Host–Virus Interactions in Japanese Encephalitis Virus
Abstract
1. Introduction
2. Materials and Methods
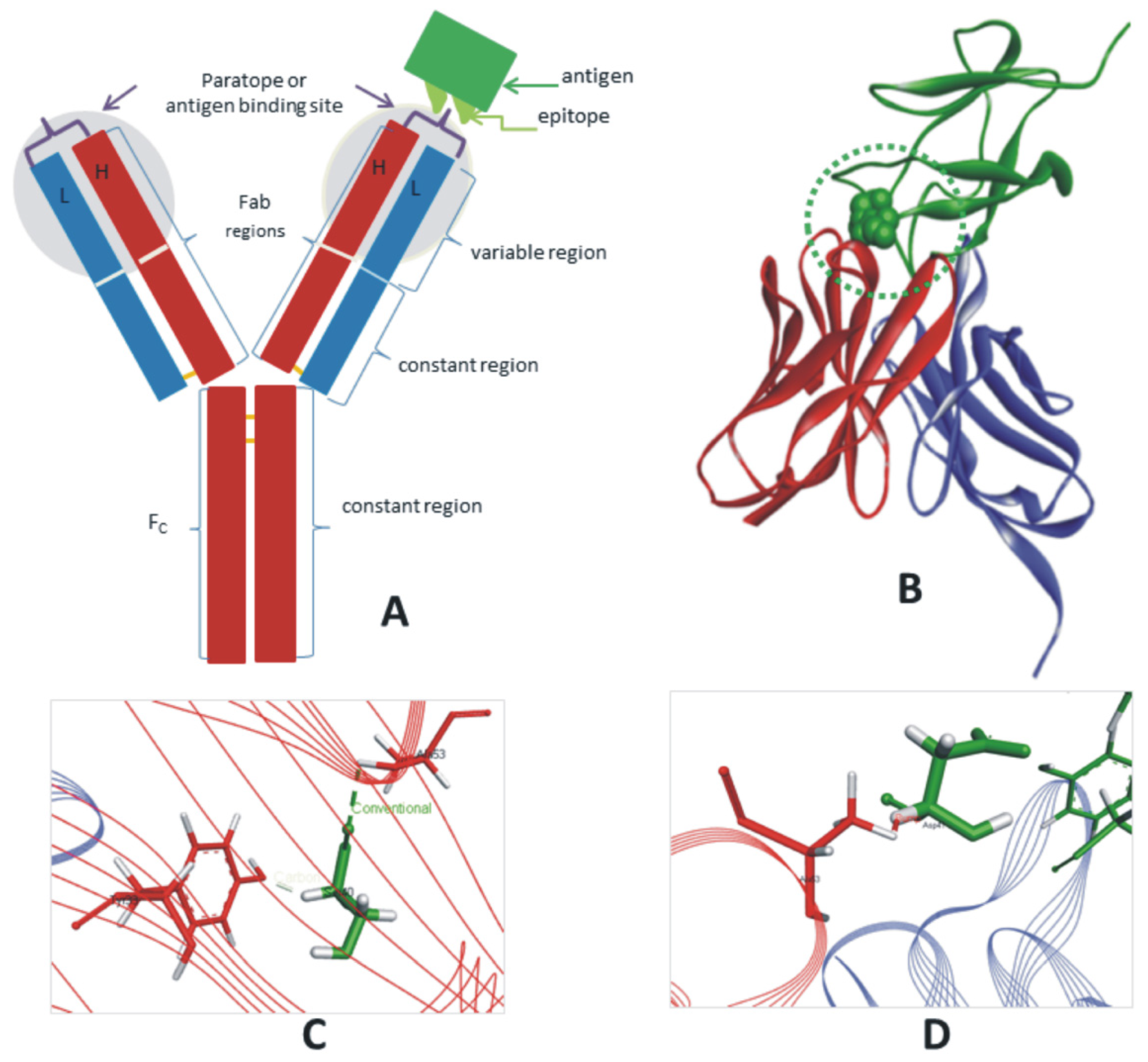
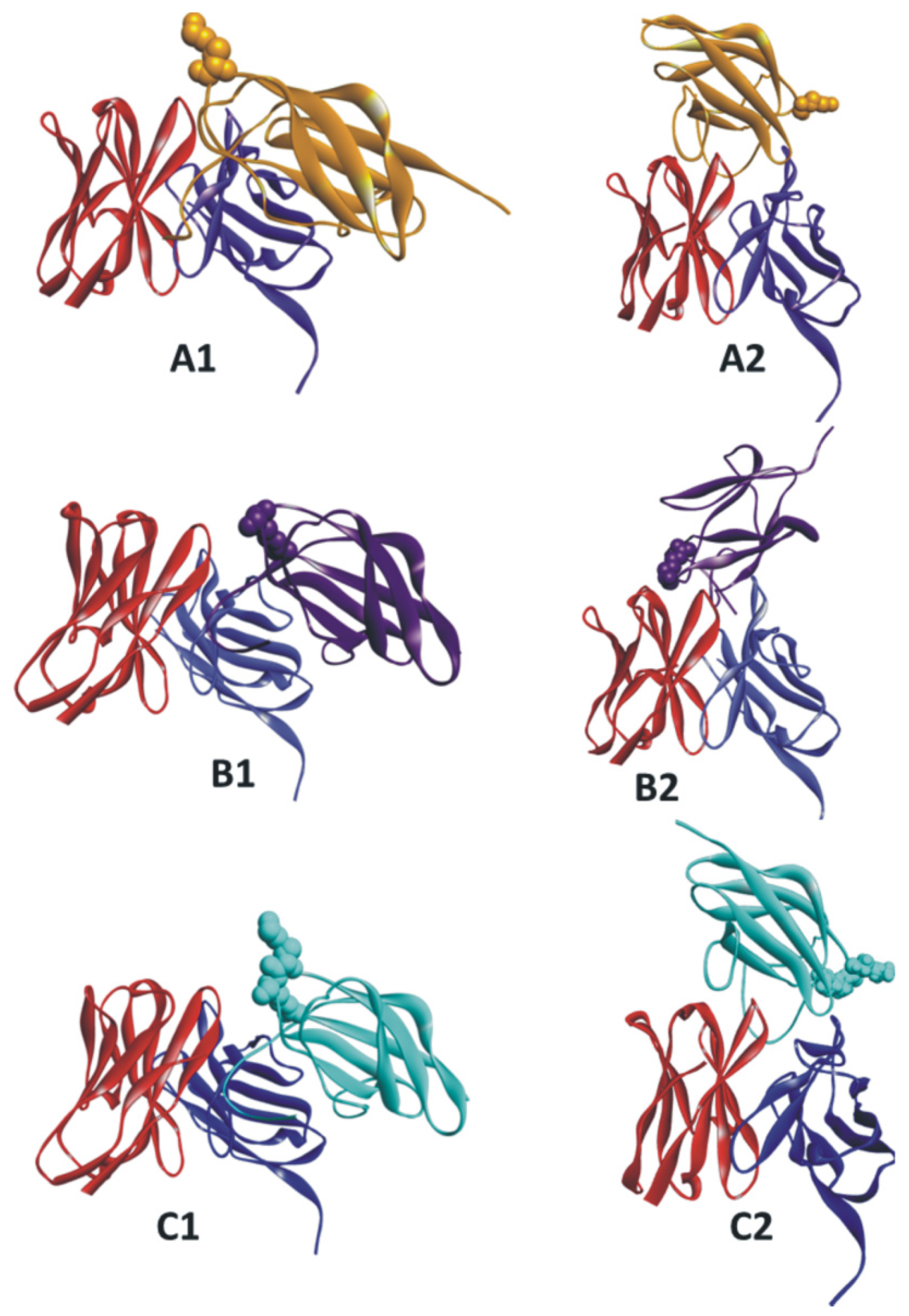
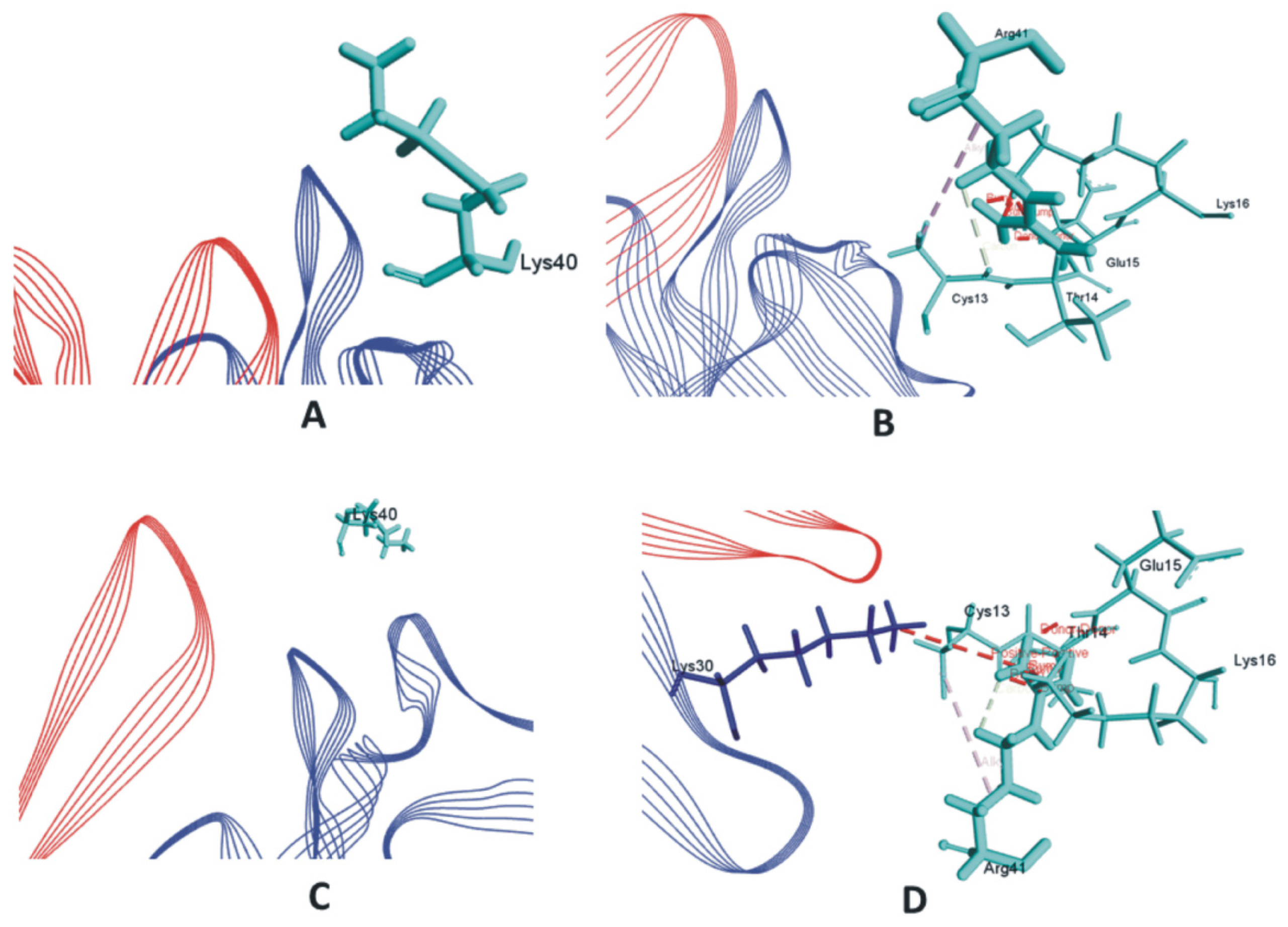
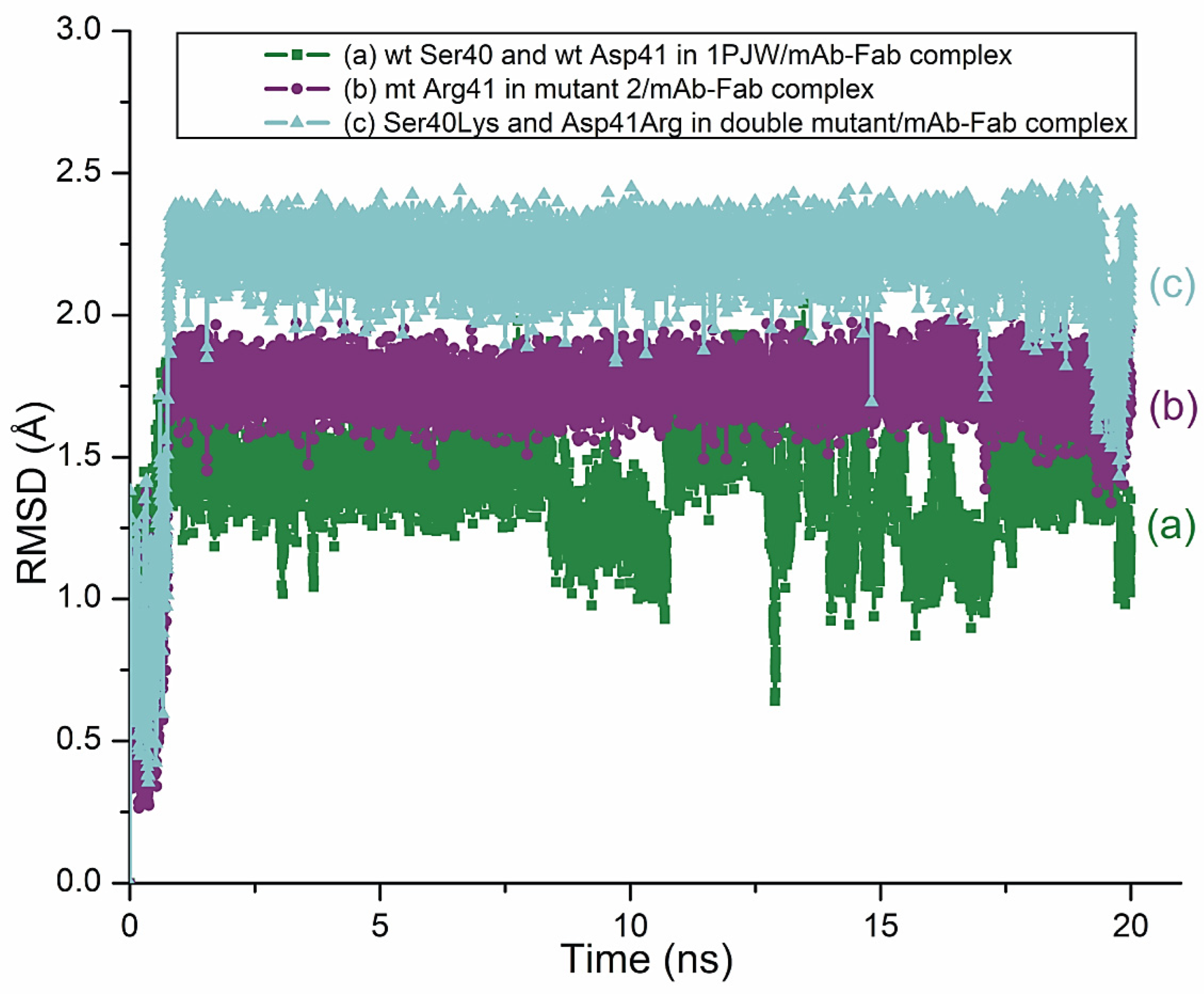
3. Results and Discussions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Al-Abdulla, O.; Kallström, A.; Valderrama, C.; Kauhanen, J. Simulation of the Progression of the COVID-19 Outbreak in Northwest Syria Using a Basic and Adjusted SIR Model. Zoonotic Dis. 2022, 2, 44–58. [Google Scholar] [CrossRef]
- Moczydlowski, E. Ion Channels. In Methods in Enzymology; Rudy, B., Iverson, L., Eds.; Elsevier Inc.: Amsterdam, The Netherlands, 1992; Volume 207, pp. 791–806. [Google Scholar]
- Head, J.F.; Swamy, N.; Ray, R. Crystal structure of the complex between actin and human vitamin D-binding protein at 2.5 A resolution. Biochemistry 2002, 41, 9015–9020. [Google Scholar] [CrossRef] [PubMed]
- Wikel, S. Ticks and tick-borne pathogens at the cutaneous interface: Host defenses, tick countermeasures, and a suitable environment for pathogen establishment. Front. Microbiol. 2013, 4, 337. [Google Scholar] [CrossRef] [PubMed]
- Khayat, R.; Russell, R.S. It is all about the Structure. Viral Immunol. 2021, 34, 1–2. [Google Scholar] [CrossRef]
- Beck, C.; Hamel, R.; Dumarest, M.; Gonzalez, G.; Lecollinet, S. Chapter 42—Nonhuman occurrence of Zika virus infection: Implications for control. In Zika Virus Biology, Transmission, and Pathology; Martin, C.R., Martin, C.J.H., Preedy, V.R., Rajendram, R., Eds.; Academic Press: Cambridge, MA, USA, 2021; pp. 453–465. [Google Scholar]
- Cáceres, C.J.; Rajao, D.S.; Perez, D.R. Airborne Transmission of Avian Origin H9N2 Influenza A Viruses in Mammals. Viruses 2021, 13, 1919. [Google Scholar] [CrossRef]
- Rupprecht, C.E.; Van Pelt, L.I.; Davis, A.D.; Chipman, R.B.; Bergman, D.L. Use of a Direct, Rapid Immunohistochemical Test for Diagnosis of Rabies Virus in Bats. Zoonotic Dis. 2022, 2, 1–8. [Google Scholar] [CrossRef]
- Talactac, M.R.; Hernandez, E.P.; Hatta, T.; Yoshii, K.; Kusakisako, K.; Tsuji, N.; Tanaka, T. The antiviral immunity of ticks against transmitted viral pathogens. Dev. Comp. Immunol. 2021, 119, 104012. [Google Scholar] [CrossRef]
- Hai, R.; García-Sastre, A.; Swayne, D.E.; Palese, P. A reassortment-incompetent live attenuated influenza virus vaccine for protection against pandemic virus strains. J. Virol. 2011, 85, 6832–6843. [Google Scholar] [CrossRef]
- Yamayoshi, S.; Kawaoka, Y. Emergence of SARS-CoV-2 and its outlook. Glob. Health Med. 2020, 2, 1–2. [Google Scholar] [CrossRef]
- CDC. Centers for Disease Control and Prevention (CDC), Japanese Encephalitis. Available online: https://www.cdc.gov/japaneseencephalitis/ (accessed on 1 July 2022).
- WHO. World Health Organization, Japanese Encephalitis. Available online: https://www.who.int/en/news-room/fact-sheets/detail/japanese-encephalitis (accessed on 21 May 2022).
- Lin, C.-W.; Wu, S.-C. A functional epitope determinant on domain III of the Japanese encephalitis virus envelope protein interacted with neutralizing-antibody combining sites. J. Virol. 2003, 77, 2600–2606. [Google Scholar] [CrossRef][Green Version]
- Luca, V.C.; AbiMansour, J.; Nelson, C.A.; Fremont, D.H. Crystal structure of the Japanese encephalitis virus envelope protein. J. Virol. 2012, 86, 2337–2346. [Google Scholar] [CrossRef] [PubMed]
- Roy, U. Structural and molecular analyses of functional epitopes and escape mutants in Japanese encephalitis virus envelope protein domain III. Immunol. Res. 2020, 68, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Rey, F.A.; Stiasny, K.; Vaney, M.C.; Dellarole, M.; Heinz, F.X. The bright and the dark side of human antibody responses to flaviviruses: Lessons for vaccine design. EMBO Rep. 2018, 19, 206–224. [Google Scholar] [CrossRef] [PubMed]
- Goncalvez, A.P.; Chien, C.H.; Tubthong, K.; Gorshkova, I.; Roll, C.; Donau, O.; Schuck, P.; Yoksan, S.; Wang, S.D.; Purcell, R.H.; et al. Humanized monoclonal antibodies derived from chimpanzee Fabs protect against Japanese encephalitis virus in vitro and in vivo. J. Virol. 2008, 82, 7009–7021. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.P.; Wu, C.W.; Tsao, Y.P.; Kuo, T.W.; Lou, Y.C.; Lin, C.W.; Wu, S.C.; Cheng, J.W. Structural basis of a flavivirus recognized by its neutralizing antibody: Solution structure of the domain III of the Japanese encephalitis virus envelope protein. J. Biol. Chem. 2003, 278, 46007–46013. [Google Scholar] [CrossRef]
- Tovchigrechko, A.; Vakser, I.A. GRAMM-X public web server for protein-protein docking. Nucleic Acids Res. 2006, 34, W310–W314. [Google Scholar] [CrossRef]
- Tovchigrechko, A.; Vakser, I.A. Development and testing of an automated approach to protein docking. Proteins 2005, 60, 296–301. [Google Scholar] [CrossRef]
- Duhovny, D.; Nussinov, R.; Wolfson, H.J. Efficient Unbound Docking of Rigid Molecules. In Proceedings of the 2nd Workshop on Algorithms in Bioinformatics (WABI), Rome, Italy, 17–21 September 2002; Gusfield, E., Ed.; Springer: Berlin/Heidelberg, Germany, 2002; Volume 2452, pp. 185–200. [Google Scholar]
- Schneidman-Duhovny, D.; Inbar, Y.; Nussinov, R.; Wolfson, H.J. PatchDock and SymmDock: Servers for rigid and symmetric docking. Nucleic Acids Res. 2005, 33, W363–W367. [Google Scholar] [CrossRef]
- Andrusier, N.; Nussinov, R.; Wolfson, H.J. FireDock: Fast interaction refinement in molecular docking. Proteins 2007, 69, 139–159. [Google Scholar] [CrossRef] [PubMed]
- Mashiach, E.; Schneidman-Duhovny, D.; Andrusier, N.; Nussinov, R.; Wolfson, H.J. FireDock: A web server for fast interaction refinement in molecular docking. Nucl. Acids Res. 2008, 36, W229–W232. [Google Scholar] [CrossRef]
- Tormo, J.; Blaas, D.; Parry, N.R.; Rowlands, D.; Stuart, D.; Fita, I. Crystal structure of a human rhinovirus neutralizing antibody complexed with a peptide derived from viral capsid protein VP2. Embo J. 1994, 13, 2247–2256. [Google Scholar] [CrossRef]
- Phillips, J.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.; Kale, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef]
- Ribeiro, J.V.; Bernardi, R.C.; Rudack, T.; Stone, J.E.; Phillips, J.C.; Freddolino, P.L.; Schulten, K. QwikMD -integrative molecular dynamics toolkit for novices and experts. Sci. Rep. 2016, 6, 26536. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Tanner, D.E.; Chan, K.Y.; Phillips, J.C.; Schulten, K. Parallel Generalized Born implicit solvent calculations with NAMD. J. Chem. Theory Comput. 2011, 7, 3635–3642. [Google Scholar] [CrossRef]
- MacKerell, A.D.; Bashford, D.; Bellott, M.; Dunbrack, R.L.; Evanseck, J.D.; Field, M.J.; Fischer, S.; Gao, J.; Guo, H.; Ha, S.; et al. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J. Phys. Chem. B 1998, 102, 3586–3616. [Google Scholar] [CrossRef]
- Roy, U. Structural characterizations of the fas Receptor and the fas-associated protein with death domain interactions. Protein J. 2016, 35, 51–60. [Google Scholar] [CrossRef]
- Roy, U. Structural modeling of tumor necrosis factor: A protein of immunological importance. Biotechnol. Appl. Biochem. 2017, 64, 454–463. [Google Scholar] [CrossRef]
- Roy, U. Role of N501Y Mutation in SARS-CoV-2 Spike Protein Structure. Preprints 2021, 2021060238. [Google Scholar] [CrossRef]
- DSV; Discovery Studio Modeling Environment. Dassault Systèmes BIOVIA Discovery Studio Modeling Environment; Dassault Systèmes: San Diego, CA, USA, 2015. [Google Scholar]
- Chiou, S.S.; Fan, Y.C.; Crill, W.D.; Chang, R.Y.; Chang, G.J. Mutation analysis of the cross-reactive epitopes of Japanese encephalitis virus envelope glycoprotein. J. Gen. Virol. 2012, 93, 1185–1192. [Google Scholar] [CrossRef]
- Guex, N.; Peitsch, M.C. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis 1997, 18, 2714–2723. [Google Scholar] [CrossRef]
- Scott, W.R.P.; Hünenberger, P.H.; Tironi, I.G.; Mark, A.E.; Billeter, S.R.; Fennen, J.; Torda, A.E.; Huber, T.; Krüger, P.; van Gunsteren, W.F. The GROMOS Biomolecular Simulation Program Package. J. Phys. Chem. A 1999, 103, 3596–3607. [Google Scholar] [CrossRef]
- Yuan, S.; Ye, Z.W.; Liang, R.; Tang, K.; Zhang, A.J.; Lu, G.; Ong, C.P.; Man Poon, V.K.; Chan, C.C.; Mok, B.W.; et al. Pathogenicity, transmissibility, and fitness of SARS-CoV-2 Omicron in Syrian hamsters. Science (N. Y.) 2022, 377, 428–433. [Google Scholar] [CrossRef]
- Hashem, A.M.; Algaissi, A.; Almahboub, S.A.; Alfaleh, M.A.; Abujamel, T.S.; Alamri, S.S.; Alluhaybi, K.A.; Hobani, H.I.; AlHarbi, R.H.; Alsulaiman, R.M.; et al. Early Humoral Response Correlates with Disease Severity and Outcomes in COVID-19 Patients. Viruses 2020, 12, 1390. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roy, U. Host–Virus Interactions in Japanese Encephalitis Virus. Zoonotic Dis. 2022, 2, 117-125. https://doi.org/10.3390/zoonoticdis2030012
Roy U. Host–Virus Interactions in Japanese Encephalitis Virus. Zoonotic Diseases. 2022; 2(3):117-125. https://doi.org/10.3390/zoonoticdis2030012
Chicago/Turabian StyleRoy, Urmi. 2022. "Host–Virus Interactions in Japanese Encephalitis Virus" Zoonotic Diseases 2, no. 3: 117-125. https://doi.org/10.3390/zoonoticdis2030012
APA StyleRoy, U. (2022). Host–Virus Interactions in Japanese Encephalitis Virus. Zoonotic Diseases, 2(3), 117-125. https://doi.org/10.3390/zoonoticdis2030012





