The Dynamic 3D Genome in Gametogenesis and Early Embryonic Development
Abstract
:1. Introduction
2. Low-Input Hi-C Methods and Analysis
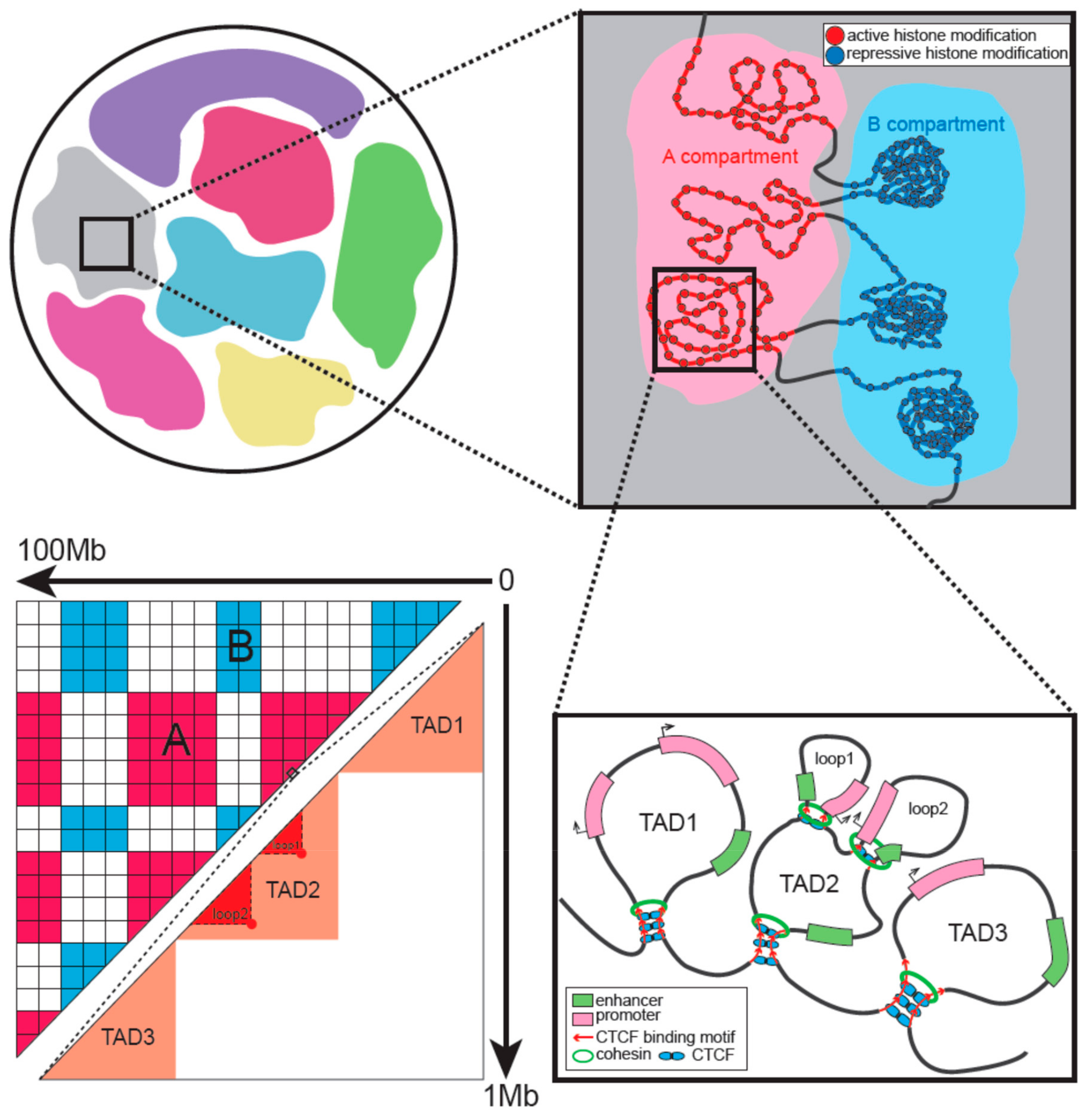
3. Hierarchical Organization of Interphase Chromatin
4. Chromatin Remodeling in Gametogenesis and Early Embryonic Development
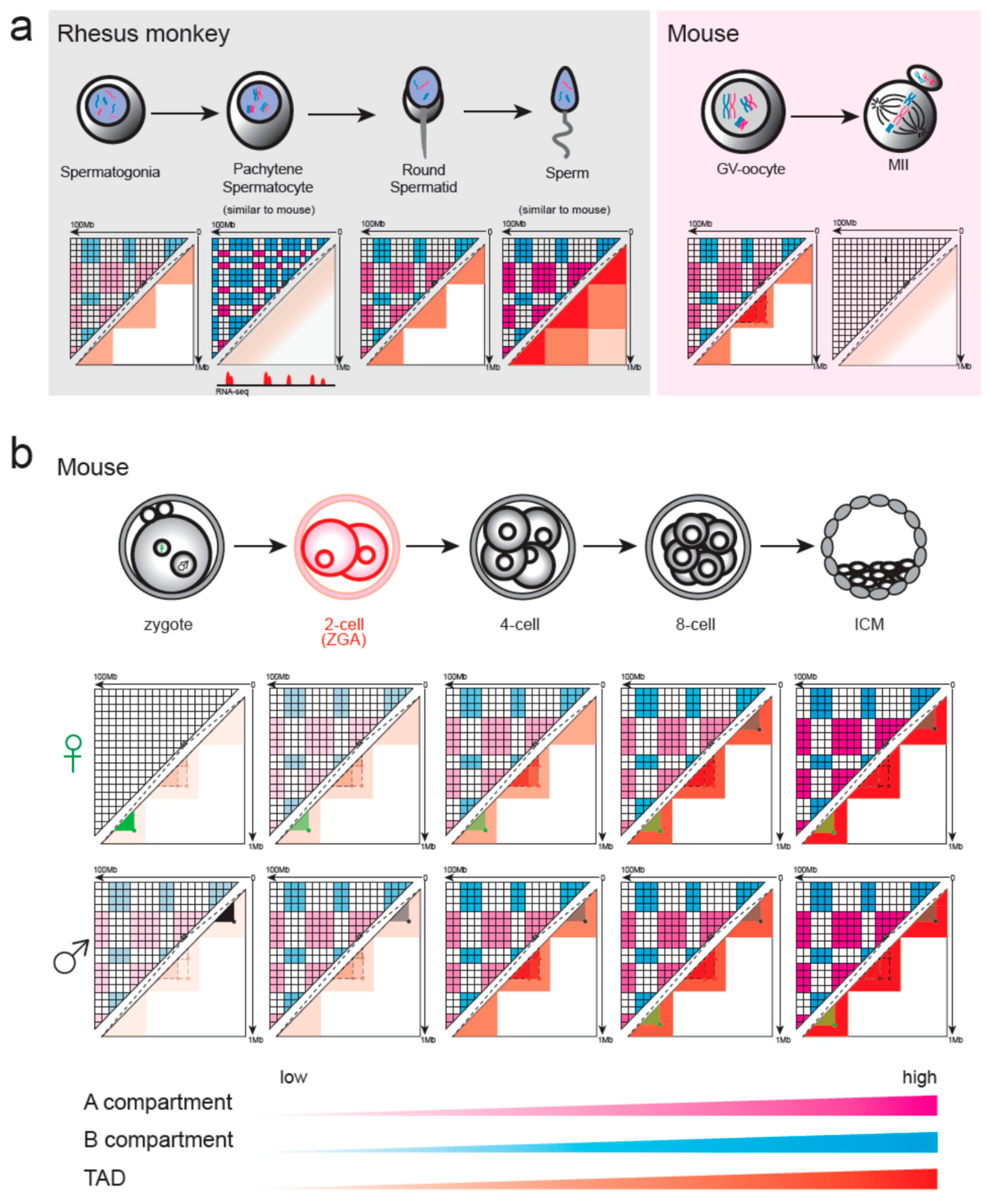
4.1. Chromatin Remodeling in Gametogenesis and Pre-Implantation Development in Mammals
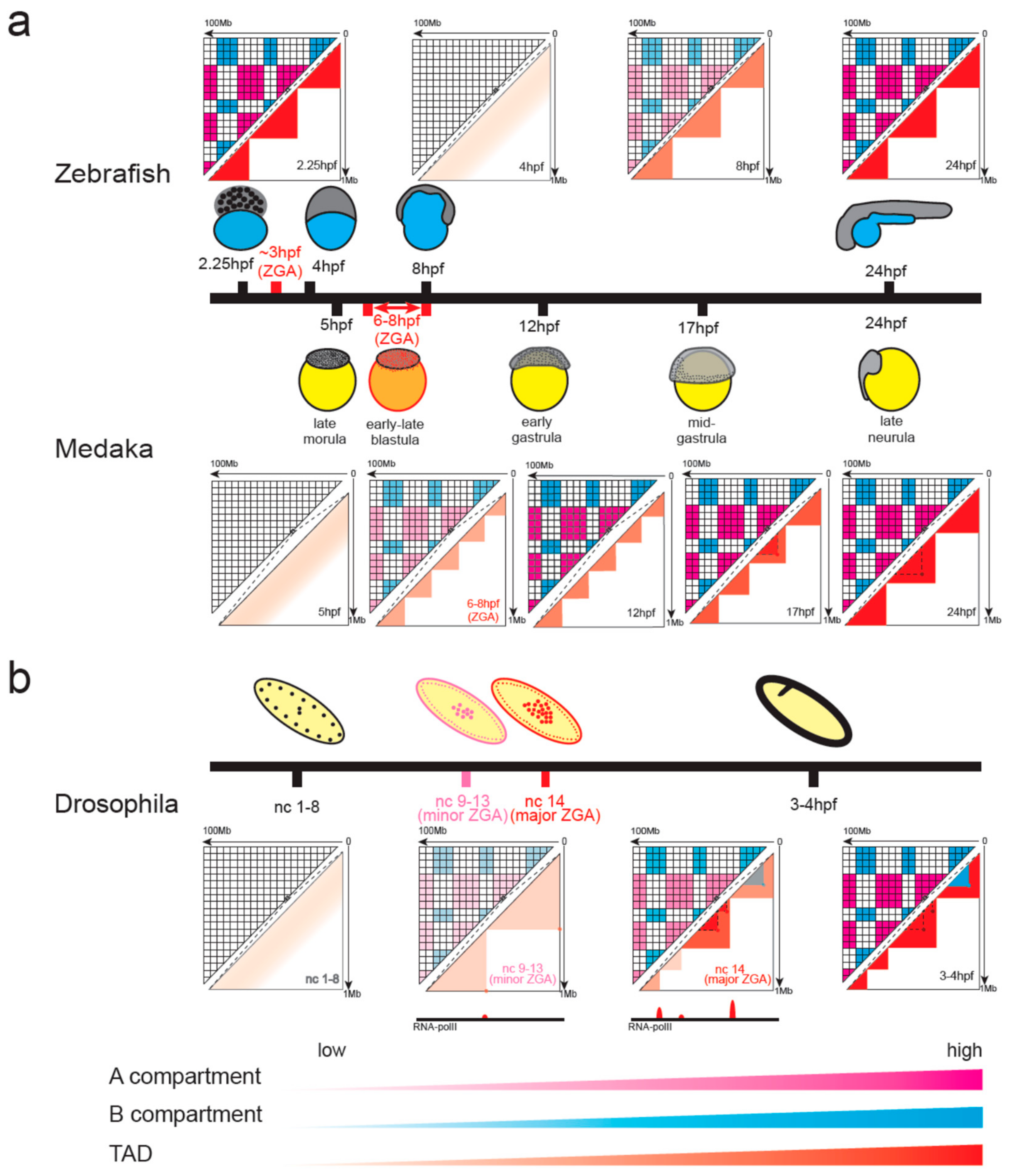
4.2. Chromatin Organization in Non-Mammalian Vertebrates during Early Embryonic Development
4.3. Emergence of Chromatin Organization in Insect Embryos
4.4. Similarities and Differences between Species
5. Mechanisms of 3D Genome Formation in Early Embryonic Development
5.1. Architectural Proteins
5.2. Transcription and Establishment of 3D Genome
5.3. Transposable Elements (TEs) and 3D Genome Folding
5.4. Phase Separation
6. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Hales, B.F.; Grenier, L.; Lalancette, C.; Robaire, B. Epigenetic programming: From gametes to blastocyst. Birth Defects Res. Part A Clin. Mol. Teratol. 2011, 91, 652–665. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Xie, W. Epigenome in Early Mammalian Development: Inheritance, Reprogramming and Establishment. Trends Cell Biol. 2018, 28, 237–253. [Google Scholar] [CrossRef] [PubMed]
- Langley, A.R.; Smith, J.C.; Stemple, D.L.; Harvey, S.A. New insights into the maternal to zygotic transition. Development 2014, 141, 3834–3841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eckersley-Maslin, M.A.; Alda-Catalinas, C.; Reik, W. Dynamics of the epigenetic landscape during the maternal-to-zygotic transition. Nat. Rev. Mol. Cell Biol. 2018, 19, 436–450. [Google Scholar] [CrossRef] [PubMed]
- Bonev, B.; Cavalli, G. Organization and function of the 3D genome. Nat. Rev. Genet. 2016, 17, 772. [Google Scholar] [CrossRef] [PubMed]
- Rowley, M.J.; Corces, V.G. Organizational principles of 3D genome architecture. Nat. Rev. Genet. 2018, 19, 789–800. [Google Scholar] [CrossRef] [PubMed]
- Mourad, R.; Cuvier, O. Predicting the spatial organization of chromosomes using epigenetic data. Genome Biol. 2015, 16, 182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Chen, Z.; Zhang, K.; Wang, M.; Medovoy, D.; Whitaker, J.W.; Ding, B.; Li, N.; Zheng, L.; Wang, W. Constructing 3D interaction maps from 1D epigenomes. Nat. Commun. 2016, 7, 10812. [Google Scholar] [CrossRef] [PubMed]
- Denker, A.; de Laat, W. The second decade of 3C technologies: Detailed insights into nuclear organization. Genes Dev. 2016, 30, 1357–1382. [Google Scholar] [CrossRef]
- Lakadamyali, M.; Cosma, M.P. Advanced microscopy methods for visualizing chromatin structure. FEBS Lett. 2015, 589, 3023–3030. [Google Scholar] [CrossRef] [Green Version]
- Lieberman-Aiden, E.; van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.S.; Huntley, M.H.; Durand, N.C.; Stamenova, E.K.; Bochkov, I.D.; Robinson, J.T.; Sanborn, A.L.; Machol, I.; Omer, A.D.; Lander, E.S.; et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 2014, 159, 1665–1680. [Google Scholar] [CrossRef] [PubMed]
- Sati, S.; Cavalli, G. Chromosome conformation capture technologies and their impact in understanding genome function. Chromosoma 2017, 126, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Zheng, H.; Huang, B.; Ma, R.; Wu, J.; Zhang, X.; He, J.; Xiang, Y.; Wang, Q.; Li, Y.; et al. Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature 2017, 547, 232–235. [Google Scholar] [CrossRef] [PubMed]
- Ke, Y.; Xu, Y.; Chen, X.; Feng, S.; Liu, Z.; Sun, Y.; Yao, X.; Li, F.; Zhu, W.; Gao, L.; et al. 3D Chromatin Structures of Mature Gametes and Structural Reprogramming during Mammalian Embryogenesis. Cell 2017, 170, 367–381.e20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diaz, N.; Kruse, K.; Erdmann, T.; Staiger, A.M.; Ott, G.; Lenz, G.; Vaquerizas, J.M. Chromatin conformation analysis of primary patient tissue using a low input Hi-C method. Nat. Commun. 2018, 9, 4938. [Google Scholar] [CrossRef] [Green Version]
- Nagano, T.; Lubling, Y.; Stevens, T.J.; Schoenfelder, S.; Yaffe, E.; Dean, W.; Laue, E.D.; Tanay, A.; Fraser, P. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 2013, 502, 59–64. [Google Scholar] [CrossRef] [Green Version]
- Nagano, T.; Lubling, Y.; Yaffe, E.; Wingett, S.W.; Dean, W.; Tanay, A.; Fraser, P. Single-cell Hi-C for genome-wide detection of chromatin interactions that occur simultaneously in a single cell. Nat. Protoc. 2015, 10, 1986–2003. [Google Scholar] [CrossRef]
- Ramani, V.; Deng, X.; Qiu, R.; Gunderson, K.L.; Steemers, F.J.; Disteche, C.M.; Noble, W.S.; Duan, Z.; Shendure, J. Massively multiplex single-cell Hi-C. Nat. Methods 2017, 14, 263–266. [Google Scholar] [CrossRef] [Green Version]
- Ramani, V.; Deng, X.; Qiu, R.; Lee, C.; Disteche, C.M.; Noble, W.S.; Duan, Z.; Shendure, J. Sci-Hi-C: A single-cell Hi-C method for mapping 3D genome organization in large number of single cells. bioRxiv 2019. [Google Scholar] [CrossRef]
- Stevens, T.J.; Lando, D.; Basu, S.; Atkinson, L.P.; Cao, Y.; Lee, S.F.; Leeb, M.; Wohlfahrt, K.J.; Boucher, W.; O’Shaughnessy-Kirwan, A.; et al. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature 2017, 544, 59–64. [Google Scholar] [CrossRef] [Green Version]
- Flyamer, I.M.; Gassler, J.; Imakaev, M.; Brandao, H.B.; Ulianov, S.V.; Abdennur, N.; Razin, S.V.; Mirny, L.A.; Tachibana-Konwalski, K. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature 2017, 544, 110–114. [Google Scholar] [CrossRef] [Green Version]
- Nagano, T.; Lubling, Y.; Varnai, C.; Dudley, C.; Leung, W.; Baran, Y.; Mendelson Cohen, N.; Wingett, S.; Fraser, P.; Tanay, A. Cell-cycle dynamics of chromosomal organization at single-cell resolution. Nature 2017, 547, 61–67. [Google Scholar] [CrossRef] [Green Version]
- Tan, L.; Xing, D.; Chang, C.H.; Li, H.; Xie, X.S. Three-dimensional genome structures of single diploid human cells. Science 2018, 361, 924–928. [Google Scholar] [CrossRef] [Green Version]
- Lando, D.; Stevens, T.J.; Basu, S.; Laue, E.D. Calculation of 3D genome structures for comparison of chromosome conformation capture experiments with microscopy: An evaluation of single-cell Hi-C protocols. Nucleus 2018, 9, 190–201. [Google Scholar] [CrossRef] [Green Version]
- Dixon, J.R.; Selvaraj, S.; Yue, F.; Kim, A.; Li, Y.; Shen, Y.; Hu, M.; Liu, J.S.; Ren, B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012, 485, 376–380. [Google Scholar] [CrossRef] [Green Version]
- Yaffe, E.; Tanay, A. Probabilistic modeling of Hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture. Nat. Genet. 2011, 43, 1059–1065. [Google Scholar] [CrossRef]
- Li, R.; Liu, Y.; Hou, Y.; Gan, J.; Wu, P.; Li, C. 3D genome and its disorganization in diseases. Cell Biol. Toxicol. 2018, 34, 351–365. [Google Scholar] [CrossRef]
- Forcato, M.; Nicoletti, C.; Pal, K.; Livi, C.M.; Ferrari, F.; Bicciato, S. Comparison of computational methods for Hi-C data analysis. Nat. Methods 2017, 14, 679–685. [Google Scholar] [CrossRef]
- Chakraborty, A.; Ay, F. The role of 3D genome organization in disease: From compartments to single nucleotides. Semin. Cell Dev. Biol. 2019, 90, 104–113. [Google Scholar] [CrossRef]
- Nozaki, T.; Imai, R.; Tanbo, M.; Nagashima, R.; Tamura, S.; Tani, T.; Joti, Y.; Tomita, M.; Hibino, K.; Kanemaki, M.T.; et al. Dynamic Organization of Chromatin Domains Revealed by Super-Resolution Live-Cell Imaging. Mol. Cell. 2017, 67, 282–293.e7. [Google Scholar] [CrossRef] [Green Version]
- Fraser, J.; Ferrai, C.; Chiariello, A.M.; Schueler, M.; Rito, T.; Laudanno, G.; Barbieri, M.; Moore, B.L.; Kraemer, D.C.; Aitken, S.; et al. Hierarchical folding and reorganization of chromosomes are linked to transcriptional changes in cellular differentiation. Mol. Syst. Biol. 2015, 11, 852. [Google Scholar] [CrossRef]
- Li, L.; Lyu, X.; Hou, C.; Takenaka, N.; Nguyen, H.Q.; Ong, C.T.; Cubenas-Potts, C.; Hu, M.; Lei, E.P.; Bosco, G.; et al. Widespread rearrangement of 3D chromatin organization underlies polycomb-mediated stress-induced silencing. Mol. Cell 2015, 58, 216–231. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, H.; Zhang, Y.; Du, Z.; Si, W.; Fan, S.; Qin, D.; Wang, M.; Duan, Y.; Li, L.; et al. Reprogramming of Meiotic Chromatin Architecture during Spermatogenesis. Mol. Cell 2019, 73, 547–561.e6. [Google Scholar] [CrossRef] [Green Version]
- Jung, Y.H.; Kremsky, I.; Gold, H.B.; Rowley, M.J.; Punyawai, K.; Buonanotte, A.; Lyu, X.; Bixler, B.J.; Chan, A.W.S.; Corces, V.G. Maintenance of CTCF—and Transcription Factor-Mediated Interactions from the Gametes to the Early Mouse Embryo. Mol. Cell 2019. [Google Scholar] [CrossRef]
- Jung, Y.H.; Sauria, M.E.G.; Lyu, X.; Cheema, M.S.; Ausio, J.; Taylor, J.; Corces, V.G. Chromatin States in Mouse Sperm Correlate with Embryonic and Adult Regulatory Landscapes. Cell Rep. 2017, 18, 1366–1382. [Google Scholar] [CrossRef]
- Von Stetina, J.R.; Orr-Weaver, T.L. Developmental control of oocyte maturation and egg activation in metazoan models. Cold Spring Harb. Perspect. Biol. 2011, 3, a005553. [Google Scholar] [CrossRef]
- Naumova, N.; Imakaev, M.; Fudenberg, G.; Zhan, Y.; Lajoie, B.R.; Mirny, L.A.; Dekker, J. Organization of the mitotic chromosome. Science 2013, 342, 948–953. [Google Scholar] [CrossRef]
- Gibcus, J.H.; Samejima, K.; Goloborodko, A.; Samejima, I.; Naumova, N.; Nuebler, J.; Kanemaki, M.T.; Xie, L.; Paulson, J.R.; Earnshaw, W.C.; et al. A pathway for mitotic chromosome formation. Science 2018, 359. [Google Scholar] [CrossRef] [Green Version]
- Gassler, J.; Brandao, H.B.; Imakaev, M.; Flyamer, I.M.; Ladstatter, S.; Bickmore, W.A.; Peters, J.M.; Mirny, L.A.; Tachibana, K. A mechanism of cohesin-dependent loop extrusion organizes zygotic genome architecture. EMBO J. 2017, 36, 3600–3618. [Google Scholar] [CrossRef]
- Kaaij, L.J.T.; van der Weide, R.H.; Ketting, R.F.; de Wit, E. Systemic Loss and Gain of Chromatin Architecture throughout Zebrafish Development. Cell Rep. 2018, 24, 1–10.e4. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, R.; Motai, Y.; Kumagai, M.; Nishiyama, H.; Durand, N.C.; Kondo, K.; Kondo, T.; Tsukahara, T.; Shimada, A.; Aiden, E.L.; et al. CTCF looping is established during gastrulation in medaka embryos. bioRxiv 2018. [Google Scholar] [CrossRef]
- Hug, C.B.; Grimaldi, A.G.; Kruse, K.; Vaquerizas, J.M. Chromatin Architecture Emerges during Zygotic Genome Activation Independent of Transcription. Cell 2017, 169, 216–228.e19. [Google Scholar] [CrossRef] [Green Version]
- Ogiyama, Y.; Schuettengruber, B.; Papadopoulos, G.L.; Chang, J.M.; Cavalli, G. Polycomb-Dependent Chromatin Looping Contributes to Gene Silencing during Drosophila Development. Mol. Cell 2018, 71, 73–88.e5. [Google Scholar] [CrossRef] [Green Version]
- Sanborn, A.L.; Rao, S.S.; Huang, S.C.; Durand, N.C.; Huntley, M.H.; Jewett, A.I.; Bochkov, I.D.; Chinnappan, D.; Cutkosky, A.; Li, J.; et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl. Acad. Sci. USA 2015, 112, E6456–E6465. [Google Scholar] [CrossRef] [Green Version]
- Fudenberg, G.; Imakaev, M.; Lu, C.; Goloborodko, A.; Abdennur, N.; Mirny, L.A. Formation of Chromosomal Domains by Loop Extrusion. Cell Rep. 2016, 15, 2038–2049. [Google Scholar] [CrossRef] [Green Version]
- Weintraub, A.S.; Li, C.H.; Zamudio, A.V.; Sigova, A.A.; Hannett, N.M.; Day, D.S.; Abraham, B.J.; Cohen, M.A.; Nabet, B.; Buckley, D.L.; et al. YY1 Is a Structural Regulator of Enhancer-Promoter Loops. Cell 2017, 171, 1573–1588.e28. [Google Scholar] [CrossRef] [Green Version]
- Ye, B.Y.; Shen, W.L.; Wang, D.; Li, P.; Zhang, Z.; Shi, M.L.; Zhang, Y.; Zhang, F.X.; Zhao, Z.H. ZNF143 is involved in CTCF-mediated chromatin interactions by cooperation with cohesin and other partners. Mol. Biol. 2016, 50, 496–503. [Google Scholar] [CrossRef]
- Meier, M.; Grant, J.; Dowdle, A.; Thomas, A.; Gerton, J.; Collas, P.; O’Sullivan, J.M.; Horsfield, J.A. Cohesin facilitates zygotic genome activation in zebrafish. Development 2018, 145, dev156521. [Google Scholar] [CrossRef]
- Wang, Q.; Sun, Q.; Czajkowsky, D.M.; Shao, Z. Sub-kb Hi-C in D. melanogaster reveals conserved characteristics of TADs between insect and mammalian cells. Nat. Commun. 2018, 9, 188. [Google Scholar]
- McDaniel, S.L.; Gibson, T.J.; Schulz, K.N.; Fernandez Garcia, M.; Nevil, M.; Jain, S.U.; Lewis, P.W.; Zaret, K.S.; Harrison, M.M. Continued Activity of the Pioneer Factor Zelda Is Required to Drive Zygotic Genome Activation. Mol. Cell. 2019, 74, 185–195.e4. [Google Scholar] [CrossRef] [Green Version]
- Racko, D.; Benedetti, F.; Dorier, J.; Stasiak, A. Transcription-induced supercoiling as the driving force of chromatin loop extrusion during formation of TADs in interphase chromosomes. Nucleic Acids Res. 2018, 46, 1648–1660. [Google Scholar] [CrossRef]
- Busslinger, G.A.; Stocsits, R.R.; van der Lelij, P.; Axelsson, E.; Tedeschi, A.; Galjart, N.; Peters, J.M. Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl. Nature 2017, 544, 503–507. [Google Scholar] [CrossRef]
- Lengronne, A.; Katou, Y.; Mori, S.; Yokobayashi, S.; Kelly, G.P.; Itoh, T.; Watanabe, Y.; Shirahige, K.; Uhlmann, F. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 2004, 430, 573–578. [Google Scholar] [CrossRef]
- Heinz, S.; Texari, L.; Hayes, M.G.B.; Urbanowski, M.; Chang, M.W.; Givarkes, N.; Rialdi, A.; White, K.M.; Albrecht, R.A.; Pache, L.; et al. Transcription Elongation Can Affect Genome 3D Structure. Cell 2018, 174, 1522–1536.e22. [Google Scholar] [CrossRef] [Green Version]
- Rowley, M.J.; Nichols, M.H.; Lyu, X.; Ando-Kuri, M.; Rivera, I.S.M.; Hermetz, K.; Wang, P.; Ruan, Y.; Corces, V.G. Evolutionarily Conserved Principles Predict 3D Chromatin Organization. Mol. Cell. 2017, 67, 837–852.e7. [Google Scholar] [CrossRef] [Green Version]
- De Koning, A.P.; Gu, W.; Castoe, T.A.; Batzer, M.A.; Pollock, D.D. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011, 7, e1002384. [Google Scholar] [CrossRef]
- Erwin, J.A.; Marchetto, M.C.; Gage, F.H. Mobile DNA elements in the generation of diversity and complexity in the brain. Nat. Rev. Neurosci. 2014, 15, 497–506. [Google Scholar] [CrossRef]
- Larsen, P.A. Transposable elements and the multidimensional genome. Chromosome Res. 2018, 26, 1–3. [Google Scholar] [CrossRef]
- Cao, Y.; Chen, G.; Wu, G.; Zhang, X.; McDermott, J.; Chen, X.; Xu, C.; Jiang, Q.; Chen, Z.; Zeng, Y.; et al. Widespread roles of enhancer-like transposable elements in cell identity and long-range genomic interactions. Genome Res. 2019, 29, 40–52. [Google Scholar] [CrossRef]
- Chuong, E.B.; Elde, N.C.; Feschotte, C. Regulatory activities of transposable elements: From conflicts to benefits. Nat. Rev. Genet. 2017, 18, 71–86. [Google Scholar] [CrossRef]
- Raviram, R.; Rocha, P.P.; Luo, V.M.; Swanzey, E.; Miraldi, E.R.; Chuong, E.B.; Feschotte, C.; Bonneau, R.; Skok, J.A. Analysis of 3D genomic interactions identifies candidate host genes that transposable elements potentially regulate. Genome Biol. 2018, 19, 216. [Google Scholar] [CrossRef]
- Darrow, E.M.; Huntley, M.H.; Dudchenko, O.; Stamenova, E.K.; Durand, N.C.; Sun, Z.; Huang, S.C.; Sanborn, A.L.; Machol, I.; Shamim, M.; et al. Deletion of DXZ4 on the human inactive X chromosome alters higher-order genome architecture. Proc. Natl. Acad. Sci. USA 2016, 113, E4504–E4512. [Google Scholar] [CrossRef]
- Winter, D.J.; Ganley, A.R.D.; Young, C.A.; Liachko, I.; Schardl, C.L.; Dupont, P.Y.; Berry, D.; Ram, A.; Scott, B.; Cox, M.P. Repeat elements organise 3D genome structure and mediate transcription in the filamentous fungus Epichloe festucae. PLoS Genet. 2018, 14, e1007467. [Google Scholar] [CrossRef]
- Wang, J.; Vicente-Garcia, C.; Seruggia, D.; Molto, E.; Fernandez-Minan, A.; Neto, A.; Lee, E.; Gomez-Skarmeta, J.L.; Montoliu, L.; Lunyak, V.V.; et al. MIR retrotransposon sequences provide insulators to the human genome. Proc. Natl. Acad. Sci. USA 2015, 112, E4428–E4437. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, D.; Schwalie, P.C.; Wilson, M.D.; Ballester, B.; Goncalves, A.; Kutter, C.; Brown, G.D.; Marshall, A.; Flicek, P.; Odom, D.T. Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell 2012, 148, 335–348. [Google Scholar] [CrossRef]
- Rodriguez-Terrones, D.; Torres-Padilla, M.E. Nimble and Ready to Mingle: Transposon Outbursts of Early Development. Trends Genet. TIG 2018, 34, 806–820. [Google Scholar] [CrossRef]
- Wu, J.; Huang, B.; Chen, H.; Yin, Q.; Liu, Y.; Xiang, Y.; Zhang, B.; Liu, B.; Wang, Q.; Xia, W.; et al. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 2016, 534, 652–657. [Google Scholar] [CrossRef]
- Kruse, K.; Díaz, N.; Enriquez-Gasca, R.; Gaume, X.; Torres-Padilla, M.-E.; Vaquerizas, J.M. Transposable elements drive reorganisation of 3D chromatin during early embryogenesis. bioRxiv 2019. [Google Scholar] [CrossRef]
- Schwarzer, W.; Abdennur, N.; Goloborodko, A.; Pekowska, A.; Fudenberg, G.; Loe-Mie, Y.; Fonseca, N.A.; Huber, W.; Haering, C.H.; Mirny, L.; et al. Two independent modes of chromatin organization revealed by cohesin removal. Nature 2017, 551, 51–56. [Google Scholar] [CrossRef] [Green Version]
- Rao, S.S.P.; Huang, S.C.; Glenn St Hilaire, B.; Engreitz, J.M.; Perez, E.M.; Kieffer-Kwon, K.R.; Sanborn, A.L.; Johnstone, S.E.; Bascom, G.D.; Bochkov, I.D.; et al. Cohesin Loss Eliminates All Loop Domains. Cell 2017, 171, 305–320.e24. [Google Scholar] [CrossRef] [Green Version]
- Nora, E.P.; Goloborodko, A.; Valton, A.L.; Gibcus, J.H.; Uebersohn, A.; Abdennur, N.; Dekker, J.; Mirny, L.A.; Bruneau, B.G. Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell 2017, 169, 930–944.e22. [Google Scholar] [CrossRef] [Green Version]
- Haarhuis, J.H.I.; van der Weide, R.H.; Blomen, V.A.; Yanez-Cuna, J.O.; Amendola, M.; van Ruiten, M.S.; Krijger, P.H.L.; Teunissen, H.; Medema, R.H.; van Steensel, B.; et al. The Cohesin Release Factor WAPL Restricts Chromatin Loop Extension. Cell 2017, 169, 693–707.e14. [Google Scholar] [CrossRef] [Green Version]
- Erdel, F.; Rippe, K. Formation of Chromatin Subcompartments by Phase Separation. Biophys. J. 2018, 114, 2262–2270. [Google Scholar] [CrossRef] [Green Version]
- Larson, A.G.; Narlikar, G.J. The Role of Phase Separation in Heterochromatin Formation, Function, and Regulation. Biochemistry 2018, 57, 2540–2548. [Google Scholar] [CrossRef]
- Sabari, B.R.; Dall’Agnese, A.; Boija, A.; Klein, I.A.; Coffey, E.L.; Shrinivas, K.; Abraham, B.J.; Hannett, N.M.; Zamudio, A.V.; Manteiga, J.C.; et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 2018, 361. [Google Scholar] [CrossRef]
- Nuebler, J.; Fudenberg, G.; Imakaev, M.; Abdennur, N.; Mirny, L.A. Chromatin organization by an interplay of loop extrusion and compartmental segregation. Proc. Natl. Acad. Sci. USA 2018, 115, E6697–E6706. [Google Scholar] [CrossRef] [Green Version]
| Methods | Full Name | Procedure | Characteristics |
|---|---|---|---|
| Hi-C [11] (in situ Hi-C) [12] | Chromosome conformation capture by high-throughput sequencing | Crosslinking, restriction enzyme digestion, end filling with biotinylated dNTP and proximity ligation (ligation performed in intact nuclei in an in situ Hi-C), reverse crosslinking, sonication and streptavidin enrichment, and sequencing. | Widely used genome-wide method |
| Single-cell Hi-C [17,18] | Single-cell Hi-C | Similar to in situ Hi-C, individual nuclei selected using microscopy after proximity ligation. Remaining steps done in single cells separately. Sonication replaced with a second restriction enzyme to fragment ligation products. | The first single-cell chromatin structure method, relatively low throughput |
| Sci-Hi-C [19,20] | Single-cell combinatorial indexed Hi-C | Crosslinking, restriction digestion, distributed to 96 wells and barcoded bridge-adaptor ligation, nuclei pooled and proximity ligation, redistribution to 96 wells and barcoded sequencing-adaptor ligation, sequencing. | A larger number of single cells with fewer interactions per cell |
| Single-cell Hi-C [21] | Single-cell Hi-C | Crosslinking, single nuclei sorting with FACS, nuclei imaging, overlaid nuclei with low melting agarose. Remaining steps similar to in situ Hi-C but done in single cells. | Combination of imaging with determination of genome structure |
| Sn Hi-C [22] | Single-nucleus Hi-C | Similar to in situ Hi-C but omitting biotin incorporation. Single nuclei sorted by FACS after proximity ligation and then whole genome amplification was done to single nuclei. | More contacts per single cell |
| Improved multiplexed single-cell Hi-C [23] | Improved multiplexed single-cell Hi-C | Improved from [17], with flow cytometry sorting, Tn5 transposase library preparation, and an automation scheme. | Moderate contacts per single cell |
| Dip-C [24] | Single-cell Hi-C of diploid cells | Similar to Sn Hi-C [22]. Whole-genome amplification done with multiplex end-tagging amplification. | Distinguishes two haplotypes of each chromosome |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, F.; An, Z.; Zhang, Z. The Dynamic 3D Genome in Gametogenesis and Early Embryonic Development. Cells 2019, 8, 788. https://doi.org/10.3390/cells8080788
Li F, An Z, Zhang Z. The Dynamic 3D Genome in Gametogenesis and Early Embryonic Development. Cells. 2019; 8(8):788. https://doi.org/10.3390/cells8080788
Chicago/Turabian StyleLi, Feifei, Ziyang An, and Zhihua Zhang. 2019. "The Dynamic 3D Genome in Gametogenesis and Early Embryonic Development" Cells 8, no. 8: 788. https://doi.org/10.3390/cells8080788
APA StyleLi, F., An, Z., & Zhang, Z. (2019). The Dynamic 3D Genome in Gametogenesis and Early Embryonic Development. Cells, 8(8), 788. https://doi.org/10.3390/cells8080788




