Nuclear Organization in Stress and Aging
Abstract
1. Introduction
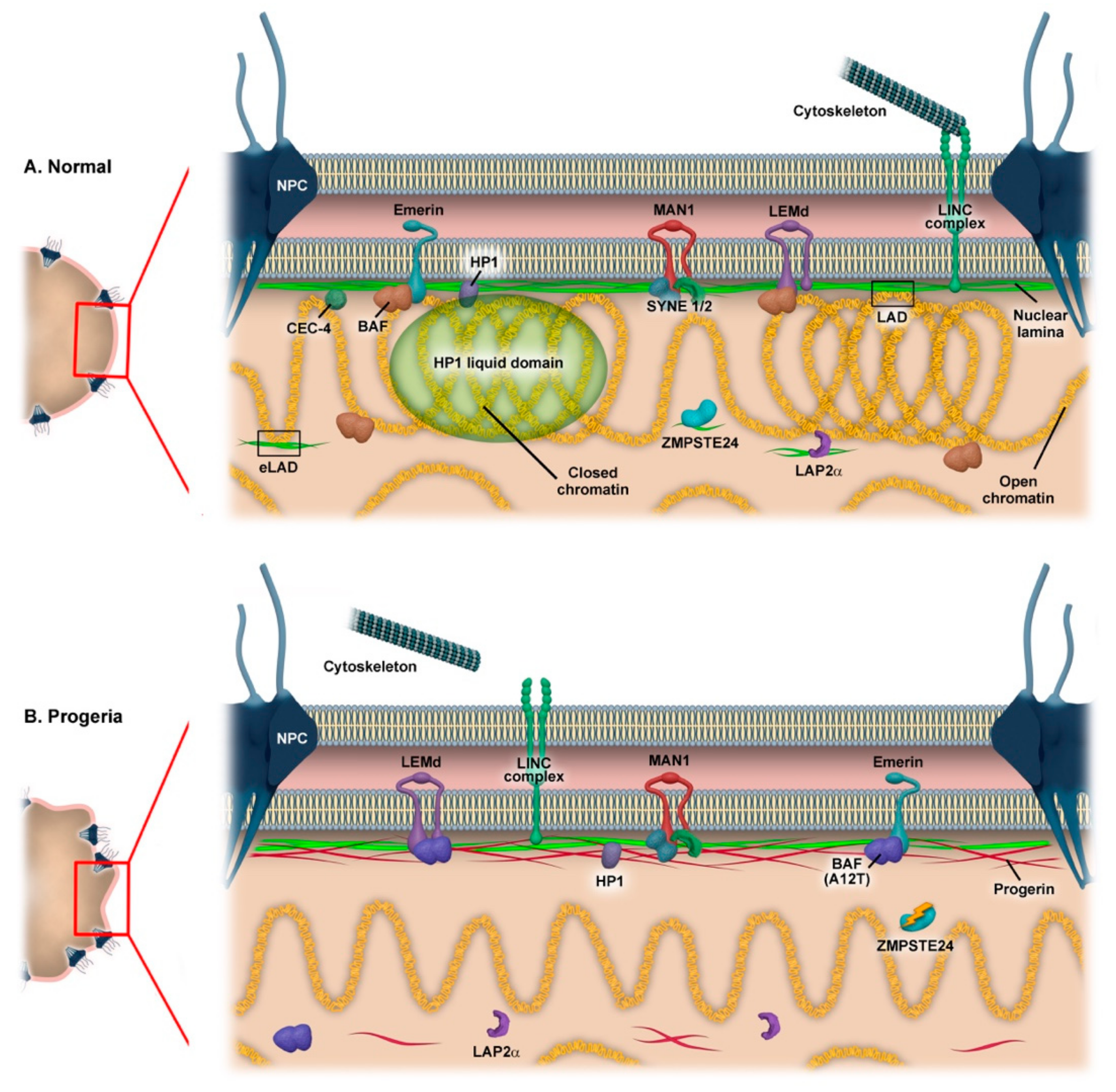
2. The Nuclear Envelope as Chromatin Organizer
3. Mutations in Nuclear Envelope Genes Causing Progeria
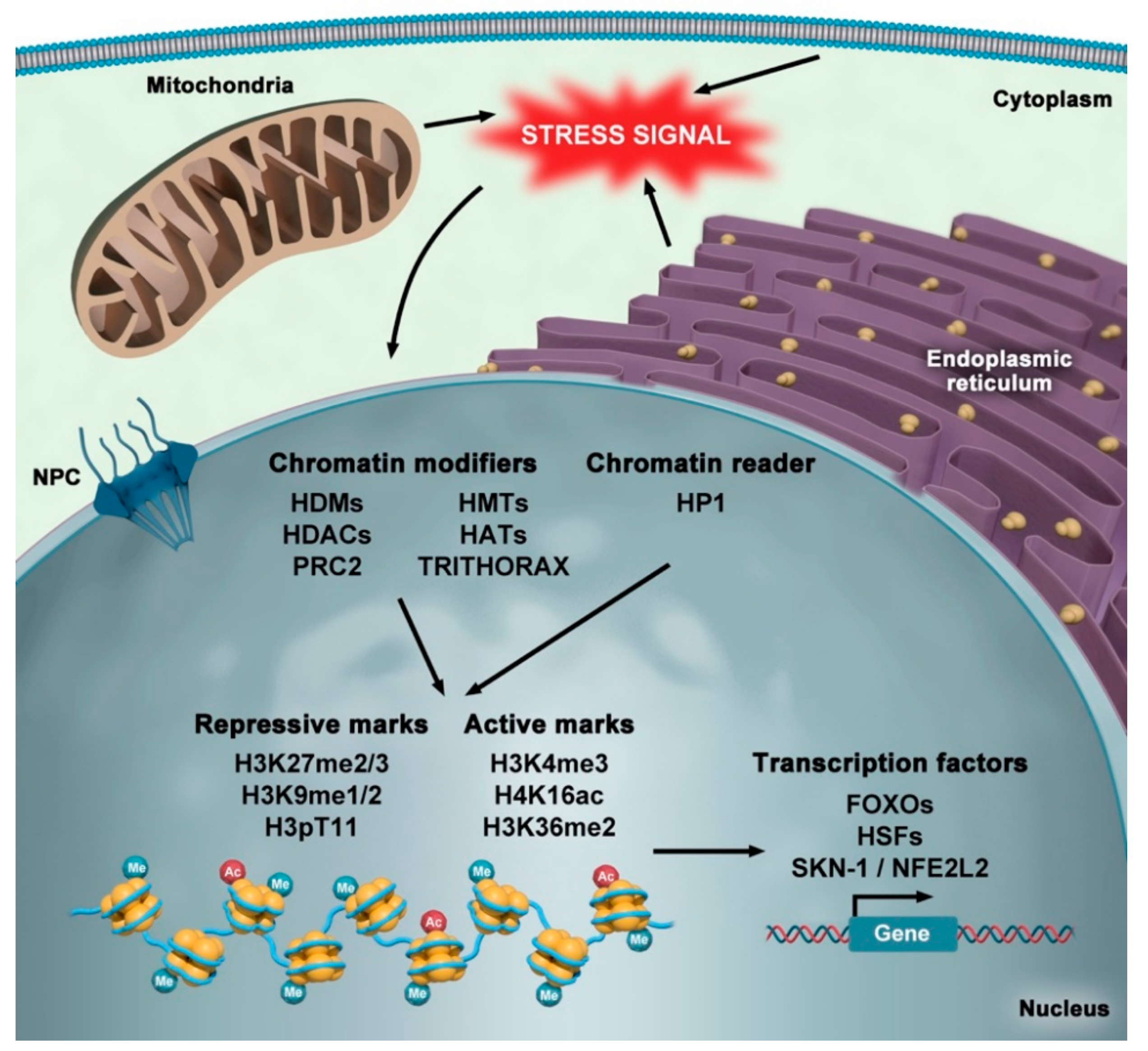
4. Alterations in Nuclear Organization during Aging
5. Stress and Epigenetics
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- De Las Heras, J.I.; Meinke, P.; Batrakou, D.G.; Srsen, V.; Zuleger, N.; Kerr, A.R.; Schirmer, E.C. Tissue specificity in the nuclear envelope supports its functional complexity. Nucleus 2013, 4, 460–477. [Google Scholar] [CrossRef] [PubMed]
- Buchwalter, A.; Kaneshiro, J.M.; Hetzer, M.W. Coaching from the sidelines: the nuclear periphery in genome regulation. Nat. Rev. Genet. 2019, 20, 39–50. [Google Scholar] [CrossRef]
- Burke, B.; Stewart, C.L. The nuclear lamins: flexibility in function. Nat. Rev. Mol. Cell Biol. 2013, 14, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Cavazos, J.S.; Hetzer, M.W. Outfits for different occasions: tissue-specific roles of Nuclear Envelope proteins. Curr. Opin. Cell Biol. 2012, 24, 775–783. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gruenbaum, Y.; Foisner, R. Lamins: nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annu. Rev. Biochem. 2015, 84, 131–164. [Google Scholar] [CrossRef] [PubMed]
- Cohen-Fix, O.; Askjaer, P. Cell Biology of the Caenorhabditis elegans Nucleus. Genetics 2017, 205, 25–59. [Google Scholar] [CrossRef] [PubMed]
- Starr, D.A. A nuclear-envelope bridge positions nuclei and moves chromosomes. J. Cell Sci. 2009, 122, 577–586. [Google Scholar] [CrossRef] [PubMed]
- Ahringer, J.; Gasser, S.M. Repressive Chromatin in Caenorhabditis elegans: Establishment, Composition, and Function. Genetics 2018, 208, 491–511. [Google Scholar] [CrossRef]
- Chandra, T.; Kirschner, K. Chromosome organisation during ageing and senescence. Curr. Opin. Cell Biol. 2016, 40, 161–167. [Google Scholar] [CrossRef]
- Gruenbaum, Y.; Medalia, O. Lamins: the structure and protein complexes. Curr. Opin. Cell Biol. 2015, 32, 7–12. [Google Scholar] [CrossRef]
- Dechat, T.; Adam, S.A.; Goldman, R.D. Nuclear lamins and chromatin: When structure meets function. Adv. Enzyme Regul. 2009, 49, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Miroshnikova, Y.A.; Nava, M.M.; Wickstrom, S.A. Emerging roles of mechanical forces in chromatin regulation. J. Cell Sci. 2017, 130, 2243–2250. [Google Scholar] [CrossRef]
- Uhler, C.; Shivashankar, G.V. Regulation of genome organization and gene expression by nuclear mechanotransduction. Nat. Rev. Mol. Cell Biol. 2017, 18, 717–727. [Google Scholar] [CrossRef]
- Towbin, B.D.; Gonzalez-Aguilera, C.; Sack, R.; Gaidatzis, D.; Kalck, V.; Meister, P.; Askjaer, P.; Gasser, S.M. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell 2012, 150, 934–947. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Sandoval, A.; Towbin, B.D.; Kalck, V.; Cabianca, D.S.; Gaidatzis, D.; Hauer, M.H.; Geng, L.; Wang, L.; Yang, T.; Wang, X.; et al. Perinuclear Anchoring of H3K9-Methylated Chromatin Stabilizes Induced Cell Fate in C. elegans Embryos. Cell 2015, 163, 1333–1347. [Google Scholar] [CrossRef] [PubMed]
- Cabianca, D.S.; Munoz-Jimenez, C.; Kalck, V.; Gaidatzis, D.; Padeken, J.; Seeber, A.; Askjaer, P.; Gasser, S.M. Active chromatin marks drive spatial sequestration of heterochromatin in C. elegans nuclei. Nature 2019, 569, 734–739. [Google Scholar] [CrossRef]
- Lee, D.C.; Welton, K.L.; Smith, E.D.; Kennedy, B.K. A-type nuclear lamins act as transcriptional repressors when targeted to promoters. Exp. Cell Res. 2009, 315, 996–1007. [Google Scholar] [CrossRef]
- Lund, E.; Oldenburg, A.R.; Delbarre, E.; Freberg, C.T.; Duband-Goulet, I.; Eskeland, R.; Buendia, B.; Collas, P. Lamin A/C-promoter interactions specify chromatin state-dependent transcription outcomes. Genome Res. 2013, 23, 1580–1589. [Google Scholar] [CrossRef]
- Meister, P.; Towbin, B.D.; Pike, B.L.; Ponti, A.; Gasser, S.M. The spatial dynamics of tissue-specific promoters during C. elegans development. Genes Dev. 2010, 24, 766–782. [Google Scholar] [CrossRef]
- Milon, B.C.; Cheng, H.; Tselebrovsky, M.V.; Lavrov, S.A.; Nenasheva, V.V.; Mikhaleva, E.A.; Shevelyov, Y.Y.; Nurminsky, D.I. Role of histone deacetylases in gene regulation at nuclear lamina. PLoS ONE 2012, 7, e49692. [Google Scholar] [CrossRef]
- Kosak, S.T.; Skok, J.A.; Medina, K.L.; Riblet, R.; Le Beau, M.M.; Fisher, A.G.; Singh, H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science 2002, 296, 158–162. [Google Scholar] [CrossRef] [PubMed]
- Zeller, P.; Padeken, J.; van Schendel, R.; Kalck, V.; Tijsterman, M.; Gasser, S.M. Histone H3K9 methylation is dispensable for Caenorhabditis elegans development but suppresses RNA:DNA hybrid-associated repeat instability. Nat. Genet. 2016, 48, 1385–1395. [Google Scholar] [CrossRef] [PubMed]
- Peric-Hupkes, D.; Meuleman, W.; Pagie, L.; Bruggeman, S.W.; Solovei, I.; Brugman, W.; Graf, S.; Flicek, P.; Kerkhoven, R.M.; van Lohuizen, M.; et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol. Cell 2010, 38, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Van Steensel, B.; Belmont, A.S. Lamina-Associated Domains: Links with Chromosome Architecture, Heterochromatin, and Gene Repression. Cell 2017, 169, 780–791. [Google Scholar] [CrossRef] [PubMed]
- Pascual-Reguant, L.; Blanco, E.; Galan, S.; Le Dily, F.; Cuartero, Y.; Serra-Bardenys, G.; Di Carlo, V.; Iturbide, A.; Cebria-Costa, J.P.; Nonell, L.; et al. Lamin B1 mapping reveals the existence of dynamic and functional euchromatin lamin B1 domains. Nat. Commun. 2018, 9, 3420. [Google Scholar] [CrossRef] [PubMed]
- Gesson, K.; Rescheneder, P.; Skoruppa, M.P.; von Haeseler, A.; Dechat, T.; Foisner, R. A-type lamins bind both hetero- and euchromatin, the latter being regulated by lamina-associated polypeptide 2 alpha. Genome Res. 2016, 26, 462–473. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Sandoval, A.; Gasser, S.M. Mechanism of chromatin segregation to the nuclear periphery in C. elegans embryos. Worm 2016, 5, e1190900. [Google Scholar] [CrossRef]
- Zheng, X.; Hu, J.; Yue, S.; Kristiani, L.; Kim, M.; Sauria, M.; Taylor, J.; Kim, Y.; Zheng, Y. Lamins Organize the Global Three-Dimensional Genome from the Nuclear Periphery. Mol. Cell 2018, 71, 802–815.e7. [Google Scholar] [CrossRef]
- Paulsen, J.; Liyakat Ali, T.M.; Nekrasov, M.; Delbarre, E.; Baudement, M.O.; Kurscheid, S.; Tremethick, D.; Collas, P. Long-range interactions between topologically associating domains shape the four-dimensional genome during differentiation. Nat. Genet. 2019, 51, 835–843. [Google Scholar] [CrossRef]
- Leemans, C.; van der Zwalm, M.C.H.; Brueckner, L.; Comoglio, F.; van Schaik, T.; Pagie, L.; van Arensbergen, J.; van Steensel, B. Promoter-Intrinsic and Local Chromatin Features Determine Gene Repression in LADs. Cell 2019, 177, 852–864. [Google Scholar] [CrossRef]
- Davidson, P.M.; Lammerding, J. Broken nuclei—lamins, nuclear mechanics, and disease. Trends Cell Biol. 2014, 24, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Gonzalo, S.; Kreienkamp, R.; Askjaer, P. Hutchinson-Gilford Progeria Syndrome: A premature aging disease caused by LMNA gene mutations. Ageing Res. Rev. 2017, 33, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Worman, H.J. Nuclear lamins and laminopathies. J. Pathol. 2012, 226, 316–325. [Google Scholar] [CrossRef] [PubMed]
- Paulsen, J.; Sekelja, M.; Oldenburg, A.R.; Barateau, A.; Briand, N.; Delbarre, E.; Shah, A.; Sorensen, A.L.; Vigouroux, C.; Buendia, B.; et al. Chrom3D: Three-dimensional genome modeling from Hi-C and nuclear lamin-genome contacts. Genome Biol. 2017, 18, 21. [Google Scholar] [CrossRef] [PubMed]
- Bank, E.M.; Gruenbaum, Y. Caenorhabditis elegans as a model system for studying the nuclear lamina and laminopathic diseases. Nucleus 2011, 2, 350–357. [Google Scholar] [CrossRef] [PubMed]
- Dialynas, G.; Speese, S.; Budnik, V.; Geyer, P.K.; Wallrath, L.L. The role of Drosophila Lamin C in muscle function and gene expression. Development 2010, 137, 3067–3077. [Google Scholar] [CrossRef] [PubMed]
- Dobrzynska, A.; Askjaer, P.; Gruenbaum, Y. Lamin-Binding Proteins in Caenorhabditis elegans. Methods Enzymol. 2016, 569, 455–483. [Google Scholar] [CrossRef] [PubMed]
- Mattout, A.; Pike, B.L.; Towbin, B.D.; Bank, E.M.; Gonzalez-Sandoval, A.; Stadler, M.B.; Meister, P.; Gruenbaum, Y.; Gasser, S.M. An EDMD mutation in C. elegans lamin blocks muscle-specific gene relocation and compromises muscle integrity. Curr. Biol. 2011, 21, 1603–1614. [Google Scholar] [CrossRef]
- Morales-Martinez, A.; Dobrzynska, A.; Askjaer, P. Inner nuclear membrane protein LEM-2 is required for correct nuclear separation and morphology in C. elegans. J. Cell Sci. 2015, 128, 1090–1096. [Google Scholar] [CrossRef]
- Lee, M.S.; Craigie, R. A previously unidentified host protein protects retroviral DNA from autointegration. Proc. Natl. Acad. Sci. USA 1998, 95, 1528–1533. [Google Scholar] [CrossRef]
- Jamin, A.; Wiebe, M.S. Barrier to Autointegration Factor (BANF1): Interwoven roles in nuclear structure, genome integrity, innate immunity, stress responses and progeria. Curr. Opin. Cell Biol. 2015, 34, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Gorjanacz, M.; Klerkx, E.P.; Galy, V.; Santarella, R.; Lopez-Iglesias, C.; Askjaer, P.; Mattaj, I.W. Caenorhabditis elegans BAF-1 and its kinase VRK-1 participate directly in post-mitotic nuclear envelope assembly. EMBO J. 2007, 26, 132–143. [Google Scholar] [CrossRef] [PubMed]
- Samwer, M.; Schneider, M.W.G.; Hoefler, R.; Schmalhorst, P.S.; Jude, J.G.; Zuber, J.; Gerlich, D.W. DNA Cross-Bridging Shapes a Single Nucleus from a Set of Mitotic Chromosomes. Cell 2017, 170, 956–972.e23. [Google Scholar] [CrossRef] [PubMed]
- Margalit, A.; Neufeld, E.; Feinstein, N.; Wilson, K.L.; Podbilewicz, B.; Gruenbaum, Y. Barrier to autointegration factor blocks premature cell fusion and maintains adult muscle integrity in C. elegans. J. Cell Biol. 2007, 178, 661–673. [Google Scholar] [CrossRef] [PubMed]
- Berk, J.M.; Simon, D.N.; Jenkins-Houk, C.R.; Westerbeck, J.W.; Gronning-Wang, L.M.; Carlson, C.R.; Wilson, K.L. The molecular basis of emerin-emerin and emerin-BAF interactions. J. Cell Sci. 2014, 127, 3956–3969. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Stoops, E.; Cp, U.; Markus, B.; Reuveny, A.; Ordan, E.; Volk, T. Mechanotransduction via the LINC complex regulates DNA replication in myonuclei. J. Cell Biol. 2018, 217, 2005–2018. [Google Scholar] [CrossRef]
- Puente, X.S.; Quesada, V.; Osorio, F.G.; Cabanillas, R.; Cadinanos, J.; Fraile, J.M.; Ordonez, G.R.; Puente, D.A.; Gutierrez-Fernandez, A.; Fanjul-Fernandez, M.; et al. Exome sequencing and functional analysis identifies BANF1 mutation as the cause of a hereditary progeroid syndrome. Am. J. Hum. Genet. 2011, 88, 650–656. [Google Scholar] [CrossRef]
- Eissenberg, J.C. Molecular biology of the chromo domain: An ancient chromatin module comes of age. Gene 2001, 275, 19–29. [Google Scholar] [CrossRef]
- Eissenberg, J.C.; Elgin, S.C. HP1a: A structural chromosomal protein regulating transcription. Trends Genet. 2014, 30, 103–110. [Google Scholar] [CrossRef]
- James, T.C.; Elgin, S.C. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol. Cell Biol. 1986, 6, 3862–3872. [Google Scholar] [CrossRef]
- Ye, Q.; Callebaut, I.; Pezhman, A.; Courvalin, J.C.; Worman, H.J. Domain-specific interactions of human HP1-type chromodomain proteins and inner nuclear membrane protein LBR. J. Biol. Chem. 1997, 272, 14983–14989. [Google Scholar] [CrossRef] [PubMed]
- Poleshko, A.; Mansfield, K.M.; Burlingame, C.C.; Andrake, M.D.; Shah, N.R.; Katz, R.A. The human protein PRR14 tethers heterochromatin to the nuclear lamina during interphase and mitotic exit. Cell Rep. 2013, 5, 292–301. [Google Scholar] [CrossRef] [PubMed]
- Larson, A.G.; Elnatan, D.; Keenen, M.M.; Trnka, M.J.; Johnston, J.B.; Burlingame, A.L.; Agard, D.A.; Redding, S.; Narlikar, G.J. Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin. Nature 2017, 547, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Strom, A.R.; Emelyanov, A.V.; Mir, M.; Fyodorov, D.V.; Darzacq, X.; Karpen, G.H. Phase separation drives heterochromatin domain formation. Nature 2017, 547, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Hiragami-Hamada, K.; Soeroes, S.; Nikolov, M.; Wilkins, B.; Kreuz, S.; Chen, C.; De La Rosa-Velazquez, I.A.; Zenn, H.M.; Kost, N.; Pohl, W.; et al. Dynamic and flexible H3K9me3 bridging via HP1beta dimerization establishes a plastic state of condensed chromatin. Nat. Commun. 2016, 7, 11310. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, S.; D’Angelo, M.A. The roles of the nuclear pore complex in cellular dysfunction, aging and disease. Semin. Cell Dev. Biol. 2017, 68, 72–84. [Google Scholar] [CrossRef]
- Kuhn, T.; Capelson, M. Nuclear Pore and Genome Organization and Gene Expression in Drosophila. In Nuclear Pore Complexes in Genome Organization, Function and Maintenance; D’Angelo, M., Ed.; Springer: Cham, Switzerland, 2018; pp. 111–135. [Google Scholar]
- Muñoz Jiménez, C.; Askjaer, P. Caenorhabditis elegans Nuclear Pore Complexes in Genome Organization and Gene Expression. In Nuclear Pore Complexes in Genome Organization, Function and Maintenance; D’Angelo, M., Ed.; Springer: Cham, Switzerland, 2018; pp. 137–158. [Google Scholar]
- Raices, M.; D’Angelo, M.A. Nuclear Pore Complexes in the Organization and Regulation of the Mammalian Genome. In Nuclear Pore Complexes in Genome Organization, Function and Maintenance; D’Angelo, M., Ed.; Springer: Cham, Switzerland, 2018; pp. 159–182. [Google Scholar]
- Randise-Hinchliff, C.; Brickner, J. Nuclear Pore Complex in Genome Organization and Gene Expression in Yeast. In Nuclear Pore Complexes in Genome Organization, Function and Maintenance; D’Angelo, M., Ed.; Springer: Cham, Switzerland, 2018; pp. 87–109. [Google Scholar]
- Fabrizio, P.; Garvis, S.; Palladino, F. Histone Methylation and Memory of Environmental Stress. Cells 2019, 8, 339. [Google Scholar] [CrossRef]
- Franks, T.M.; Benner, C.; Narvaiza, I.; Marchetto, M.C.; Young, J.M.; Malik, H.S.; Gage, F.H.; Hetzer, M.W. Evolution of a transcriptional regulator from a transmembrane nucleoporin. Genes Dev. 2016, 30, 1155–1171. [Google Scholar] [CrossRef]
- Kalverda, B.; Pickersgill, H.; Shloma, V.V.; Fornerod, M. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell 2010, 140, 360–371. [Google Scholar] [CrossRef]
- Toyama, B.H.; Savas, J.N.; Park, S.K.; Harris, M.S.; Ingolia, N.T.; Yates, J.R., 3rd; Hetzer, M.W. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell 2013, 154, 971–982. [Google Scholar] [CrossRef]
- D’Angelo, M.A.; Raices, M.; Panowski, S.H.; Hetzer, M.W. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell 2009, 136, 284–295. [Google Scholar] [CrossRef] [PubMed]
- Burtner, C.R.; Kennedy, B.K. Progeria syndromes and ageing: What is the connection? Nat. Rev. Mol. Cell Biol. 2010, 11, 567–578. [Google Scholar] [CrossRef] [PubMed]
- Doubaj, Y.; De Sandre-Giovannoli, A.; Vera, E.V.; Navarro, C.L.; Elalaoui, S.C.; Tajir, M.; Levy, N.; Sefiani, A. An inherited LMNA gene mutation in atypical Progeria syndrome. Am. J. Med. Genet. A 2012, 158A, 2881–2887. [Google Scholar] [CrossRef] [PubMed]
- Scaffidi, P.; Misteli, T. Lamin A-dependent nuclear defects in human aging. Science 2006, 312, 1059–1063. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.S.; Wu, S.C.; Hou, J.W.; Chu, C.P.; Tseng, L.L.; Lue, H.C. Restrictive dermopathy: Report of two siblings. Pediatr. Neonatol. 2013, 54, 198–201. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Novelli, G.; Muchir, A.; Sangiuolo, F.; Helbling-Leclerc, A.; D’Apice, M.R.; Massart, C.; Capon, F.; Sbraccia, P.; Federici, M.; Lauro, R.; et al. Mandibuloacral dysplasia is caused by a mutation in LMNA-encoding lamin A/C. Am. J. Hum. Genet. 2002, 71, 426–431. [Google Scholar] [CrossRef]
- Dobrzynska, A.; Gonzalo, S.; Shanahan, C.; Askjaer, P. The nuclear lamina in health and disease. Nucleus 2016, 7, 233–248. [Google Scholar] [CrossRef] [PubMed]
- Janin, A.; Bauer, D.; Ratti, F.; Millat, G.; Mejat, A. Nuclear envelopathies: a complex LINC between nuclear envelope and pathology. Orphanet J. Rare Dis. 2017, 12, 147. [Google Scholar] [CrossRef]
- Navarro, C.L.; Cadinanos, J.; De Sandre-Giovannoli, A.; Bernard, R.; Courrier, S.; Boccaccio, I.; Boyer, A.; Kleijer, W.J.; Wagner, A.; Giuliano, F.; et al. Loss of ZMPSTE24 (FACE-1) causes autosomal recessive restrictive dermopathy and accumulation of Lamin A precursors. Hum. Mol. Genet. 2005, 14, 1503–1513. [Google Scholar] [CrossRef]
- Agarwal, A.K.; Fryns, J.P.; Auchus, R.J.; Garg, A. Zinc metalloproteinase, ZMPSTE24, is mutated in mandibuloacral dysplasia. Hum. Mol. Genet. 2003, 12, 1995–2001. [Google Scholar] [CrossRef]
- Reunert, J.; Wentzell, R.; Walter, M.; Jakubiczka, S.; Zenker, M.; Brune, T.; Rust, S.; Marquardt, T. Neonatal progeria: Increased ratio of progerin to lamin A leads to progeria of the newborn. Eur. J. Hum. Genet. 2012, 20, 933–937. [Google Scholar] [CrossRef] [PubMed]
- Kubben, N.; Misteli, T. Shared molecular and cellular mechanisms of premature ageing and ageing-associated diseases. Nat. Rev. Mol. Cell Biol. 2017, 18, 595–609. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Otin, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef] [PubMed]
- Arancio, W.; Pizzolanti, G.; Genovese, S.I.; Pitrone, M.; Giordano, C. Epigenetic involvement in Hutchinson-Gilford progeria syndrome: A mini-review. Gerontology 2014, 60, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Columbaro, M.; Capanni, C.; Mattioli, E.; Novelli, G.; Parnaik, V.K.; Squarzoni, S.; Maraldi, N.M.; Lattanzi, G. Rescue of heterochromatin organization in Hutchinson-Gilford progeria by drug treatment. Cell Mol. Life Sci. 2005, 62, 2669–2678. [Google Scholar] [CrossRef]
- Heyn, H.; Moran, S.; Esteller, M. Aberrant DNA methylation profiles in the premature aging disorders Hutchinson-Gilford Progeria and Werner syndrome. Epigenetics 2013, 8, 28–33. [Google Scholar] [CrossRef]
- Zhang, J.; Lian, Q.; Zhu, G.; Zhou, F.; Sui, L.; Tan, C.; Mutalif, R.A.; Navasankari, R.; Zhang, Y.; Tse, H.F.; et al. A human iPSC model of Hutchinson Gilford Progeria reveals vascular smooth muscle and mesenchymal stem cell defects. Cell Stem Cell 2011, 8, 31–45. [Google Scholar] [CrossRef]
- Vidak, S.; Kubben, N.; Dechat, T.; Foisner, R. Proliferation of progeria cells is enhanced by lamina-associated polypeptide 2alpha (LAP2alpha) through expression of extracellular matrix proteins. Genes Dev. 2015, 29, 2022–2036. [Google Scholar] [CrossRef]
- Chojnowski, A.; Ong, P.F.; Wong, E.S.; Lim, J.S.; Mutalif, R.A.; Navasankari, R.; Dutta, B.; Yang, H.; Liow, Y.Y.; Sze, S.K.; et al. Progerin reduces LAP2alpha-telomere association in Hutchinson-Gilford progeria. Elife 2015, 4, e07759. [Google Scholar] [CrossRef]
- Loi, M.; Cenni, V.; Duchi, S.; Squarzoni, S.; Lopez-Otin, C.; Foisner, R.; Lattanzi, G.; Capanni, C. Barrier-to-autointegration factor (BAF) involvement in prelamin A-related chromatin organization changes. Oncotarget 2016, 7, 15662–15677. [Google Scholar] [CrossRef]
- Fong, L.G.; Frost, D.; Meta, M.; Qiao, X.; Yang, S.H.; Coffinier, C.; Young, S.G. A protein farnesyltransferase inhibitor ameliorates disease in a mouse model of progeria. Science 2006, 311, 1621–1623. [Google Scholar] [CrossRef] [PubMed]
- Moiseeva, O.; Lopes-Paciencia, S.; Huot, G.; Lessard, F.; Ferbeyre, G. Permanent farnesylation of lamin A mutants linked to progeria impairs its phosphorylation at serine 22 during interphase. Aging 2016, 8, 366–381. [Google Scholar] [CrossRef] [PubMed]
- Snider, N.T.; Omary, M.B. Post-translational modifications of intermediate filament proteins: Mechanisms and functions. Nat. Rev. Mol. Cell Biol. 2014, 15, 163–177. [Google Scholar] [CrossRef] [PubMed]
- Kubben, N.; Zhang, W.; Wang, L.; Voss, T.C.; Yang, J.; Qu, J.; Liu, G.H.; Misteli, T. Repression of the Antioxidant NRF2 Pathway in Premature Aging. Cell 2016, 165, 1361–1374. [Google Scholar] [CrossRef] [PubMed]
- Kim, P.H.; Luu, J.; Heizer, P.; Tu, Y.; Weston, T.A.; Chen, N.; Lim, C.; Li, R.L.; Lin, P.Y.; Dunn, J.C.Y.; et al. Disrupting the LINC complex in smooth muscle cells reduces aortic disease in a mouse model of Hutchinson-Gilford progeria syndrome. Sci. Transl. Med. 2018, 10, eaat7163. [Google Scholar] [CrossRef] [PubMed]
- Cao, K.; Blair, C.D.; Faddah, D.A.; Kieckhaefer, J.E.; Olive, M.; Erdos, M.R.; Nabel, E.G.; Collins, F.S. Progerin and telomere dysfunction collaborate to trigger cellular senescence in normal human fibroblasts. J. Clin. Investig. 2011, 121, 2833–2844. [Google Scholar] [CrossRef] [PubMed]
- Brunauer, R.; Kennedy, B.K. Medicine. Progeria accelerates adult stem cell aging. Science 2015, 348, 1093–1094. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Zhang, W.; Song, M.; Wang, W.; Wei, G.; Li, W.; Lei, J.; Huang, Y.; Sang, Y.; Chan, P.; et al. Differential stem cell aging kinetics in Hutchinson-Gilford progeria syndrome and Werner syndrome. Protein Cell 2018, 9, 333–350. [Google Scholar] [CrossRef] [PubMed]
- Cabanillas, R.; Cadinanos, J.; Villameytide, J.A.; Perez, M.; Longo, J.; Richard, J.M.; Alvarez, R.; Duran, N.S.; Illan, R.; Gonzalez, D.J.; et al. Nestor-Guillermo progeria syndrome: A novel premature aging condition with early onset and chronic development caused by BANF1 mutations. Am. J. Med. Genet. A 2011, 155A, 2617–2625. [Google Scholar] [CrossRef] [PubMed]
- Paquet, N.; Box, J.K.; Ashton, N.W.; Suraweera, A.; Croft, L.V.; Urquhart, A.J.; Bolderson, E.; Zhang, S.D.; O’Byrne, K.J.; Richard, D.J. Nestor-Guillermo Progeria Syndrome: A biochemical insight into Barrier-to-Autointegration Factor 1, alanine 12 threonine mutation. BMC Mol. Biol. 2014, 15, 27. [Google Scholar] [CrossRef]
- Capanni, C.; Squarzoni, S.; Cenni, V.; D’Apice, M.R.; Gambineri, A.; Novelli, G.; Wehnert, M.; Pasquali, R.; Maraldi, N.M.; Lattanzi, G. Familial partial lipodystrophy, mandibuloacral dysplasia and restrictive dermopathy feature barrier-to-autointegration factor (BAF) nuclear redistribution. Cell Cycle 2012, 11, 3568–3577. [Google Scholar] [CrossRef] [PubMed]
- Osorio, F.G.; Navarro, C.L.; Cadinanos, J.; Lopez-Mejia, I.C.; Quiros, P.M.; Bartoli, C.; Rivera, J.; Tazi, J.; Guzman, G.; Varela, I.; et al. Splicing-directed therapy in a new mouse model of human accelerated aging. Sci. Transl. Med. 2011, 3, 106ra107. [Google Scholar] [CrossRef] [PubMed]
- Mehta, I.S.; Eskiw, C.H.; Arican, H.D.; Kill, I.R.; Bridger, J.M. Farnesyltransferase inhibitor treatment restores chromosome territory positions and active chromosome dynamics in Hutchinson-Gilford progeria syndrome cells. Genome Biol. 2011, 12, R74. [Google Scholar] [CrossRef] [PubMed]
- Bikkul, M.U.; Clements, C.S.; Godwin, L.S.; Goldberg, M.W.; Kill, I.R.; Bridger, J.M. Farnesyltransferase inhibitor and rapamycin correct aberrant genome organisation and decrease DNA damage respectively, in Hutchinson-Gilford progeria syndrome fibroblasts. Biogerontology 2018, 19, 579–602. [Google Scholar] [CrossRef] [PubMed]
- Driscoll, M.K.; Albanese, J.L.; Xiong, Z.M.; Mailman, M.; Losert, W.; Cao, K. Automated image analysis of nuclear shape: what can we learn from a prematurely aged cell? Aging 2012, 4, 119–132. [Google Scholar] [CrossRef] [PubMed][Green Version]
- DuBose, A.J.; Lichtenstein, S.T.; Petrash, N.M.; Erdos, M.R.; Gordon, L.B.; Collins, F.S. Everolimus rescues multiple cellular defects in laminopathy-patient fibroblasts. Proc. Natl. Acad. Sci. USA 2018, 115, 4206–4211. [Google Scholar] [CrossRef] [PubMed]
- Adam, S.A.; Butin-Israeli, V.; Cleland, M.M.; Shimi, T.; Goldman, R.D. Disruption of lamin B1 and lamin B2 processing and localization by farnesyltransferase inhibitors. Nucleus 2013, 4, 142–150. [Google Scholar] [CrossRef]
- Varela, I.; Pereira, S.; Ugalde, A.P.; Navarro, C.L.; Suarez, M.F.; Cau, P.; Cadinanos, J.; Osorio, F.G.; Foray, N.; Cobo, J.; et al. Combined treatment with statins and aminobisphosphonates extends longevity in a mouse model of human premature aging. Nat. Med. 2008, 14, 767–772. [Google Scholar] [CrossRef]
- Ibrahim, M.X.; Sayin, V.I.; Akula, M.K.; Liu, M.; Fong, L.G.; Young, S.G.; Bergo, M.O. Targeting isoprenylcysteine methylation ameliorates disease in a mouse model of progeria. Science 2013, 340, 1330–1333. [Google Scholar] [CrossRef]
- Kubben, N.; Brimacombe, K.R.; Donegan, M.; Li, Z.; Misteli, T. A high-content imaging-based screening pipeline for the systematic identification of anti-progeroid compounds. Methods 2016, 96, 46–58. [Google Scholar] [CrossRef]
- Song, S.; Johnson, F.B. Epigenetic Mechanisms Impacting Aging: A Focus on Histone Levels and Telomeres. Genes 2018, 9, 201. [Google Scholar] [CrossRef] [PubMed]
- Sogabe, Y.; Seno, H.; Yamamoto, T.; Yamada, Y. Unveiling epigenetic regulation in cancer, aging, and rejuvenation with in vivo reprogramming technology. Cancer Sci. 2018, 109, 2641–2650. [Google Scholar] [CrossRef] [PubMed]
- Pu, M.; Wang, M.; Wang, W.; Velayudhan, S.S.; Lee, S.S. Unique patterns of trimethylation of histone H3 lysine 4 are prone to changes during aging in Caenorhabditis elegans somatic cells. PLoS Genet. 2018, 14, e1007466. [Google Scholar] [CrossRef] [PubMed]
- Greer, E.L.; Maures, T.J.; Hauswirth, A.G.; Green, E.M.; Leeman, D.S.; Maro, G.S.; Han, S.; Banko, M.R.; Gozani, O.; Brunet, A. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature 2010, 466, 383–387. [Google Scholar] [CrossRef] [PubMed]
- Benayoun, B.A.; Pollina, E.A.; Brunet, A. Epigenetic regulation of ageing: Linking environmental inputs to genomic stability. Nat. Rev. Mol. Cell Biol. 2015, 16, 593–610. [Google Scholar] [CrossRef]
- Li, L.; Greer, C.; Eisenman, R.N.; Secombe, J. Essential functions of the histone demethylase lid. PLoS Genet. 2010, 6, e1001221. [Google Scholar] [CrossRef]
- Cruz, C.; Della Rosa, M.; Krueger, C.; Gao, Q.; Horkai, D.; King, M.; Field, L.; Houseley, J. Tri-methylation of histone H3 lysine 4 facilitates gene expression in ageing cells. Elife 2018, 7, e34081. [Google Scholar] [CrossRef]
- Sarg, B.; Koutzamani, E.; Helliger, W.; Rundquist, I.; Lindner, H.H. Postsynthetic trimethylation of histone H4 at lysine 20 in mammalian tissues is associated with aging. J. Biol. Chem. 2002, 277, 39195–39201. [Google Scholar] [CrossRef]
- Shumaker, D.K.; Dechat, T.; Kohlmaier, A.; Adam, S.A.; Bozovsky, M.R.; Erdos, M.R.; Eriksson, M.; Goldman, A.E.; Khuon, S.; Collins, F.S.; et al. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc. Natl. Acad. Sci. USA 2006, 103, 8703–8708. [Google Scholar] [CrossRef]
- Denisenko, O.; Mar, D.; Trawczynski, M.; Bomsztyk, K. Chromatin changes trigger laminin genes dysregulation in aging kidneys. Aging (Albany NY) 2018, 10, 1133–1145. [Google Scholar] [CrossRef]
- McColl, G.; Killilea, D.W.; Hubbard, A.E.; Vantipalli, M.C.; Melov, S.; Lithgow, G.J. Pharmacogenetic analysis of lithium-induced delayed aging in Caenorhabditis elegans. J. Biol. Chem. 2008, 283, 350–357. [Google Scholar] [CrossRef]
- Maures, T.J.; Greer, E.L.; Hauswirth, A.G.; Brunet, A. The H3K27 demethylase UTX-1 regulates C. elegans lifespan in a germline-independent, insulin-dependent manner. Aging Cell 2011, 10, 980–990. [Google Scholar] [CrossRef] [PubMed]
- Greer, E.L.; Becker, B.; Latza, C.; Antebi, A.; Shi, Y. Mutation of C. elegans demethylase spr-5 extends transgenerational longevity. Cell Res. 2016, 26, 229–238. [Google Scholar] [CrossRef] [PubMed]
- De Vaux, V.; Pfefferli, C.; Passannante, M.; Belhaj, K.; von Essen, A.; Sprecher, S.G.; Muller, F.; Wicky, C. The Caenorhabditis elegans LET-418/Mi2 plays a conserved role in lifespan regulation. Aging Cell 2013, 12, 1012–1020. [Google Scholar] [CrossRef] [PubMed]
- Riedel, C.G.; Dowen, R.H.; Lourenco, G.F.; Kirienko, N.V.; Heimbucher, T.; West, J.A.; Bowman, S.K.; Kingston, R.E.; Dillin, A.; Asara, J.M.; et al. DAF-16 employs the chromatin remodeller SWI/SNF to promote stress resistance and longevity. Nat. Cell Biol. 2013, 15, 491–501. [Google Scholar] [CrossRef]
- Kaeberlein, M.; McVey, M.; Guarente, L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999, 13, 2570–2580. [Google Scholar] [CrossRef]
- Ikeda, T.; Uno, M.; Honjoh, S.; Nishida, E. The MYST family histone acetyltransferase complex regulates stress resistance and longevity through transcriptional control of DAF-16/FOXO transcription factors. EMBO Rep. 2017, 18, 1716–1726. [Google Scholar] [CrossRef]
- Woodhouse, R.M.; Buchmann, G.; Hoe, M.; Harney, D.J.; Low, J.K.K.; Larance, M.; Boag, P.R.; Ashe, A. Chromatin Modifiers SET-25 and SET-32 Are Required for Establishment but Not Long-Term Maintenance of Transgenerational Epigenetic Inheritance. Cell Rep. 2018, 25, 2259–2272. [Google Scholar] [CrossRef]
- Siebold, A.P.; Banerjee, R.; Tie, F.; Kiss, D.L.; Moskowitz, J.; Harte, P.J. Polycomb Repressive Complex 2 and Trithorax modulate Drosophila longevity and stress resistance. Proc. Natl. Acad. Sci. USA 2010, 107, 169–174. [Google Scholar] [CrossRef]
- Su, L.; Li, H.; Huang, C.; Zhao, T.; Zhang, Y.; Ba, X.; Li, Z.; Zhang, Y.; Huang, B.; Lu, J.; et al. Muscle-Specific Histone H3K36 Dimethyltransferase SET-18 Shortens Lifespan of Caenorhabditis elegans by Repressing daf-16a Expression. Cell Rep. 2018, 22, 2716–2729. [Google Scholar] [CrossRef]
- Wang, W.; Chaturbedi, A.; Wang, M.; An, S.; Santhi Velayudhan, S.; Lee, S.S. SET-9 and SET-26 are H3K4me3 readers and play critical roles in germline development and longevity. Elife 2018, 7, e34970. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yuan, Q.; Xie, L. Histone Modifications in Aging: The Underlying Mechanisms and Implications. Curr. Stem Cell Res. Ther. 2018, 13, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Dang, W.; Steffen, K.K.; Perry, R.; Dorsey, J.A.; Johnson, F.B.; Shilatifard, A.; Kaeberlein, M.; Kennedy, B.K.; Berger, S.L. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature 2009, 459, 802–807. [Google Scholar] [CrossRef] [PubMed]
- Feser, J.; Truong, D.; Das, C.; Carson, J.J.; Kieft, J.; Harkness, T.; Tyler, J.K. Elevated histone expression promotes life span extension. Mol. Cell 2010, 39, 724–735. [Google Scholar] [CrossRef]
- Li, Y.; Jin, M.; O’Laughlin, R.; Bittihn, P.; Tsimring, L.S.; Pillus, L.; Hasty, J.; Hao, N. Multigenerational silencing dynamics control cell aging. Proc. Natl. Acad. Sci. USA 2017, 114, 11253–11258. [Google Scholar] [CrossRef] [PubMed]
- Larson, K.; Yan, S.J.; Tsurumi, A.; Liu, J.; Zhou, J.; Gaur, K.; Guo, D.; Eickbush, T.H.; Li, W.X. Heterochromatin formation promotes longevity and represses ribosomal RNA synthesis. PLoS Genet. 2012, 8, e1002473. [Google Scholar] [CrossRef]
- Mouchiroud, L.; Houtkooper, R.H.; Moullan, N.; Katsyuba, E.; Ryu, D.; Canto, C.; Mottis, A.; Jo, Y.S.; Viswanathan, M.; Schoonjans, K.; et al. The NAD(+)/Sirtuin Pathway Modulates Longevity through Activation of Mitochondrial UPR and FOXO Signaling. Cell 2013, 154, 430–441. [Google Scholar] [CrossRef]
- Moroz, N.; Carmona, J.J.; Anderson, E.; Hart, A.C.; Sinclair, D.A.; Blackwell, T.K. Dietary restriction involves NAD(+) -dependent mechanisms and a shift toward oxidative metabolism. Aging Cell 2014, 13, 1075–1085. [Google Scholar] [CrossRef] [PubMed]
- Haithcock, E.; Dayani, Y.; Neufeld, E.; Zahand, A.J.; Feinstein, N.; Mattout, A.; Gruenbaum, Y.; Liu, J. Age-related changes of nuclear architecture in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2005, 102, 16690–16695. [Google Scholar] [CrossRef]
- Perez-Jimenez, M.M.; Rodriguez-Palero, M.J.; Rodenas, E.; Askjaer, P.; Munoz, M.J. Age-dependent changes of nuclear morphology are uncoupled from longevity in Caenorhabditis elegans IGF/insulin receptor daf-2 mutants. Biogerontology 2014, 15, 279–288. [Google Scholar] [CrossRef]
- Xue, Y.; Wong, J.; Moreno, G.T.; Young, M.K.; Cote, J.; Wang, W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell 1998, 2, 851–861. [Google Scholar] [CrossRef]
- Zhang, Y.; LeRoy, G.; Seelig, H.P.; Lane, W.S.; Reinberg, D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 1998, 95, 279–289. [Google Scholar] [CrossRef]
- Pegoraro, G.; Kubben, N.; Wickert, U.; Gohler, H.; Hoffmann, K.; Misteli, T. Ageing-related chromatin defects through loss of the NURD complex. Nat. Cell Biol. 2009, 11, 1261–1267. [Google Scholar] [CrossRef] [PubMed]
- Peleg, S.; Sananbenesi, F.; Zovoilis, A.; Burkhardt, S.; Bahari-Javan, S.; Agis-Balboa, R.C.; Cota, P.; Wittnam, J.L.; Gogol-Doering, A.; Opitz, L.; et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science 2010, 328, 753–756. [Google Scholar] [CrossRef] [PubMed]
- Ryu, S.H.; Kang, K.; Yoo, T.; Joe, C.O.; Chung, J.H. Transcriptional repression of repeat-derived transcripts correlates with histone hypoacetylation at repetitive DNA elements in aged mice brain. Exp. Gerontol. 2011, 46, 811–818. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Chen, K.; Xia, Z.; Chavez, M.; Pal, S.; Seol, J.H.; Chen, C.C.; Li, W.; Tyler, J.K. Nucleosome loss leads to global transcriptional up-regulation and genomic instability during yeast aging. Genes Dev. 2014, 28, 396–408. [Google Scholar] [CrossRef] [PubMed]
- Ni, Z.; Ebata, A.; Alipanahiramandi, E.; Lee, S.S. Two SET domain containing genes link epigenetic changes and aging in Caenorhabditis elegans. Aging Cell 2012, 11, 315–325. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, R.J.; Kubicek, S.; Schreiber, S.L.; Karlseder, J. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat. Struct. Mol. Biol. 2010, 17, 1218–1225. [Google Scholar] [CrossRef] [PubMed]
- Maze, I.; Wenderski, W.; Noh, K.M.; Bagot, R.C.; Tzavaras, N.; Purushothaman, I.; Elsasser, S.J.; Guo, Y.; Ionete, C.; Hurd, Y.L.; et al. Critical Role of Histone Turnover in Neuronal Transcription and Plasticity. Neuron 2015, 87, 77–94. [Google Scholar] [CrossRef] [PubMed]
- Tvardovskiy, A.; Schwammle, V.; Kempf, S.J.; Rogowska-Wrzesinska, A.; Jensen, O.N. Accumulation of histone variant H3.3 with age is associated with profound changes in the histone methylation landscape. Nucleic Acids Res. 2017, 45, 9272–9289. [Google Scholar] [CrossRef] [PubMed]
- Piazzesi, A.; Papic, D.; Bertan, F.; Salomoni, P.; Nicotera, P.; Bano, D. Replication-Independent Histone Variant H3.3 Controls Animal Lifespan through the Regulation of Pro-longevity Transcriptional Programs. Cell Rep. 2016, 17, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Weiner, A.; Chen, H.V.; Liu, C.L.; Rahat, A.; Klien, A.; Soares, L.; Gudipati, M.; Pfeffner, J.; Regev, A.; Buratowski, S.; et al. Systematic dissection of roles for chromatin regulators in a yeast stress response. PLoS Biol. 2012, 10, e1001369. [Google Scholar] [CrossRef] [PubMed]
- Shaughnessy, D.T.; McAllister, K.; Worth, L.; Haugen, A.C.; Meyer, J.N.; Domann, F.E.; Van Houten, B.; Mostoslavsky, R.; Bultman, S.J.; Baccarelli, A.A.; et al. Mitochondria, energetics, epigenetics, and cellular responses to stress. Environ. Health Perspect. 2014, 122, 1271–1278. [Google Scholar] [CrossRef] [PubMed]
- Kishimoto, S.; Uno, M.; Okabe, E.; Nono, M.; Nishida, E. Environmental stresses induce transgenerationally inheritable survival advantages via germline-to-soma communication in Caenorhabditis elegans. Nat. Commun. 2017, 8, 14031. [Google Scholar] [CrossRef] [PubMed]
- Kozlowski, L.; Garvis, S.; Bedet, C.; Palladino, F. The Caenorhabditis elegans HP1 family protein HPL-2 maintains ER homeostasis through the UPR and hormesis. Proc. Natl. Acad. Sci. USA 2014, 111, 5956–5961. [Google Scholar] [CrossRef]
- Merkwirth, C.; Jovaisaite, V.; Durieux, J.; Matilainen, O.; Jordan, S.D.; Quiros, P.M.; Steffen, K.K.; Williams, E.G.; Mouchiroud, L.; Tronnes, S.U.; et al. Two Conserved Histone Demethylases Regulate Mitochondrial Stress-Induced Longevity. Cell 2016, 165, 1209–1223. [Google Scholar] [CrossRef]
- Tian, Y.; Garcia, G.; Bian, Q.; Steffen, K.K.; Joe, L.; Wolff, S.; Meyer, B.J.; Dillin, A. Mitochondrial Stress Induces Chromatin Reorganization to Promote Longevity and UPR(mt). Cell 2016, 165, 1197–1208. [Google Scholar] [CrossRef]
- Ma, C.; Niu, R.; Huang, T.; Shao, L.W.; Peng, Y.; Ding, W.; Wang, Y.; Jia, G.; He, C.; Li, C.Y.; et al. N6-methyldeoxyadenine is a transgenerational epigenetic signal for mitochondrial stress adaptation. Nat. Cell Biol. 2019, 21, 319–327. [Google Scholar] [CrossRef]
- Dillin, A.; Hsu, A.L.; Arantes-Oliveira, N.; Lehrer-Graiwer, J.; Hsin, H.; Fraser, A.G.; Kamath, R.S.; Ahringer, J.; Kenyon, C. Rates of behavior and aging specified by mitochondrial function during development. Science 2002, 298, 2398–2401. [Google Scholar] [CrossRef]
- Houtkooper, R.H.; Mouchiroud, L.; Ryu, D.; Moullan, N.; Katsyuba, E.; Knott, G.; Williams, R.W.; Auwerx, J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature 2013, 497, 451–457. [Google Scholar] [CrossRef]
- Lee, S.S.; Lee, R.Y.; Fraser, A.G.; Kamath, R.S.; Ahringer, J.; Ruvkun, G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat. Genet. 2003, 33, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Quiros, P.M.; Mottis, A.; Auwerx, J. Mitonuclear communication in homeostasis and stress. Nat. Rev. Mol. Cell Biol. 2016, 17, 213–226. [Google Scholar] [CrossRef] [PubMed]
- Bosch-Presegue, L.; Raurell-Vila, H.; Marazuela-Duque, A.; Kane-Goldsmith, N.; Valle, A.; Oliver, J.; Serrano, L.; Vaquero, A. Stabilization of Suv39H1 by SirT1 is part of oxidative stress response and ensures genome protection. Mol. Cell 2011, 42, 210–223. [Google Scholar] [CrossRef] [PubMed]
- Frost, B.; Hemberg, M.; Lewis, J.; Feany, M.B. Tau promotes neurodegeneration through global chromatin relaxation. Nat. Neurosci. 2014, 17, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Tauffenberger, A.; Parker, J.A. Heritable transmission of stress resistance by high dietary glucose in Caenorhabditis elegans. PLoS Genet. 2014, 10, e1004346. [Google Scholar] [CrossRef] [PubMed]
- Greer, E.L.; Maures, T.J.; Ucar, D.; Hauswirth, A.G.; Mancini, E.; Lim, J.P.; Benayoun, B.A.; Shi, Y.; Brunet, A. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature 2011, 479, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Tran, K.; Jethmalani, Y.; Jaiswal, D.; Green, E.M. Set4 is a chromatin-associated protein, promotes survival during oxidative stress, and regulates stress response genes in yeast. J. Biol Chem. 2018, 293, 14429–14443. [Google Scholar] [CrossRef]
- Sharma, V.; Kohli, S.; Brahmachari, V. Correlation between desiccation stress response and epigenetic modifications of genes in Drosophila melanogaster: An example of environment-epigenome interaction. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 1058–1068. [Google Scholar] [CrossRef]
- Klosin, A.; Casas, E.; Hidalgo-Carcedo, C.; Vavouri, T.; Lehner, B. Transgenerational transmission of environmental information in C. elegans. Science 2017, 356, 320–323. [Google Scholar] [CrossRef]
- Raurell-Vila, H.; Bosch-Presegue, L.; Gonzalez, J.; Kane-Goldsmith, N.; Casal, C.; Brown, J.P.; Marazuela-Duque, A.; Singh, P.B.; Serrano, L.; Vaquero, A. An HP1 isoform-specific feedback mechanism regulates Suv39h1 activity under stress conditions. Epigenetics 2017, 12, 166–175. [Google Scholar] [CrossRef]
- McMurchy, A.N.; Stempor, P.; Gaarenstroom, T.; Wysolmerski, B.; Dong, Y.; Aussianikava, D.; Appert, A.; Huang, N.; Kolasinska-Zwierz, P.; Sapetschnig, A.; et al. A team of heterochromatin factors collaborates with small RNA pathways to combat repetitive elements and germline stress. Elife 2017, 6, e21666. [Google Scholar] [CrossRef] [PubMed]
- Schott, S.; Coustham, V.; Simonet, T.; Bedet, C.; Palladino, F. Unique and redundant functions of C. elegans HP1 proteins in post-embryonic development. Dev. Biol. 2006, 298, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Meister, P.; Schott, S.; Bedet, C.; Xiao, Y.; Rohner, S.; Bodennec, S.; Hudry, B.; Molin, L.; Solari, F.; Gasser, S.M.; et al. Caenorhabditis elegans Heterochromatin protein 1 (HPL-2) links developmental plasticity, longevity and lipid metabolism. Genome Biol. 2011, 12, R123. [Google Scholar] [CrossRef] [PubMed]
- Mills, B.B.; Thomas, A.D.; Riddle, N.C. HP1B is a euchromatic Drosophila HP1 homolog with links to metabolism. PLoS ONE 2018, 13, e0205867. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.; Suganuma, T.; Gogol, M.M.; Workman, J.L. Histone H3 threonine 11 phosphorylation by Sch9 and CK2 regulates chronological lifespan by controlling the nutritional stress response. Elife 2018, 7, e36157. [Google Scholar] [CrossRef]
- Molina-Serrano, D.; Schiza, V.; Demosthenous, C.; Stavrou, E.; Oppelt, J.; Kyriakou, D.; Liu, W.; Zisser, G.; Bergler, H.; Dang, W.; et al. Loss of Nat4 and its associated histone H4 N-terminal acetylation mediates calorie restriction-induced longevity. EMBO Rep. 2016, 17, 1829–1843. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tollefsbol, T.O. p16(INK4a) suppression by glucose restriction contributes to human cellular lifespan extension through SIRT1-mediated epigenetic and genetic mechanisms. PLoS ONE 2011, 6, e17421. [Google Scholar] [CrossRef]
- Singh, A.; Kumar, N.; Matai, L.; Jain, V.; Garg, A.; Mukhopadhyay, A. A chromatin modifier integrates insulin/IGF-1 signalling and dietary restriction to regulate longevity. Aging Cell 2016, 15, 694–705. [Google Scholar] [CrossRef]
- Hahn, O.; Gronke, S.; Stubbs, T.M.; Ficz, G.; Hendrich, O.; Krueger, F.; Andrews, S.; Zhang, Q.; Wakelam, M.J.; Beyer, A.; et al. Dietary restriction protects from age-associated DNA methylation and induces epigenetic reprogramming of lipid metabolism. Genome Biol. 2017, 18, 56. [Google Scholar] [CrossRef]
- Maegawa, S.; Lu, Y.; Tahara, T.; Lee, J.T.; Madzo, J.; Liang, S.; Jelinek, J.; Colman, R.J.; Issa, J.J. Caloric restriction delays age-related methylation drift. Nat. Commun. 2017, 8, 539. [Google Scholar] [CrossRef]
- Chouliaras, L.; van den Hove, D.L.; Kenis, G.; Draanen, M.; Hof, P.R.; van Os, J.; Steinbusch, H.W.; Schmitz, C.; Rutten, B.P. Histone deacetylase 2 in the mouse hippocampus: attenuation of age-related increase by caloric restriction. Curr. Alzheimer Res. 2013, 10, 868–876. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nie, L.; Shuai, L.; Zhu, M.; Liu, P.; Xie, Z.F.; Jiang, S.; Jiang, H.W.; Li, J.; Zhao, Y.; Li, J.Y.; et al. The Landscape of Histone Modifications in a High-Fat Diet-Induced Obese (DIO) Mouse Model. Mol. Cell Proteom. 2017, 16, 1324–1334. [Google Scholar] [CrossRef] [PubMed]
- Carrer, A.; Parris, J.L.; Trefely, S.; Henry, R.A.; Montgomery, D.C.; Torres, A.; Viola, J.M.; Kuo, Y.M.; Blair, I.A.; Meier, J.L.; et al. Impact of a High-fat Diet on Tissue Acyl-CoA and Histone Acetylation Levels. J. Biol. Chem. 2017, 292, 3312–3322. [Google Scholar] [CrossRef] [PubMed]
- Siersbaek, M.; Varticovski, L.; Yang, S.; Baek, S.; Nielsen, R.; Mandrup, S.; Hager, G.L.; Chung, J.H.; Grontved, L. High fat diet-induced changes of mouse hepatic transcription and enhancer activity can be reversed by subsequent weight loss. Sci. Rep. 2017, 7, 40220. [Google Scholar] [CrossRef] [PubMed]
- Sohi, G.; Marchand, K.; Revesz, A.; Arany, E.; Hardy, D.B. Maternal protein restriction elevates cholesterol in adult rat offspring due to repressive changes in histone modifications at the cholesterol 7alpha-hydroxylase promoter. Mol. Endocrinol. 2011, 25, 785–798. [Google Scholar] [CrossRef] [PubMed]
- Strakovsky, R.S.; Zhang, X.; Zhou, D.; Pan, Y.X. Gestational high fat diet programs hepatic phosphoenolpyruvate carboxykinase gene expression and histone modification in neonatal offspring rats. J. Physiol. 2011, 589, 2707–2717. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Lyu, X.; Hou, C.; Takenaka, N.; Nguyen, H.Q.; Ong, C.T.; Cubenas-Potts, C.; Hu, M.; Lei, E.P.; Bosco, G.; et al. Widespread rearrangement of 3D chromatin organization underlies polycomb-mediated stress-induced silencing. Mol. Cell 2015, 58, 216–231. [Google Scholar] [CrossRef]
- Amat, R.; Bottcher, R.; Le Dily, F.; Vidal, E.; Quilez, J.; Cuartero, Y.; Beato, M.; de Nadal, E.; Posas, F. Rapid reversible changes in compartments and local chromatin organization revealed by hyperosmotic shock. Genome Res. 2019, 29, 18–28. [Google Scholar] [CrossRef]
- Delaney, K.; Mailler, J.; Wenda, J.M.; Gabus, C.; Steiner, F.A. Differential Expression of Histone H3.3 Genes and Their Role in Modulating Temperature Stress Response in Caenorhabditis elegans. Genetics 2018, 209, 551–565. [Google Scholar] [CrossRef]
| Disease | Gene | Reference | |
|---|---|---|---|
| HGPS | Hutchinson–Gilford progeria syndrome | LMNA | [3] |
| APSs | Atypical progeria syndromes | LMNA | [67] |
| MADA | Mandibuloacral dysplasia | LMNA | [68] |
| RD | Restrictive dermopathy | ZMPSTE24 | [69] |
| MADB | Mandibuloacral dysplasia | ZMPSTE24 | [70] |
| NGPS | Nestor–Guillermo progeria syndrome | BANF1 | [46] |
| Type of Enzyme | Protein | Intervention | Targets | Species | Reference |
|---|---|---|---|---|---|
| histone demethylase (HDM) | LSD-1 | RNAi | H3K9me/ H3K4me | C. elegans | [115] |
| UTX-1 | deletion | H3K27me3 | C. elegans | [116] | |
| SPR-5 | deletion | H3K4me2 | C. elegans | [117] | |
| histone deacetylase/ remodeling complex (HDAC) | LET-418 (NuRD 2) | loss-of-function allele | Nucleosome | C. elegans | [118] |
| SWI/SNF | RNAi | Nucleosome | C. elegans | [119] | |
| SIR2 | overexpression | H4K16ac | S. cerevisiae | [120] | |
| histone acetyltransferase (HAT) | MYS-1 & TRR-1 | RNAi | H4K16ac | C. elegans | [121] |
| histone methyltransferase (HMT) | Trithorax (WDR-5, SET-2 and ASH-2) | deletion | H3K4me3 | C. elegans | [108] |
| SET-32 | deletion | H3K9me3 | C. elegans | [108,122] | |
| PRC2 3 | deletion | H3K27me3 | D. melanogaster | [123] | |
| SET-18 | deletion | H3K36me2 | C. elegans | [124] | |
| SET-26 | deletion | H3K4me3 | C. elegans | [125] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romero-Bueno, R.; Ruiz, P.d.l.C.; Artal-Sanz, M.; Askjaer, P.; Dobrzynska, A. Nuclear Organization in Stress and Aging. Cells 2019, 8, 664. https://doi.org/10.3390/cells8070664
Romero-Bueno R, Ruiz PdlC, Artal-Sanz M, Askjaer P, Dobrzynska A. Nuclear Organization in Stress and Aging. Cells. 2019; 8(7):664. https://doi.org/10.3390/cells8070664
Chicago/Turabian StyleRomero-Bueno, Raquel, Patricia de la Cruz Ruiz, Marta Artal-Sanz, Peter Askjaer, and Agnieszka Dobrzynska. 2019. "Nuclear Organization in Stress and Aging" Cells 8, no. 7: 664. https://doi.org/10.3390/cells8070664
APA StyleRomero-Bueno, R., Ruiz, P. d. l. C., Artal-Sanz, M., Askjaer, P., & Dobrzynska, A. (2019). Nuclear Organization in Stress and Aging. Cells, 8(7), 664. https://doi.org/10.3390/cells8070664






