Inflammatory Bowel Diseases and Gut Microbiota
Abstract
:1. Introduction
2. Gut Microbiota and IBD
3. Effect of Probiotics, Prebiotics, and Symbiotics on IBD
4. FMT for IBD
5. Microbial Metabolites and IBD
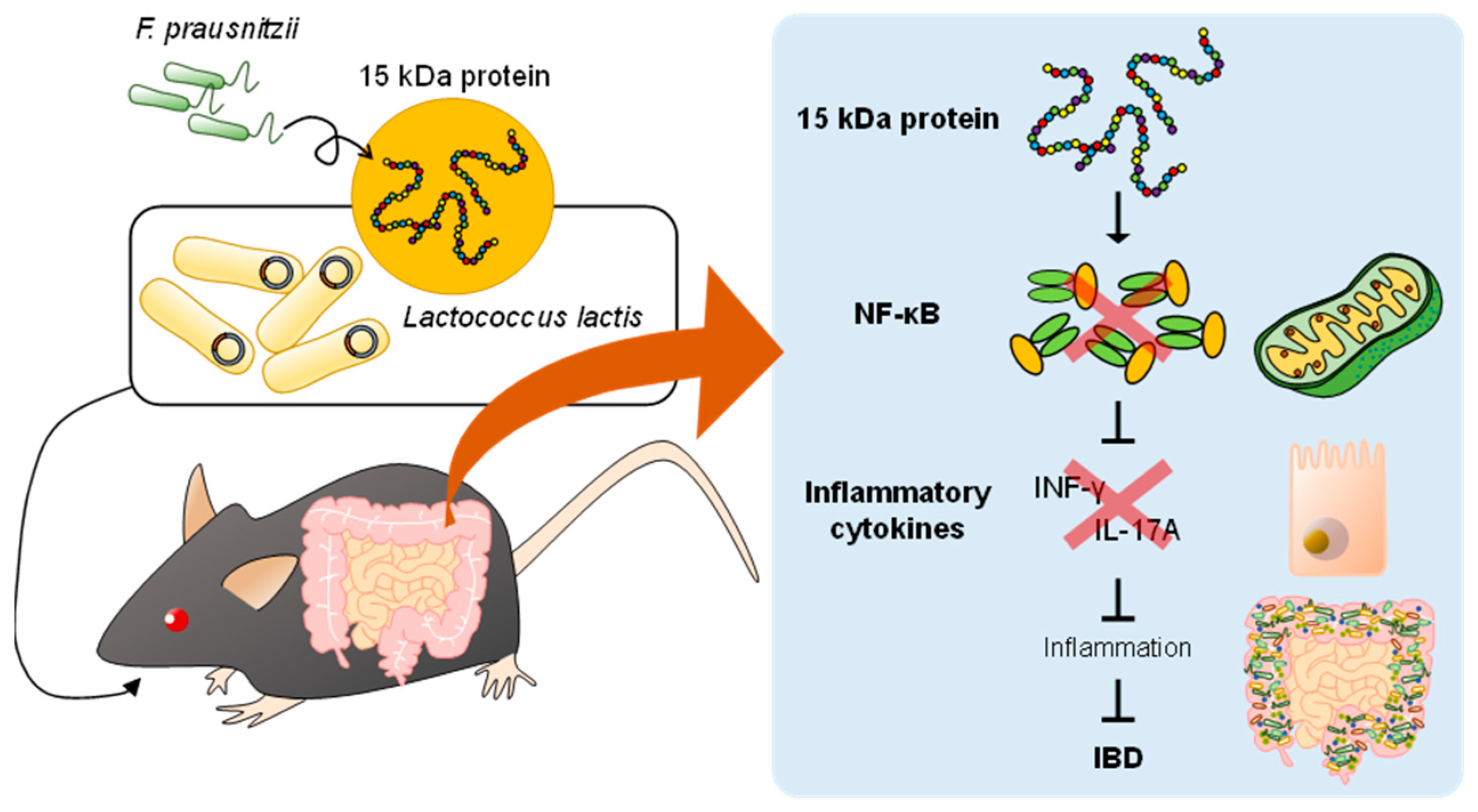
6. A Potential Innovative Strategy by Postbiotic Treatment
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Prescott, S.L. Early-life environmental determinants of allergic diseases and the wider pandemic of inflammatory noncommunicable diseases. J. Allergy Clin. Immunol. 2013, 131, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Al Bander, Z.; Nitert, M.D.; Mousa, A.; Naderpoor, N. The Gut Microbiota and Inflammation: An Overview. Int. J. Environ. Res. Public Health 2020, 17, 7618. [Google Scholar] [CrossRef] [PubMed]
- Aron-Wisnewsky, J.; Warmbrunn, M.V.; Nieuwdorp, M.; Clément, K. Metabolism and Metabolic Disorders and the Microbiome: The Intestinal Microbiota Associated With Obesity, Lipid Metabolism, and Metabolic Health-Pathophysiology and Therapeutic Strategies. Gastroenterology 2021, 160, 573–599. [Google Scholar] [CrossRef] [PubMed]
- Sekirov, I.; Russell, S.L.; Antunes, L.C.; Finlay, B.B. Gut microbiota in health and disease. Physiol. Rev. 2010, 90, 859–904. [Google Scholar] [CrossRef] [PubMed]
- Maloy, K.J.; Powrie, F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature 2011, 474, 298–306. [Google Scholar] [CrossRef]
- Honda, K.; Littman, D.R. The microbiota in adaptive immune homeostasis and disease. Nature 2016, 535, 75–84. [Google Scholar] [CrossRef]
- Hooper, L.V.; Littman, D.R.; Macpherson, A.J. Interactions between the microbiota and the immune system. Science 2012, 336, 1268–1273. [Google Scholar] [CrossRef]
- Nieuwdorp, M.; Gilijamse, P.W.; Pai, N.; Kaplan, L.M. Role of the microbiome in energy regulation and metabolism. Gastroenterology 2014, 146, 1525–1533. [Google Scholar] [CrossRef]
- Gomes, A.C.; Hoffmann, C.; Mota, J.F. The human gut microbiota: Metabolism and perspective in obesity. Gut Microbes 2018, 9, 308–325. [Google Scholar] [CrossRef]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef] [Green Version]
- Brestoff, J.R.; Artis, D. Commensal bacteria at the interface of host metabolism and the immune system. Nat. Immunol. 2013, 14, 676–684. [Google Scholar] [CrossRef] [PubMed]
- Ridaura, V.K.; Faith, J.J.; Rey, F.E.; Cheng, J.; Duncan, A.E.; Kau, A.L.; Griffin, N.W.; Lombard, V.; Henrissat, B.; Bain, J.R.; et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 2013, 341, 1241214. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Baker, S.S.; Gill, C.; Liu, W.; Alkhouri, R.; Baker, R.D.; Gill, S.R. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: A connection between endogenous alcohol and NASH. Hepatology 2013, 57, 601–609. [Google Scholar] [CrossRef]
- Ivanov, I.I.; Honda, K. Intestinal commensal microbes as immune modulators. Cell Host Microbe 2012, 12, 496–508. [Google Scholar] [CrossRef] [PubMed]
- Wen, L.; Ley, R.E.; Volchkov, P.Y.; Stranges, P.B.; Avanesyan, L.; Stonebraker, A.C.; Hu, C.; Wong, F.S.; Szot, G.L.; Bluestone, J.A.; et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature 2008, 455, 1109–1113. [Google Scholar] [CrossRef] [PubMed]
- Shimokawa, C.; Kato, T.; Takeuchi, T.; Ohshima, N.; Furuki, T.; Ohtsu, Y.; Suzue, K.; Imai, T.; Obi, S.; Olia, A.; et al. CD8+ Regulatory T Cells Are Critical in Prevention of Autoimmune-Mediated Diabetes. Nat. Commun. 2020, 11, 1922. [Google Scholar] [CrossRef]
- Cignarella, F.; Cantoni, C.; Ghezzi, L.; Salter, A.; Dorsett, Y.; Chen, L.; Phillips, D.; Weinstock, G.M.; Fontana, L.; Cross, A.H.; et al. Intermittent Fasting Confers Protection in CNS Autoimmunity by Altering the Gut Microbiota. Cell Metab. 2018, 27, 1222–1235.e6. [Google Scholar] [CrossRef]
- Berer, K.; Gerdes, L.A.; Cekanaviciute, E.; Jia, X.; Xiao, L.; Xia, Z.; Liu, C.; Klotz, L.; Stauffer, U.; Baranzini, S.E.; et al. Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proc. Natl. Acad. Sci. USA 2017, 114, 10719–10724. [Google Scholar] [CrossRef]
- Miyauchi, E.; Kim, S.W.; Suda, W.; Kawasumi, M.; Onawa, S.; Taguchi-Atarashi, N.; Morita, H.; Taylor, T.D.; Hattori, M.; Ohno, H. Gut microorganisms act together to exacerbate inflammation in spinal cords. Nature 2020, 585, 102–106. [Google Scholar] [CrossRef]
- Chen, J.; Wright, K.; Davis, J.M.; Jeraldo, P.; Marietta, E.V.; Murray, J.; Nelson, H.; Matteson, E.L.; Taneja, V. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. 2016, 8, 43. [Google Scholar] [CrossRef] [Green Version]
- Leonard, M.M.; Valitutti, F.; Karathia, H.; Pujolassos, M.; Kenyon, V.; Fanelli, B.; Troisi, J.; Subramanian, P.; Camhi, S.; Colucci, A.; et al. Microbiome signatures of progression toward celiac disease onset in at-risk children in a longitudinal prospective cohort study. Proc. Natl. Acad. Sci. USA 2021, 118, e2020322118. [Google Scholar] [CrossRef] [PubMed]
- Windsor, J.W.; Kaplan, G.G. Evolving Epidemiology of IBD. Curr. Gastroenterol. Rep. 2019, 21, 40. [Google Scholar] [CrossRef] [PubMed]
- Kaser, A.; Zeissig, S.; Blumberg, R.S. Genes and environment: How will our concepts on the pathophysiology of IBD develop in the future? Dig. Dis. 2010, 28, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Knowles, S.R.; Graff, L.A.; Wilding, H.; Hewitt, C.; Keefer, L.; Mikocka-Walus, A. Quality of Life in Inflammatory Bowel Disease: A Systematic Review and Meta-analyses-Part I. Inflamm. Bowel Dis. 2018, 24, 742–751. [Google Scholar] [CrossRef] [PubMed]
- Dorofeyev, A.E.; Vasilenko, I.V.; Rassokhina, O.A.; Kondratiuk, R.B. Mucosal barrier in ulcerative colitis and Crohn’s disease. Gastroenterol. Res. Pract. 2013, 2013, 431231. [Google Scholar] [CrossRef]
- Peterson, D.A.; Frank, D.N.; Pace, N.R.; Gordon, J.I. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe 2008, 3, 417–427. [Google Scholar] [CrossRef]
- Yeshi, K.; Ruscher, R.; Hunter, L.; Daly, N.L.; Loukas, A.; Wangchuk, P. Revisiting Inflammatory Bowel Disease: Pathology, Treatments, Challenges and Emerging Therapeutics Including Drug Leads from Natural Products. J. Clin. Med. 2020, 9, 1273. [Google Scholar] [CrossRef]
- Santana, P.T.; Rosas, S.L.B.; Ribeiro, B.E.; Marinho, Y.; de Souza, H.S.P. Dysbiosis in Inflammatory Bowel Disease: Pathogenic Role and Potential Therapeutic Targets. Int. J. Mol. Sci. 2022, 23, 3464. [Google Scholar] [CrossRef]
- Loftus, E.V., Jr. Clinical epidemiology of inflammatory bowel disease: Incidence, prevalence, and environmental influences. Gastroenterology 2004, 126, 1504–1517. [Google Scholar] [CrossRef]
- Glassner, K.L.; Abraham, B.P.; Quigley, E.M.M. The microbiome and inflammatory bowel disease. J. Allergy Clin. Immunol. 2020, 145, 16–27. [Google Scholar] [CrossRef] [Green Version]
- Aniwan, S.; Tremaine, W.J.; Raffals, L.E.; Kane, S.V.; Loftus, E.V., Jr. Antibiotic Use and New-Onset Inflammatory Bowel Disease in Olmsted County, Minnesota: A Population-Based Case-Control Study. J. Crohns Colitis 2018, 12, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Rohr, M.W.; Narasimhulu, C.A.; Rudeski-Rohr, T.A.; Parthasarathy, S. Negative Effects of a High-Fat Diet on Intestinal Permeability: A Review. Adv. Nutr. 2020, 11, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Belkaid, Y.; Hand, T.W. Role of the Microbiota in Immunity and Inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef] [PubMed]
- Khor, B.; Gardet, A.; Xavier, R.J. Genetics and pathogenesis of inflammatory bowel disease. Nature 2011, 474, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Fava, F.; Danese, S. Intestinal microbiota in inflammatory bowel disease: Friend of foe? World J. Gastroenterol. 2011, 17, 557–566. [Google Scholar] [CrossRef]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef]
- Morgan, X.C.; Tickle, T.L.; Sokol, H.; Gevers, D.; Devaney, K.L.; Ward, D.V.; Reyes, J.A.; Shah, S.A.; LeLeiko, N.; Snapper, S.B.; et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012, 13, R79. [Google Scholar] [CrossRef]
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.; Yamasaki, S.; Saito, T.; Ohba, Y.; et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011, 331, 337–341. [Google Scholar] [CrossRef]
- Watanabe, Y.; Nagai, F.; Morotomi, M. Characterization of Phascolarctobacterium succinatutens sp. nov., an asaccharolytic, succinate-utilizing bacterium isolated from human feces. Appl. Environ. Microbiol. 2012, 78, 511–518. [Google Scholar] [CrossRef]
- Eslick, S.; Thompson, C.; Berthon, B.; Wood, L. Short-chain fatty acids as anti-inflammatory agents in overweight and obesity: A systematic review and meta-analysis. Nutr. Rev. 2022, 80, 838–856. [Google Scholar] [CrossRef]
- Kabeerdoss, J.; Sankaran, V.; Pugazhendhi, S.; Ramakrishna, B.S. Clostridium leptum group bacteria abundance and diversity in the fecal microbiota of patients with inflammatory bowel disease: A case-control study in India. BMC Gastroenterol. 2013, 13, 20. [Google Scholar] [CrossRef]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermúdez-Humarán, L.G.; Gratadoux, J.J.; Blugeon, S.; Bridonneau, C.; Furet, J.P.; Corthier, G.; et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef]
- Willing, B.P.; Dicksved, J.; Halfvarson, J.; Andersson, A.F.; Lucio, M.; Zheng, Z.; Järnerot, G.; Tysk, C.; Jansson, J.K.; Engstrand, L. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 2010, 139, 1844–1854.e1. [Google Scholar] [CrossRef]
- Lloyd-Price, J.; Arze, C.; Ananthakrishnan, A.N.; Schirmer, M.; Avila-Pacheco, J.; Poon, T.W.; Andrews, E.; Ajami, N.J.; Bonham, K.S.; Brislawn, C.J.; et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019, 569, 655–662. [Google Scholar] [CrossRef]
- Selinger, C.P.; Bell, A.; Cairns, A.; Lockett, M.; Sebastian, S.; Haslam, N. Probiotic VSL#3 prevents antibiotic-associated diarrhoea in a double-blind, randomized, placebo-controlled clinical trial. J. Hosp. Infect. 2013, 84, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.S.; Li, W.B.; Wang, H.Y.; Ma, Y.M.; Zhao, X.H.; Yang, H.; Qian, J.M.; Li, J.N. VSL#3 can prevent ulcerative colitis-associated carcinogenesis in mice. World J. Gastroenterol. 2018, 24, 4254–4262. [Google Scholar] [CrossRef] [PubMed]
- Rossi, G.; Pengo, G.; Caldin, M.; Palumbo Piccionello, A.; Steiner, J.M.; Cohen, N.D.; Jergens, A.E.; Suchodolski, J.S. Comparison of microbiological, histological, and immunomodulatory parameters in response to treatment with either combination therapy with prednisone and metronidazole or probiotic VSL#3 strains in dogs with idiopathic inflammatory bowel disease. PLoS ONE 2014, 9, e94699. [Google Scholar] [CrossRef]
- Uronis, J.M.; Arthur, J.C.; Keku, T.; Fodor, A.; Carroll, I.M.; Cruz, M.L.; Appleyard, C.B.; Jobin, C. Gut microbial diversity is reduced by the probiotic VSL#3 and correlates with decreased TNBS-induced colitis. Inflamm. Bowel Dis. 2011, 17, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Carnero-Gregorio, M.; Molares-Vila, A.; Corbalán-Rivas, A.; Villaverde-Taboada, C.; Rodríguez-Cerdeira, C. Effect of VSL#3 Probiotic in a Patient with Glycogen Storage Disease Type Ia and Irritable Bowel Disease-like Disease. Probiotics Antimicrob. Proteins 2019, 11, 143–149. [Google Scholar] [CrossRef]
- Gionchetti, P.; Rizzello, F.; Helwig, U.; Venturi, A.; Lammers, K.M.; Brigidi, P.; Vitali, B.; Poggioli, G.; Miglioli, M.; Campieri, M. Prophylaxis of pouchitis onset with probiotic therapy: A double-blind, placebo-controlled trial. Gastroenterology 2003, 124, 1202–1209. [Google Scholar] [CrossRef] [PubMed]
- Gionchetti, P.; Rizzello, F.; Venturi, A.; Brigidi, P.; Matteuzzi, D.; Bazzocchi, G.; Poggioli, G.; Miglioli, M.; Campieri, M. Oral bacteriotherapy as maintenance treatment in patients with chronic pouchitis: A double-blind, placebo-controlled trial. Gastroenterology 2000, 119, 305–309. [Google Scholar] [CrossRef] [PubMed]
- Kühbacher, T.; Ott, S.J.; Helwig, U.; Mimura, T.; Rizzello, F.; Kleessen, B.; Gionchetti, P.; Blaut, M.; Campieri, M.; Fölsch, U.R.; et al. Bacterial and fungal microbiota in relation to probiotic therapy (VSL#3) in pouchitis. Gut 2006, 55, 833–841. [Google Scholar] [CrossRef] [PubMed]
- Dang, X.; Xu, M.; Liu, D.; Zhou, D.; Yang, W. Assessing the efficacy and safety of fecal microbiota transplantation and probiotic VSL#3 for active ulcerative colitis: A systematic review and meta-analysis. PLoS ONE 2020, 15, e0228846. [Google Scholar] [CrossRef]
- Quévrain, E.; Maubert, M.A.; Michon, C.; Chain, F.; Marquant, R.; Tailhades, J.; Miquel, S.; Carlier, L.; Bermúdez-Humarán, L.G.; Pigneur, B.; et al. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut 2016, 65, 415–425. [Google Scholar] [CrossRef] [PubMed]
- Kruis, W.; Fric, P.; Pokrotnieks, J.; Lukás, M.; Fixa, B.; Kascák, M.; Kamm, M.A.; Weismueller, J.; Beglinger, C.; Stolte, M.; et al. Maintaining remission of ulcerative colitis with the probiotic Escherichia coli Nissle 1917 is as effective as with standard mesalazine. Gut 2004, 53, 1617–1623. [Google Scholar] [CrossRef]
- Dieleman, L.A.; Goerres, M.S.; Arends, A.; Sprengers, D.; Torrice, C.; Hoentjen, F.; Grenther, W.B.; Sartor, R.B. Lactobacillus GG prevents recurrence of colitis in HLA-B27 transgenic rats after antibiotic treatment. Gut 2003, 52, 370–376. [Google Scholar] [CrossRef]
- Li, Q.; Sun, X.; Yu, K.; Lv, J.; Miao, C.; Yang, J.; Wang, S.; Fu, Z.; Sun, Y.; Zhang, H.; et al. Enterobacter ludwigii protects DSS-induced colitis through choline-mediated immune tolerance. Cell Rep. 2022, 40, 111308. [Google Scholar] [CrossRef]
- Hedin, C.R.; McCarthy, N.E.; Louis, P.; Farquharson, F.M.; McCartney, S.; Stagg, A.J.; Lindsay, J.O.; Whelan, K. Prebiotic Fructans Have Greater Impact on Luminal Microbiology and CD3+ T Cells in Healthy Siblings than Patients with Crohn’s Disease: A Pilot Study Investigating the Potential for Primary Prevention of Inflammatory Bowel Disease. Clin. Nutr. 2021, 40, 5009–5019. [Google Scholar] [CrossRef]
- Zhang, X.F.; Guan, X.X.; Tang, Y.J.; Sun, J.F.; Wang, X.K.; Wang, W.D.; Fan, J.M. Clinical Effects and Gut Microbiota Changes of Using Probiotics, Prebiotics or Synbiotics in Inflammatory Bowel Disease: A Systematic Review and Meta-Analysis. Eur. J. Nutr. 2021, 60, 2855–2875. [Google Scholar] [CrossRef]
- Colman, R.J.; Rubin, D.T. Fecal microbiota transplantation as therapy for inflammatory bowel disease: A systematic review and meta-analysis. J. Crohns Colitis 2014, 8, 1569–1581. [Google Scholar] [CrossRef] [Green Version]
- Nishida, A.; Imaeda, H.; Ohno, M.; Inatomi, O.; Bamba, S.; Sugimoto, M.; Andoh, A. Efficacy and safety of single fecal microbiota transplantation for Japanese patients with mild to moderately active ulcerative colitis. J. Gastroenterol. 2017, 52, 476–482. [Google Scholar] [CrossRef] [PubMed]
- Goyal, A.; Yeh, A.; Bush, B.R.; Firek, B.A.; Siebold, L.M.; Rogers, M.B.; Kufen, A.D.; Morowitz, M.J. Safety, Clinical Response, and Microbiome Findings Following Fecal Microbiota Transplant in Children With Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2018, 24, 410–421. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.D.; Sandler, R.S.; Brotherton, C.; Brensinger, C.; Li, H.; Kappelman, M.D.; Daniel, S.G.; Bittinger, K.; Albenberg, L.; Valentine, J.F.; et al. A Randomized Trial Comparing the Specific Carbohydrate Diet to a Mediterranean Diet in Adults With Crohn’s Disease. Gastroenterology 2021, 161, 837–852.e9. [Google Scholar] [CrossRef] [PubMed]
- Burrello, C.; Giuffrè, M.R.; Macandog, A.D.; Diaz-Basabe, A.; Cribiù, F.M.; Lopez, G.; Borgo, F.; Nezi, L.; Caprioli, F.; Vecchi, M.; et al. Fecal Microbiota Transplantation Controls Murine Chronic Intestinal Inflammation by Modulating Immune Cell Functions and Gut Microbiota Composition. Cells 2019, 8, 517. [Google Scholar] [CrossRef]
- Wen, X.; Wang, H.G.; Zhang, M.N.; Zhang, M.H.; Wang, H.; Yang, X.Z. Fecal microbiota transplantation ameliorates experimental colitis via gut microbiota and T-cell modulation. World J. Gastroenterol. 2021, 27, 2834–2849. [Google Scholar] [CrossRef]
- Zhang, W.; Zou, G.; Li, B.; Du, X.; Sun, Z.; Sun, Y.; Jiang, X. Fecal Microbiota Transplantation (FMT) Alleviates Experimental Colitis in Mice by Gut Microbiota Regulation. J. Microbiol. Biotechnol. 2020, 30, 1132–1141. [Google Scholar] [CrossRef]
- Maslowski, K.M.; Vieira, A.T.; Ng, A.; Kranich, J.; Sierro, F.; Yu, D.; Schilter, H.C.; Rolph, M.S.; Mackay, F.; Artis, D.; et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 2009, 461, 1282–1286. [Google Scholar] [CrossRef]
- Allegretti, J.R.; Kassam, Z.; Carrellas, M.; Mullish, B.H.; Marchesi, J.R.; Pechlivanis, A.; Smith, M.; Gerardin, Y.; Timberlake, S.; Pratt, D.S.; et al. Fecal Microbiota Transplantation in Patients With Primary Sclerosing Cholangitis: A Pilot Clinical Trial. Am. J. Gastroenterol. 2019, 114, 1071–1079. [Google Scholar] [CrossRef]
- Caruso, R.; Lo, B.C.; Núñez, G. Host-microbiota interactions in inflammatory bowel disease. Nat. Rev. Immunol. 2020, 20, 411–426. [Google Scholar] [CrossRef]
- Schmidt, T.S.B.; Li, S.S.; Maistrenko, O.M.; Akanni, W.; Coelho, L.P.; Dolai, S.; Fullam, A.; Glazek, A.M.; Hercog, R.; Herrema, H.; et al. Drivers and determinants of strain dynamics following fecal microbiota transplantation. Nat. Med. 2022, 28, 1902–1912. [Google Scholar] [CrossRef]
- Paramsothy, S.; Nielsen, S.; Kamm, M.A.; Deshpande, N.P.; Faith, J.J.; Clemente, J.C.; Paramsothy, R.; Walsh, A.J.; van den Bogaerde, J.; Samuel, D.; et al. Specific Bacteria and Metabolites Associated With Response to Fecal Microbiota Transplantation in Patients With Ulcerative Colitis. Gastroenterology 2019, 156, 1440–1454.e2. [Google Scholar] [CrossRef] [PubMed]
- Vaughn, B.P.; Vatanen, T.; Allegretti, J.R.; Bai, A.; Xavier, R.J.; Korzenik, J.; Gevers, D.; Ting, A.; Robson, S.C.; Moss, A.C. Increased Intestinal Microbial Diversity Following Fecal Microbiota Transplant for Active Crohn’s Disease. Inflamm. Bowel Dis. 2016, 22, 2182–2190. [Google Scholar] [CrossRef] [PubMed]
- Christ, A.; Lauterbach, M.; Latz, E. Western Diet and the Immune System: An Inflammatory Connection. Immunity 2019, 51, 794–811. [Google Scholar] [CrossRef]
- Moszak, M.; Szulińska, M.; Bogdański, P. You Are What You Eat—The Relationship between Diet, Microbiota, and Metabolic Disorders—A Review. Nutrients 2020, 12, 1096. [Google Scholar] [CrossRef] [PubMed]
- Mentella, M.C.; Scaldaferri, F.; Pizzoferrato, M.; Gasbarrini, A.; Miggiano, G.A.D. Nutrition, IBD and Gut Microbiota: A Review. Nutrients 2020, 12, 944. [Google Scholar] [CrossRef]
- Sugihara, K.; Kamada, N. Diet–Microbiota Interactions in Inflammatory Bowel Disease. Nutrients 2021, 13, 1533. [Google Scholar] [CrossRef]
- Tursi, A.; Brandimarte, G. The symptomatic and histologic response to a gluten-free diet in patients with borderline enteropathy. J. Clin. Gastroenterol. 2003, 36, 13–17. [Google Scholar] [CrossRef]
- Adolph, T.E.; Zhang, J. Diet fuelling inflammatory bowel diseases: Preclinical and clinical concepts. Gut 2022, 71, 2574–2586. [Google Scholar] [CrossRef]
- Roberts, C.L.; Keita, A.V.; Duncan, S.H.; O’Kennedy, N.; Söderholm, J.D.; Rhodes, J.M.; Campbell, B.J. Translocation of Crohn’s disease Escherichia coli across M-cells: Contrasting effects of soluble plant fibres and emulsifiers. Gut 2010, 59, 1331–1339. [Google Scholar] [CrossRef] [PubMed]
- Chassaing, B.; Koren, O.; Goodrich, J.K.; Poole, A.C.; Srinivasan, S.; Ley, R.E.; Gewirtz, A.T. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature 2015, 519, 92–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Chen, W.D.; Wang, Y.D. Nuclear Receptors: A Bridge Linking the Gut Microbiome and the Host. Mol. Med. 2021, 27, 144. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Chen, Z.; Xu, C.; Kan, S.; Chen, D. Disturbances of the Gut Microbiota and Microbiota-Derived Metabolites in Inflammatory Bowel Disease. Nutrients 2022, 14, 5140. [Google Scholar] [CrossRef] [PubMed]
- Huda-Faujan, N.; Abdulamir, A.S.; Fatimah, A.B.; Anas, O.M.; Shuhaimi, M.; Yazid, A.M.; Loong, Y.Y. The Impact of the Level of the Intestinal Short Chain Fatty Acids in Inflammatory Bowel Disease Patients Versus Healthy Subjects. Open Biochem. J. 2010, 4, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Kumari, R.; Ahuja, V.; Paul, J. Fluctuations in butyrate-producing bacteria in ulcerative colitis patients of North India. World J. Gastroenterol. 2013, 19, 3404–3414. [Google Scholar] [CrossRef]
- Ji, J.; Shu, D.; Zheng, M.; Wang, J.; Luo, C.; Wang, Y.; Guo, F.; Zou, X.; Lv, X.; Li, Y.; et al. Microbial metabolite butyrate facilitates M2 macrophage polarization and function. Sci. Rep. 2016, 6, 24838. [Google Scholar] [CrossRef]
- Bian, X.; Yang, L.; Wu, W.; Lv, L.; Jiang, X.; Wang, Q.; Wu, J.; Li, Y.; Ye, J.; Fang, D.; et al. Pediococcus pentosaceus LI05 alleviates DSS-induced colitis by modulating immunological profiles, the gut microbiota and short-chain fatty acid levels in a mouse model. Microb. Biotechnol. 2020, 13, 1228–1244. [Google Scholar] [CrossRef]
- Gudi, R.; Suber, J.; Brown, R.; Johnson, B.M.; Vasu, C. Pretreatment with Yeast-Derived Complex Dietary Polysaccharides Suppresses Gut Inflammation, Alters the Microbiota Composition, and Increases Immune Regulatory Short-Chain Fatty Acid Production in C57BL/6 Mice. J. Nutr. 2020, 150, 1291–1302. [Google Scholar] [CrossRef]
- Devkota, S.; Wang, Y.; Musch, M.W.; Leone, V.; Fehlner-Peach, H.; Nadimpalli, A.; Antonopoulos, D.A.; Jabri, B.; Chang, E.B. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature 2012, 487, 104–108. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]
- Duboc, H.; Rajca, S.; Rainteau, D.; Benarous, D.; Maubert, M.A.; Quervain, E.; Thomas, G.; Barbu, V.; Humbert, L.; Despras, G.; et al. Connecting Dysbiosis, Bile-Acid Dysmetabolism and Gut Inflammation in Inflammatory Bowel Diseases. Gut 2013, 62, 531–539. [Google Scholar] [CrossRef]
- Paik, D.; Yao, L.; Zhang, Y.; Bae, S.; D’Agostino, G.D.; Zhang, M.; Kim, E.; Franzosa, E.A.; Avila-Pacheco, J.; Bisanz, J.E.; et al. Human Gut Bacteria Produce TH17-Modulating Bile Acid Metabolites. Nature 2022, 603, 907–912. [Google Scholar] [CrossRef] [PubMed]
- Nikolaus, S.; Schulte, B.; Al-Massad, N.; Thieme, F.; Schulte, D.M.; Bethge, J.; Rehman, A.; Tran, F.; Aden, K.; Häsler, R.; et al. Increased Tryptophan Metabolism Is Associated With Activity of Inflammatory Bowel Diseases. Gastroenterology 2017, 153, 1504–1516.e2. [Google Scholar] [CrossRef] [PubMed]
- Naganuma, M.; Sugimoto, S.; Mitsuyama, K.; Kobayashi, T.; Yoshimura, N.; Ohi, H.; Tanaka, S.; Andoh, A.; Ohmiya, N.; Saigusa, K.; et al. Efficacy of Indigo Naturalis in a Multicenter Randomized Controlled Trial of Patients With Ulcerative Colitis. Gastroenterology 2018, 154, 935–947. [Google Scholar] [CrossRef] [PubMed]
- Mayne, C.G.; Williams, C.B. Induced and natural regulatory T cells in the development of inflammatory bowel disease. Inflamm. Bowel Dis. 2013, 19, 1772–1788. [Google Scholar] [CrossRef] [PubMed]
- Lê, A.; Mantel, M.; Marchix, J.; Bodinier, M.; Jan, G.; Rolli-Derkinderen, M. Inflammatory bowel disease therapeutic strategies by modulation of the microbiota: How and when to introduce pre-, pro-, syn-, or postbiotics? Am. J. Physiol. Gastrointest. Liver Physiol. 2022, 323, G523–G553. [Google Scholar] [CrossRef]
- Aguilar-Toalá, J.E.; Garcia-Varela, R.; Garcia, H.S.; Mata-Haro, V.; González-Córdova, A.F.; Vallejo-Cordoba, B.; Hernández-Mendoza, A. Postbiotics: An Evolving Term within the Functional Foods Field. Trends Food Sci. Technol. 2018, 75, 105–114. [Google Scholar] [CrossRef]
- Lebeer, S.; Vanderleyden, J.; De Keersmaecker, S.C. Host interactions of probiotic bacterial surface molecules: Comparison with commensals and pathogens. Nat. Rev. Microbiol. 2010, 8, 171–184. [Google Scholar] [CrossRef]
- Hynönen, U.; Palva, A. Lactobacillus surface layer proteins: Structure, function and applications. Appl. Microbiol. Biotechnol. 2013, 97, 5225–5243. [Google Scholar] [CrossRef]
- Klotz, C.; Goh, Y.J.; O’Flaherty, S.; Barrangou, R. S-layer associated proteins contribute to the adhesive and immunomodulatory properties of Lactobacillus acidophilus NCFM. BMC Microbiol. 2020, 20, 248. [Google Scholar] [CrossRef]
- Lathrop, S.K.; Bloom, S.M.; Rao, S.M.; Nutsch, K.; Lio, C.W.; Santacruz, N.; Peterson, D.A.; Stappenbeck, T.S.; Hsieh, C.S. Peripheral education of the immune system by colonic commensal microbiota. Nature 2011, 478, 250–254. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Zhang, L.; Xu, S.; Pan, J.; Zhang, Q.; Lu, R. Surface-Layer Protein from Lactobacillus acidophilus NCFM Inhibits Lipopolysaccharide-Induced Inflammation through MAPK and NF-κB Signaling Pathways in RAW264.7 Cells. J. Agric. Food Chem. 2018, 66, 7655–7662. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Xu, P.; Wu, Z.; Pan, D. Anti-Inflammatory Activity of Surface Layer Protein SlpA of Lactobacillus Acidophilus CICC 6074 in LPS-Induced RAW 264.7 Cells and DSS-Induced Mice Colitis. J. Funct. Foods 2018, 51, 16–27. [Google Scholar] [CrossRef]
- Ge, Y.; Gong, M.; Zadeh, M.; Li, J.; Abbott, J.R.; Li, W.; Morel, L.; Sonon, R.; Supekar, N.T.; Azadi, P.; et al. Regulating colonic dendritic cells by commensal glycosylated large surface layer protein A to sustain gut homeostasis against pathogenic inflammation. Mucosal. Immunol. 2020, 13, 34–46. [Google Scholar] [CrossRef]
- Aldars-García, L.; Chaparro, M.; Gisbert, J.P. Systematic Review: The Gut Microbiome and Its Potential Clinical Application in Inflammatory Bowel Disease. Microorganisms 2021, 9, 977. [Google Scholar] [CrossRef]
- Sudhakar, P.; Andrighetti, T.; Verstockt, S.; Caenepeel, C.; Ferrante, M.; Sabino, J.; Verstockt, B.; Vermeire, S. Integrated Analysis of Microbe-Host Interactions in Crohn’s Disease Reveals Potential Mechanisms of Microbial Proteins on Host Gene Expression. iScience 2022, 25, 103963. [Google Scholar] [CrossRef]
- Aleksandrova, K.; Romero-Mosquera, B.; Hernandez, V. Diet, Gut Microbiome and Epigenetics: Emerging Links with Inflammatory Bowel Diseases and Prospects for Management and Prevention. Nutrients 2017, 9, 962. [Google Scholar] [CrossRef]
- Khorsand, B.; Asadzadeh Aghdaei, H.; Nazemalhosseini-Mojarad, E.; Nadalian, B.; Nadalian, B.; Houri, H. Overrepresentation of Enterobacteriaceae and Escherichia Coli Is the Major Gut Microbiome Signature in Crohn’s Disease and Ulcerative Colitis; A Comprehensive Metagenomic Analysis of IBDMDB Datasets. Front. Cell. Infect. Microbiol. 2022, 12, 1015890. [Google Scholar] [CrossRef]

| Diseases | Gut Microbiota | References |
|---|---|---|
| Type 1 diabetes | Lactobacillaceae ↓ Rikenellaceae ↓ Porphoromadaceae ↓ | [15] |
| Ruminococcaceae ↓ Ruminococcus ↓ | [16] | |
| Multiple sclerosis | Lactobacillaceae ↓ Bacteroidaceae ↓ Prevotellaceae ↓ | [17] |
| Akkermansia ↑ | [18,19] | |
| Erysipelotrichaceae ↑ Lactobacillus reuteri ↑ | ||
| Rheumatoid arthritis | Collinsella ↑ Faecalibacterium ↑ Eggerthella ↑ | [20] |
| Celiac disease | Dialister invisus ↑ Parabacteroides ↑ Lachnospiraceae bacterium ↑ | [21] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haneishi, Y.; Furuya, Y.; Hasegawa, M.; Picarelli, A.; Rossi, M.; Miyamoto, J. Inflammatory Bowel Diseases and Gut Microbiota. Int. J. Mol. Sci. 2023, 24, 3817. https://doi.org/10.3390/ijms24043817
Haneishi Y, Furuya Y, Hasegawa M, Picarelli A, Rossi M, Miyamoto J. Inflammatory Bowel Diseases and Gut Microbiota. International Journal of Molecular Sciences. 2023; 24(4):3817. https://doi.org/10.3390/ijms24043817
Chicago/Turabian StyleHaneishi, Yuri, Yuma Furuya, Mayu Hasegawa, Antonio Picarelli, Mauro Rossi, and Junki Miyamoto. 2023. "Inflammatory Bowel Diseases and Gut Microbiota" International Journal of Molecular Sciences 24, no. 4: 3817. https://doi.org/10.3390/ijms24043817






