588-597g 798-812i? | 32-47 mouse [67] | - |
| hNPAS1 UniProtKB - Q99742 | 45-98 bHLH 135-207 PAS-1 293-359 PAS-2 365-408 PAC | 33-76abe 89-128ab 250-284ab 456-487abf | 64-78gi 87-101h?i 273-287i 300-320ghi | - | 310-317 [71] |
| hNPAS2 UniProtKB - Q99743 | 9-59 bHLH 82-152 PAS-1 237-307 PAS-2 311-354 PAC | 4-47abcde 214-265a 502-532a 698-730a 803-824b | 25-29i 88-102hi 492-501g 525-540fgi 545-559i? | - | - |
| hNPAS3 UniProtKB - Q8IXF0 | 51-104 bHLH 147-217 PAS-1 319-389 PAS-2 363-406 PAC | 1-18e 29-78ab 130-161ab 266-297ab 578-635abcdf 645-674a 727—773ab | 70-84gi? 88-103ghi 284-298gi 333-347gi? 871-885gi? 903-921i? | 568-585 [76] | NES in bHLH [77] |
| hNPAS4 UniProtKB - Q8IUM7 | 1-53 bHLH 70-144 PAS-1 203-273 PAS-2 278-317 PAC | 8-39ab 158-191ab 193-211b 220-251a 283-314a 593-621a | 20-46 ghi 230-245h?i 394-408i? 590-604ghi? 664-678gi | 10-52 rat 158-191 285-316? 593-622? [21] | 29-45 rat 227-2424 60-580? 591-600 [21] |
| Protein Name | Domains According to UniProt | Predicted NLS aa Area | Predicted NES aa Area | EXPERIMENTAL NLS | EXPERIMENTAL NES |
|---|---|---|---|---|---|
| hARNT HIF-1β UniProtKB - P27540 | 89-142 bHLH 161-235 PAS-1 349-419 PAS-2 424-467 PAC | 34-61abdef 83-130ab 151-182a 260-291a 310-339a 390-421a | 159-178hi 274-288i 336-345g 417-431i 671-685i? | 39-61 [93] | - |
| hARNT2 HIF-2β UniProtKB - Q9HBZ2 | 63-116 bHLH 134-209 PAS-1 323-393 PAS-2 398-441 PAC | 15-29e 34-85b 39-66aef 73-104acd 256-269abf 364-394a 591-619a | 133-150h 310-319gh? 702-711g | 42-64 by similarity to ARNT [93] | 132-144 by similarity to BMAL1 [87] |
| hBMAL1 ARNTL, ARNT3 UniProtKB - O00327 | 78-131 bHLH 146-213 PAS-1 328-394 PAS-2 401-444 PAC | 23-45acdef 37-91b 82-113acd 174205a 244-279ab 291-322a 471-502a | 105-121ghi 142-159hi? | 36-41 active 81-87 not active mouse [94] | 109-116 not active 142-152 active 361-369 active mouse [94] |
| hBMAL2 ARNTL2 ARNT4 UniProtKB - Q8WYA1 | 107-160 bHLH 178-250 PAS-1 357-475 PAS-2 432-475 PAC | 1-10e 19-26e 35-60abcdef 103-127abcd 209-240a 279-322ab 444-474a 554-586a | 140-156ghi 238-252g 463-477f | 13-16 32-35 mouse [90] | - |
| Protein Name | Domains According to UniProt | Predicted NLS | Predicted NES | EXPERIMENTAL NLS | EXPERIMENTAL NES |
|---|---|---|---|---|---|
| MET UniProtKB - Q9VYW2 | 36-89 bHLH 143-185 PAS-1 403-508 PAS-2 Domains according to Uniprot and [19] | 34-72abe 68-115b 98-128acdf 252-285ab 296—330a 489-519a 532-554a 628-662ab | 130-142g 262-276gi 425-443ghi? 446-460i? 542-556i? L603h? | 98-102 absent in GCE 482-498 present in GCE [19] | 126-139 present in GCE 446-456 present in GCE 598-631 present in GCE [19] |
| GCE UniProtKB - Q9VXW7 | 277-330 bHLH 341-416 PAS-1 Uniprot 272-324 bHLH339-425 PAS-1 531-660 PAS-2 According to [20] * | 12-25e? 39-56e? 278-323ab 424-458ab 469-499a 521-554a 625-656abf 838-860adf | 44-50h 347-361hi 567-576g 582-596i? 650-659g 778-792i? | 618-634 ** present in MET 840-859** absent in MET [20] | 343-359** present in Met 580-610** present in MET 762-795** present in MET [20] |
| SIMA UniProtKB - Q24167 | 72-125 bHLH 167-249 PAS-1 307-377 PAS-2 381-422 PAC | 46-91bde 150-180a 347-374a 383-414a 545-546b 554-591abd 1102-1131a 1212-1232af 1288-1316a 1408-1417af | 90-105fghi? 114-130gh?i 173-189fh?i 477-491i 636-650i? 1006-1020i 1064-1078 1125-1139gi? | 537-568 not active? 1210-1230 activeConserved with HIF-1-3α 1406-1409 not active? [55] | 92-101 active 115-124 active 1011-1020 not active? 1131-1140 not active? [55] |
| TANGO UniProtKB - O15945 | 13-66 bHLH 85-156 PAS-1 271-341 PAS-2 346-389 PAC | 11-52b 23-54acd 167-196a? 207-237a? 288-320a | 83-97hi 259-272ghi? 624-638h?i | - | - |
| CYCLE (BMAL1) UniProtKB - O61734 | 30-83 bHLH 104-175 PAS-1 297-367 PAS-2 372-413 PAC | 25-46 bcd 41-71ae 210-240a 229-262abf | 50-59g 63-75gh? 102-111h? | - | - |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Greb-Markiewicz, B.; Kolonko, M. Subcellular Localization Signals of bHLH-PAS Proteins: Their Significance, Current State of Knowledge and Future Perspectives. Int. J. Mol. Sci. 2019, 20, 4746. https://doi.org/10.3390/ijms20194746
Greb-Markiewicz B, Kolonko M. Subcellular Localization Signals of bHLH-PAS Proteins: Their Significance, Current State of Knowledge and Future Perspectives. International Journal of Molecular Sciences. 2019; 20(19):4746. https://doi.org/10.3390/ijms20194746
Chicago/Turabian StyleGreb-Markiewicz, Beata, and Marta Kolonko. 2019. "Subcellular Localization Signals of bHLH-PAS Proteins: Their Significance, Current State of Knowledge and Future Perspectives" International Journal of Molecular Sciences 20, no. 19: 4746. https://doi.org/10.3390/ijms20194746
APA StyleGreb-Markiewicz, B., & Kolonko, M. (2019). Subcellular Localization Signals of bHLH-PAS Proteins: Their Significance, Current State of Knowledge and Future Perspectives. International Journal of Molecular Sciences, 20(19), 4746. https://doi.org/10.3390/ijms20194746





