Processing and Maturation of Cathepsin C Zymogen: A Biochemical and Molecular Modeling Analysis
Abstract
1. Introduction
2. Results
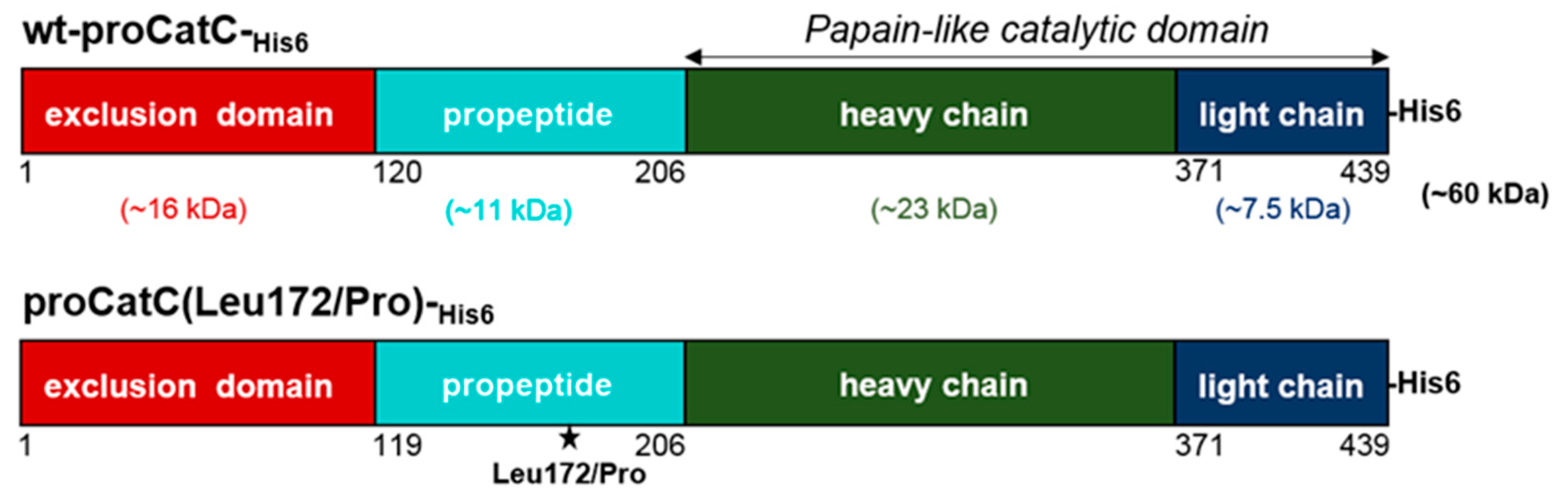
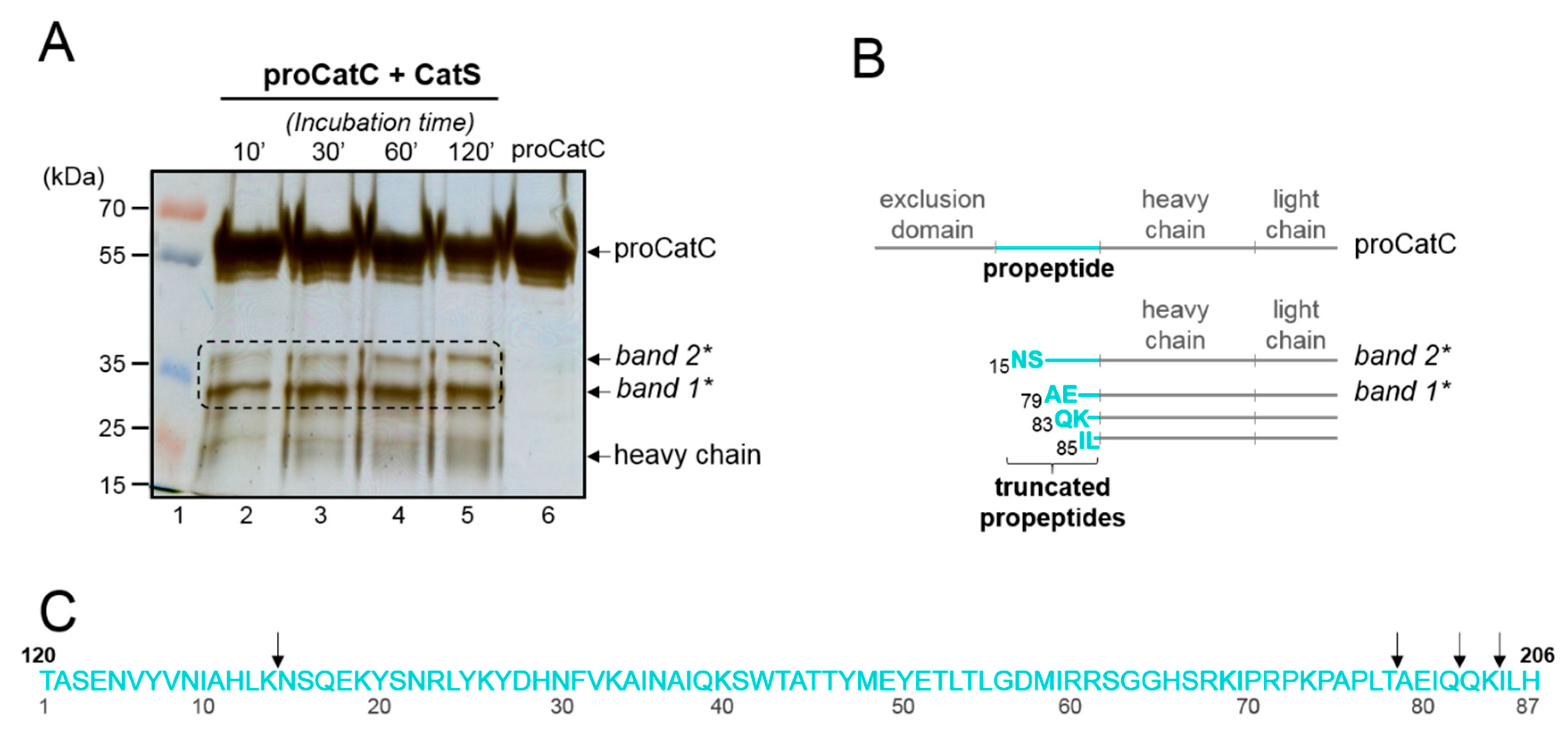
2.1. Processing and Maturation of Wt-proCatC by CatK, CatV, or CatF
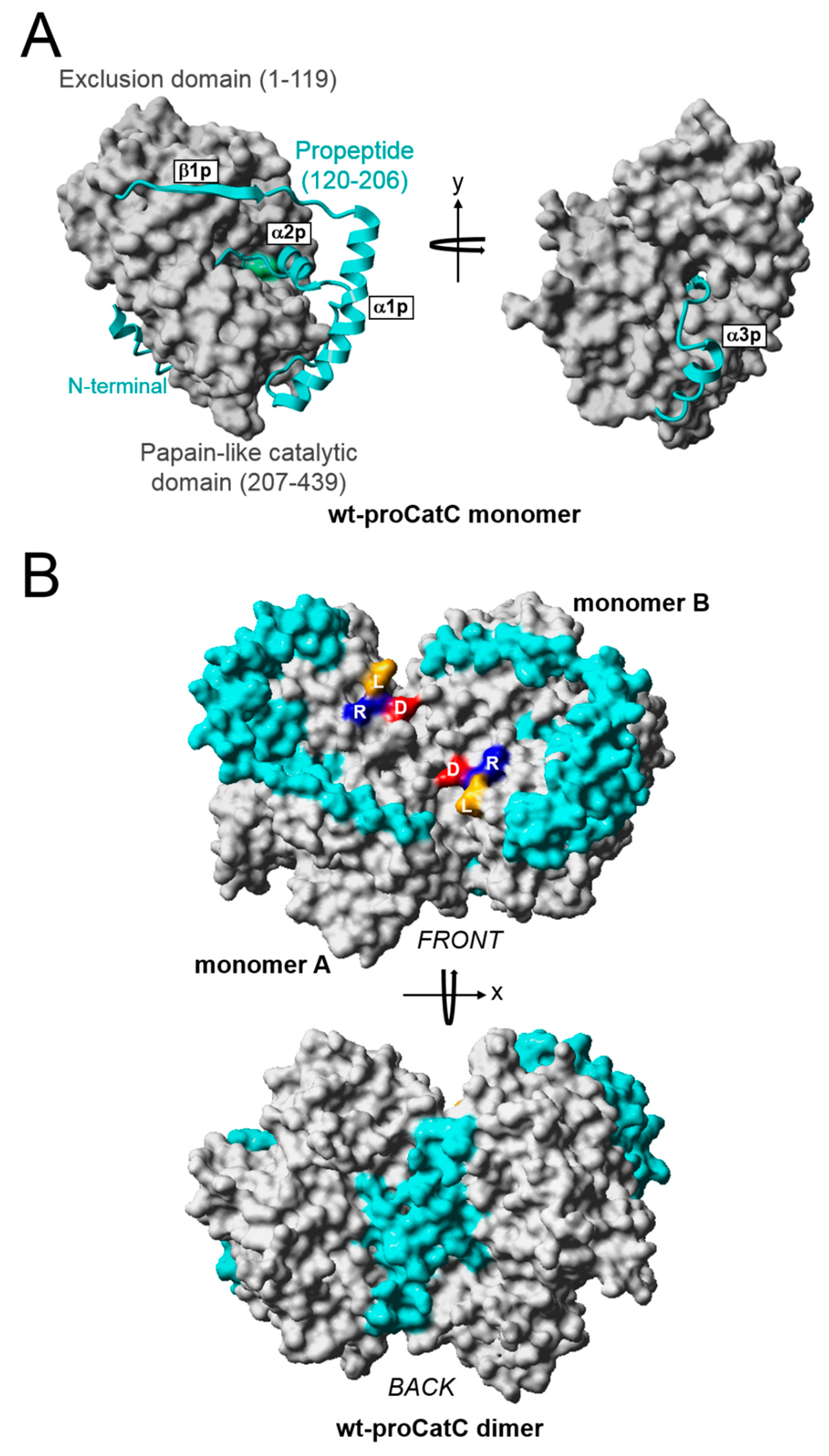
2.2. Structural Modeling of Wt-proCatC
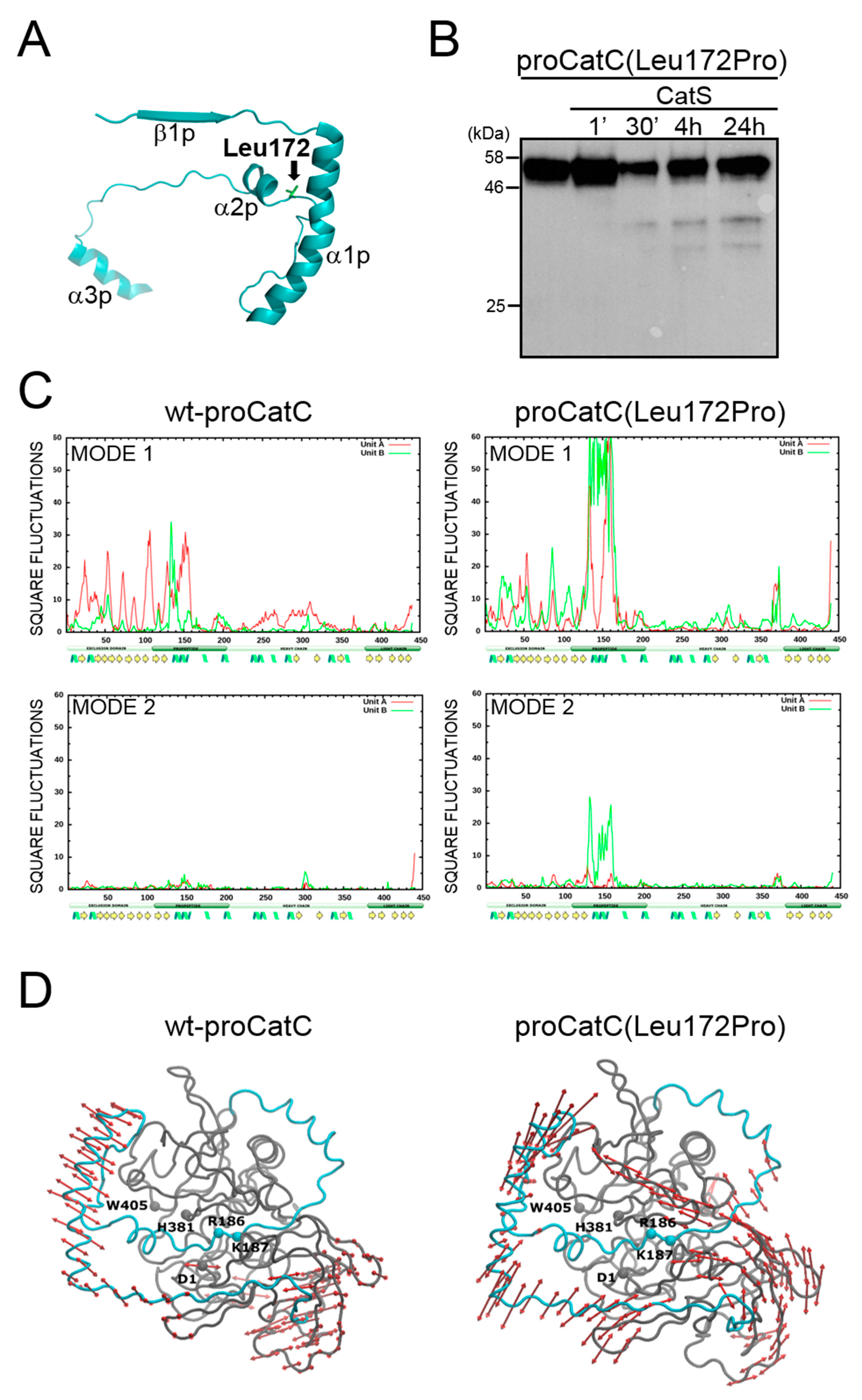
2.3. Characterization and Structural Modeling of ProCatC(Leu172Pro)
3. Discussion
4. Materials and Methods
4.1. Enzymes and Reagents
4.2. Structural Modeling of Human Wt-proCatC and ProCatC(Leu172Pro)
4.3. Principal Component Analysis (PCA) of Human Wt-proCatC and ProCatC(Leu172Pro)
4.4. Cloning, Production, and Purification of Recombinant ProCatC Proteins
4.5. Processing of Human ProCatC Proteins
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| Cat | Cathepsin |
| HMS | Haim–Munk syndrome |
| HEK | HEK, human embryonic kidney |
| PLS | PLS, Papillon–Lefèvre syndrome |
| PDB | PDB, Protein Data Bank |
| Enzymes | Cathepsin C (EC 3.4.14.1), cathepsin F (EC 3.4.22.41), cathepsin K (EC 3.4.22.38), cathepsin L (EC 3.4.22.15), cathepsin S (EC 3.4.22.27), cathepsin V (EC 3.4.22.43) |
References
- Novinec, M.; Lenarcic, B. Papain-like peptidases: Structure, function, and evolution. Biomol. Concepts 2013, 4, 287–308. [Google Scholar] [CrossRef] [PubMed]
- Turk, V.; Stoka, V.; Vasiljeva, O.; Renko, M.; Sun, T.; Turk, B.; Turk, D. Cysteine cathepsins: From structure, function and regulation to new frontiers. Biochim. Biophys. Acta. 2012, 1824, 68–88. [Google Scholar] [CrossRef] [PubMed]
- Kramer, L.; Turk, D.; Turk, B. The Future of Cysteine Cathepsins in Disease Management. Trends Pharm. Sci. 2017, 38, 873–898. [Google Scholar] [CrossRef] [PubMed]
- Dahl, S.W.; Halkier, T.; Lauritzen, C.; Dolenc, I.; Pedersen, J.; Turk, V.; Turk, B. Human recombinant pro-dipeptidyl peptidase I (cathepsin C) can be activated by cathepsins L and S but not by autocatalytic processing. Biochemistry 2001, 40, 1671–1678. [Google Scholar] [CrossRef] [PubMed]
- Turk, D.; Janjic, V.; Stern, I.; Podobnik, M.; Lamba, D.; Dahl, S.W.; Lauritzen, C.; Pedersen, J.; Turk, V.; Turk, B. Structure of human dipeptidyl peptidase I (cathepsin C): Exclusion domain added to an endopeptidase framework creates the machine for activation of granular serine proteases. EMBO J. 2001, 20, 6570–6582. [Google Scholar] [CrossRef] [PubMed]
- Toomes, C.; James, J.; Wood, A.J.; Wu, C.L.; McCormick, D.; Lench, N.; Hewitt, C.; Moynihan, L.; Roberts, E.; Woods, C.G.; et al. Loss-of-function mutations in the cathepsin C gene result in periodontal disease and palmoplantar keratosis. Nat. Genet. 1991, 23, 421–424. [Google Scholar] [CrossRef] [PubMed]
- Hart, T.C.; Hart, P.S.; Bowden, D.W.; Michalec, M.D.; Callison, S.A.; Walker, S.J.; Zhang, Y.; Firatli, E. Mutations of the cathepsin C gene are responsible for Papillon-Lefevre syndrome. J. Med. Genet. 1999, 36, 881–887. [Google Scholar] [PubMed]
- Hart, T.C.; Hart, P.S.; Michalec, M.D.; Zhang, Y.; Firatli, E.; Van Dyke, T.E.; Stabholz, A.; Zlotogorski, A.; Shapira, L.; Soskolne, W.A. Haim–Munk syndrome and Papillon-Lefevre syndrome are allelic mutations in cathepsin C. J. Med. Genet. 2000, 37, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Adkison, A.M.; Raptis, S.Z.; Kelley, D.G.; Pham, C.T. Dipeptidyl peptidase I activates neutrophil-derived serine proteases and regulates the development of acute experimental arthritis. J. Clin. Investig. 2002, 109, 363–371. [Google Scholar] [CrossRef]
- Perera, N.C.; Wiesmuller, K.H.; Larsen, M.T.; Schacher, B.; Eickholz, P.; Borregaard, N.; Jenne, D.E. NSP4 is stored in azurophil granules and released by activated neutrophils as active endoprotease with restricted specificity. J. Immunol. 2013, 191, 2700–2707. [Google Scholar] [CrossRef]
- Korkmaz, B.; Horwitz, M.; Jenne, D.E.; Gauthier, F. Neutrophil elastase, proteinase 3 and cathepsin G as therapeutic targets in human diseases. Pharm. Rev. 2010, 62, 726–759. [Google Scholar] [CrossRef] [PubMed]
- Seren, S.; Rashed Abouzaid, M.; Eulenberg-Gustavus, C.; Hirschfeld, J.; Nasr Soliman, H.; Jerke, U.; N’Guessan, K.; Dallet-Choisy, S.; Lesner, A.; Lauritzen, C.; et al. Consequences of cathepsin C inactivation for membrane exposure of proteinase 3, the target antigen in autoimmune vasculitis. J. Biol. Chem. 2018, 293, 12415–12428. [Google Scholar] [CrossRef] [PubMed]
- Pham, C.T.; Ivanovich, J.L.; Raptis, S.Z.; Zehnbauer, B.; Ley, T.J. Papillon-Lefevre syndrome: Correlating the molecular, cellular, and clinical consequences of cathepsin C/dipeptidyl peptidase I deficiency in humans. J. Immunol. 2004, 173, 7277–7281. [Google Scholar] [CrossRef] [PubMed]
- Korkmaz, B.; Lesner, A.; Letast, S.; Mahdi, Y.K.; Jourdan, M.L.; Dallet-Choisy, S.; Marchand-Adam, S.; Kellenberger, C.; Viaud-Massuard, M.C.; Jenne, D.E.; et al. Neutrophil proteinase 3 and dipeptidyl peptidase I (cathepsin C) as pharmacological targets in granulomatosis with polyangiitis (Wegener granulomatosis). Semin Immunopathol. 2013, 35, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Guay, D.; Beaulieu, C.; Jagadeeswar Reddy, T.; Zamboni, R.; Methot, N.; Rubin, J.; Ethier, D.; David Percival, M. Design and synthesis of dipeptidyl nitriles as potent, selective, and reversible inhibitors of cathepsin C. Bioorg. Med. Chem. Lett. 2009, 19, 5392–5396. [Google Scholar] [CrossRef] [PubMed]
- Korkmaz, B.; Caughey, G.H.; Chapple, I.; Gauthier, F.; Hirschfeld, J.; Jenne, D.E.; Kettritz, R.; Lalmanach, G.; Lamort, A.S.; Lauritzen, C.; et al. Therapeutic targeting of cathepsin C: From pathophysiology to treatment. Pharm. Ther. 2018, 190, 202–236. [Google Scholar] [CrossRef] [PubMed]
- Mallen-St Clair, J.; Shi, G.P.; Sutherland, R.E.; Chapman, H.A.; Caughey, G.H.; Wolters, P.J. Cathepsins L and S are not required for activation of dipeptidyl peptidase I (cathepsin C) in mice. Biol. Chem. 2006, 387, 1143–1146. [Google Scholar] [CrossRef] [PubMed]
- Hamon, Y.; Legowska, M.; Herve, V.; Dallet-Choisy, S.; Marchand-Adam, S.; Vanderlynden, L.; Demonte, M.; Williams, R.; Scott, C.J.; Si-Tahar, M.; et al. Neutrophilic cathepsin C is maturated by a multi-step proteolytic process and secreted by activated cells during inflammatory lung diseases. J. Biol. Chem. 2016, 291, 8486–8499. [Google Scholar] [CrossRef]
- Cury, V.F.; Costa, J.E.; Gomez, R.S.; Boson, W.L.; Loures, C.G.; De Marco, L. A Novel Mutation of the Cathepsin C Gene in Papillon-Lefevre Syndrome. J. Periodontol. 2002, 73, 307–312. [Google Scholar] [CrossRef]
- Cury, V.F.; Gomez, R.S.; Costa, J.E.; Friedman, E.; Boson, W.; De Marco, L. A homozygous cathepsin C mutation associated with Haim-Munk syndrome. Br. J. Dermatol. 2005, 152, 353–356. [Google Scholar] [CrossRef]
- Redecke, L.; Nass, K.; DePonte, D.P.; White, T.A.; Rehders, D.; Barty, A.; Stellato, F.; Liang, M.; Barends, T.R.M.; Boutet, S.; et al. Natively inhibited Trypanosoma brucei cathepsin B structure determined by using an X-ray laser. Science 2013, 339, 227–230. [Google Scholar] [CrossRef] [PubMed]
- Coulombe, R.; Grochulski, P.; Sivaraman, J.; Menard, R.; Mort, J.S.; Cygler, M. Structure of human procathepsin L reveals the molecular basis of inhibition by the prosegment. EMBO J. 1996, 15, 5492–5503. [Google Scholar] [CrossRef] [PubMed]
- Sivaraman, J.; Lalumiere, M.; Menard, R.; Cygler, M. Crystal structure of wild-type human procathepsin K. Protein Sci. 1999, 8, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Somoza, J.R.; Palmer, J.T.; Ho, J.D. The crystal structure of human cathepsin F and its implications for the development of novel immunomodulators. J. Mol. Biol. 2002, 322, 559–568. [Google Scholar] [CrossRef]
- Wang, B.; Shi, G.P.; Yao, P.M.; Li, Z.; Chapman, H.A.; Bromme, D. Human cathepsin F. Molecular cloning, functional expression, tissue localization, and enzymatic characterization. J. Biol. Chem. 1998, 273, 32000–32008. [Google Scholar] [CrossRef] [PubMed]
- Nagler, D.K.; Menard, R. Family C1 cysteine proteases: Biological diversity or redundancy? Biol. Chem. 2003, 384, 837–843. [Google Scholar] [CrossRef] [PubMed]
- Rebernik, M.; Lenarcic, B.; Novinec, M. The catalytic domain of cathepsin C (dipeptidyl-peptidase I) alone is a fully functional endoprotease. Protein Expr. Purif. 2019, 157, 21–27. [Google Scholar] [CrossRef]
- Nagy, N.; Valyi, P.; Csoma, Z.; Sulak, A.; Tripolszki, K.; Farkas, K.; Paschali, E.; Papp, F.; Toth, L.; Fabos, B.; et al. CTSC and Papillon-Lefevre syndrome: Detection of recurrent mutations in Hungarian patients, a review of published variants and database update. Mol. Genet. Genomic Med. 2014, 2, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Sorensen, O.E.; Clemmensen, S.N.; Dahl, S.L.; Ostergaard, O.; Heegaard, N.H.; Glenthoj, A.; Nielsen, F.C.; Borregaard, N. Papillon-Lefevre syndrome patient reveals species-dependent requirements for neutrophil defenses. J. Clin. Investig. 2014, 124, 4539–4548. [Google Scholar] [CrossRef]
- Hamon, Y.; Legowska, M.; Fergelot, P.; Dallet-Choisy, S.; Newell, L.; Vanderlynden, L.; Kord Valeshabad, A.; Acrich, K.; Kord, H.; Charalampos, T. Analysis of urinary cathepsin C for diagnosing Papillon-Lefevre syndrome. FEBS J. 2016, 283, 498–509. [Google Scholar] [CrossRef]
- Laine, D.; Palovich, M.; McCleland, B.; Petitjean, E.; Delhom, I.; Xie, H.; Deng, J.; Lin, G.; Davis, R.; Jolit, A.; et al. Discovery of novel cyanamide-based inhibitors of cathepsin C. ACS Med. Chem. Lett. 2011, 2, 142–147. [Google Scholar] [CrossRef] [PubMed]
- Liwo, A.; Oldziej, S.; Czaplewski, C.; Kleinerman, D.S.; Blood, P.; Scheraga, H.A. Implementation of molecular dynamics and its extensions with the coarse-grained UNRES force field on massively parallel systems; towards millisecond-scale simulations of protein structure, dynamics, and thermodynamics. J. Chem. Theory Comput. 2010, 6, 890–909. [Google Scholar] [CrossRef] [PubMed]
- Pikora, M.; Gieldon, A. RASMOL AB—New functionalities in the program for structure analysis. Acta. Biochim. Pol. 2015, 62, 629–631. [Google Scholar] [CrossRef] [PubMed]
- Bakan, A.; Meireles, L.M.; Bahar, I. ProDy: Protein dynamics inferred from theory and experiments. Bioinformatics 2011, 27, 1575–1577. [Google Scholar] [CrossRef] [PubMed]


| Chain A/B | Chain B/A |
|---|---|
| Y8 | D371, N374 |
| L9 | D371 |
| Q45 | N332, E333 |
| K46 | G325 (backbone) |
| L47 | L47, F327, N374 |
| T49 | L335 |
| Y51 | E333, A334 |
| N56 | L205 |
| S57 | Q201 |
| H59 | T197 |
| L196 | L196 |
| F327 | F327 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lamort, A.-S.; Hamon, Y.; Czaplewski, C.; Gieldon, A.; Seren, S.; Coquet, L.; Lecaille, F.; Lesner, A.; Lalmanach, G.; Gauthier, F.; et al. Processing and Maturation of Cathepsin C Zymogen: A Biochemical and Molecular Modeling Analysis. Int. J. Mol. Sci. 2019, 20, 4747. https://doi.org/10.3390/ijms20194747
Lamort A-S, Hamon Y, Czaplewski C, Gieldon A, Seren S, Coquet L, Lecaille F, Lesner A, Lalmanach G, Gauthier F, et al. Processing and Maturation of Cathepsin C Zymogen: A Biochemical and Molecular Modeling Analysis. International Journal of Molecular Sciences. 2019; 20(19):4747. https://doi.org/10.3390/ijms20194747
Chicago/Turabian StyleLamort, Anne-Sophie, Yveline Hamon, Cezary Czaplewski, Artur Gieldon, Seda Seren, Laurent Coquet, Fabien Lecaille, Adam Lesner, Gilles Lalmanach, Francis Gauthier, and et al. 2019. "Processing and Maturation of Cathepsin C Zymogen: A Biochemical and Molecular Modeling Analysis" International Journal of Molecular Sciences 20, no. 19: 4747. https://doi.org/10.3390/ijms20194747
APA StyleLamort, A.-S., Hamon, Y., Czaplewski, C., Gieldon, A., Seren, S., Coquet, L., Lecaille, F., Lesner, A., Lalmanach, G., Gauthier, F., Jenne, D., & Korkmaz, B. (2019). Processing and Maturation of Cathepsin C Zymogen: A Biochemical and Molecular Modeling Analysis. International Journal of Molecular Sciences, 20(19), 4747. https://doi.org/10.3390/ijms20194747






