Kujigamberol Inhibits IFN-γ and IL-2 mRNA Expression and NFATc2 Binding to Their Promoters in Response to a Phorbol Ester and Ionomycin Stimulation
Abstract
1. Introduction
2. Results
2.1. Kujigamberol Inhibited IFN-γ and IL-2 mRNA Expression in Murine T-Cell Lymphoma BW5147 Cells
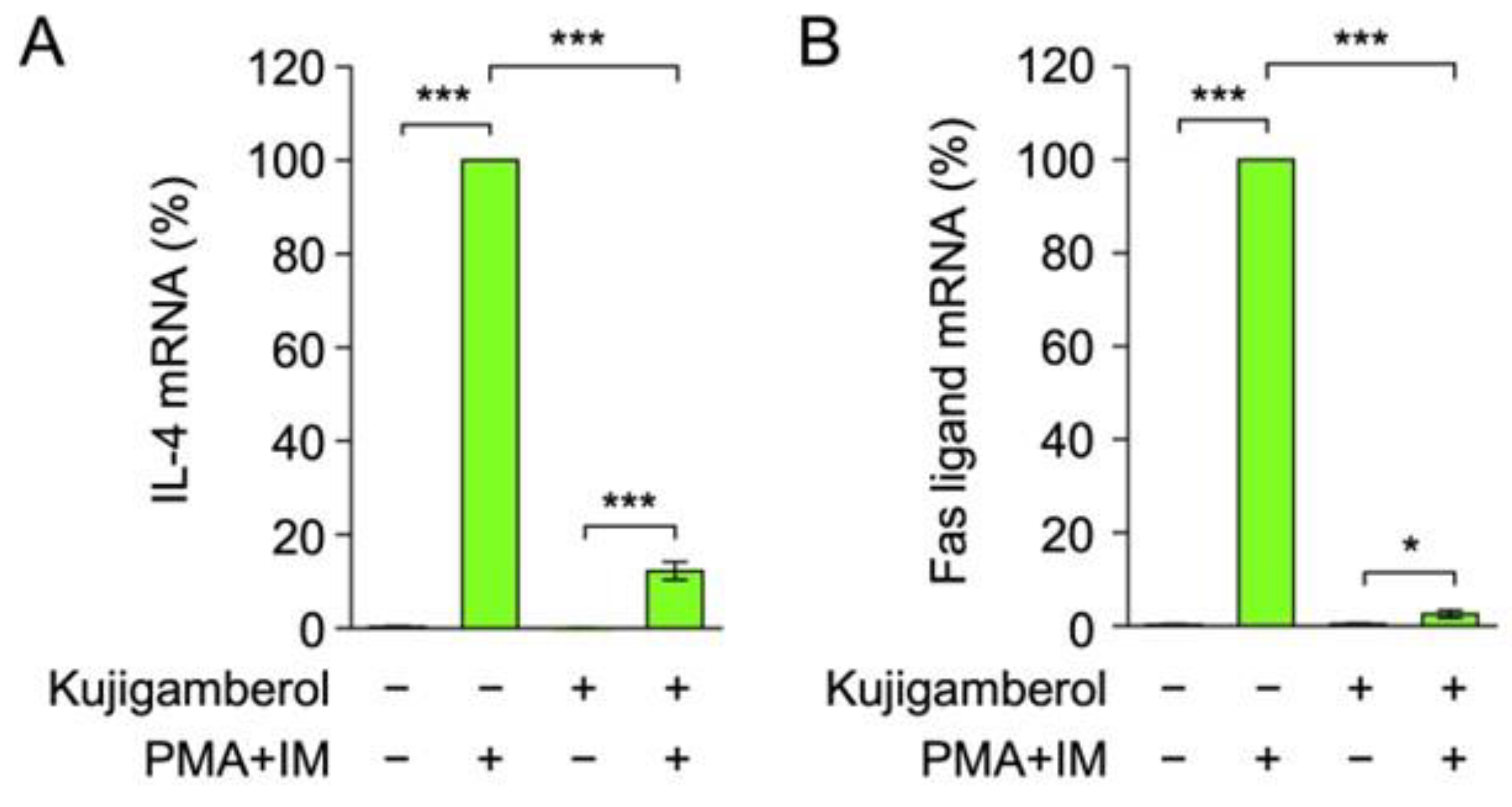
2.2. Kujigamberol Inhibited IL-4 and Fas Ligand mRNA Expression in Eomes-Transfected BW5147 Cells
2.3. Kujigamberol Inhibited IFN-γ mRNA Expression in the Murine Cytotoxic T-Cell Line CTLL-2
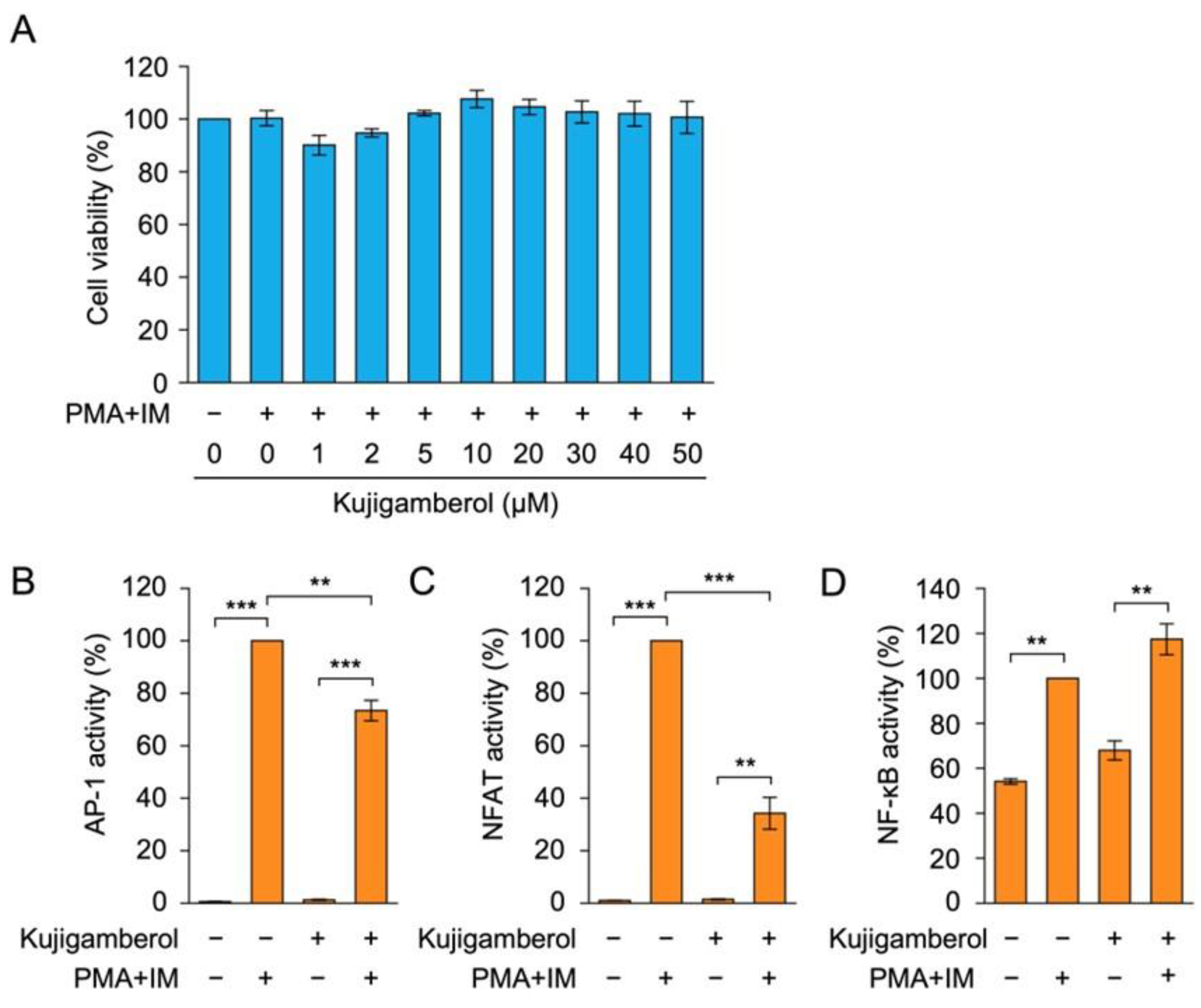
2.4. Kujigamberol Suppressed NFAT-Dependent Transcriptional Activity in Human Embryonic Kidney 293T Cells
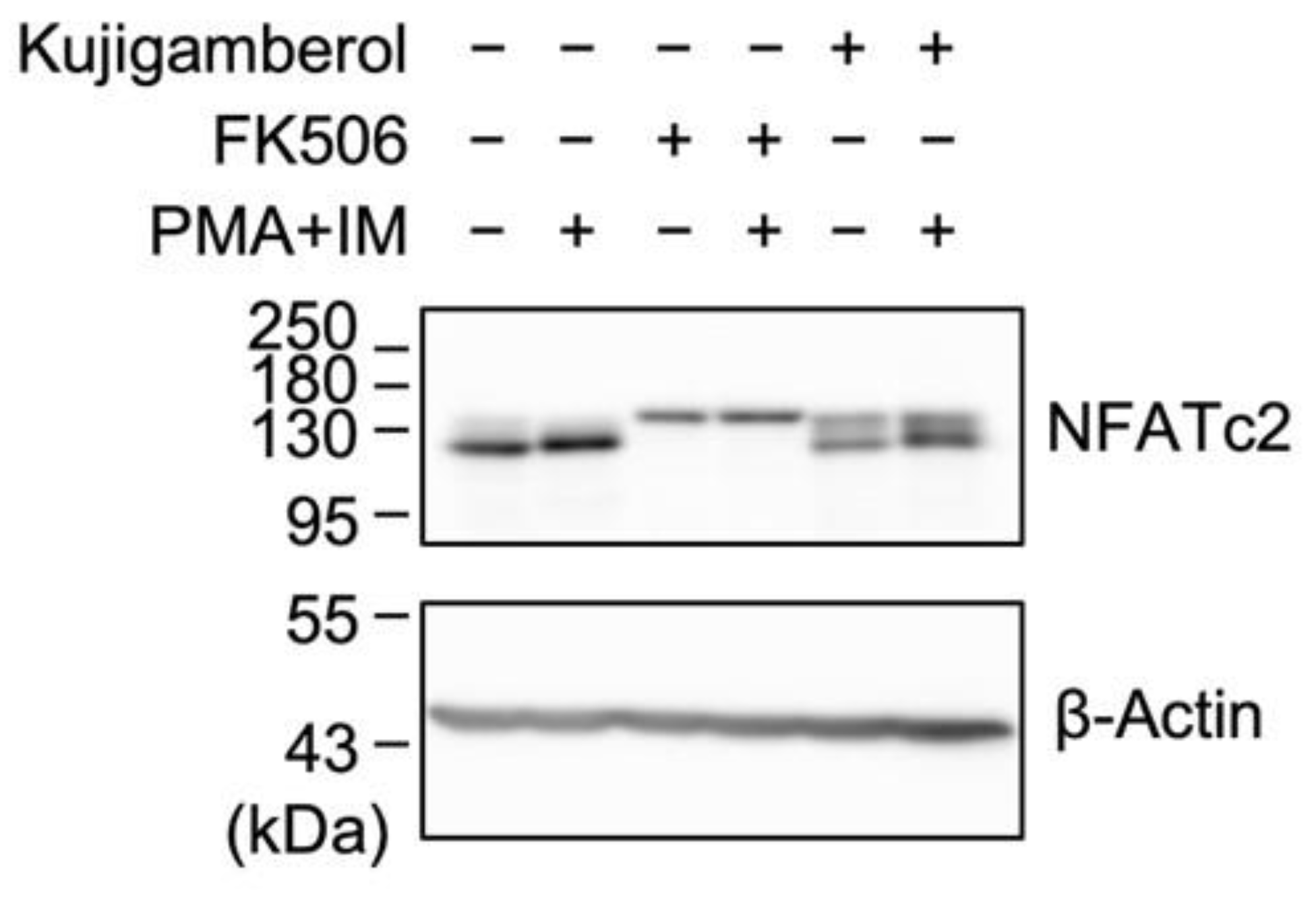
2.5. Kujigamberol Did Not Affect NFATc2 Protein Expression in Eomes-Transfected BW5147 Cells
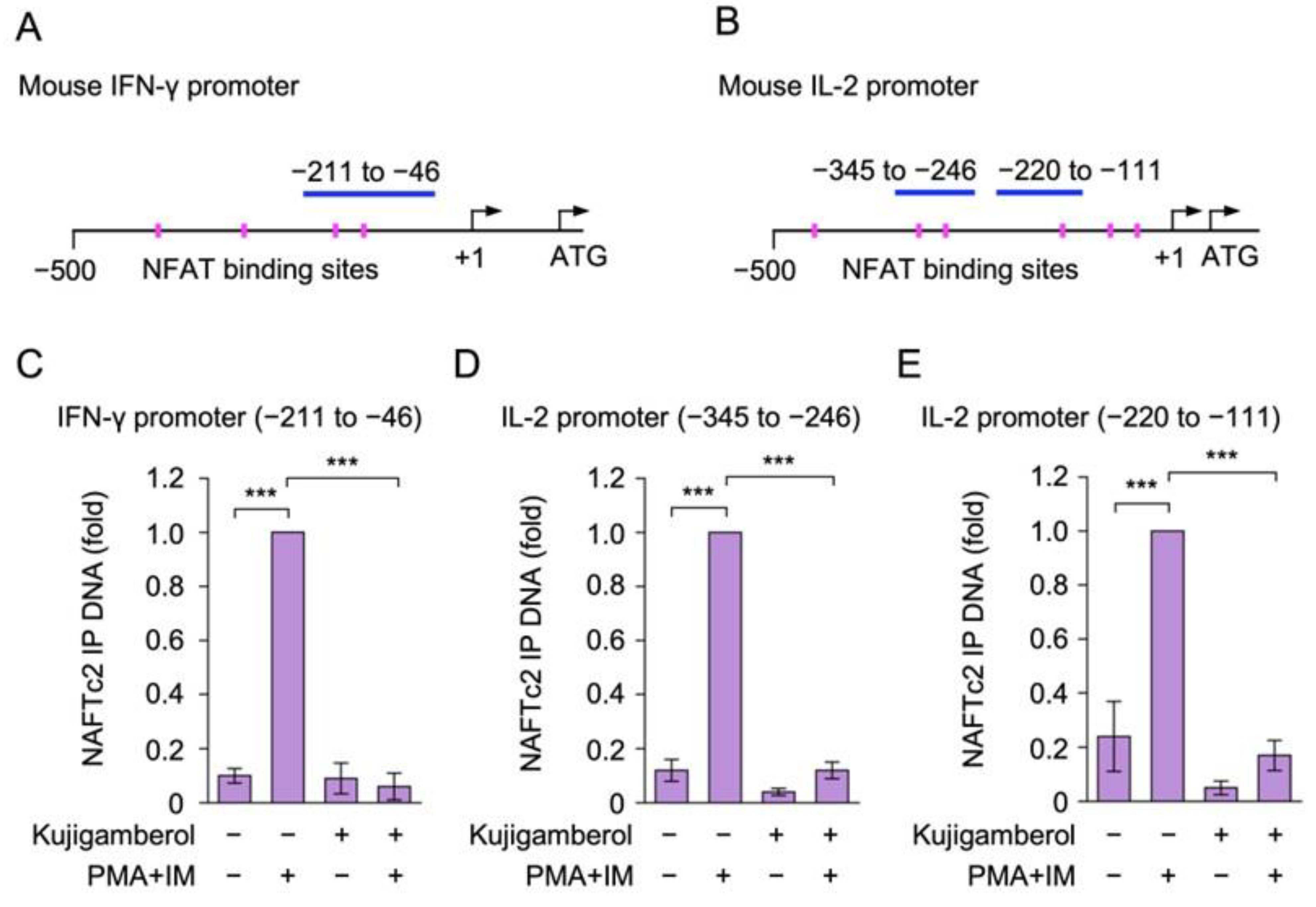
2.6. Kujigamberol Interfered with the Binding of NFATc2 to IFN-γ and IL-2 Promoters in Eomes-Transfected BW5147 Cells
3. Discussion
4. Materials and Methods
4.1. Cells
4.2. Reagents
4.3. Antibodies
4.4. Plasmid Vectors
4.5. Cell Viability Assay
4.6. RT-qPCR
4.7. Reporter Assay
4.8. Western Blotting
4.9. ChIP Assay
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, W.; Zhu, C. Mechanical regulation of T-cell functions. Immunol. Rev. 2013, 256, 160–176. [Google Scholar] [CrossRef] [PubMed]
- Raphael, I.; Nalawade, S.; Eagar, T.N.; Forsthuber, T.G. T cell subsets and their signature cytokines in autoimmune and inflammatory diseases. Cytokine 2015, 74, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Dong, C. Cytokine Regulation and Function in T Cells. Annu. Rev. Immunol. 2021, 39, 51–76. [Google Scholar] [CrossRef]
- Turner, M.D.; Nedjai, B.; Hurst, T.; Pennington, D.J. Cytokines and chemokines: At the crossroads of cell signalling and inflammatory disease. Biochim. Biophys. Acta. 2014, 1843, 2563–2582. [Google Scholar] [CrossRef]
- Liu, C.; Chu, D.; Kalantar-Zadeh, K.; George, J.; Young, H.A.; Liu, G. Cytokines: From clinical significance to quantification. Adv. Sci. 2021, 8, e2004433. [Google Scholar] [CrossRef]
- Rosenberg, S.A. IL-2: The first effective immunotherapy for human cancer. J. Immunol. 2014, 192, 5451–5458. [Google Scholar] [CrossRef]
- Damoiseaux, J. The IL-2—IL-2 receptor pathway in health and disease: The role of the soluble IL-2 receptor. Clin. Immunol. 2020, 218, 108515. [Google Scholar] [CrossRef]
- Liao, W.; Lin, J.X.; Leonard, W.J. IL-2 family cytokines: New insights into the complex roles of IL-2 as a broad regulator of T helper cell differentiation. Curr. Opin. Immunol. 2011, 23, 598–604. [Google Scholar] [CrossRef]
- Shouse, A.N.; LaPorte, K.M.; Malek, T.R. Interleukin-2 signaling in the regulation of T cell biology in autoimmunity and cancer. Immunity 2024, 57, 414–428. [Google Scholar] [CrossRef]
- Bunting, K.; Wang, J.; Shannon, M.F. Control of interleukin-2 gene transcription: A paradigm for inducible, tissue-specific gene expression. Vitam. Horm. 2006, 74, 105–145. [Google Scholar]
- Crispín, J.C.; Tsokos, G.C. Transcriptional regulation of IL-2 in health and autoimmunity. Autoimmun. Rev. 2009, 8, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Shah, K.; Al-Haidari, A.; Sun, J.; Kazi, J.U. T cell receptor (TCR) signaling in health and disease. Signal Transduct. Target Ther. 2021, 6, 412. [Google Scholar] [CrossRef] [PubMed]
- Hermann-Kleiter, N.; Baier, G. NFAT pulls the strings during CD4+ T helper cell effector functions. Blood 2010, 115, 2989–2997. [Google Scholar] [CrossRef] [PubMed]
- Schroder, K.; Hertzog, P.J.; Ravasi, T.; Hume, D.A. Interferon-γ: An overview of signals, mechanisms and functions. J. Leukoc. Biol. 2004, 75, 163–189. [Google Scholar] [CrossRef] [PubMed]
- Jorgovanovic, D.; Song, M.; Wang, L.; Zhang, Y. Roles of IFN-γ in tumor progression and regression: A review. Biomark. Res. 2020, 8, 49. [Google Scholar] [CrossRef]
- Rožman, P.; Švajger, U. The tolerogenic role of IFN-γ. Cytokine Growth Factor Rev. 2018, 41, 40–53. [Google Scholar] [CrossRef]
- Burke, J.D.; Young, H.A. IFN-γ: A cytokine at the right time, is in the right place. Semin. Immunol. 2019, 43, 101280. [Google Scholar] [CrossRef]
- Balasubramani, A.; Mukasa, R.; Hatton, R.D.; Weaver, C.T. Regulation of the Ifng locus in the context of T-lineage specification and plasticity. Immunol. Rev. 2010, 238, 216–232. [Google Scholar] [CrossRef]
- Wilson, C.B.; Rowell, E.; Sekimata, M. Epigenetic control of T-helper-cell differentiation. Nat. Rev. Immunol. 2009, 9, 91–105. [Google Scholar] [CrossRef]
- Aune, T.M.; Collins, P.L.; Collier, S.P.; Henderson, M.A.; Chang, S. Epigenetic activation and silencing of the gene that encodes IFN-γ. Front. Immunol. 2013, 4, 112. [Google Scholar] [CrossRef]
- Li, H.; Rao, A.; Hogan, P.G. Interaction of calcineurin with substrates and targeting proteins. Trends Cell Biol. 2011, 21, 91–103. [Google Scholar] [CrossRef]
- Qin, J.J.; Nag, S.; Wang, W.; Zhou, J.; Zhang, W.D.; Wang, H.; Zhang, R. NFAT as cancer target: Mission possible? Biochim. Biophys. Acta. 2014, 1846, 297–311. [Google Scholar] [CrossRef] [PubMed]
- Dewenter, M.; von der Lieth, A.; Katus, H.A.; Backs, J. Calcium signaling and transcriptional regulation in cardiomyocytes. Circ. Res. 2017, 121, 1000–1020. [Google Scholar] [CrossRef] [PubMed]
- Fric, J.; Zelante, T.; Wong, A.Y.W.; Mertes, A.; Yu, H.B.; Ricciardi-Castagnoli, P. NFAT control of innate immunity. Blood 2012, 120, 1380–1389. [Google Scholar] [CrossRef]
- Hodge, M.R.; Ranger, A.M.; Charles de la Brousse, F.; Hoey, T.; Grusby, M.J.; Glimcher, L.H. Hyperproliferation and dysregulation of IL-4 expression in NF-ATp-deficient mice. Immunity 1996, 4, 397–405. [Google Scholar] [CrossRef]
- Kiani, A.; García-Cózar, F.J.; Habermann, I.; Laforsch, S.; Aebischer, T.; Ehninger, G.; Rao, A. Regulation of interferon-γ gene expression by nuclear factor of activated T cells. Blood 2001, 98, 1480–1488. [Google Scholar] [CrossRef]
- Teixeira, L.K.; Fonseca, B.P.F.; Vieira-de-Abreu, A.; Barboza, B.A.; Robbs, B.K.; Bozza, P.T.; Viola, J.P. IFN-γ production by CD8+ T cells depends on NFAT1 transcription factor and regulates Th differentiation. J. Immunol. 2005, 175, 5931–5939. [Google Scholar] [CrossRef]
- Zhang, J.; Marotel, M.; Fauteux-Daniel, S.; Mathieu, A.L.; Viel, S.; Marçais, A.; Walzer, T. T-bet and Eomes govern differentiation and function of mouse and human NK cells and ILC1. Eur. J. Immunol. 2018, 48, 738–750. [Google Scholar] [CrossRef]
- Pritchard, G.H.; Kedl, R.M.; Hunter, C.A. The evolving role of T-bet in resistance to infection. Nat. Rev. Immunol. 2019, 19, 398–410. [Google Scholar] [CrossRef]
- Fukuoka, N.; Harada, M.; Nishida, A.; Ito, Y.; Shiota, H.; Kataoka, T. Eomesodermin promotes interferon-γ expression and binds to multiple conserved noncoding sequences across the Ifng locus in mouse thymoma cell lines. Genes Cells 2016, 21, 146–162. [Google Scholar] [CrossRef]
- Harada, M.; Vo, N.T.; Nakao, A.; Tanigaki, R.; Fukuoka, N.; Nishida, A.; Kataoka, T. Eomesodermin promotes interaction of RelA and NFATc2 with the Ifng promoter and multiple conserved noncoding sequences across the Ifng locus in mouse lymphoma BW5147 cells. Immunol. Lett. 2020, 225, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Kimura, K.; Minamikawa, Y.; Ogasawara, Y.; Yoshida, J.; Saitoh, K.; Shinden, H.; Ye, Y.Q.; Takahashi, S.; Miyakawa, T.; Koshino, H. Kujigamberol, a new dinorlabdane diterpenoid isolated from 85 million years old Kuji amber using a biotechnological assay. Fitoterapia 2012, 83, 907–912. [Google Scholar] [CrossRef]
- Fukuhara, S.; Tanigaki, R.; Kimura, K.; Kataoka, T. Kujigamberol interferes with pro-inflammatory cytokine-induced expression of and N-glycan modifications to cell adhesion molecules at different stages in human umbilical vein endothelial cells. Int. Immunopharmacol. 2018, 62, 313–325. [Google Scholar] [CrossRef]
- Maruyama, M.; Kobayashi, M.; Uchida, T.; Shimizu, E.; Higashio, H.; Ohno, M.; Uesugi, S.; Kimura, K. Anti-allergy activities of Kuji amber extract and kujigamberol. Fitoterapia 2018, 127, 263–270. [Google Scholar] [CrossRef]
- Hagiwara, H.; Yokota, T.; Luh, J.; Lee, F.; Arai, K.; Arai, N.; Zlotnik, A. The AKR thymoma BW5147 is able to produce lymphokines when stimulated with calcium ionophore and phorbol ester. J. Immunol. 1988, 140, 1561–1565. [Google Scholar] [CrossRef]
- Li, Y.Q.; Kobayashi, M.; Yuan, L.; Wang, J.; Matsushita, K.; Hamada, J.; Kimura, K.; Yagita, H.; Okumura, K.; Hosokawa, M. Protein kinase C mediates the signal for interferon-γ mRNA expression in cytotoxic T cells after their adhesion to laminin. Immunology 1998, 93, 455–461. [Google Scholar] [CrossRef]
- Lv, S.; Yi, P.F.; Shen, H.Q.; Zhang, L.Y.; Dong, H.B.; Wu, S.C.; Xia, F.; Guo, X.; Wei, X.B.; Fu, B.D. Ginsenoside Rh2-B1 stimulates cell proliferation and IFN-γ production by activating the p38 MAPK and ERK-dependent signaling pathways in CTLL-2 cells. Immunopharmacol. Immunotoxicol. 2014, 36, 43–51. [Google Scholar] [CrossRef]
- Sieber, M.; Baumgrass, R. Novel inhibitors of the calcineurin/NFATc hub–Alternatives to CsA and FK506? Cell Commun. Signal. 2009, 7, 25. [Google Scholar] [CrossRef]
- Rauluseviciute, I.; Riudavets-Puig, R.; Blanc-Mathieu, R.; Castro-Mondragon, J.A.; Ferenc, K.; Kumar, V.; Lemma, R.B.; Lucas, J.; Chèneby, J.; Baranasic, D.; et al. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2024, 52, D174–D182. [Google Scholar] [CrossRef]
- Benbijja, M.; Mellouk, A.; Bobé, P. Sensitivity of leukemic T-cell lines to arsenic trioxide cytotoxicity is dependent on the induction of phosphatase B220/CD45R expression at the cell surface. Mol. Cancer. 2014, 13, 251. [Google Scholar] [CrossRef]
- Pearce, E.L.; Mullen, A.C.; Martins, G.A.; Krawczyk, C.M.; Hutchins, A.S.; Zediak, V.P.; Banica, M.; DiCioccio, C.B.; Gross, D.A.; Mao, C.A.; et al. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science 2003, 302, 1041–1043. [Google Scholar] [CrossRef]
- Eshima, K.; Chiba, S.; Suzuki, H.; Kokubo, K.; Kobayashi, H.; Iizuka, M.; Iwabuchi, K.; Shinohara, N. Ectopic expression of a T-box transcription factor, eomesodermin, renders CD4+ Th cells cytotoxic by activating both perforin- and FasL-pathways. Immunol. Lett. 2012, 144, 7–15. [Google Scholar] [CrossRef]
- Yagi, R.; Junttila, I.S.; Wei, G.; Urban, J.F., Jr.; Zhao, K.; Paul, W.E.; Zhu, J. The transcription factor GATA3 actively represses RUNX3 protein-regulated production of interferon-γ. Immunity 2010, 23, 507–517. [Google Scholar] [CrossRef]
- Endo, Y.; Iwamura, C.; Kuwahara, M.; Suzuki, A.; Sugaya, K.; Tumes, D.J.; Tokoyoda, K.; Hosokawa, H.; Yamashita, M.; Nakayama, T. Eomesodermin controls interleukin-5 production in memory T helper 2 cells through inhibition of activity of the transcription factor GATA3. Immunity 2011, 35, 733–745. [Google Scholar] [CrossRef]
- Aune, T.M.; Penix, L.A.; Rincón, M.R.; Flavell, R.A. Differential transcription directed by discrete gamma interferon promoter elements in naive and memory (effector) CD4 T cells and CD8 T cells. Mol. Cell. Biol. 1997, 17, 199–208. [Google Scholar] [CrossRef]
- Sica, A.; Dorman, L.; Viggiano, V.; Cippitelli, M.; Ghosh, P.; Rice, N.; Young, H.A. Interaction of NF-κB and NFAT with the interferon-γ promoter. J. Biol. Chem. 1997, 272, 30412–30420. [Google Scholar] [CrossRef]
- Rooney, J.W.; Sun, Y.L.; Glimcher, L.H.; Hoey, T. Novel NFAT sites that mediate activation of the interleukin-2 promoter in response to T-cell receptor stimulation. Mol. Cell. Biol. 1995, 15, 6299–6310. [Google Scholar] [CrossRef]
- Walters, R.D.; Drullinger, L.F.; Kugel, J.F.; Goodrich, J.A. NFATc2 recruits cJun homodimers to an NFAT site to synergistically activate interleukin-2 transcription. Mol. Immunol. 2013, 56, 48–56. [Google Scholar] [CrossRef]
- Li-Weber, M.; Giaisi, M.; Baumann, S.; Pálfi, K.; Krammer, P.H. NF-κB synergizes with NF-AT and NF-IL6 in activation of the IL-4 gene in T cells. Eur. J. Immunol. 2004, 34, 1111–1118. [Google Scholar] [CrossRef]
- Kavurma, M.M.; Khachigian, L.M. Signaling and transcriptional control of Fas ligand gene expression. Cell Death Differ. 2003, 10, 36–44. [Google Scholar] [CrossRef]
- Peng, S.L.; Gerth, A.J.; Ranger, A.M.; Glimcher, L.H. NFATc1 and NFATc2 together control both T and B cell activation and differentiation. Immunity 2001, 14, 13–20. [Google Scholar] [CrossRef]
- Wu, Y.; Borde, M.; Heissmeyer, V.; Feuerer, M.; Lapan, A.D.; Stroud, J.C.; Bates, D.L.; Guo, L.; Han, A.; Ziegler, S.F.; et al. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell 2006, 126, 375–387. [Google Scholar] [CrossRef]
- Ishihara, S.; Schwartz, R.H. Two-step binding of transcription factors causes sequential chromatin structural changes at the activated IL-2 promoter. J. Immunol. 2011, 187, 3292–3299. [Google Scholar] [CrossRef]
- Abe, T.; Kobayashi, M.; Okawa, Y.; Inui, T.; Yoshida, J.; Higashio, H.; Shinden, H.; Uesugi, S.; Koshino, H.; Kimura, K. Yeast Ca2+-signal transduction inhibitors isolated from Dominican amber prevent the degranulation of RBL-2H3 cells through the inhibition of Ca2+-influx. Fitoterapia 2016, 113, 188–194. [Google Scholar] [CrossRef]
- Dohrman, A.; Kataoka, T.; Cuenin, S.; Russell, J.Q.; Tschopp, J.; Budd, R.C. Cellular FLIP (long form) regulates CD8+ T cell activation through caspase-8-dependent NF-κB activation. J. Immunol. 2005, 174, 5270–5278. [Google Scholar] [CrossRef]
- Matsuda, I.; Matsuo, K.; Matsushita, Y.; Haruna, Y.; Niwa, M.; Kataoka, T. The C-terminal domain of the long form of cellular FLICE-inhibitory protein (c-FLIPL) inhibits the interaction of the caspase 8 prodomain with the receptor-interacting protein 1 (RIP1) death domain and regulates caspase 8-dependent nuclear factor κB (NF-κB) activation. J. Biol. Chem. 2014, 289, 3876–3887. [Google Scholar]
- Casteels, K.M.; Mathieu, C.; Waer, M.; Valckx, D.; Overbergh, L.; Laureys, J.M.; Bouillon, R. Prevention of type I diabetes in nonobese diabetic mice by late intervention with nonhypercalcemic analogs of 1,25-dihydroxyvitamin D3 in combination with a short induction course of cyclosporin A. Endocrinology 1998, 139, 95–102. [Google Scholar] [CrossRef]
- Orsatti, C.L.; Missima, F.; Pagliarone, A.C.; Sforcin, J.M. Th1/Th2 cytokines’ expression and production by propolis-treated mice. J. Ethnopharmacol. 2010, 129, 314–318. [Google Scholar] [CrossRef]
- Pinhu, L.; Qin, Y.; Xiong, B.; You, Y.; Li, J.; Sooranna, S.R. Overexpression of Fas and FasL is associated with infectious complications and severity of experimental severe acute pancreatitis by promoting apoptosis of lymphocytes. Inflammation 2014, 37, 1202–1212. [Google Scholar] [CrossRef]
- Tanaka, Y.; Nakao, A.; Miyake, Y.; Higashi, Y.; Tanigaki, R.; Kataoka, T. Small molecule inhibitors targeting nuclear factor κB activation markedly reduce expression of interleukin-2, but not interferon-γ, induced by phorbol esters and calcium ionophores. Int. J. Mol. Sci. 2021, 22, 13098. [Google Scholar] [CrossRef]
- Chang, S.; Aune, T.M. Dynamic changes in histone-methylation “marks” across the locus encoding interferon-γ during the differentiation of T helper type 2 cells. Nat. Immunol. 2007, 8, 723–731. [Google Scholar] [CrossRef] [PubMed]
- Dheer, D.; Jyoti; Gupta, P.N.; Shankar, R. Tacrolimus: An updated review on delivering strategies for multifarious diseases. Eur. J. Pharm. Sci. 2018, 114, 217–227. [Google Scholar] [CrossRef]
- Ong, S.C.; Gaston, R.S. Thirty years of tacrolimus in clinical practice. Transplantation 2021, 105, 484–495. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yodweerapong, T.; Ueno, Y.; Yamaguchi, R.; Yarangsee, P.; Kimura, K.-i.; Kataoka, T. Kujigamberol Inhibits IFN-γ and IL-2 mRNA Expression and NFATc2 Binding to Their Promoters in Response to a Phorbol Ester and Ionomycin Stimulation. Molecules 2025, 30, 2214. https://doi.org/10.3390/molecules30102214
Yodweerapong T, Ueno Y, Yamaguchi R, Yarangsee P, Kimura K-i, Kataoka T. Kujigamberol Inhibits IFN-γ and IL-2 mRNA Expression and NFATc2 Binding to Their Promoters in Response to a Phorbol Ester and Ionomycin Stimulation. Molecules. 2025; 30(10):2214. https://doi.org/10.3390/molecules30102214
Chicago/Turabian StyleYodweerapong, Tanpitcha, Yuto Ueno, Rikako Yamaguchi, Piimwara Yarangsee, Ken-ichi Kimura, and Takao Kataoka. 2025. "Kujigamberol Inhibits IFN-γ and IL-2 mRNA Expression and NFATc2 Binding to Their Promoters in Response to a Phorbol Ester and Ionomycin Stimulation" Molecules 30, no. 10: 2214. https://doi.org/10.3390/molecules30102214
APA StyleYodweerapong, T., Ueno, Y., Yamaguchi, R., Yarangsee, P., Kimura, K.-i., & Kataoka, T. (2025). Kujigamberol Inhibits IFN-γ and IL-2 mRNA Expression and NFATc2 Binding to Their Promoters in Response to a Phorbol Ester and Ionomycin Stimulation. Molecules, 30(10), 2214. https://doi.org/10.3390/molecules30102214






