Altered Faecal Microbiota Composition and Structure of Ghanaian Children with Acute Gastroenteritis
Abstract
1. Introduction
2. Results
2.1. Demographic and Clinical Characteristics of Study Participants
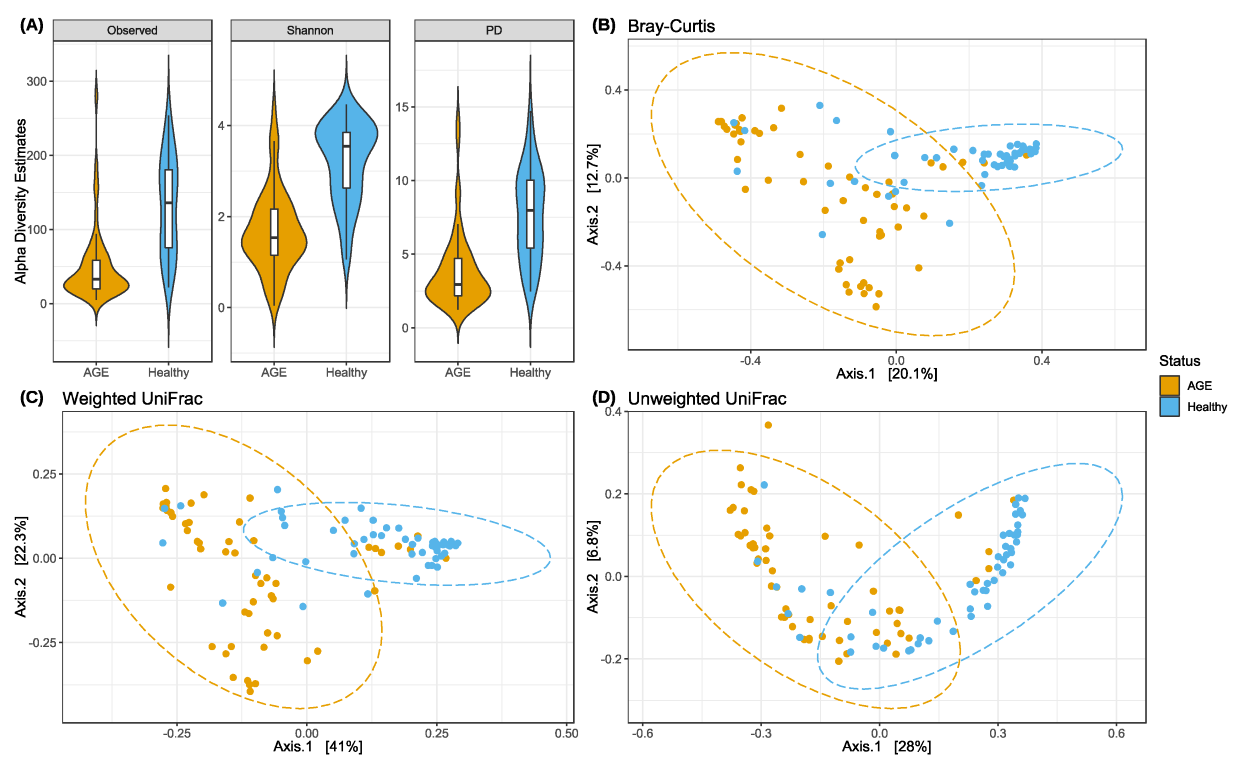
2.2. AGE Cases Have Lower Alpha Diversity and Distinct Beta Diversity Profiles
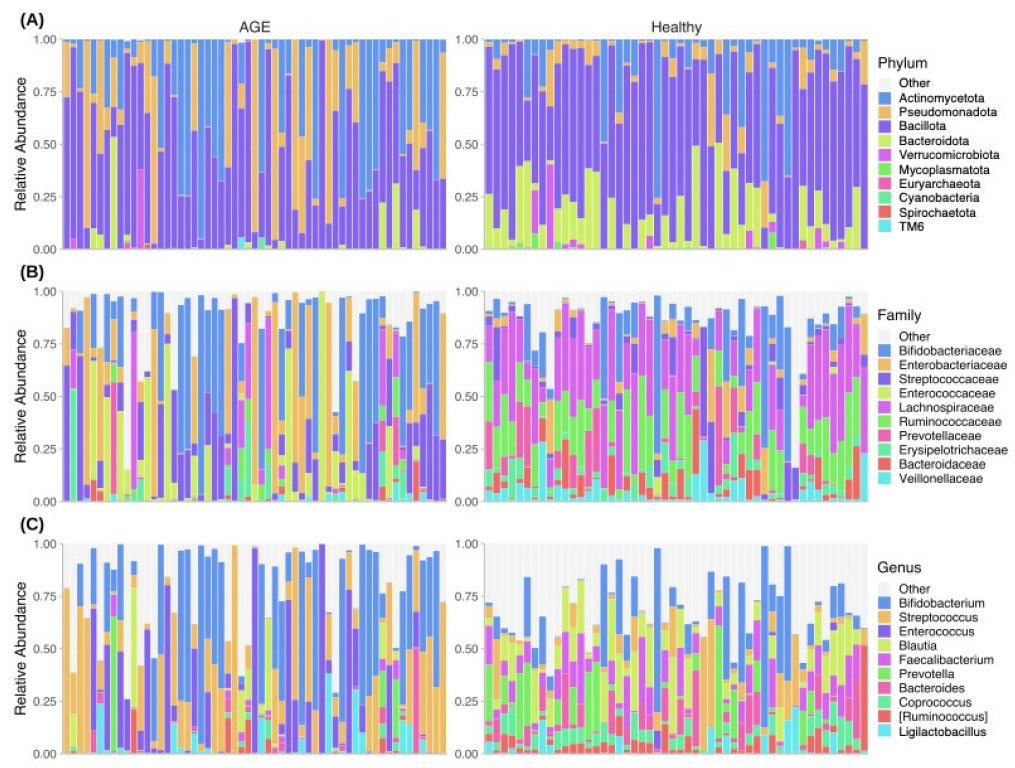
2.3. Taxonomic Profiles Differ between AGE Cases and Healthy Controls
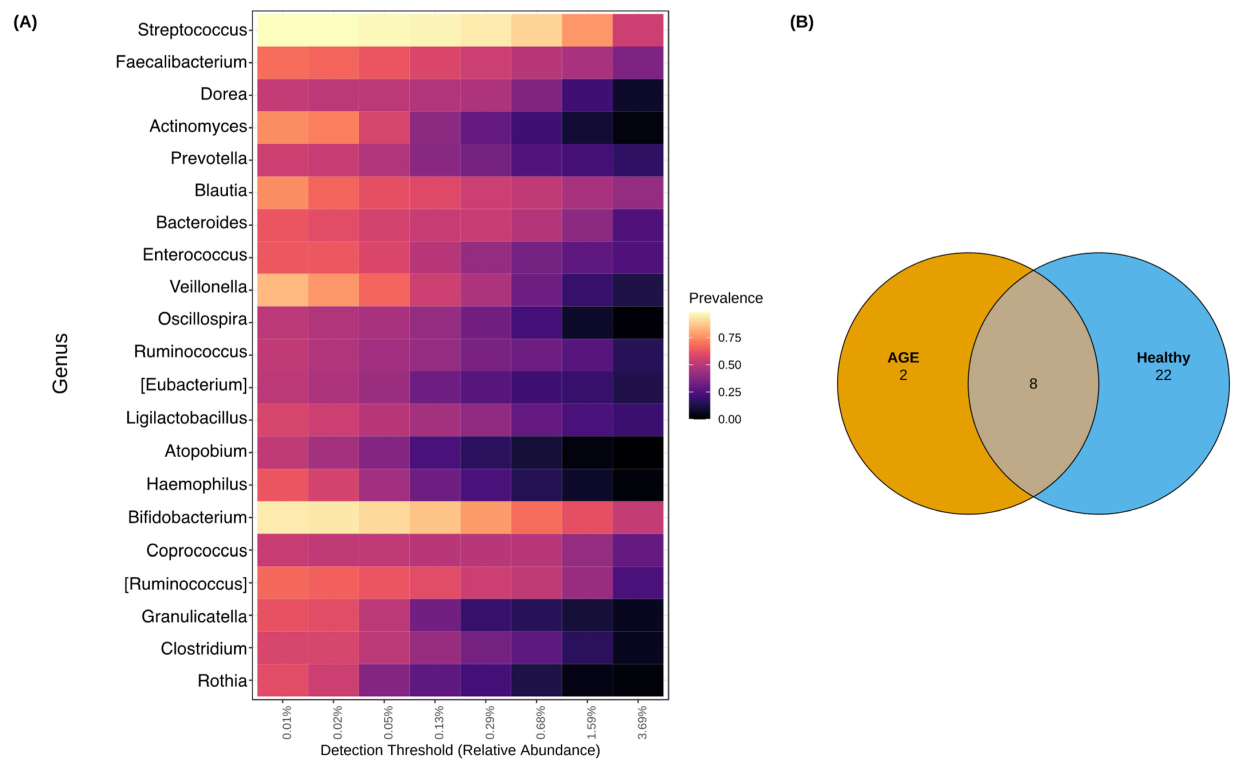
2.4. Shared and Unique Core Genera between AGE Cases and Healthy Controls
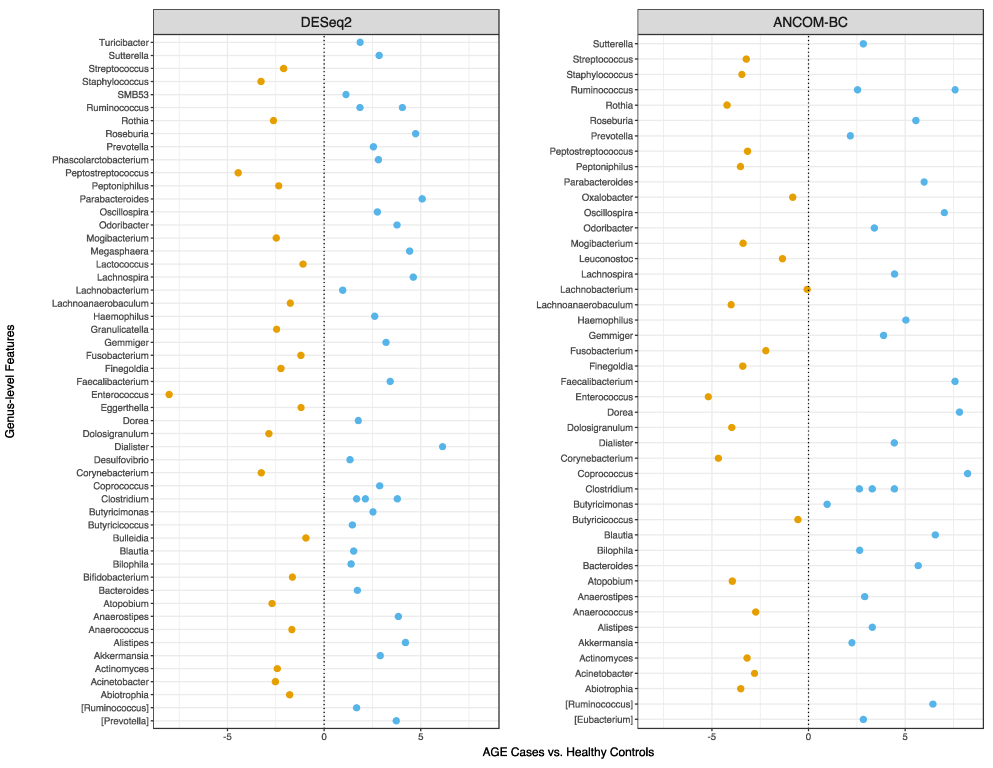
2.5. Faecal Microbial Sequence Profiles Based on Differential Abundance Testing
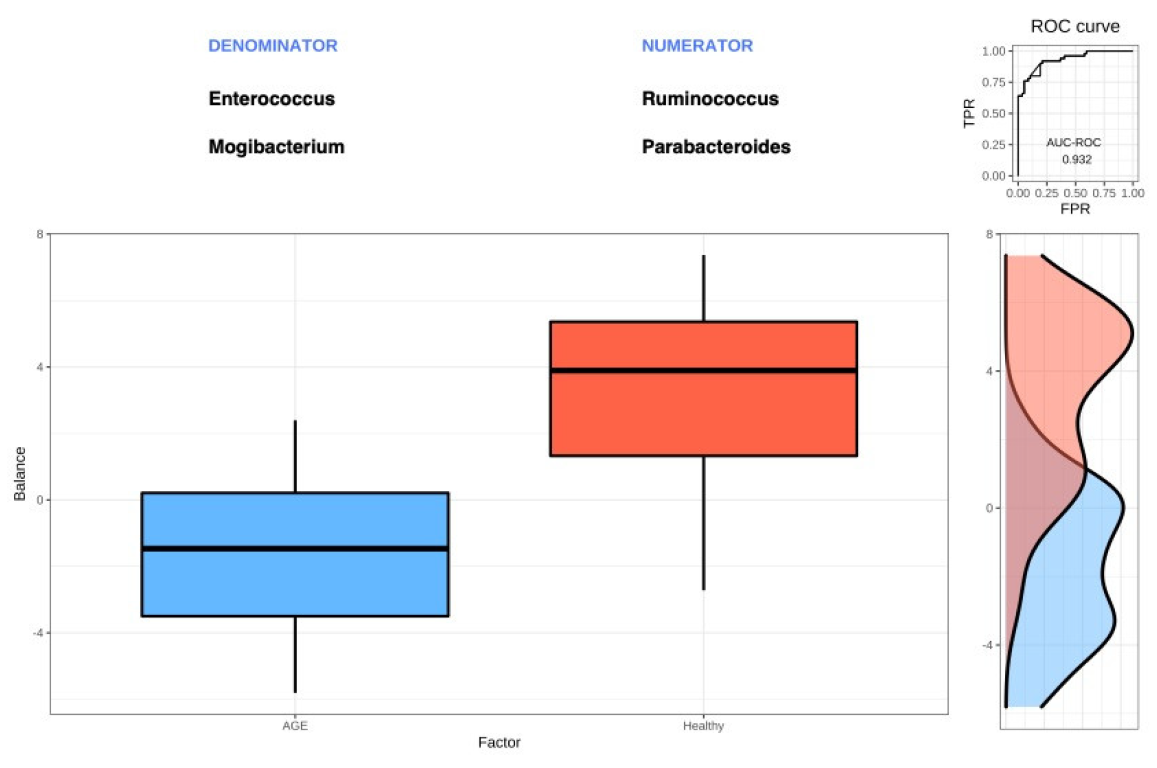
2.6. Selbal Identifies Balances Associated with AGE Cases and Healthy Controls
2.7. Network Analysis further Reveals Differences in Faecal Microbiota Structure
3. Discussion
4. Materials and Methods
4.1. Study Design and Participant Recruitment Criteria
4.2. Sample Size, Sample Collection, and Processing
4.3. DNA Extraction and 16S rRNA Gene Amplicon Sequencing
4.4. 16S rRNA Gene Sequence Processing
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vos, T.; Lim, S.S.; Abbafati, C.; Abbasifard, C.; Abbasi-Kangevari, M.; Abbastabar, H.; Abd-Allah, F.; Abdelalim, A.; Abdollahi, M.; Abdollahpour, I.; et al. Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet 2020, 396, 1204–1222. [Google Scholar] [CrossRef] [PubMed]
- WHO. Diarrhoeal Disease. 2017. Available online: https://www.who.int/news-room/fact-sheets/detail/diarrhoeal-disease (accessed on 7 June 2019).
- Oppong, T.B.; Yang, H.; Amponsem-Boateng, C.; Kyere, E.K.D.; Abdulai, T.; Duan, G.; Opolot, G. Enteric pathogens associated with gastroenteritis among children under 5 years in sub-Saharan Africa: A systematic review and meta-analysis. Epidemiology Infect. 2020, 148, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Vega, R.M.; Avva, U. Pediatric Dehydration. Available online: https://www.ncbi.nlm.nih.gov/books/NBK436022/ (accessed on 3 August 2022).
- Liu, J.; Platts-Mills, J.A.; Juma, J.; Kabir, F.; Nkeze, J.; Okoi, C.; Operario, D.J.; Uddin, J.; Ahmed, S.; Alonso, P.L.; et al. Use of quantitative molecular diagnostic methods to identify causes of diarrhoea in children: A reanalysis of the GEMS case-control study. Lancet 2016, 388, 1291–1301. [Google Scholar] [CrossRef] [PubMed]
- Belkaid, Y.; Hand, T.W. Role of the Microbiota in Immunity and Inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef] [PubMed]
- Ferretti, P.; Pasolli, E.; Tett, A.; Asnicar, F.; Gorfer, V.; Fedi, S.; Armanini, F.; Truong, D.T.; Manara, S.; Zolfo, M.; et al. Mother-to-Infant Microbial Transmission from Different Body Sites Shapes the Developing Infant Gut Microbiome. Cell Host Microbe 2018, 24, 133–145.e5. [Google Scholar] [CrossRef] [PubMed]
- Aagaard, K.; Stewart, C.J.; Chu, D. Una destinatio, viae diversae: Does exposure to the vaginal microbiota confer health benefits to the infant, and does lack of exposure confer disease risk? EMBO Rep. 2016, 17, 1679–1684. [Google Scholar] [CrossRef]
- Medlock, G.L.; Carey, M.A.; McDuffie, D.G.; Mundy, M.B.; Giallourou, N.; Swann, J.R.; Kolling, G.L.; Papin, J.A. Inferring Metabolic Mechanisms of Interaction within a Defined Gut Microbiota. Cell Syst. 2018, 7, 245–257.e7. [Google Scholar] [CrossRef]
- Whitney, J.C.; Peterson, S.B.; Kim, J.; Pazos, M.; Verster, A.J.; Radey, M.C.; Kulasekara, H.D.; Ching, M.Q.; Bullen, N.P.; Bryant, D.; et al. A broadly distributed toxin family mediates contact-dependent antagonism between gram-positive bacteria. Elife 2017, 6, e26938. [Google Scholar] [CrossRef]
- Round, J.L.; Palm, N.W. Causal effects of the microbiota on immune-mediated diseases. Sci. Immunol. 2018, 3, eaao1603. [Google Scholar] [CrossRef]
- Round, J.L.; Mazmanian, S.K. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009, 9, 313–323. [Google Scholar] [CrossRef]
- Vatanen, T.; Kostic, A.D.; D’Hennezel, E.; Siljander, H.; Franzosa, E.A.; Yassour, M.; Kolde, R.; Vlamakis, H.; Arthur, T.D.; Hämäläinen, A.-M.; et al. Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell 2016, 165, 842–853. [Google Scholar] [CrossRef]
- Monaghan, T.M.; Sloan, T.J.; Stockdale, S.R.; Blanchard, A.M.; Emes, R.D.; Wilcox, M.; Biswas, R.; Nashine, R.; Manke, S.; Gandhi, J.; et al. Metagenomics reveals impact of geography and acute diarrheal disease on the Central Indian human gut microbiome. Gut Microbes 2020, 12, 1752605. [Google Scholar] [CrossRef]
- David, L.A.; Weil, A.; Ryan, E.T.; Calderwood, S.B.; Harris, J.B.; Chowdhury, F.; Begum, Y.; Qadri, F.; LaRocque, R.C.; Turnbaugh, P.J. Gut Microbial Succession Follows Acute Secretory Diarrhea in Humans. Mbio 2015, 6, e00381-15. [Google Scholar] [CrossRef]
- Pop, M.; Walker, A.W.; Paulson, J.; Lindsay, B.; Antonio, M.; Hossain, M.A.; Oundo, J.; Tamboura, B.; Mai, V.; Astrovskaya, I.; et al. Diarrhea in young children from low-income countries leads to large-scale alterations in intestinal microbiota composition. Genome Biol. 2014, 15, R76. [Google Scholar] [CrossRef]
- The, H.C.; De Sessions, P.F.; Jie, S.; Thanh, D.P.; Thompson, C.N.; Minh, C.N.N.; Chung, H.; Tran, T.-A.; Thomson, N.R.; Thwaites, G.E.; et al. Assessing gut microbiota perturbations during the early phase of infectious diarrhea in Vietnamese children. Gut Microbes 2017, 9, 38–54. [Google Scholar] [CrossRef]
- Castaño-Rodríguez, N.; Underwood, A.; Merif, J.; Riordan, S.M.; Rawlinson, W.D.; Mitchell, H.M.; Kaakoush, N.O. Gut Microbiome Analysis Identifies Potential Etiological Factors in Acute Gastroenteritis. Infect. Immun. 2018, 86, e00060-18. [Google Scholar] [CrossRef]
- Iddrisu, I.; Monteagudo-Mera, A.; Poveda, C.; Pyle, S.; Shahzad, M.; Andrews, S.; Walton, G. Malnutrition and Gut Microbiota in Children. Nutrients 2021, 13, 2727. [Google Scholar] [CrossRef]
- Mizutani, T.; Aboagye, S.Y.; Ishizaka, A.; Afum, T.; Mensah, G.I.; Asante-Poku, A.; Asandem, D.A.; Parbie, P.K.; Abana, C.Z.-Y.; Kushitor, D.; et al. Gut microbiota signature of pathogen-dependent dysbiosis in viral gastroenteritis. Sci. Rep. 2021, 11, 13945. [Google Scholar] [CrossRef]
- Rivera-Pinto, J.; Egozcue, J.J.; Pawlowsky-Glahn, V.; Paredes, R.; Noguera-Julian, M.; Calle, M.L. Balances: A New Perspective for Microbiome Analysis. Msystems 2018, 3, e00053-18. [Google Scholar] [CrossRef]
- Martin, R.; Nauta, A.; Ben Amor, K.; Knippels, L.; Knol, J.; Garssen, J. Early life: Gut microbiota and immune development in infancy. Benef. Microbes 2010, 1, 367–382. [Google Scholar] [CrossRef]
- Rouhani, S.; Griffin, N.W.; Yori, P.P.; Gehrig, J.L.; Olortegui, M.P.; Salas, M.S.; Trigoso, D.R.; Moulton, L.H.; Houpt, E.R.; Barratt, M.J.; et al. Diarrhea as a Potential Cause and Consequence of Reduced Gut Microbial Diversity Among Undernourished Children in Peru. Clin. Infect. Dis. 2019, 71, 989–999. [Google Scholar] [CrossRef] [PubMed]
- Saffouri, G.B.; Shields-Cutler, R.R.; Chen, J.; Yang, Y.; Lekatz, H.R.; Hale, V.L.; Cho, J.M.; Battaglioli, E.J.; Bhattarai, Y.; Thompson, K.J.; et al. Small intestinal microbial dysbiosis underlies symptoms associated with functional gastrointestinal disorders. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Stewart, C.J.; Ajami, N.J.; O’Brien, J.L.; Hutchinson, D.S.; Smith, D.P.; Wong, M.C.; Ross, M.C.; Lloyd, R.E.; Doddapaneni, H.; Metcalf, G.A.; et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 2018, 562, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Adadey, S.M.; Quaye, O. The Burden of Gastroenteritis in the Post-Rotavirus Vaccine Era in Ghana: A Hospital Diagnoses-Based Study. Int. J. Med. Res. Health Sci. 2017, 6, 45–49. [Google Scholar]
- Enweronu-Laryea, C.C.; Sagoe, K.W.C.; Mwenda, J.M.; Armah, G.E. Severe Acute Rotavirus Gastroenteritis in Children Less Than 5 Years in Southern Ghana. Pediatr. Infect. Dis. J. 2014, 33, S9–S13. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Chung, J.; Battaglia, T.; Henderson, N.; Jay, M.; Li, H.; Lieber, A.D.; Wu, F.; Perez-Perez, G.I.; Chen, Y.; et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci. Transl. Med. 2016, 8, 343ra82. [Google Scholar] [CrossRef]
- Reyman, M.; van Houten, M.A.; Watson, R.L.; Chu, M.L.J.N.; Arp, K.; de Waal, W.J.; Schiering, I.; Plötz, F.B.; Willems, R.J.L.; van Schaik, W.; et al. Effects of early-life antibiotics on the developing infant gut microbiome and resistome: A randomized trial. Nat. Commun. 2022, 13, 1–12. [Google Scholar] [CrossRef]
- Knights, D.; Ward, T.L.; McKinlay, C.E.; Miller, H.; Gonzalez, A.; McDonald, D.; Knight, R. Rethinking “Enterotypes”. Cell Host Microbe 2014, 16, 433–437. [Google Scholar] [CrossRef]
- Lozupone, C.; Lladser, M.E.; Knights, D.; Stombaugh, J.; Knight, R. UniFrac: An effective distance metric for microbial community comparison. ISME J. 2011, 5, 169–172. [Google Scholar] [CrossRef]
- Duvallet, C.; Gibbons, S.M.; Gurry, T.; Irizarry, R.A.; Alm, E.J. Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat. Commun. 2017, 8, 1–10. [Google Scholar] [CrossRef]
- Lackey, K.A.; Williams, J.E.; Meehan, C.L.; Zachek, J.A.; Benda, E.D.; Price, W.J.; Foster, J.A.; Sellen, D.W.; Kamau-Mbuthia, E.W.; Kamundia, E.W.; et al. What's Normal? Microbiomes in Human Milk and Infant Feces Are Related to Each Other but Vary Geographically: The INSPIRE Study. Front. Nutr. 2019, 6, 45. [Google Scholar] [CrossRef]
- Ruiz, L.; Alba, C.; García-Carral, C.; Jiménez, E.A.; Lackey, K.A.; McGuire, M.K.; Meehan, C.L.; Foster, J.; Sellen, D.W.; Kamau-Mbuthia, E.W.; et al. Comparison of Two Approaches for the Metataxonomic Analysis of the Human Milk Microbiome. Front. Cell Infect. Microbiol. 2021, 11, 622550. [Google Scholar] [CrossRef]
- Neu, A.T.; Allen, E.E.; Roy, K. Defining and quantifying the core microbiome: Challenges and prospects. Proc. Natl. Acad. Sci. USA 2021, 118, e2104429118. [Google Scholar] [CrossRef]
- Abt, M.C. Acute Gastroenteritis Leaves a Lasting Impression. Cell Host Microbe 2016, 19, 3–5. [Google Scholar] [CrossRef]
- Vieira, S.M.; Hiltensperger, M.; Kumar, V.; Zegarra-Ruiz, D.; Dehner, C.; Khan, N.; Costa, F.R.C.; Tiniakou, E.; Greiling, T.; Ruff, W.; et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science 2018, 359, 1156–1161. [Google Scholar] [CrossRef]
- Liss, B.J.; Vehreschild, J.J.; Cornely, O.A.; Hallek, M.; Fätkenheuer, G.; Wisplinghoff, H.; Seifert, H.; Vehreschild, M.J.G.T. Intestinal colonisation and blood stream infections due to vancomycin-resistant enterococci (VRE) and extended-spectrum beta-lactamase-producing Enterobacteriaceae (ESBLE) in patients with haematological and oncological malignancies. Infection 2012, 40, 613–619. [Google Scholar] [CrossRef]
- Ayobami, O.; Willrich, N.; Reuss, A.; Eckmanns, T.; Markwart, R. The ongoing challenge of vancomycin-resistant Enterococcus faecium and Enterococcus faecalis in Europe: An epidemiological analysis of bloodstream infections. Emerg. Microbes Infect. 2020, 9, 1180–1193. [Google Scholar] [CrossRef]
- Becker-Dreps, S.; Rogawski, E.T.; Monteagudo, A.; Carroll, I.M.; Vilchez, S.; Zambrana, L.E.; Azcarate-Peril, M.A.; Hudgens, M.G.; Allali, I.; Espinoza, F. Gut Microbiome Composition in Young Nicaraguan Children During Diarrhea Episodes and Recovery. Am. J. Trop. Med. Hyg. 2015, 93, 1187–1193. [Google Scholar] [CrossRef]
- Schmidt, T.S.; Hayward, M.R.; Coelho, L.P.; Li, S.S.; Costea, P.I.; Voigt, A.Y.; Wirbel, J.; Maistrenko, O.M.; Alves, R.J.; Bergsten, E.; et al. Extensive transmission of microbes along the gastrointestinal tract. eLife 2019, 8, e42693. [Google Scholar] [CrossRef]
- Rosier, B.; Takahashi, N.; Zaura, E.; Krom, B.; Martínez-Espinosa, R.; van Breda, S.; Marsh, P.; Mira, A. The Importance of Nitrate Reduction for Oral Health. J. Dent. Res. 2022, 101, 887–897. [Google Scholar] [CrossRef] [PubMed]
- Vonaesch, P.; Anderson, M.; Sansonetti, P.J. Pathogens, microbiome and the host: Emergence of the ecological Koch's postulates. FEMS Microbiol. Rev. 2018, 42, 273–292. [Google Scholar] [CrossRef] [PubMed]
- Rojas-Tapias, D.F.; Brown, E.M.; Temple, E.R.; Onyekaba, M.A.; Mohamed, A.M.T.; Duncan, K.; Schirmer, M.; Walker, R.L.; Mayassi, T.; Pierce, K.A.; et al. Inflammation-associated nitrate facilitates ectopic colonization of oral bacterium Veillonella parvula in the intestine. Nat. Microbiol. 2022, 7, 1673–1685. [Google Scholar] [CrossRef] [PubMed]
- Martel, J.; Chang, S.-H.; Ko, Y.-F.; Hwang, T.-L.; Young, J.D.; Ojcius, D.M. Gut barrier disruption and chronic disease. Trends Endocrinol. Metab. 2022, 33, 247–265. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Nguyen, L.H.; Song, M.; Wang, D.D.; Franzosa, E.A.; Cao, Y.; Joshi, A.; Drew, D.A.; Mehta, R.; Ivey, K.L.; et al. Dietary fiber intake, the gut microbiome, and chronic systemic inflammation in a cohort of adult men. Genome Med. 2021, 13, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Schulthess, J.; Pandey, S.; Capitani, M.; Rue-Albrecht, K.C.; Arnold, I.; Franchini, F.; Chomka, A.; Ilott, N.E.; Johnston, D.G.W.; Pires, E.; et al. The Short Chain Fatty Acid Butyrate Imprints an Antimicrobial Program in Macrophages. Immunity 2019, 50, 432–445.e437. [Google Scholar] [CrossRef]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef]
- Vonaesch, P.; Morien, E.; Andrianonimiadana, L.; Sanke, H.; Mbecko, J.-R.; Huus, K.E.; Naharimanananirina, T.; Gondje, B.P.; Nigatoloum, S.N.; Vondo, S.S.; et al. Stunted childhood growth is associated with decompartmentalization of the gastrointestinal tract and overgrowth of oropharyngeal taxa. Proc. Natl. Acad. Sci. USA 2018, 115, E8489–E8498. [Google Scholar] [CrossRef]
- Tamashiro, R.; Strange, L.; Schnackenberg, K.; Santos, J.; Gadalla, H.; Zhao, L.; Li, E.C.; Hill, E.; Hill, B.; Sidhu, G.; et al. Stability of healthy subgingival microbiome across space and time. Sci. Rep. 2021, 11, 23987. [Google Scholar] [CrossRef]
- McCann, K.S. The diversity–stability debate. Nature 2000, 405, 228–233. [Google Scholar] [CrossRef]
- Parente, E.; Zotta, T.; Ricciardi, A. Microbial association networks in cheese: A meta-analysis. bioRxiv 2021, arXiv:2021.2007.2021.453196. [Google Scholar] [CrossRef]
- Chen, S.-Y.; Tsai, C.-N.; Lee, Y.-S.; Lin, C.-Y.; Huang, K.-Y.; Chao, H.-C.; Lai, M.-W.; Chiu, C.-H. Intestinal microbiome in children with severe and complicated acute viral gastroenteritis. Sci. Rep. 2017, 7, srep46130. [Google Scholar] [CrossRef]
- Mathew, S.; Smatti, M.K.; Al Ansari, K.; Nasrallah, G.K.; Al Thani, A.A.; Yassine, H.M. Mixed Viral-Bacterial Infections and Their Effects on Gut Microbiota and Clinical Illnesses in Children. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Sohail, M.U.; Al Khatib, H.A.; Al Thani, A.A.; Al Ansari, K.; Yassine, H.M.; Al-Asmakh, M. Microbiome profiling of rotavirus infected children suffering from acute gastroenteritis. Gut Pathog. 2021, 13, 1–9. [Google Scholar] [CrossRef]
- Kaplan, I.; Bokulich, N.A.; Caporaso, J.G.; Enders, L.S.; Ghanem, W.; Ingerslew, K.S. Phylogenetic farming: Can evolutionary history predict crop rotation via the soil microbiome? Evol. Appl. 2020, 13, 1984–1999. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 1), 4516–4522. [Google Scholar] [CrossRef]
- Cock, P.J.A.; Fields, C.J.; Goto, N.; Heuer, M.L.; Rice, P.M. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2009, 38, 1767–1771. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Janssen, S.; McDonald, D.; Gonzalez, A.; Navas-Molina, J.A.; Jiang, L.; Xu, Z.Z.; Winker, K.; Kado, D.M.; Orwoll, E.; Manary, M.; et al. Phylogenetic Placement of Exact Amplicon Sequences Improves Associations with Clinical Information. mSystems 2018, 3, e00021-18. [Google Scholar] [CrossRef] [PubMed]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Estaki, M.; Jiang, L.; Bokulich, N.A.; McDonald, D.; González, A.; Kosciolek, T.; Martino, C.; Zhu, Q.; Birmingham, A.; Vázquez-Baeza, Y.; et al. QIIME 2 Enables Comprehensive End-to-End Analysis of Diverse Microbiome Data and Comparative Studies with Publicly Available Data. Curr. Protoc. Bioinform. 2020, 70, e100. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Werner, J.J.; Koren, O.; Hugenholtz, P.; DeSantis, T.Z.; Walters, W.A.; Caporaso, J.G.; Angenent, L.; Knight, R.; Ley, R. Impact of training sets on classification of high-throughput bacterial 16s rRNA gene surveys. ISME J. 2011, 6, 94–103. [Google Scholar] [CrossRef]
- Oren, A.; Garrity, G.M. Valid publication of the names of forty-two phyla of prokaryotes. Int. J. Syst. Evol. Microbiol. 2021, 71, 5056. [Google Scholar] [CrossRef]
- Lappan, R.; Imbrogno, K.; Sikazwe, C.; Anderson, D.; Mok, D.; Coates, H.; Vijayasekaran, S.; Bumbak, P.; Blyth, C.C.; Jamieson, S.E.; et al. A microbiome case-control study of recurrent acute otitis media identified potentially protective bacterial genera. BMC Microbiol. 2018, 18, 1–20. [Google Scholar] [CrossRef]
- Walker, R.E.; Walker, C.G.; Camargo, C.A., Jr.; Bartley, J.; Flint, D.; Thompson, J.M.D.; Mitchell, E.A. Nasal microbial composition and chronic otitis media with effusion: A case-control study. PLoS ONE 2019, 14, e0212473. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Davis, N.M.; Proctor, D.M.; Holmes, S.P.; Relman, D.A.; Callahan, B.J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 2018, 6, 226. [Google Scholar] [CrossRef]
- Weiss, S.; Xu, Z.Z.; Peddada, S.; Amir, A.; Bittinger, K.; Gonzalez, A.; Lozupone, C.; Zaneveld, J.R.; Vázquez-Baeza, Y.; Birmingham, A.; et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 2017, 5, 27. [Google Scholar] [CrossRef]
- Bray, J.R.; Curtis, J.T. An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecol. Monogr. 1957, 27, 325–349. [Google Scholar] [CrossRef]
- Lozupone, C.; Knight, R. UniFrac: A New Phylogenetic Method for Comparing Microbial Communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and Qualitative β Diversity Measures Lead to Different Insights into Factors That Structure Microbial Communities. Appl. Environ. Microbiol. 2007, 73, 1576–1585. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.; O’hara, R.; Simpson, G.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Community ecology package. R Package Version 2013, 2, 321–326. [Google Scholar]
- Anderson, M.J. Permutational Multivariate Analysis of Variance (PERMANOVA); Balakrishnan, N., Colton, T., Everitt, B., Piegorsch, W., Ruggeri, F., Teugels, J.L., Eds.; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2017; pp. 1–15. [Google Scholar]
- Lahti, L.; Shetty, S. Introduction to the Microbiome R Package. 2018. Available online: http://microbiome.github.io/microbiome (accessed on 25 July 2021).
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Lin, H.; Das Peddada, S. Analysis of compositions of microbiomes with bias correction. Nat. Commun. 2020, 11, 3514. [Google Scholar] [CrossRef]
- Nearing, J.T.; Douglas, G.M.; Hayes, M.G.; MacDonald, J.; Desai, D.K.; Allward, N.; Jones, C.M.A.; Wright, R.J.; Dhanani, A.S.; Comeau, A.M.; et al. Microbiome differential abundance methods produce different results across 38 datasets. Nat. Commun. 2022, 13, 1–16. [Google Scholar] [CrossRef]
- Shaffer, M.; Thurimella, K.; Sterrett, J.D.; Lozupone, C.A. SCNIC: Sparse correlation network investigation for compositional data. Mol. Ecol. Resour. 2022, 23, 312–325. [Google Scholar] [CrossRef]
- Friedman, J.; Alm, E.J. Inferring Correlation Networks from Genomic Survey Data. PLOS Comput. Biol. 2012, 8, e1002687. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Team, R.C. R: A Language and Environment for Statistical Computing. 2013. Available online: http://www.r-project.org/ (accessed on 15 September 2021).
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.A.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Wickham, H. Package ‘ggplot2’: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Garnier, S.; Ross, N.; Rudis, R.; Camargo, A.P.; Sciaini, M.; Scherer, C. Rvision-Colorblind-Friendly Color Maps for R. R Package Version 0.6. 2021. Available online: https://rdrr.io/cran/viridis/ (accessed on 15 September 2021).
- Neuwirth, E.; Brewer, R.C. ColorBrewer Palettes. R Package Version 2014, 1, 4. [Google Scholar]
- Teunisse, G. Fantaxtic: Fantaxtic Plots for Phyloseq Data. 2021. Available online: https://github.com/gmteunisse/Fantaxtic (accessed on 2 June 2022).
- Shetty, S.A.; Lahti, L.; de Vos, W.M.; Smidt, H. Microbiomeutilities: An R Package for Utilities to Guide in-depth Marker Gene Amplicon Data Analysis. Ecophysiological Insights into the Human Intestinal Microbiota: From Single Strains to Defined Consortia. 2018, Volume 95. Available online: https://zenodo.org/record/1471685#.Y-ZgzHbMJPY (accessed on 25 July 2021).
- Larsson, J. Eulerr: Area-proportional Euler Diagrams with Ellipses. 2018. Available online: https://jolars.github.io/eulerr/ (accessed on 25 July 2021).
- Kassambara, A.; Kassambara, M.A. Package ‘ggpubr’. R Package Version 0.1. 2020, Volume 6. Available online: https://r-pkgs.org/lifecycle.html (accessed on 25 July 2021).
- Qiu, Y. showtext: Using System Fonts in R Graphics. R J. 2015, 7, 99–108. [Google Scholar] [CrossRef]
- Bache, S.M.; Wickham, H. Magrittr: A Forward-Pipe Operator for R. R Package Version 2.0.3. 2014. Available online: https://rdrr.io/cran/magrittr/ (accessed on 25 July 2021).
- Robinson, D.; Hayes, A.; Couch, S. Broom: Convert statistical objects into tidy tibbles. R Package Version 2022, 5. [Google Scholar]
- Harrell, F. CRAN-Package Hmisc. Hmisc: Harrell Miscellaneous. 2019. Available online: https://cran.r-project.org/web/packages/Hmisc/index.html (accessed on 25 July 2021).
- Xie, Y. knitr: A General-Purpose Package for Dynamic Report Generation in R. R Package Version 1.28. 2020. Available online: https://cran.r-project.org/web/packages/knitr/index.html (accessed on 25 July 2021).
- Wickham, H.; Seidel, D. scales: Scale functions for visualization. R Package Version 2020, 1, 678. [Google Scholar]

| Number, n | AGE Cases | Healthy Controls | p-Value |
|---|---|---|---|
| 57 | 50 | ||
| Age (Months) | <0.0001 | ||
| Mean ± SD | 15.6 ± 14.2 | 29.6 ± 16.0 | |
| (Range) | (1.5–60.0) | (5.0–59.0) | |
| Sex (%) | 0.9884 | ||
| Female | 25 (43.9) | 22 (44.0) | |
| Male | 32 (56.1) | 28 (56.0) | |
| Vomiting (%) | <0.0001 | ||
| Yes | 30 (52.6) | 0 (0) | |
| No | 23 (40.4) | 50 (100) | |
| Missing * | 4 (7.0) | - | |
| Fever (%) | <0.0001 | ||
| Yes | 44 (77.2) | 0 (0) | |
| No | 13 (22.8) | 50 (100) | |
| Mode of Feeding, Yes (%) | |||
| Breastfeeding | 19 (33.3) | 16 (32.0) | 0.8834 |
| Artificial Milk | 12 (21.1) | 2 (4.0) | 0.0091 |
| Formula | 0 (0) | 2 (4.0) | 0.1274 |
| Family Meal | 32 (56.1) | 43 (86.0) | 0.0008 |
| Rotavirus Vaccination (%) | 0.3244 | ||
| Yes | 51 (89.5) | 50 (100) | |
| No | 1 (1.7) | 0 (0) | |
| Missing * | 5 (8.8) | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quaye, E.K.; Adjei, R.L.; Isawumi, A.; Allen, D.J.; Caporaso, J.G.; Quaye, O. Altered Faecal Microbiota Composition and Structure of Ghanaian Children with Acute Gastroenteritis. Int. J. Mol. Sci. 2023, 24, 3607. https://doi.org/10.3390/ijms24043607
Quaye EK, Adjei RL, Isawumi A, Allen DJ, Caporaso JG, Quaye O. Altered Faecal Microbiota Composition and Structure of Ghanaian Children with Acute Gastroenteritis. International Journal of Molecular Sciences. 2023; 24(4):3607. https://doi.org/10.3390/ijms24043607
Chicago/Turabian StyleQuaye, Emmanuel Kofi, Raymond Lovelace Adjei, Abiola Isawumi, David J. Allen, J. Gregory Caporaso, and Osbourne Quaye. 2023. "Altered Faecal Microbiota Composition and Structure of Ghanaian Children with Acute Gastroenteritis" International Journal of Molecular Sciences 24, no. 4: 3607. https://doi.org/10.3390/ijms24043607
APA StyleQuaye, E. K., Adjei, R. L., Isawumi, A., Allen, D. J., Caporaso, J. G., & Quaye, O. (2023). Altered Faecal Microbiota Composition and Structure of Ghanaian Children with Acute Gastroenteritis. International Journal of Molecular Sciences, 24(4), 3607. https://doi.org/10.3390/ijms24043607







