Role of Histone H3 Lysine 4 Methylation in Chromatin Biology
Abstract
1. Introduction
2. Writers and Erasers of H3K4 Methylation
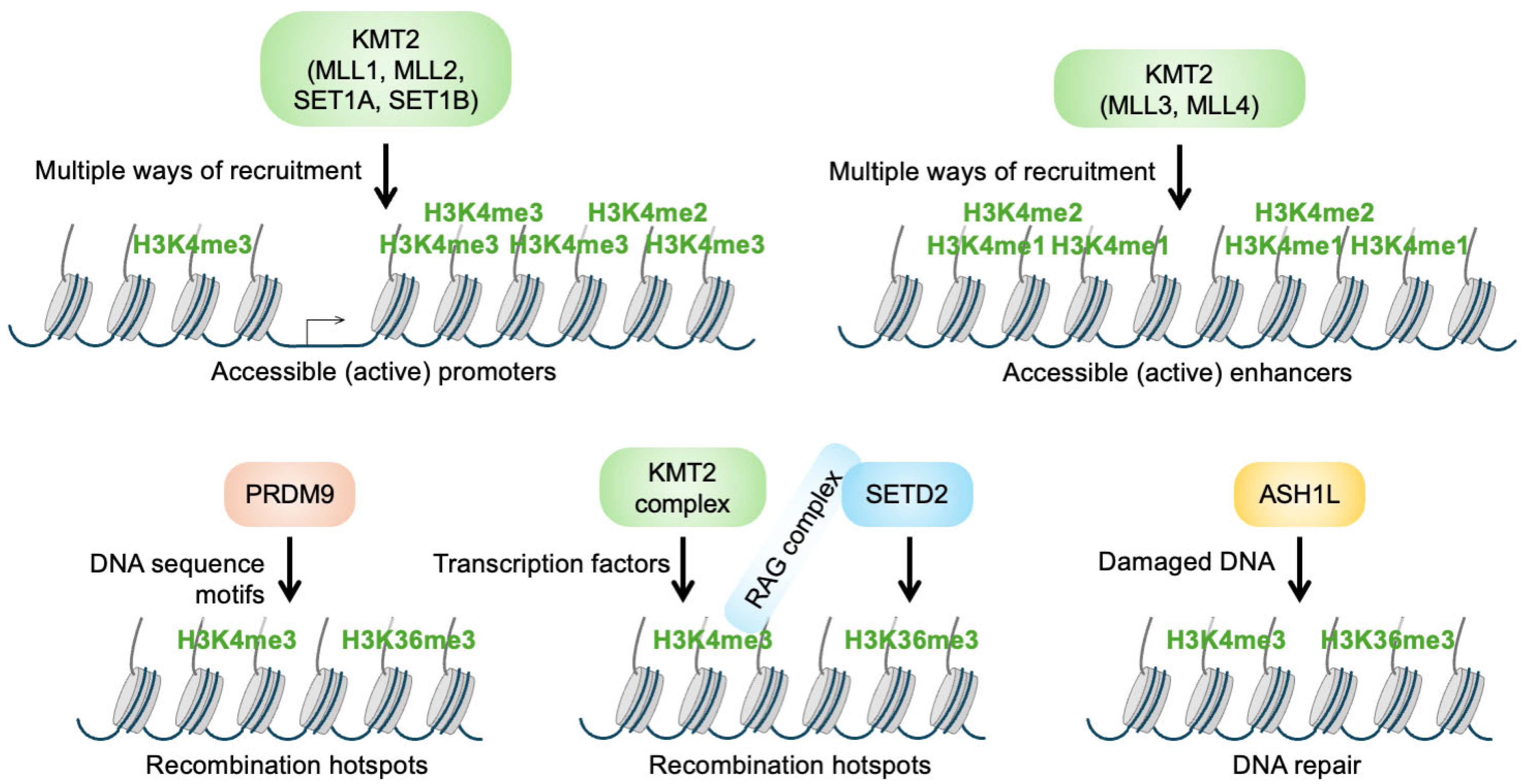
2.1. H3K4 Methyltransferases
2.2. H3K4 Demethylases
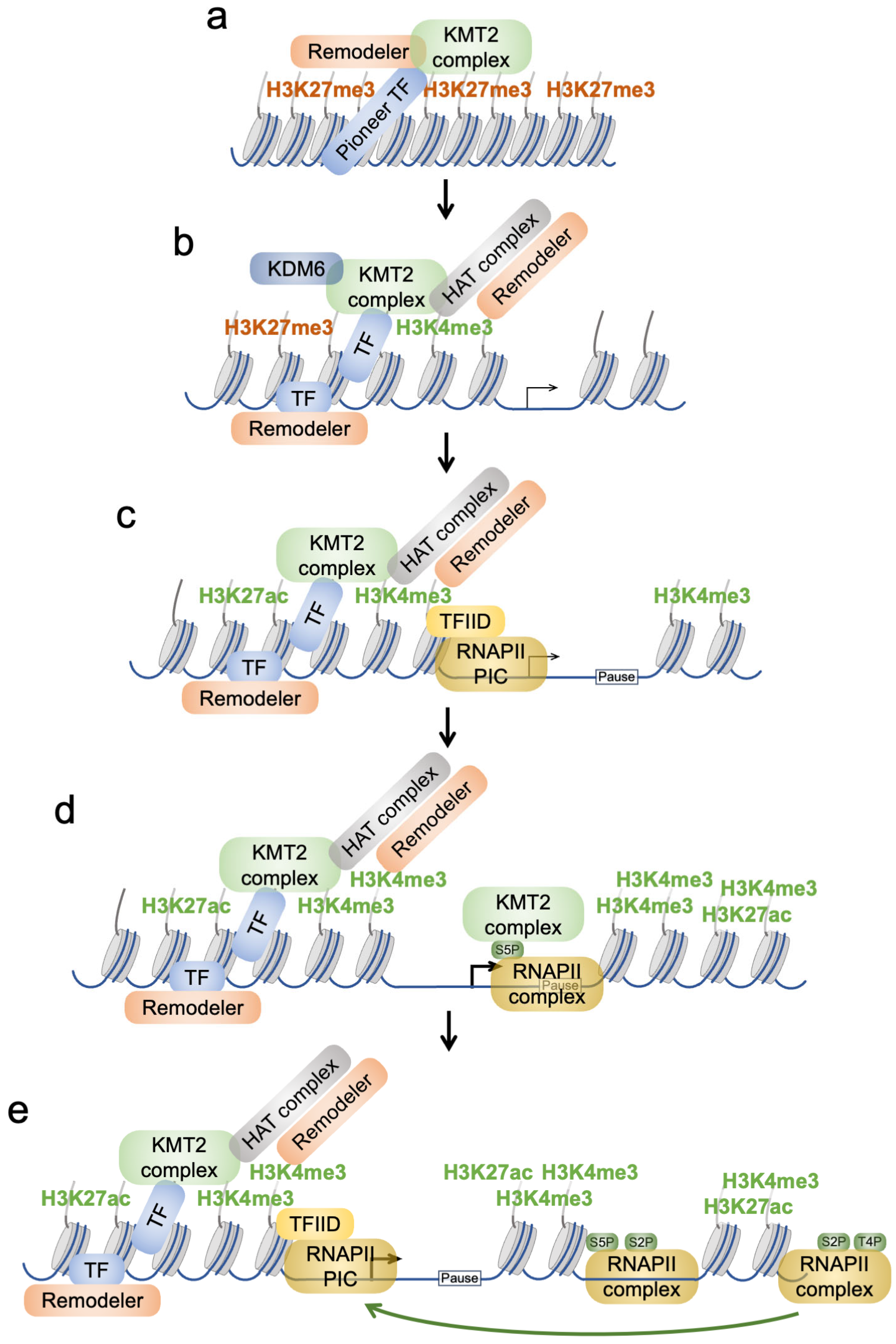
3. Locating KMT2 Complexes to Distinct Chromatin Sites
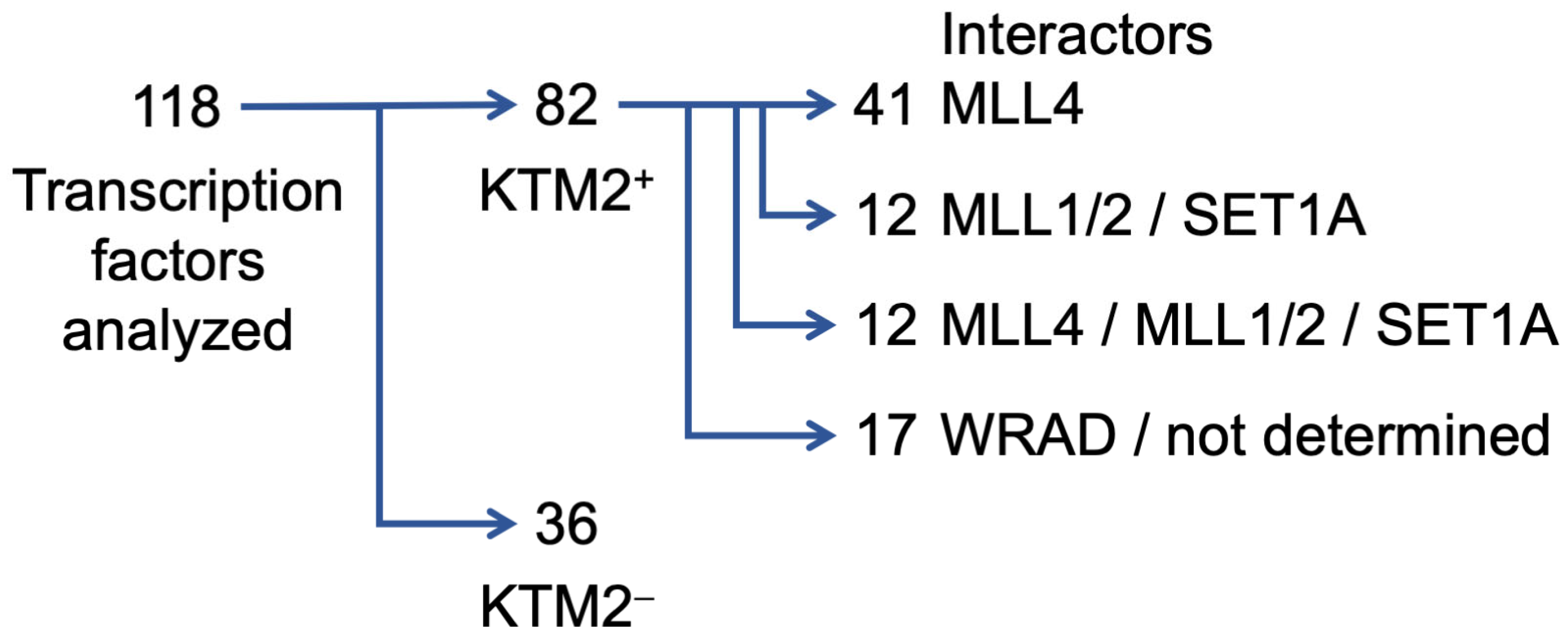
3.1. Sequence-Specific Transcription Factors Interact with KMT2 Complexes
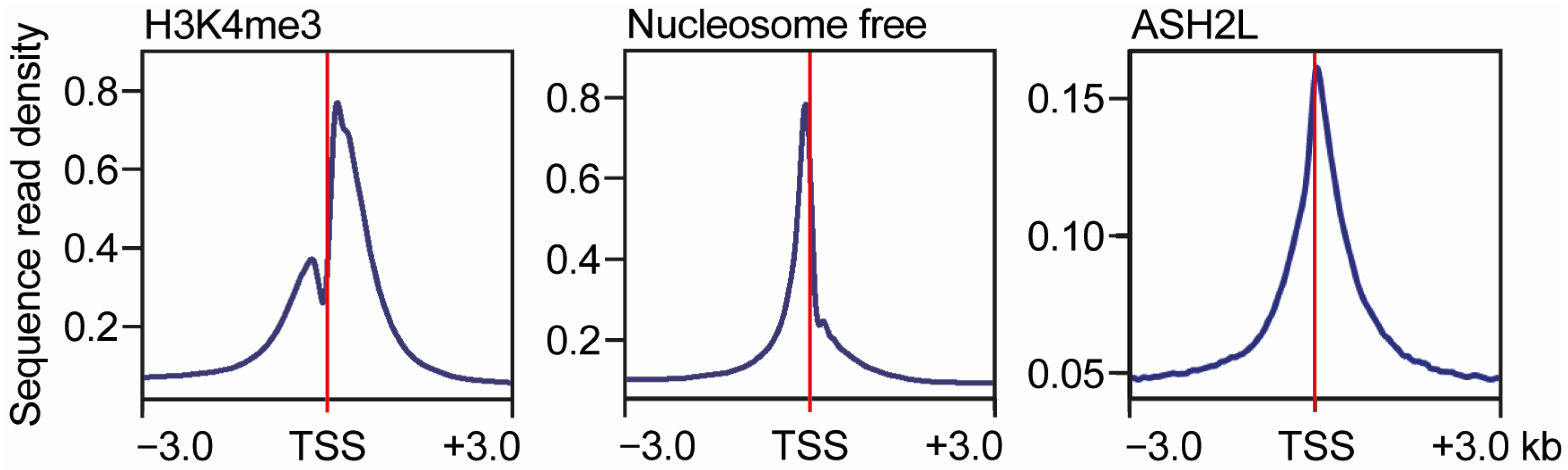
3.2. Positioning of KMT2 Complexes near Promoters
3.3. Positioning of KMT2 Complexes by Binding to CpG Island Promoters
4. Readers of H3K4 Methylation and Functional Consequences
4.1. Recruiting Cofactors by H3K4me3
4.2. Interactors at Enhancers and Response Elements
4.3. Recruitment and Regulation of the RNAPII Complex
4.4. Interplay of H3K4 Methylation with Other Histone Marks
5. Manipulating H3K4 Methylation and Their Consequences on Gene Expression
6. PROTAC Systems to Rapidly Control KMT2 Complex Functions
7. Is H3K4me3 Sufficient to Activate Gene Expression?
8. Does H3K4 Methylation Have a Role in Chromatin Organization?
9. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ac | Acetylation |
| ATAC-seq | Assay for Transposase accessible chromatin with high-throughput sequencing |
| BAH | Bromo-adjacent homology domain |
| CFP1 | CxxC zinc finger protein 1 |
| CGI | CpG island |
| ChIP-seq | Chromatin immunoprecipitation with high-throughput sequencing |
| COMPASS | Complex of proteins associated with SET1 |
| CTD | C-terminal domain of RNAPII |
| FAD | Flavin adenine dinucleotide |
| H3K4 | Histone H3 lysine 4 |
| IDR | Intrinsically disordered region |
| ING | Inhibitor of growth protein |
| KDM | Lysine demethylase |
| KMT | Lysine methyltransferase |
| me1-3 | Mono-, di- and tri-methylation |
| Rme2s | Symmetric di-methylation of arginine |
| Rme2a | Asymmetric di-methylation of arginine |
| MLL | Mixed lineage leukemia |
| NDR | Nucleosome-depleted regions |
| NURF | Nucleosome remodeling factor |
| PIC | RNAPII pre-initiation complex |
| PoI | Protein of interest |
| PROTAC | Proteolysis targeting chimera |
| PTM | Post-translational modification |
| RNAPII | DNA-dependent RNA polymerase II |
| S2P/S5P/T3P/T6P | Phosphorylated serine 2 or 5 or threonine 3 or 5 |
| SAGA | Spt-Ada-Gcn5 acetyltransferase complex |
| SAM | S-adenosyl-L-methionine |
| SET | Su(var)3-9, Enhancer-of-zeste and Trithorax domain |
| SMC | Structural maintenance of chromosome complex |
| SWI/SNF | SWItch/Sucrose Non-Fermentable remodeling complexes |
| TAD | Topologically associated domains |
| TAF | TATA-box binding protein associated factor |
| TF | Transcription factor |
References
- Lambert, S.A.; Jolma, A.; Campitelli, L.F.; Das, P.K.; Yin, Y.; Albu, M.; Chen, X.; Taipale, J.; Hughes, T.R.; Weirauch, M.T. The Human Transcription Factors. Cell 2018, 172, 650–665. [Google Scholar] [CrossRef]
- Andersson, R.; Sandelin, A. Determinants of Enhancer and Promoter Activities of Regulatory Elements. Nat. Rev. Genet. 2020, 21, 71–87. [Google Scholar] [CrossRef]
- Morgan, M.A.J.; Shilatifard, A. Reevaluating the Roles of Histone-Modifying Enzymes and Their Associated Chromatin Modifications in Transcriptional Regulation. Nat. Genet. 2020, 52, 1271–1281. [Google Scholar] [CrossRef]
- Cenik, B.K.; Shilatifard, A. Compass and Swi/Snf Complexes in Development and Disease. Nat. Rev. Genet. 2021, 22, 38–58. [Google Scholar] [CrossRef]
- Isbel, L.; Grand, R.S.; Schubeler, D. Generating Specificity in Genome Regulation through Transcription Factor Sensitivity to Chromatin. Nat. Rev. Genet. 2022, 23, 728–740. [Google Scholar] [CrossRef]
- Kornberg, R.D.; Lorch, Y. Perspective Primary Role of the Nucleosome. Mol. Cell 2020, 79, 371–375. [Google Scholar] [CrossRef]
- Luger, K.; Dechassa, M.L.; Tremethick, D.J. New Insights into Nucleosome and Chromatin Structure: An Ordered State or a Disordered Affair? Nat. Rev. Mol. Cell Biol. 2012, 13, 436–447. [Google Scholar] [CrossRef]
- Ahmad, K.; Henikoff, S.; Ramachandran, S. Managing the Steady State Chromatin Landscape by Nucleosome Dynamics. Annu. Rev. Biochem. 2022, 91, 183–195. [Google Scholar] [CrossRef]
- Janssen, S.M.; Lorincz, M.C. Interplay between Chromatin Marks in Development and Disease. Nat. Rev. Genet. 2022, 23, 137–153. [Google Scholar] [CrossRef]
- Millan-Zambrano, G.; Burton, A.; Bannister, A.J.; Schneider, R. Histone Post-Translational Modifications—Cause and Consequence of Genome Function. Nat. Rev. Genet. 2022, 23, 563–580. [Google Scholar] [CrossRef]
- McCarthy, R.L.; Zhang, J.; Zaret, K.S. Diverse Heterochromatin States Restricting Cell Identity and Reprogramming. Trends Biochem. Sci. 2023, 48, 513–526. [Google Scholar] [CrossRef]
- Chen, Y.C.; Koutelou, E.; Dent, S.Y.R. Now Open: Evolving Insights to the Roles of Lysine Acetylation in Chromatin Organization and Function. Mol. Cell 2022, 82, 716–727. [Google Scholar] [CrossRef]
- Rose, N.R.; Klose, R.J. Understanding the Relationship between DNA Methylation and Histone Lysine Methylation. Biochim. Biophys. Acta 2014, 1839, 1362–1372. [Google Scholar] [CrossRef]
- Wang, Z.A.; Cole, P.A. The Chemical Biology of Reversible Lysine Post-Translational Modifications. Cell Chem. Biol. 2020, 27, 953–969. [Google Scholar] [CrossRef]
- Jiang, H. The Complex Activities of the Set1/Mll Complex Core Subunits in Development and Disease. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194560. [Google Scholar] [CrossRef]
- Rao, R.C.; Dou, Y. Hijacked in Cancer: The Kmt2 (Mll) Family of Methyltransferases. Nat. Rev. Cancer 2015, 15, 334–346. [Google Scholar] [CrossRef]
- Wang, H.; Helin, K. Roles of H3k4 Methylation in Biology and Disease. Trends Cell Biol. 2025, 35, 115–128. [Google Scholar] [CrossRef]
- Yancoskie, M.N.; Maritz, C.; van Eijk, P.; Reed, S.H.; Naegeli, H. To Incise or Not and Where: Set-Domain Methyltransferases Know. Trends Biochem. Sci. 2023, 48, 321–330. [Google Scholar] [CrossRef]
- Damm, E.; Odenthal-Hesse, L. Orchestrating Recombination Initiation in Mice and Men. Curr. Top. Dev. Biol. 2023, 151, 27–42. [Google Scholar] [CrossRef]
- Murn, J.; Shi, Y. The Winding Path of Protein Methylation Research: Milestones and New Frontiers. Nat. Rev. Mol. Cell Biol. 2017, 18, 517–527. [Google Scholar] [CrossRef]
- Allfrey, V.G.; Faulkner, R.; Mirsky, A.E. Acetylation and Methylation of Histones and Their Possible Role in Regulation of Rna Synthesis. Proc. Natl. Acad. Sci. USA 1964, 51, 786–794. [Google Scholar] [CrossRef]
- Strahl, B.D.; Ohba, R.; Cook, R.G.; Allis, C.D. Methylation of Histone H3 at Lysine 4 Is Highly Conserved and Correlates with Transcriptionally Active Nuclei in Tetrahymena. Proc. Natl. Acad. Sci. USA 1999, 96, 14967–14972. [Google Scholar] [CrossRef]
- Strahl, B.D.; Allis, C.D. The Language of Covalent Histone Modifications. Nature 2000, 403, 41–45. [Google Scholar] [CrossRef]
- Jenuwein, T.; Allis, C.D. Translating the Histone Code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef]
- Cosgrove, M.S.; Boeke, J.D.; Wolberger, C. Regulated Nucleosome Mobility and the Histone Code. Nat. Struct. Mol. Biol. 2004, 11, 1037–1043. [Google Scholar] [CrossRef]
- Briggs, S.D.; Bryk, M.; Strahl, B.D.; Cheung, W.L.; Davie, J.K.; Dent, S.Y.; Winston, F.; Allis, C.D. Histone H3 Lysine 4 Methylation Is Mediated by Set1 and Required for Cell Growth and Rdna Silencing in Saccharomyces Cerevisiae. Genes Dev. 2001, 15, 3286–3295. [Google Scholar] [CrossRef]
- Santos-Rosa, H.; Schneider, R.; Bannister, A.J.; Sherriff, J.; Bernstein, B.E.; Emre, N.C.; Schreiber, S.L.; Mellor, J.; Kouzarides, T. Active Genes Are Tri-Methylated at K4 of Histone H3. Nature 2002, 419, 407–411. [Google Scholar] [CrossRef]
- Allis, C.D.; Berger, S.L.; Cote, J.; Dent, S.; Jenuwien, T.; Kouzarides, T.; Pillus, L.; Reinberg, D.; Shi, Y.; Shiekhattar, R.; et al. New Nomenclature for Chromatin-Modifying Enzymes. Cell 2007, 131, 633–636. [Google Scholar] [CrossRef]
- Miller, T.; Krogan, N.J.; Dover, J.; Erdjument-Bromage, H.; Tempst, P.; Johnston, M.; Greenblatt, J.F.; Shilatifard, A. Compass: A Complex of Proteins Associated with a Trithorax-Related Set Domain Protein. Proc. Natl. Acad. Sci. USA 2001, 98, 12902–12907. [Google Scholar] [CrossRef]
- Bochynska, A.; Luscher-Firzlaff, J.; Luscher, B. Modes of Interaction of Kmt2 Histone H3 Lysine 4 Methyltransferase/Compass Complexes with Chromatin. Cells 2018, 7, 17. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, K.; Yin, L.; Yu, Y.; Qi, J.; Shen, W.H.; Zhu, J.; Zhang, Y.; Dong, A. H3k4me2 Functions as a Repressive Epigenetic Mark in Plants. Epigenetics Chromatin 2019, 12, 40. [Google Scholar] [CrossRef]
- Hurton, M.D.; Miller, J.M.; Lee, M.T. H3k4me2 Distinguishes a Distinct Class of Enhancers During the Maternal-to-Zygotic Transition. PLoS Biol. 2025, 23, e3003239. [Google Scholar] [CrossRef]
- Kim, T.; Buratowski, S. Dimethylation of H3k4 by Set1 Recruits the Set3 Histone Deacetylase Complex to 5′ Transcribed Regions. Cell 2009, 137, 259–272. [Google Scholar] [CrossRef]
- Di Tullio, F.; Schwarz, M.; Zorgati, H.; Mzoughi, S.; Guccione, E. The Duality of Prdm Proteins: Epigenetic and Structural Perspectives. FEBS J. 2022, 289, 1256–1275. [Google Scholar] [CrossRef]
- Grey, C.; Baudat, F.; de Massy, B. Prdm9, a Driver of the Genetic Map. PLoS Genet. 2018, 14, e1007479. [Google Scholar] [CrossRef]
- Shimazaki, N.; Lieber, M.R. Histone Methylation and V(D)J Recombination. Int. J. Hematol. 2014, 100, 230–237. [Google Scholar] [CrossRef]
- Ji, Z.; Sheng, Y.; Miao, J.; Li, X.; Zhao, H.; Wang, J.; Cheng, C.; Wang, X.; Liu, K.; Zhang, K.; et al. The Histone Methyltransferase Setd2 Is Indispensable for V(D)J Recombination. Nat. Commun. 2019, 10, 3353. [Google Scholar] [CrossRef]
- Matthews, A.G.; Kuo, A.J.; Ramon-Maiques, S.; Han, S.; Champagne, K.S.; Ivanov, D.; Gallardo, M.; Carney, D.; Cheung, P.; Ciccone, D.N.; et al. Rag2 Phd Finger Couples Histone H3 Lysine 4 Trimethylation with V(D)J Recombination. Nature 2007, 450, 1106–1110. [Google Scholar] [CrossRef]
- Poreba, E.; Lesniewicz, K.; Durzynska, J. Histone-Lysine N-Methyltransferase 2 (Kmt2) Complexes—A New Perspective. Mutat. Res. Rev. Mutat. Res. 2022, 790, 108443. [Google Scholar] [CrossRef]
- Shi, Y.; Lan, F.; Matson, C.; Mulligan, P.; Whetstine, J.R.; Cole, P.A.; Casero, R.A.; Shi, Y. Histone Demethylation Mediated by the Nuclear Amine Oxidase Homolog Lsd1. Cell 2004, 119, 941–953. [Google Scholar] [CrossRef]
- Gray, Z.H.; Honer, M.A.; Ghatalia, P.; Shi, Y.; Whetstine, J.R. 20 Years of Histone Lysine Demethylases: From Discovery to the Clinic and Beyond. Cell 2025, 188, 1747–1783. [Google Scholar] [CrossRef]
- Rasmussen, P.B.; Staller, P. The Kdm5 Family of Histone Demethylases as Targets in Oncology Drug Discovery. Epigenomics 2014, 6, 277–286. [Google Scholar] [CrossRef]
- Hu, S.; Song, A.; Peng, L.; Tang, N.; Qiao, Z.; Wang, Z.; Lan, F.; Chen, F.X. H3k4me2/3 Modulate the Stability of Rna Polymerase Ii Pausing. Cell Res. 2023, 33, 403–406. [Google Scholar] [CrossRef]
- Wang, H.; Fan, Z.; Shliaha, P.V.; Miele, M.; Hendrickson, R.C.; Jiang, X.; Helin, K. H3k4me3 Regulates Rna Polymerase Ii Promoter-Proximal Pause-Release. Nature 2023, 615, 339–348. [Google Scholar] [CrossRef]
- Gold, S.; Shilatifard, A. Epigenetic Therapies Targeting Histone Lysine Methylation: Complex Mechanisms and Clinical Challenges. J. Clin. Investig. 2024, 134, e183391. [Google Scholar] [CrossRef]
- Yu, H.; Lesch, B.J. Functional Roles of H3k4 Methylation in Transcriptional Regulation. Mol. Cell Biol. 2024, 44, 505–515. [Google Scholar] [CrossRef]
- Guenther, M.G.; Levine, S.S.; Boyer, L.A.; Jaenisch, R.; Young, R.A. A Chromatin Landmark and Transcription Initiation at Most Promoters in Human Cells. Cell 2007, 130, 77–88. [Google Scholar] [CrossRef]
- Mikkelsen, T.S.; Ku, M.C.; Jaffe, D.B.; Issac, B.; Lieberman, E.; Giannoukos, G.; Alvarez, P.; Brockman, W.; Kim, T.K.; Koche, R.P.; et al. Genome-Wide Maps of Chromatin State in Pluripotent and Lineage-Committed Cells. Nature 2007, 448, 553–560. [Google Scholar] [CrossRef]
- Lloret-Llinares, M.; Perez-Lluch, S.; Rossell, D.; Moran, T.; Ponsa-Cobas, J.; Auer, H.; Corominas, M.; Azorin, F. Dkdm5/Lid Regulates H3k4me3 Dynamics at the Transcription-Start Site (Tss) of Actively Transcribed Developmental Genes. Nucleic Acids Res. 2012, 40, 9493–9505. [Google Scholar] [CrossRef]
- Heikamp, E.B.; Henrich, J.A.; Perner, F.; Wong, E.M.; Hatton, C.; Wen, Y.; Barwe, S.P.; Gopalakrishnapillai, A.; Xu, H.; Uckelmann, H.J.; et al. The Menin-Mll1 Interaction Is a Molecular Dependency in Nup98-Rearranged Aml. Blood 2022, 139, 894–906. [Google Scholar] [CrossRef]
- Barsoum, M.; Sayadi-Boroujeni, R.; Stenzel, A.T.; Bussmann, P.; Luscher-Firzlaff, J.; Luscher, B. Sequential Deregulation of Histone Marks, Chromatin Accessibility and Gene Expression in Response to Protac-Induced Degradation of Ash2l. Sci. Rep. 2023, 13, 22565. [Google Scholar] [CrossRef]
- Hughes, A.L.; Szczurek, A.T.; Kelley, J.R.; Lastuvkova, A.; Turberfield, A.H.; Dimitrova, E.; Blackledge, N.P.; Klose, R.J. A Cpg Island-Encoded Mechanism Protects Genes from Premature Transcription Termination. Nat. Commun. 2023, 14, 726. [Google Scholar] [CrossRef]
- Park, S.; Kim, G.W.; Kwon, S.H.; Lee, J.S. Broad Domains of Histone H3 Lysine 4 Trimethylation in Transcriptional Regulation and Disease. FEBS J. 2020, 287, 2891–2902. [Google Scholar] [CrossRef]
- Beacon, T.H.; Delcuve, G.P.; Lopez, C.; Nardocci, G.; Kovalchuk, I.; van Wijnen, A.J.; Davie, J.R. The Dynamic Broad Epigenetic (H3k4me3, H3k27ac) Domain as a Mark of Essential Genes. Clin. Epigenetics 2021, 13, 138. [Google Scholar] [CrossRef]
- Bai, L.; Morozov, A.V. Gene Regulation by Nucleosome Positioning. Trends Genet. 2010, 26, 476–483. [Google Scholar] [CrossRef]
- Muller, F.; Tora, L. Chromatin and DNA Sequences in Defining Promoters for Transcription Initiation. Biochim. Biophys. Acta 2014, 1839, 118–128. [Google Scholar] [CrossRef]
- Pokholok, D.K.; Harbison, C.T.; Levine, S.; Cole, M.; Hannett, N.M.; Lee, T.I.; Bell, G.W.; Walker, K.; Rolfe, P.A.; Herbolsheimer, E.; et al. Genome-Wide Map of Nucleosome Acetylation and Methylation in Yeast. Cell 2005, 122, 517–527. [Google Scholar] [CrossRef]
- Barsoum, M.; Stenzel, A.T.; Bochynska, A.; Kuo, C.C.; Tsompanidis, A.; Sayadi-Boroujeni, R.; Bussmann, P.; Luscher-Firzlaff, J.; Costa, I.G.; Luscher, B. Loss of the Ash2l Subunit of Histone H3k4 Methyltransferase Complexes Reduces Chromatin Accessibility at Promoters. Sci. Rep. 2022, 12, 21506. [Google Scholar] [CrossRef]
- Fields, J.K.; Hicks, C.W.; Wolberger, C. Diverse Modes of Regulating Methyltransferase Activity by Histone Ubiquitination. Curr. Opin. Struct. Biol. 2023, 82, 102649. [Google Scholar] [CrossRef]
- Chan, J.; Kumar, A.; Kono, H. Rnapii Driven Post-Translational Modifications of Nucleosomal Histones. Trends Genet. 2022, 38, 1076–1095. [Google Scholar] [CrossRef]
- Barral, A.; Zaret, K.S. Pioneer Factors: Roles and Their Regulation in Development. Trends Genet. 2024, 40, 134–148. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, K.; Brahma, S.; Henikoff, S. Epigenetic Pioneering by Swi/Snf Family Remodelers. Mol. Cell 2024, 84, 194–201. [Google Scholar] [CrossRef]
- Goos, H.; Kinnunen, M.; Salokas, K.; Tan, Z.; Liu, X.; Yadav, L.; Zhang, Q.; Wei, G.H.; Varjosalo, M. Human Transcription Factor Protein Interaction Networks. Nat. Commun. 2022, 13, 766. [Google Scholar] [CrossRef]
- Mohammadparast, S.; Chang, C. Ash2l, an Obligatory Component of H3k4 Methylation Complexes, Regulates Neural Crest Development. Dev. Biol. 2022, 492, 14–24. [Google Scholar] [CrossRef]
- Ullius, A.; Luscher-Firzlaff, J.; Costa, I.G.; Walsemann, G.; Forst, A.H.; Gusmao, E.G.; Kapelle, K.; Kleine, H.; Kremmer, E.; Vervoorts, J.; et al. The Interaction of Myc with the Trithorax Protein Ash2l Promotes Gene Transcription by Regulating H3k27 Modification. Nucleic Acids Res. 2014, 42, 6901–6920. [Google Scholar] [CrossRef] [PubMed]
- Luscher, B.; Vervoorts, J. Regulation of Gene Transcription by the Oncoprotein Myc. Gene 2012, 494, 145–160. [Google Scholar] [CrossRef]
- Thomas, L.R.; Wang, Q.; Grieb, B.C.; Phan, J.; Foshage, A.M.; Sun, Q.; Olejniczak, E.T.; Clark, T.; Dey, S.; Lorey, S.; et al. Interaction with Wdr5 Promotes Target Gene Recognition and Tumorigenesis by Myc. Mol. Cell 2015, 58, 440–452. [Google Scholar] [CrossRef]
- Staller, M.V. Transcription Factors Perform a 2-Step Search of the Nucleus. Genetics 2022, 222, iyac111. [Google Scholar] [CrossRef]
- Brodsky, S.; Jana, T.; Barkai, N. Order through Disorder: The Role of Intrinsically Disordered Regions in Transcription Factor Binding Specificity. Curr. Opin. Struct. Biol. 2021, 71, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Soto, L.F.; Li, Z.; Santoso, C.S.; Berenson, A.; Ho, I.; Shen, V.X.; Yuan, S.; Fuxman Bass, J.I. Compendium of Human Transcription Factor Effector Domains. Mol. Cell 2022, 82, 514–526. [Google Scholar] [CrossRef]
- Pei, G.; Lyons, H.; Li, P.; Sabari, B.R. Transcription Regulation by Biomolecular Condensates. Nat. Rev. Mol. Cell Biol. 2025, 26, 213–236. [Google Scholar] [CrossRef]
- Vinson, C.; Chatterjee, R.; Fitzgerald, P. Transcription Factor Binding Sites and Other Features in Human and Drosophila Proximal Promoters. Subcell. Biochem. 2011, 52, 205–222. [Google Scholar] [CrossRef]
- Koudritsky, M.; Domany, E. Positional Distribution of Human Transcription Factor Binding Sites. Nucleic Acids Res. 2008, 36, 6795–6805. [Google Scholar] [CrossRef]
- Wagh, K.; Stavreva, D.A.; Hager, G.L. Transcription Dynamics and Genome Organization in the Mammalian Nucleus: Recent Advances. Mol. Cell 2025, 85, 208–224. [Google Scholar] [CrossRef]
- Yang, Y.W.; Flynn, R.A.; Chen, Y.; Qu, K.; Wan, B.; Wang, K.C.; Lei, M.; Chang, H.Y. Essential Role of Lncrna Binding for Wdr5 Maintenance of Active Chromatin and Embryonic Stem Cell Pluripotency. eLife 2014, 3, e02046. [Google Scholar] [CrossRef] [PubMed]
- Bumpous, L.A.; Moe, K.C.; Wang, J.; Carver, L.A.; Williams, A.G.; Romer, A.S.; Scobee, J.D.; Maxwell, J.N.; Jones, C.A.; Chung, D.H.; et al. Wdr5 Facilitates Recruitment of N-Myc to Conserved Wdr5 Gene Targets in Neuroblastoma Cell Lines. Oncogenesis 2023, 12, 32. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, R.; Okuda, H.; Kanai, A.; Takahashi, S.; Kawamura, T.; Matsui, H.; Kitamura, T.; Kitabayashi, I.; Inaba, T.; Yokoyama, A. Activation of Cpg-Rich Promoters Mediated by Mll Drives Moz-Rearranged Leukemia. Cell Rep. 2020, 32, 108200. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Kanai, A.; Okuda, H.; Miyamoto, R.; Komata, Y.; Kawamura, T.; Matsui, H.; Inaba, T.; Takaori-Kondo, A.; Yokoyama, A. Hbo1-Mll Interaction Promotes Af4/Enl/P-Tefb-Mediated Leukemogenesis. eLife 2021, 10, e65872. [Google Scholar] [CrossRef]
- Denissov, S.; Hofemeister, H.; Marks, H.; Kranz, A.; Ciotta, G.; Singh, S.; Anastassiadis, K.; Stunnenberg, H.G.; Stewart, A.F. Mll2 Is Required for H3k4 Trimethylation on Bivalent Promoters in Embryonic Stem Cells, Whereas Mll1 Is Redundant. Development 2014, 141, 526–537. [Google Scholar] [CrossRef]
- Franks, T.M.; McCloskey, A.; Shokirev, M.N.; Benner, C.; Rathore, A.; Hetzer, M.W. Nup98 Recruits the Wdr82-Set1a/Compass Complex to Promoters to Regulate H3k4 Trimethylation in Hematopoietic Progenitor Cells. Genes Dev. 2017, 31, 2222–2234. [Google Scholar] [CrossRef]
- Lee, J.H.; Skalnik, D.G. Wdr82 Is a C-Terminal Domain-Binding Protein That Recruits the Setd1a Histone H3-Lys4 Methyltransferase Complex to Transcription Start Sites of Transcribed Human Genes. Mol. Cell Biol. 2008, 28, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Wang, P.F.; Lee, J.S.; Martin-Brown, S.; Florens, L.; Washburn, M.; Shilatifard, A. Molecular Regulation of H3k4 Trimethylation by Wdr82, a Component of Human Set1/Compass. Mol. Cell Biol. 2008, 28, 7337–7344. [Google Scholar] [CrossRef] [PubMed]
- Bae, H.J.; Dubarry, M.; Jeon, J.; Soares, L.M.; Dargemont, C.; Kim, J.; Geli, V.; Buratowski, S. The Set1 N-Terminal Domain and Swd2 Interact with Rna Polymerase Ii Ctd to Recruit Compass. Nat. Commun. 2020, 11, 2181. [Google Scholar] [CrossRef]
- Ng, H.H.; Robert, F.; Young, R.A.; Struhl, K. Targeted Recruitment of Set1 Histone Methylase by Elongating Pol Ii Provides a Localized Mark and Memory of Recent Transcriptional Activity. Mol. Cell 2003, 11, 709–719. [Google Scholar] [CrossRef] [PubMed]
- Eick, D.; Geyer, M. The Rna Polymerase Ii Carboxy-Terminal Domain (Ctd) Code. Chem. Rev. 2013, 113, 8456–8490. [Google Scholar] [CrossRef]
- Xu, C.; Bian, C.; Lam, R.; Dong, A.; Min, J. The Structural Basis for Selective Binding of Non-Methylated Cpg Islands by the Cfp1 Cxxc Domain. Nat. Commun. 2011, 2, 227. [Google Scholar] [CrossRef]
- Thomson, J.P.; Skene, P.J.; Selfridge, J.; Clouaire, T.; Guy, J.; Webb, S.; Kerr, A.R.; Deaton, A.; Andrews, R.; James, K.D.; et al. Cpg Islands Influence Chromatin Structure Via the Cpg-Binding Protein Cfp1. Nature 2010, 464, 1082–1086. [Google Scholar] [CrossRef]
- van de Lagemaat, L.N.; Flenley, M.; Lynch, M.D.; Garrick, D.; Tomlinson, S.R.; Kranc, K.R.; Vernimmen, D. Cpg Binding Protein (Cfp1) Occupies Open Chromatin Regions of Active Genes, Including Enhancers and Non-Cpg Islands. Epigenetics Chromatin 2018, 11, 59. [Google Scholar] [CrossRef]
- Clouaire, T.; Webb, S.; Bird, A. Cfp1 Is Required for Gene Expression-Dependent H3k4 Trimethylation and H3k9 Acetylation in Embryonic Stem Cells. Genome Biol. 2014, 15, 451. [Google Scholar] [CrossRef]
- Clouaire, T.; Webb, S.; Skene, P.; Illingworth, R.; Kerr, A.; Andrews, R.; Lee, J.H.; Skalnik, D.; Bird, A. Cfp1 Integrates Both Cpg Content and Gene Activity for Accurate H3k4me3 Deposition in Embryonic Stem Cells. Genes Dev. 2012, 26, 1714–1728. [Google Scholar] [CrossRef]
- Brown, D.A.; Di Cerbo, V.; Feldmann, A.; Ahn, J.; Ito, S.; Blackledge, N.P.; Nakayama, M.; McClellan, M.; Dimitrova, E.; Turberfield, A.H.; et al. The Set1 Complex Selects Actively Transcribed Target Genes Via Multivalent Interaction with Cpg Island Chromatin. Cell Rep. 2017, 20, 2313–2327. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Kachirskaia, I.; Walter, K.L.; Kuo, J.H.; Lake, A.; Davrazou, F.; Chan, S.M.; Martin, D.G.; Fingerman, I.M.; Briggs, S.D.; et al. Proteome-Wide Analysis in Saccharomyces Cerevisiae Identifies Several Phd Fingers as Novel Direct and Selective Binding Modules of Histone H3 Methylated at Either Lysine 4 or Lysine 36. J. Biol. Chem. 2007, 282, 2450–2455. [Google Scholar] [CrossRef]
- He, C.; Liu, N.; Xie, D.Y.; Liu, Y.H.; Xiao, Y.Z.; Li, F.D. Structural Basis for Histone H3k4me3 Recognition by the N-Terminal Domain of the Phd Finger Protein Spp1. Biochem. J. 2019, 476, 1957–1973. [Google Scholar] [CrossRef]
- Hu, D.; Gao, X.; Cao, K.; Morgan, M.A.; Mas, G.; Smith, E.R.; Volk, A.G.; Bartom, E.T.; Crispino, J.D.; Di Croce, L.; et al. Not All H3k4 Methylations Are Created Equal: Mll2/Compass Dependency in Primordial Germ Cell Specification. Mol. Cell 2017, 65, 460–475.e6. [Google Scholar] [CrossRef]
- Stroynowska-Czerwinska, A.M.; Klimczak, M.; Pastor, M.; Kazrani, A.A.; Misztal, K.; Bochtler, M. Clustered Phd Domains in Kmt2/Mll Proteins Are Attracted by H3k4me3 and H3 Acetylation-Rich Active Promoters and Enhancers. Cell Mol. Life Sci. 2023, 80, 23. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.L.; Kelley, J.R.; Klose, R.J. Understanding the Interplay between Cpg Island-Associated Gene Promoters and H3k4 Methylation. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194567. [Google Scholar] [CrossRef] [PubMed]
- Musselman, C.A.; Khorasanizadeh, S.; Kutateladze, T.G. Towards Understanding Methyllysine Readout. Biochim. Biophys. Acta 2014, 1839, 686–693. [Google Scholar] [CrossRef]
- Hyun, K.; Jeon, J.; Park, K.; Kim, J. Writing, Erasing and Reading Histone Lysine Methylations. Exp. Mol. Med. 2017, 49, e324. [Google Scholar] [CrossRef]
- Patel, D.J.; Wang, Z. Readout of Epigenetic Modifications. Annu. Rev. Biochem. 2013, 82, 81–118. [Google Scholar] [CrossRef]
- Ruthenburg, A.J.; Li, H.; Milne, T.A.; Dewell, S.; McGinty, R.K.; Yuen, M.; Ueberheide, B.; Dou, Y.; Muir, T.W.; Patel, D.J.; et al. Recognition of a Mononucleosomal Histone Modification Pattern by Bptf Via Multivalent Interactions. Cell 2011, 145, 692–706. [Google Scholar] [CrossRef]
- Li, H.; Ilin, S.; Wang, W.; Duncan, E.M.; Wysocka, J.; Allis, C.D.; Patel, D.J. Molecular Basis for Site-Specific Read-out of Histone H3k4me3 by the Bptf Phd Finger of Nurf. Nature 2006, 442, 91–95. [Google Scholar] [CrossRef]
- Wysocka, J.; Swigut, T.; Xiao, H.; Milne, T.A.; Kwon, S.Y.; Landry, J.; Kauer, M.; Tackett, A.J.; Chait, B.T.; Badenhorst, P.; et al. A Phd Finger of Nurf Couples Histone H3 Lysine 4 Trimethylation with Chromatin Remodelling. Nature 2006, 442, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Sims, R.J., 3rd; Millhouse, S.; Chen, C.F.; Lewis, B.A.; Erdjument-Bromage, H.; Tempst, P.; Manley, J.L.; Reinberg, D. Recognition of Trimethylated Histone H3 Lysine 4 Facilitates the Recruitment of Transcription Postinitiation Factors and Pre-Mrna Splicing. Mol. Cell 2007, 28, 665–676. [Google Scholar] [CrossRef]
- Alkhatib, S.G.; Landry, J.W. The Nucleosome Remodeling Factor. FEBS Lett. 2011, 585, 3197–3207. [Google Scholar] [CrossRef]
- Flanagan, J.F.; Mi, L.Z.; Chruszcz, M.; Cymborowski, M.; Clines, K.L.; Kim, Y.; Minor, W.; Rastinejad, F.; Khorasanizadeh, S. Double Chromodomains Cooperate to Recognize the Methylated Histone H3 Tail. Nature 2005, 438, 1181–1185. [Google Scholar] [CrossRef] [PubMed]
- Sims, R.J., 3rd; Chen, C.F.; Santos-Rosa, H.; Kouzarides, T.; Patel, S.S.; Reinberg, D. Human but Not Yeast Chd1 Binds Directly and Selectively to Histone H3 Methylated at Lysine 4 Via Its Tandem Chromodomains. J. Biol. Chem. 2005, 280, 41789–41792. [Google Scholar] [CrossRef]
- Bulut-Karslioglu, A.; Jin, H.; Kim, Y.K.; Cho, B.; Guzman-Ayala, M.; Williamson, A.J.K.; Hejna, M.; Stotzel, M.; Whetton, A.D.; Song, J.S.; et al. Chd1 Protects Genome Integrity at Promoters to Sustain Hypertranscription in Embryonic Stem Cells. Nat. Commun. 2021, 12, 4859. [Google Scholar] [CrossRef]
- Coles, A.H.; Jones, S.N. The Ing Gene Family in the Regulation of Cell Growth and Tumorigenesis. J. Cell. Physiol. 2009, 218, 45–57. [Google Scholar] [CrossRef]
- Taheri, M.; Hussen, B.M.; Najafi, S.; Abak, A.; Ghafouri-Fard, S.; Samsami, M.; Baniahmad, A. Molecular Mechanisms of Inhibitor of Growth (Ing) Family Members in Health and Malignancy. Cancer Cell Int. 2022, 22, 272. [Google Scholar] [CrossRef] [PubMed]
- Dantas, A.; Al Shueili, B.; Yang, Y.; Nabbi, A.; Fink, D.; Riabowol, K. Biological Functions of the Ing Proteins. Cancers 2019, 11, 1817. [Google Scholar] [CrossRef]
- Lukauskas, S.; Tvardovskiy, A.; Nguyen, N.V.; Stadler, M.; Faull, P.; Ravnsborg, T.; Ozdemir Aygenli, B.; Dornauer, S.; Flynn, H.; Lindeboom, R.G.H.; et al. Decoding Chromatin States by Proteomic Profiling of Nucleosome Readers. Nature 2024, 627, 671–679. [Google Scholar] [CrossRef]
- Vermeulen, M.; Eberl, H.C.; Matarese, F.; Marks, H.; Denissov, S.; Butter, F.; Lee, K.K.; Olsen, J.V.; Hyman, A.A.; Stunnenberg, H.G.; et al. Quantitative Interaction Proteomics and Genome-Wide Profiling of Epigenetic Histone Marks and Their Readers. Cell 2010, 142, 967–980. [Google Scholar] [CrossRef]
- Bian, C.; Xu, C.; Ruan, J.; Lee, K.K.; Burke, T.L.; Tempel, W.; Barsyte, D.; Li, J.; Wu, M.; Zhou, B.O.; et al. Sgf29 Binds Histone H3k4me2/3 and Is Required for Saga Complex Recruitment and Histone H3 Acetylation. EMBO J. 2011, 30, 2829–2842. [Google Scholar] [CrossRef]
- Helmlinger, D.; Papai, G.; Devys, D.; Tora, L. What Do the Structures of Gcn5-Containing Complexes Teach Us About Their Function? Biochim. Biophys. Acta Gene Regul. Mech. 2021, 1864, 194614. [Google Scholar] [CrossRef]
- Espinola-Lopez, J.M.; Tan, S. The Ada2/Ada3/Gcn5/Sgf29 Histone Acetyltransferase Module. Biochim. Biophys. Acta Gene Regul. Mech. 2021, 1864, 194629. [Google Scholar] [CrossRef] [PubMed]
- Grewal, S.I.S. The Molecular Basis of Heterochromatin Assembly and Epigenetic Inheritance. Mol. Cell 2023, 83, 1767–1785. [Google Scholar] [CrossRef]
- Wen, H.; Li, J.; Song, T.; Lu, M.; Kan, P.Y.; Lee, M.G.; Sha, B.; Shi, X. Recognition of Histone H3k4 Trimethylation by the Plant Homeodomain of Phf2 Modulates Histone Demethylation. J. Biol. Chem. 2010, 285, 9322–9326. [Google Scholar] [CrossRef] [PubMed]
- Local, A.; Huang, H.; Albuquerque, C.P.; Singh, N.; Lee, A.Y.; Wang, W.; Wang, C.; Hsia, J.E.; Shiau, A.K.; Ge, K.; et al. Identification of H3k4me1-Associated Proteins at Mammalian Enhancers. Nat. Genet. 2018, 50, 73–82. [Google Scholar] [CrossRef]
- Takaku, M.; Grimm, S.A.; Shimbo, T.; Perera, L.; Menafra, R.; Stunnenberg, H.G.; Archer, T.K.; Machida, S.; Kurumizaka, H.; Wade, P.A. Gata3-Dependent Cellular Reprogramming Requires Activation-Domain Dependent Recruitment of a Chromatin Remodeler. Genome Biol. 2016, 17, 36. [Google Scholar] [CrossRef] [PubMed]
- Moonen, J.R.; Chappell, J.; Shi, M.; Shinohara, T.; Li, D.; Mumbach, M.R.; Zhang, F.; Nair, R.V.; Nasser, J.; Mai, D.H.; et al. Klf4 Recruits Swi/Snf to Increase Chromatin Accessibility and Reprogram the Endothelial Enhancer Landscape under Laminar Shear Stress. Nat. Commun. 2022, 13, 4941. [Google Scholar] [CrossRef]
- King, H.W.; Klose, R.J. The Pioneer Factor Oct4 Requires the Chromatin Remodeller Brg1 to Support Gene Regulatory Element Function in Mouse Embryonic Stem Cells. eLife 2017, 6, e22631. [Google Scholar] [CrossRef]
- Cheng, S.W.; Davies, K.P.; Yung, E.; Beltran, R.J.; Yu, J.; Kalpana, G.V. C-Myc Interacts with Ini1/Hsnf5 and Requires the Swi/Snf Complex for Transactivation Function. Nat. Genet. 1999, 22, 102–105. [Google Scholar] [CrossRef] [PubMed]
- Wolf, B.K.; Zhao, Y.; McCray, A.; Hawk, W.H.; Deary, L.T.; Sugiarto, N.W.; LaCroix, I.S.; Gerber, S.A.; Cheng, C.; Wang, X. Cooperation of Chromatin Remodeling Swi/Snf Complex and Pioneer Factor Ap-1 Shapes 3d Enhancer Landscapes. Nat. Struct. Mol. Biol. 2023, 30, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Malik, S.; Roeder, R.G. Regulation of the Rna Polymerase Ii Pre-Initiation Complex by Its Associated Coactivators. Nat. Rev. Genet. 2023, 24, 767–782. [Google Scholar] [CrossRef] [PubMed]
- Vermeulen, M.; Mulder, K.W.; Denissov, S.; Pijnappel, W.W.; van Schaik, F.M.; Varier, R.A.; Baltissen, M.P.; Stunnenberg, H.G.; Mann, M.; Timmers, H.T. Selective Anchoring of Tfiid to Nucleosomes by Trimethylation of Histone H3 Lysine 4. Cell 2007, 131, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Lauberth, S.M.; Nakayama, T.; Wu, X.; Ferris, A.L.; Tang, Z.; Hughes, S.H.; Roeder, R.G. H3k4me3 Interactions with Taf3 Regulate Preinitiation Complex Assembly and Selective Gene Activation. Cell 2013, 152, 1021–1036. [Google Scholar] [CrossRef]
- van Ingen, H.; van Schaik, F.M.; Wienk, H.; Ballering, J.; Rehmann, H.; Dechesne, A.C.; Kruijzer, J.A.; Liskamp, R.M.; Timmers, H.T.; Boelens, R. Structural Insight into the Recognition of the H3k4me3 Mark by the Tfiid Subunit Taf3. Structure 2008, 16, 1245–1256. [Google Scholar] [CrossRef]
- Chen, F.X.; Smith, E.R.; Shilatifard, A. Born to Run: Control of Transcription Elongation by Rna Polymerase Ii. Nat. Rev. Mol. Cell Biol. 2018, 19, 464–478. [Google Scholar] [CrossRef]
- Harlen, K.M.; Churchman, L.S. The Code and Beyond: Transcription Regulation by the Rna Polymerase Ii Carboxy-Terminal Domain. Nat. Rev. Mol. Cell Biol. 2017, 18, 263–273. [Google Scholar] [CrossRef]
- Shandilya, J.; Roberts, S.G. The Transcription Cycle in Eukaryotes: From Productive Initiation to Rna Polymerase Ii Recycling. Biochim. Biophys. Acta 2012, 1819, 391–400. [Google Scholar] [CrossRef]
- Howe, F.S.; Fischl, H.; Murray, S.C.; Mellor, J. Is H3k4me3 Instructive for Transcription Activation? Bioessays 2017, 39, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Taylor, B.C.; Young, N.L. Combinations of Histone Post-Translational Modifications. Biochem. J. 2021, 478, 511–532. [Google Scholar] [CrossRef]
- Xue, H.; Yao, T.; Cao, M.; Zhu, G.; Li, Y.; Yuan, G.; Chen, Y.; Lei, M.; Huang, J. Structural Basis of Nucleosome Recognition and Modification by Mll Methyltransferases. Nature 2019, 573, 445–449. [Google Scholar] [CrossRef]
- Park, S.H.; Ayoub, A.; Lee, Y.T.; Xu, J.; Kim, H.; Zheng, W.; Zhang, B.; Sha, L.; An, S.; Zhang, Y.; et al. Cryo-Em Structure of the Human Mll1 Core Complex Bound to the Nucleosome. Nat. Commun. 2019, 10, 5540. [Google Scholar] [CrossRef]
- Rahman, S.; Hoffmann, N.A.; Worden, E.J.; Smith, M.L.; Namitz, K.E.W.; Knutson, B.A.; Cosgrove, M.S.; Wolberger, C. Multistate Structures of the Mll1-Wrad Complex Bound to H2b-Ubiquitinated Nucleosome. Proc. Natl. Acad. Sci. USA 2022, 119, e2205691119. [Google Scholar] [CrossRef]
- Josling, G.A.; Selvarajah, S.A.; Petter, M.; Duffy, M.F. The Role of Bromodomain Proteins in Regulating Gene Expression. Genes 2012, 3, 320–343. [Google Scholar] [CrossRef] [PubMed]
- Shema, E.; Jones, D.; Shoresh, N.; Donohue, L.; Ram, O.; Bernstein, B.E. Single-Molecule Decoding of Combinatorially Modified Nucleosomes. Science 2016, 352, 717–721. [Google Scholar] [CrossRef]
- Moriniere, J.; Rousseaux, S.; Steuerwald, U.; Soler-Lopez, M.; Curtet, S.; Vitte, A.L.; Govin, J.; Gaucher, J.; Sadoul, K.; Hart, D.J.; et al. Cooperative Binding of Two Acetylation Marks on a Histone Tail by a Single Bromodomain. Nature 2009, 461, 664–668. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Chuh, K.N.; Zhang, B.; Dul, B.E.; Thompson, R.E.; Farrelly, L.A.; Liu, X.; Xu, N.; Xue, Y.; Roeder, R.G.; et al. Histone H3q5 Serotonylation Stabilizes H3k4 Methylation and Potentiates Its Readout. Proc. Natl. Acad. Sci. USA 2021, 118, e2016742118. [Google Scholar] [CrossRef]
- Al-Kachak, A.; Di Salvo, G.; Fulton, S.L.; Chan, J.C.; Farrelly, L.A.; Lepack, A.E.; Bastle, R.M.; Kong, L.; Cathomas, F.; Newman, E.L.; et al. Histone Serotonylation in Dorsal Raphe Nucleus Contributes to Stress- and Antidepressant-Mediated Gene Expression and Behavior. Nat. Commun. 2024, 15, 5042. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.C.; He, P.; McDonald, M.; Williamson, M.R.; Varadharajan, S.; Lozzi, B.; Woo, J.; Choi, D.J.; Sardar, D.; Huang-Hobbs, E.; et al. Histone Serotonylation Regulates Ependymoma Tumorigenesis. Nature 2024, 632, 903–910. [Google Scholar] [CrossRef] [PubMed]
- Sardar, D.; Cheng, Y.T.; Woo, J.; Choi, D.J.; Lee, Z.F.; Kwon, W.; Chen, H.C.; Lozzi, B.; Cervantes, A.; Rajendran, K.; et al. Induction of Astrocytic Slc22a3 Regulates Sensory Processing through Histone Serotonylation. Science 2023, 380, eade0027. [Google Scholar] [CrossRef]
- Zhao, J.; Chen, W.; Pan, Y.; Zhang, Y.; Sun, H.; Wang, H.; Yang, F.; Liu, Y.; Shen, N.; Zhang, X.; et al. Structural Insights into the Recognition of Histone H3q5 Serotonylation by Wdr5. Sci. Adv. 2021, 7, eabf4291. [Google Scholar] [CrossRef]
- Macrae, T.A.; Fothergill-Robinson, J.; Ramalho-Santos, M. Regulation, Functions and Transmission of Bivalent Chromatin During Mammalian Development. Nat. Rev. Mol. Cell Biol. 2023, 24, 6–26. [Google Scholar] [CrossRef]
- Voigt, P.; Tee, W.W.; Reinberg, D. A Double Take on Bivalent Promoters. Genes Dev. 2013, 27, 1318–1338. [Google Scholar] [CrossRef]
- Guo, Y.; Zhao, S.; Wang, G.G. Polycomb Gene Silencing Mechanisms: Prc2 Chromatin Targeting, H3k27me3 ‘Readout’, and Phase Separation-Based Compaction. Trends Genet. 2021, 37, 547–565. [Google Scholar] [CrossRef]
- Gao, Z.; Li, Y.; Ou, Y.; Yin, M.; Chen, T.; Zeng, X.; Li, R.; He, Y. A Pair of Readers of Bivalent Chromatin Mediate Formation of Polycomb-Based “Memory of Cold” in Plants. Mol. Cell 2023, 83, 1109–1124.e4. [Google Scholar] [CrossRef] [PubMed]
- Qian, S.; Lv, X.; Scheid, R.N.; Lu, L.; Yang, Z.; Chen, W.; Liu, R.; Boersma, M.D.; Denu, J.M.; Zhong, X.; et al. Dual Recognition of H3k4me3 and H3k27me3 by a Plant Histone Reader Shl. Nat. Commun. 2018, 9, 2425. [Google Scholar] [CrossRef]
- Lee, M.G.; Villa, R.; Trojer, P.; Norman, J.; Yan, K.P.; Reinberg, D.; Di Croce, L.; Shiekhattar, R. Demethylation of H3k27 Regulates Polycomb Recruitment and H2a Ubiquitination. Science 2007, 318, 447–450. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.W.; Hong, T.; Hong, S.; Guo, H.; Yu, H.; Kim, D.; Guszczynski, T.; Dressler, G.R.; Copeland, T.D.; Kalkum, M.; et al. Ptip Associates with Mll3- and Mll4-Containing Histone H3 Lysine 4 Methyltransferase Complex. J. Biol. Chem. 2007, 282, 20395–20406. [Google Scholar] [CrossRef]
- Gozdecka, M.; Meduri, E.; Mazan, M.; Tzelepis, K.; Dudek, M.; Knights, A.J.; Pardo, M.; Yu, L.; Choudhary, J.S.; Metzakopian, E.; et al. Utx-Mediated Enhancer and Chromatin Remodeling Suppresses Myeloid Leukemogenesis through Noncatalytic Inverse Regulation of Ets and Gata Programs. Nat. Genet. 2018, 50, 883–894. [Google Scholar] [CrossRef]
- Leng, X.; Wang, J.; An, N.; Wang, X.; Sun, Y.; Chen, Z. Histone 3 Lysine-27 Demethylase Kdm6a Coordinates with Kmt2b to Play an Oncogenic Role in Nsclc by Regulating H3k4me3. Oncogene 2020, 39, 6468–6479. [Google Scholar] [CrossRef]
- Froimchuk, E.; Jang, Y.; Ge, K. Histone H3 Lysine 4 Methyltransferase Kmt2d. Gene 2017, 627, 337–342. [Google Scholar] [CrossRef]
- Varier, R.A.; Outchkourov, N.S.; de Graaf, P.; van Schaik, F.M.; Ensing, H.J.; Wang, F.; Higgins, J.M.; Kops, G.J.; Timmers, H.T. A Phospho/Methyl Switch at Histone H3 Regulates Tfiid Association with Mitotic Chromosomes. EMBO J. 2010, 29, 3967–3978. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Sultan, S.; Taylor, S.S.; Higgins, J.M. The Kinase Haspin Is Required for Mitotic Histone H3 Thr 3 Phosphorylation and Normal Metaphase Chromosome Alignment. Genes Dev. 2005, 19, 472–488. [Google Scholar] [CrossRef]
- Kang, H.; Shokhirev, M.N.; Xu, Z.; Chandran, S.; Dixon, J.R.; Hetzer, M.W. Dynamic Regulation of Histone Modifications and Long-Range Chromosomal Interactions During Postmitotic Transcriptional Reactivation. Genes Dev. 2020, 34, 913–930. [Google Scholar] [CrossRef]
- Harris, R.J.; Heer, M.; Levasseur, M.D.; Cartwright, T.N.; Weston, B.; Mitchell, J.L.; Coxhead, J.M.; Gaughan, L.; Prendergast, L.; Rico, D.; et al. Release of Histone H3k4-Reading Transcription Factors from Chromosomes in Mitosis Is Independent of Adjacent H3 Phosphorylation. Nat. Commun. 2023, 14, 7243. [Google Scholar] [CrossRef]
- Metzger, E.; Imhof, A.; Patel, D.; Kahl, P.; Hoffmeyer, K.; Friedrichs, N.; Muller, J.M.; Greschik, H.; Kirfel, J.; Ji, S.; et al. Phosphorylation of Histone H3t6 by Pkcbeta(I) Controls Demethylation at Histone H3k4. Nature 2010, 464, 792–796. [Google Scholar] [CrossRef]
- Andrews, F.H.; Gatchalian, J.; Krajewski, K.; Strahl, B.D.; Kutateladze, T.G. Regulation of Methyllysine Readers through Phosphorylation. ACS Chem. Biol. 2016, 11, 547–553. [Google Scholar] [CrossRef] [PubMed]
- Hyllus, D.; Stein, C.; Schnabel, K.; Schiltz, E.; Imhof, A.; Dou, Y.; Hsieh, J.; Bauer, U.M. Prmt6-Mediated Methylation of R2 in Histone H3 Antagonizes H3 K4 Trimethylation. Genes Dev. 2007, 21, 3369–3380. [Google Scholar] [CrossRef] [PubMed]
- Guccione, E.; Bassi, C.; Casadio, F.; Martinato, F.; Cesaroni, M.; Schuchlautz, H.; Luscher, B.; Amati, B. Methylation of Histone H3r2 by Prmt6 and H3k4 by an Mll Complex Are Mutually Exclusive. Nature 2007, 449, 933–937. [Google Scholar] [CrossRef]
- Kirmizis, A.; Santos-Rosa, H.; Penkett, C.J.; Singer, M.A.; Vermeulen, M.; Mann, M.; Bahler, J.; Green, R.D.; Kouzarides, T. Arginine Methylation at Histone H3r2 Controls Deposition of H3k4 Trimethylation. Nature 2007, 449, 928–932. [Google Scholar] [CrossRef]
- Bouchard, C.; Sahu, P.; Meixner, M.; Notzold, R.R.; Rust, M.B.; Kremmer, E.; Feederle, R.; Hart-Smith, G.; Finkernagel, F.; Bartkuhn, M.; et al. Genomic Location of Prmt6-Dependent H3r2 Methylation Is Linked to the Transcriptional Outcome of Associated Genes. Cell Rep. 2018, 24, 3339–3352. [Google Scholar] [CrossRef]
- Yuan, C.C.; Matthews, A.G.; Jin, Y.; Chen, C.F.; Chapman, B.A.; Ohsumi, T.K.; Glass, K.C.; Kutateladze, T.G.; Borowsky, M.L.; Struhl, K.; et al. Histone H3r2 Symmetric Dimethylation and Histone H3k4 Trimethylation Are Tightly Correlated in Eukaryotic Genomes. Cell Rep. 2012, 1, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Liu, Y.; Su, X.; Lee, J.E.; Song, Y.; Wang, D.; Ge, K.; Gao, J.; Zhang, M.Q.; Li, H. Molecular Basis for Histone H3 “K4me3-K9me3/2” Methylation Pattern Readout by Spindlin1. J. Biol. Chem. 2020, 295, 16877–16887. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Chen, Z.; Mao, Z.; Zhang, H.; Ding, X.; Chen, S.; Zhang, X.; Xu, R.; Zhu, B. Nucleolar Protein Spindlin1 Recognizes H3k4 Methylation and Stimulates the Expression of Rrna Genes. EMBO Rep. 2011, 12, 1160–1166. [Google Scholar] [CrossRef]
- Du, Y.; Yan, Y.; Xie, S.; Huang, H.; Wang, X.; Ng, R.K.; Zhou, M.M.; Qian, C. Structural Mechanism of Bivalent Histone H3k4me3k9me3 Recognition by the Spindlin1/C11orf84 Complex in Rrna Transcription Activation. Nat. Commun. 2021, 12, 949. [Google Scholar] [CrossRef]
- Su, X.; Zhu, G.; Ding, X.; Lee, S.Y.; Dou, Y.; Zhu, B.; Wu, W.; Li, H. Molecular Basis Underlying Histone H3 Lysine-Arginine Methylation Pattern Readout by Spin/Ssty Repeats of Spindlin1. Genes Dev. 2014, 28, 622–636. [Google Scholar] [CrossRef] [PubMed]
- Lam, U.T.F.; Tan, B.K.Y.; Poh, J.J.X.; Chen, E.S. Structural and Functional Specificity of H3k36 Methylation. Epigenetics Chromatin 2022, 15, 17. [Google Scholar] [CrossRef]
- Sharda, A.; Humphrey, T.C. The Role of Histone H3k36me3 Writers, Readers and Erasers in Maintaining Genome Stability. DNA Repair. 2022, 119, 103407. [Google Scholar] [CrossRef]
- Talbert, P.B.; Henikoff, S. The Yin and Yang of Histone Marks in Transcription. Annu. Rev. Genom. Hum. Genet. 2021, 22, 147–170. [Google Scholar] [CrossRef]
- Cavassim, M.I.A.; Baker, Z.; Hoge, C.; Schierup, M.H.; Schumer, M.; Przeworski, M. Prdm9 Losses in Vertebrates Are Coupled to Those of Paralogs Zcwpw1 and Zcwpw2. Proc. Natl. Acad. Sci. USA 2022, 119, e2114401119. [Google Scholar] [CrossRef]
- Hoolehan, W.; Harris, J.C.; Rodgers, K.K. Molecular Mechanisms of DNA Sequence Selectivity in V(D)J Recombination. ACS Omega 2023, 8, 34206–34214. [Google Scholar] [CrossRef]
- Yin, B.; Yu, F.; Wang, C.; Li, B.; Liu, M.; Ye, L. Epigenetic Control of Mesenchymal Stem Cell Fate Decision Via Histone Methyltransferase Ash1l. Stem Cells 2019, 37, 115–127. [Google Scholar] [CrossRef]
- Zhu, L.; Li, Q.; Wong, S.H.; Huang, M.; Klein, B.J.; Shen, J.; Ikenouye, L.; Onishi, M.; Schneidawind, D.; Buechele, C.; et al. Ash1l Links Histone H3 Lysine 36 Dimethylation to Mll Leukemia. Cancer Discov. 2016, 6, 770–783. [Google Scholar] [CrossRef]
- Miyazaki, H.; Higashimoto, K.; Yada, Y.; Endo, T.A.; Sharif, J.; Komori, T.; Matsuda, M.; Koseki, Y.; Nakayama, M.; Soejima, H.; et al. Ash1l Methylates Lys36 of Histone H3 Independently of Transcriptional Elongation to Counteract Polycomb Silencing. PLoS Genet. 2013, 9, e1003897. [Google Scholar] [CrossRef] [PubMed]
- Gregory, G.D.; Vakoc, C.R.; Rozovskaia, T.; Zheng, X.; Patel, S.; Nakamura, T.; Canaani, E.; Blobel, G.A. Mammalian Ash1l Is a Histone Methyltransferase That Occupies the Transcribed Region of Active Genes. Mol. Cell Biol. 2007, 27, 8466–8479. [Google Scholar] [CrossRef] [PubMed]
- Maritz, C.; Khaleghi, R.; Yancoskie, M.N.; Diethelm, S.; Brulisauer, S.; Ferreira, N.S.; Jiang, Y.; Sturla, S.J.; Naegeli, H. Ash1l-Mrg15 Methyltransferase Deposits H3k4me3 and Fact for Damage Verification in Nucleotide Excision Repair. Nat. Commun. 2023, 14, 3892. [Google Scholar] [CrossRef] [PubMed]
- Al-Harthi, S.; Li, H.; Winkler, A.; Szczepski, K.; Deng, J.; Grembecka, J.; Cierpicki, T.; Jaremko, L. Mrg15 Activates Histone Methyltransferase Activity of Ash1l by Recruiting It to the Nucleosomes. Structure 2023, 31, 1200–1207.e5. [Google Scholar] [CrossRef]
- Schneider, J.; Wood, A.; Lee, J.S.; Schuster, R.; Dueker, J.; Maguire, C.; Swanson, S.K.; Florens, L.; Washburn, M.P.; Shilatifard, A. Molecular Regulation of Histone H3 Trimethylation by Compass and the Regulation of Gene Expression. Mol. Cell 2005, 19, 849–856. [Google Scholar] [CrossRef]
- Nislow, C.; Ray, E.; Pillus, L. Set1, a Yeast Member of the Trithorax Family, Functions in Transcriptional Silencing and Diverse Cellular Processes. Mol. Biol. Cell 1997, 8, 2421–2436. [Google Scholar] [CrossRef] [PubMed]
- Margaritis, T.; Oreal, V.; Brabers, N.; Maestroni, L.; Vitaliano-Prunier, A.; Benschop, J.J.; van Hooff, S.; van Leenen, D.; Dargemont, C.; Geli, V.; et al. Two Distinct Repressive Mechanisms for Histone 3 Lysine 4 Methylation through Promoting 3′-End Antisense Transcription. PLoS Genet. 2012, 8, e1002952. [Google Scholar] [CrossRef] [PubMed]
- Hodl, M.; Basler, K. Transcription in the Absence of Histone H3.2 and H3k4 Methylation. Curr. Biol. 2012, 22, 2253–2257. [Google Scholar] [CrossRef]
- Stoller, J.Z.; Huang, L.; Tan, C.C.; Huang, F.; Zhou, D.D.; Yang, J.; Gelb, B.D.; Epstein, J.A. Ash2l Interacts with Tbx1 and Is Required During Early Embryogenesis. Exp. Biol. Med. 2010, 235, 569–576. [Google Scholar] [CrossRef]
- Liang, W.L.; Luscher-Firzlaff, J.; Ullius, A.; Schneider, U.; Longerich, T.; Luscher, B. Loss of the Epigenetic Regulator Ash2l Results in Desintegration of Hepatocytes and Liver Failure. Int. J. Clin. Exp. Patho 2016, 9, 5167–5175. [Google Scholar]
- Zhang, Z.; Yang, C.; Wang, Z.; Guo, L.; Xu, Y.; Gao, C.; Sun, Y.; Zhang, Z.; Peng, J.; Hu, M.; et al. Wdr5-Mediated H3k4me3 Coordinately Regulates Cell Differentiation, Proliferation Termination, and Survival in Digestive Organogenesis. Cell Death Discov. 2023, 9, 227. [Google Scholar] [CrossRef]
- Yang, Z.; Shah, K.; Khodadadi-Jamayran, A.; Jiang, H. Dpy30 Is Critical for Maintaining the Identity and Function of Adult Hematopoietic Stem Cells. J. Exp. Med. 2016, 213, 2349–2364. [Google Scholar] [CrossRef]
- Luscher-Firzlaff, J.; Chatain, N.; Kuo, C.C.; Braunschweig, T.; Bochynska, A.; Ullius, A.; Denecke, B.; Costa, I.G.; Koschmieder, S.; Luscher, B. Hematopoietic Stem and Progenitor Cell Proliferation and Differentiation Requires the Trithorax Protein Ash2l. Sci. Rep. 2019, 9, 8262. [Google Scholar] [CrossRef]
- Bochynska, A.; Stenzel, A.T.; Boroujeni, R.S.; Kuo, C.C.; Barsoum, M.; Liang, W.; Bussmann, P.; Costa, I.G.; Luscher-Firzlaff, J.; Luscher, B. Induction of Senescence Upon Loss of the Ash2l Core Subunit of H3k4 Methyltransferase Complexes. Nucleic Acids Res. 2022, 50, 7889–7905. [Google Scholar] [CrossRef]
- Gan, T.; Jude, C.D.; Zaffuto, K.; Ernst, P. Developmentally Induced Mll1 Loss Reveals Defects in Postnatal Haematopoiesis. Leukemia 2010, 24, 1732–1741. [Google Scholar] [CrossRef]
- Lim, D.A.; Huang, Y.C.; Swigut, T.; Mirick, A.L.; Garcia-Verdugo, J.M.; Wysocka, J.; Ernst, P.; Alvarez-Buylla, A. Chromatin Remodelling Factor Mll1 Is Essential for Neurogenesis from Postnatal Neural Stem Cells. Nature 2009, 458, 529–533. [Google Scholar] [CrossRef]
- Fang, L.; Teng, H.; Wang, Y.; Liao, G.; Weng, L.; Li, Y.; Wang, X.; Jin, J.; Jiao, C.; Chen, L.; et al. Set1a-Mediated Mono-Methylation at K342 Regulates Yap Activation by Blocking Its Nuclear Export and Promotes Tumorigenesis. Cancer Cell 2018, 34, 103–118.e9. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, L.; Tian, X.; Peng, C.; Gong, F.; Chen, Y. Crystal Structure of Mll2 Complex Guides the Identification of a Methylation Site on P53 Catalyzed by Kmt2 Family Methyltransferases. Structure 2020, 28, 1141–1148.e4. [Google Scholar] [CrossRef]
- Cho, H.S.; Shimazu, T.; Toyokawa, G.; Daigo, Y.; Maehara, Y.; Hayami, S.; Ito, A.; Masuda, K.; Ikawa, N.; Field, H.I.; et al. Enhanced Hsp70 Lysine Methylation Promotes Proliferation of Cancer Cells through Activation of Aurora Kinase B. Nat. Commun. 2012, 3, 1072. [Google Scholar] [CrossRef]
- Nikonova, A.S.; Astsaturov, I.; Serebriiskii, I.G.; Dunbrack, R.L., Jr.; Golemis, E.A. Aurora a Kinase (Aurka) in Normal and Pathological Cell Division. Cell Mol. Life Sci. 2013, 70, 661–687. [Google Scholar] [CrossRef]
- Mannervik, M. Beyond Histones: The Elusive Substrates of Chromatin Regulators. Genes Dev. 2024, 38, 357–359. [Google Scholar] [CrossRef]
- Crain, A.T.; Butler, M.B.; Hill, C.A.; Huynh, M.; McGinty, R.K.; Duronio, R.J. Drosophila Melanogaster Set8 and L(3)Mbt Function in Gene Expression Independently of Histone H4 Lysine 20 Methylation. Genes Dev. 2024, 38, 455–472. [Google Scholar] [CrossRef] [PubMed]
- Crain, A.T.; Klusza, S.; Armstrong, R.L.; Santa Rosa, P.; Temple, B.R.S.; Strahl, B.D.; McKay, D.J.; Matera, A.G.; Duronio, R.J. Distinct Developmental Phenotypes Result from Mutation of Set8/Kmt5a and Histone H4 Lysine 20 in Drosophila Melanogaster. Genetics 2022, 221, iyac054. [Google Scholar] [CrossRef] [PubMed]
- Milite, C.; Feoli, A.; Viviano, M.; Rescigno, D.; Cianciulli, A.; Balzano, A.L.; Mai, A.; Castellano, S.; Sbardella, G. The Emerging Role of Lysine Methyltransferase Setd8 in Human Diseases. Clin. Epigenetics 2016, 8, 102. [Google Scholar] [CrossRef] [PubMed]
- Ogino, J.; Dou, Y. Histone Methyltransferase Kmt2a: Developmental Regulation to Oncogenic Transformation. J. Biol. Chem. 2024, 300, 107791. [Google Scholar] [CrossRef]
- Poreba, E.; Lesniewicz, K.; Durzynska, J. Aberrant Activity of Histone-Lysine N-Methyltransferase 2 (Kmt2) Complexes in Oncogenesis. Int. J. Mol. Sci. 2020, 21, 9340. [Google Scholar] [CrossRef] [PubMed]
- Ford, D.J.; Dingwall, A.K. The Cancer Compass: Navigating the Functions of Mll Complexes in Cancer. Cancer Genet. 2015, 208, 178–191. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Li, D.; Kottur, J.; Shen, Y.; Kim, H.S.; Park, K.S.; Tsai, Y.H.; Gong, W.; Wang, J.; Suzuki, K.; et al. A Selective Wdr5 Degrader Inhibits Acute Myeloid Leukemia in Patient-Derived Mouse Models. Sci. Transl. Med. 2021, 13, eabj1578. [Google Scholar] [CrossRef] [PubMed]
- Chern, T.R.; Liu, L.; Petrunak, E.; Stuckey, J.A.; Wang, M.; Bernard, D.; Zhou, H.; Lee, S.; Dou, Y.; Wang, S. Discovery of Potent Small-Molecule Inhibitors of Mll Methyltransferase. ACS Med. Chem. Lett. 2020, 11, 1348–1352. [Google Scholar] [CrossRef]
- Barghout, S.H.; Machado, R.A.C.; Barsyte-Lovejoy, D. Chemical Biology and Pharmacology of Histone Lysine Methylation Inhibitors. Biochim. Biophys. Acta Gene Regul. Mech. 2022, 1865, 194840. [Google Scholar] [CrossRef]
- Zhai, X.; Brownell, J.E. Biochemical Perspectives on Targeting Kmt2 Methyltransferases in Cancer. Trends Pharmacol. Sci. 2021, 42, 688–699. [Google Scholar] [CrossRef]
- Perner, F.; Armstrong, S.A. Targeting Chromatin Complexes in Myeloid Malignancies and Beyond: From Basic Mechanisms to Clinical Innovation. Cells 2020, 9, 2721. [Google Scholar] [CrossRef]
- Candoni, A.; Coppola, G. A 2024 Update on Menin Inhibitors. A New Class of Target Agents against Kmt2a-Rearranged and Npm1-Mutated Acute Myeloid Leukemia. Hematol. Rep. 2024, 16, 244–254. [Google Scholar] [CrossRef]
- Cao, F.; Townsend, E.C.; Karatas, H.; Xu, J.; Li, L.; Lee, S.; Liu, L.; Chen, Y.; Ouillette, P.; Zhu, J.; et al. Targeting Mll1 H3k4 Methyltransferase Activity in Mixed-Lineage Leukemia. Mol. Cell 2014, 53, 247–261. [Google Scholar] [CrossRef]
- Jaeger, M.G.; Winter, G.E. Fast-Acting Chemical Tools to Delineate Causality in Transcriptional Control. Mol. Cell 2021, 81, 1617–1630. [Google Scholar] [CrossRef]
- Haddad, J.F.; Yang, Y.; Takahashi, Y.H.; Joshi, M.; Chaudhary, N.; Woodfin, A.R.; Benyoucef, A.; Yeung, S.; Brunzelle, J.S.; Skiniotis, G.; et al. Structural Analysis of the Ash2l/Dpy-30 Complex Reveals a Heterogeneity in H3k4 Methylation. Structure 2018, 26, 1594–1603.e4. [Google Scholar] [CrossRef]
- Patel, A.; Dharmarajan, V.; Vought, V.E.; Cosgrove, M.S. On the Mechanism of Multiple Lysine Methylation by the Human Mixed Lineage Leukemia Protein-1 (Mll1) Core Complex. J. Biol. Chem. 2009, 284, 24242–24256. [Google Scholar] [CrossRef]
- Yang, Z.; Augustin, J.; Chang, C.; Hu, J.; Shah, K.; Chang, C.W.; Townes, T.; Jiang, H. The Dpy30 Subunit in Set1/Mll Complexes Regulates the Proliferation and Differentiation of Hematopoietic Progenitor Cells. Blood 2014, 124, 2025–2033. [Google Scholar] [CrossRef] [PubMed]
- da Costa-Nunes, J.A.; Noordermeer, D. Tads: Dynamic Structures to Create Stable Regulatory Functions. Curr. Opin. Struct. Biol. 2023, 81, 102622. [Google Scholar] [CrossRef] [PubMed]
- Qi, S.; Shi, Z.; Yu, H. Genome Folding by Cohesion. Curr. Opin. Genet. Dev. 2025, 91, 102310. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Chen, S.A.; Local, A.; Liu, T.; Qiu, Y.; Dorighi, K.M.; Preissl, S.; Rivera, C.M.; Wang, C.; Ye, Z.; et al. Histone H3 Lysine 4 Monomethylation Modulates Long-Range Chromatin Interactions at Enhancers. Cell Res. 2018, 28, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Chen, Z.; Wu, D.; Zhang, L.; Lin, X.; Su, J.; Rodriguez, B.; Xi, Y.; Xia, Z.; Chen, X.; et al. Broad H3k4me3 Is Associated with Increased Transcription Elongation and Enhancer Activity at Tumor-Suppressor Genes. Nat. Genet. 2015, 47, 1149–1157. [Google Scholar] [CrossRef]
- Cao, F.; Fang, Y.; Tan, H.K.; Goh, Y.; Choy, J.Y.H.; Koh, B.T.H.; Hao Tan, J.; Bertin, N.; Ramadass, A.; Hunter, E.; et al. Super-Enhancers and Broad H3k4me3 Domains Form Complex Gene Regulatory Circuits Involving Chromatin Interactions. Sci. Rep. 2017, 7, 2186. [Google Scholar] [CrossRef]
- Thibodeau, A.; Marquez, E.J.; Shin, D.G.; Vera-Licona, P.; Ucar, D. Chromatin Interaction Networks Revealed Unique Connectivity Patterns of Broad H3k4me3 Domains and Super Enhancers in 3d Chromatin. Sci. Rep. 2017, 7, 14466. [Google Scholar] [CrossRef]
- Tang, W.; Costantino, L.; Stocsits, R.; Wutz, G.; Ladurner, R.; Hudecz, O.; Mechtler, K.; Peters, J.M. Cohesin Positions the Epigenetic Reader Phf2 within the Genome. EMBO J. 2025, 44, 736–766. [Google Scholar] [CrossRef]
- Nishiyama, T. Cohesion and Cohesin-Dependent Chromatin Organization. Curr. Opin. Cell Biol. 2019, 58, 8–14. [Google Scholar] [CrossRef]
- Hoencamp, C.; Rowland, B.D. Genome Control by Smc Complexes. Nat. Rev. Mol. Cell Biol. 2023, 24, 633–650. [Google Scholar] [CrossRef] [PubMed]
- Dekker, J.; Mirny, L.A. The Chromosome Folding Problem and How Cells Solve It. Cell 2024, 187, 6424–6450. [Google Scholar] [CrossRef] [PubMed]
- Nanni, L.; Ceri, S.; Logie, C. Spatial Patterns of Ctcf Sites Define the Anatomy of Tads and Their Boundaries. Genome Biol. 2020, 21, 197. [Google Scholar] [CrossRef] [PubMed]
- Merkenschlager, M.; Nora, E.P. Ctcf and Cohesin in Genome Folding and Transcriptional Gene Regulation. Annu. Rev. Genom. Hum. Genet. 2016, 17, 17–43. [Google Scholar] [CrossRef]
- Yuen, K.C.; Slaughter, B.D.; Gerton, J.L. Condensin Ii Is Anchored by Tfiiic and H3k4me3 in the Mammalian Genome and Supports the Expression of Active Dense Gene Clusters. Sci. Adv. 2017, 3, e1700191. [Google Scholar] [CrossRef]



| MTase (Writers) | WRAD Subunits (Common to All Complexes) | Additional Subunits | Activity (Main) | ||||
|---|---|---|---|---|---|---|---|
| Yeast | ySet1 | Cps30 (Swd3) | Cps50 (Swd1) | Cps60 (Bre2) | Cps25 (Sdc1) | Cps35 (Swd2, Wdr82) Cps40 (Spp1, Cfp1) | H3K4me1-3 |
| Drosophila melanogaster | Trithorax | WDS | RBBP5 | ASH2 | DPY30 | HCF1/2 | H3K4me(1-)3 |
| Trithorax related | PTIP PA1 NCOA6 UTX | H3K4me1 | |||||
| dSet1 | WDR82 CXXC1 HCF1/2 | H3K4me(1-)3 | |||||
| Mammals | MLL1/ KMT2A | WDR5 | RBBP5 | ASH2L | DPY30 | HCF1/2 Menin PSIP1 (LEDGF) | H3K4me(1-)3 |
| MLL2/ KMT2B | |||||||
| MLL3/ KMT2C | PTIP PA1 NCOA6 UTX | H3K4me1 | |||||
| MLL4/ KMT2D | |||||||
| SET1A/ SETD1A/ KMT2F | WDR82 CXXC1 HCF1 | H3K4me(1-)3 | |||||
| SET1B/ SETD1B/ KMT2G | |||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lüscher, B.; Bussmann, P.; Müller, J. Role of Histone H3 Lysine 4 Methylation in Chromatin Biology. Molecules 2025, 30, 4075. https://doi.org/10.3390/molecules30204075
Lüscher B, Bussmann P, Müller J. Role of Histone H3 Lysine 4 Methylation in Chromatin Biology. Molecules. 2025; 30(20):4075. https://doi.org/10.3390/molecules30204075
Chicago/Turabian StyleLüscher, Bernhard, Philip Bussmann, and Janina Müller. 2025. "Role of Histone H3 Lysine 4 Methylation in Chromatin Biology" Molecules 30, no. 20: 4075. https://doi.org/10.3390/molecules30204075
APA StyleLüscher, B., Bussmann, P., & Müller, J. (2025). Role of Histone H3 Lysine 4 Methylation in Chromatin Biology. Molecules, 30(20), 4075. https://doi.org/10.3390/molecules30204075







