Ascorbic Acid in Seeds, Priming and Beyond
Abstract
:1. Introduction
2. The AsA System
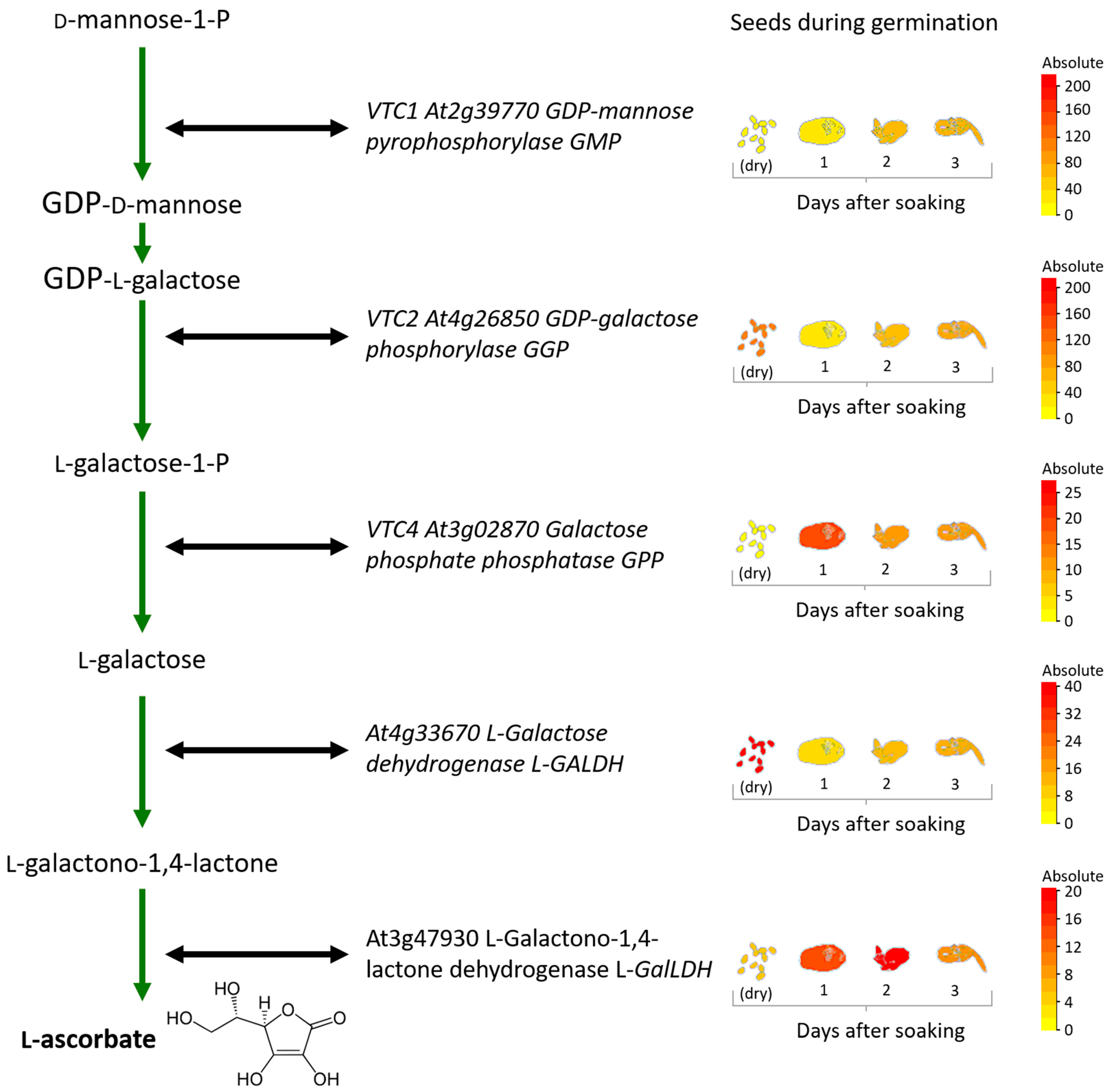
2.1. AsA Biosynthesis
2.2. AsA Transport and Intracellular Distribution
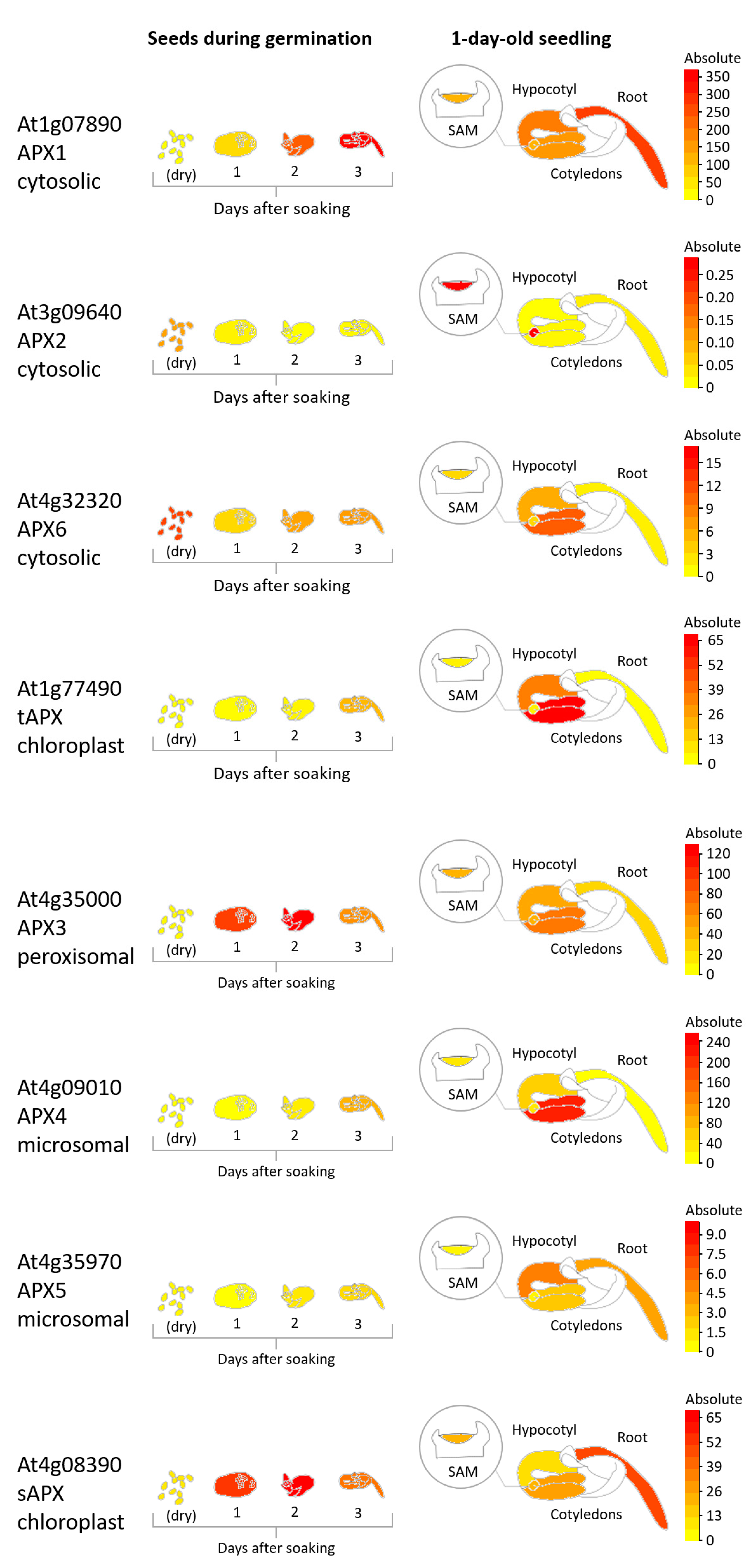
2.3. AsA Utilization
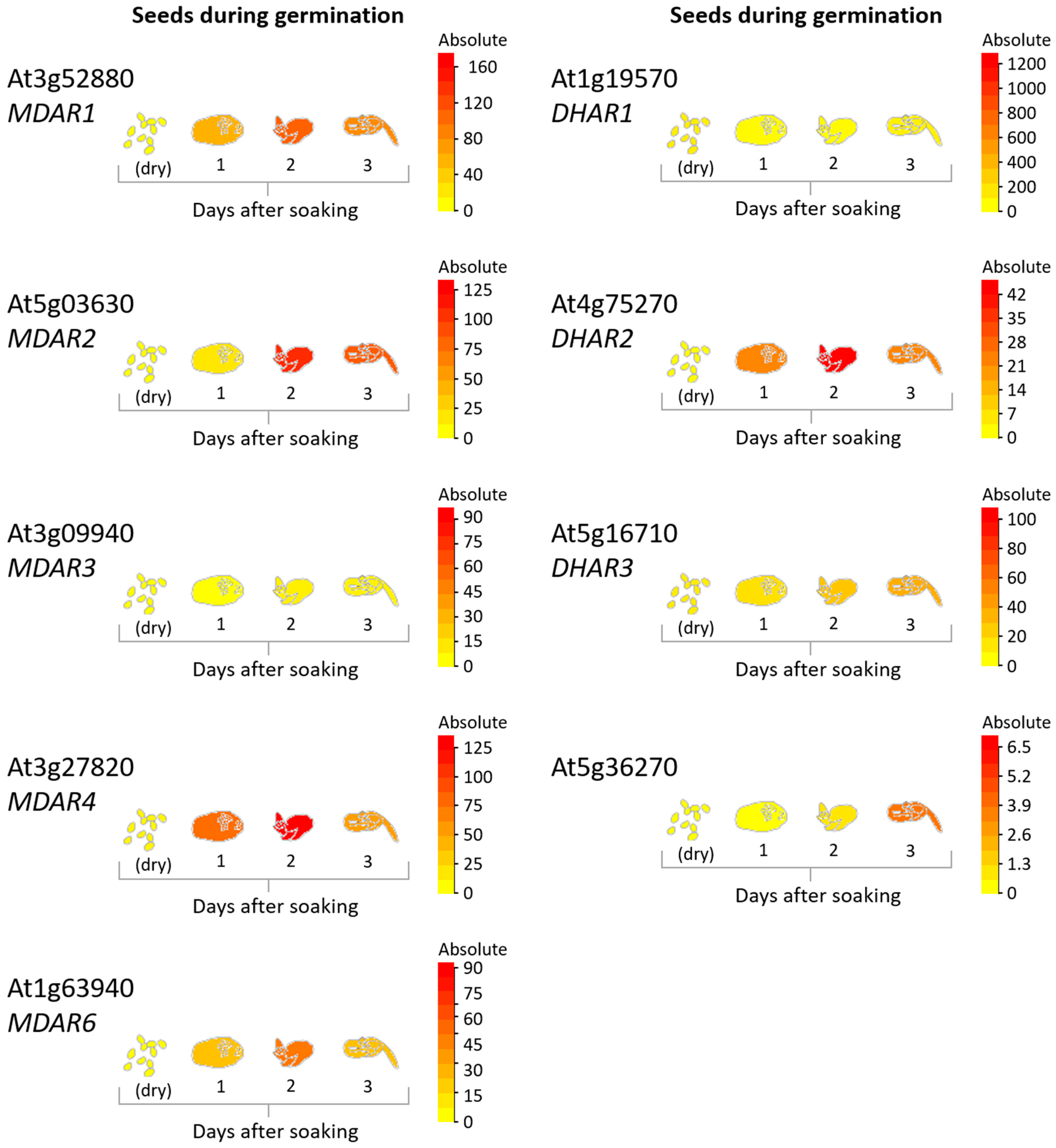
2.4. Recycling of AsA Oxidized Forms
2.5. Possible DHA Signaling and Further Catabolism
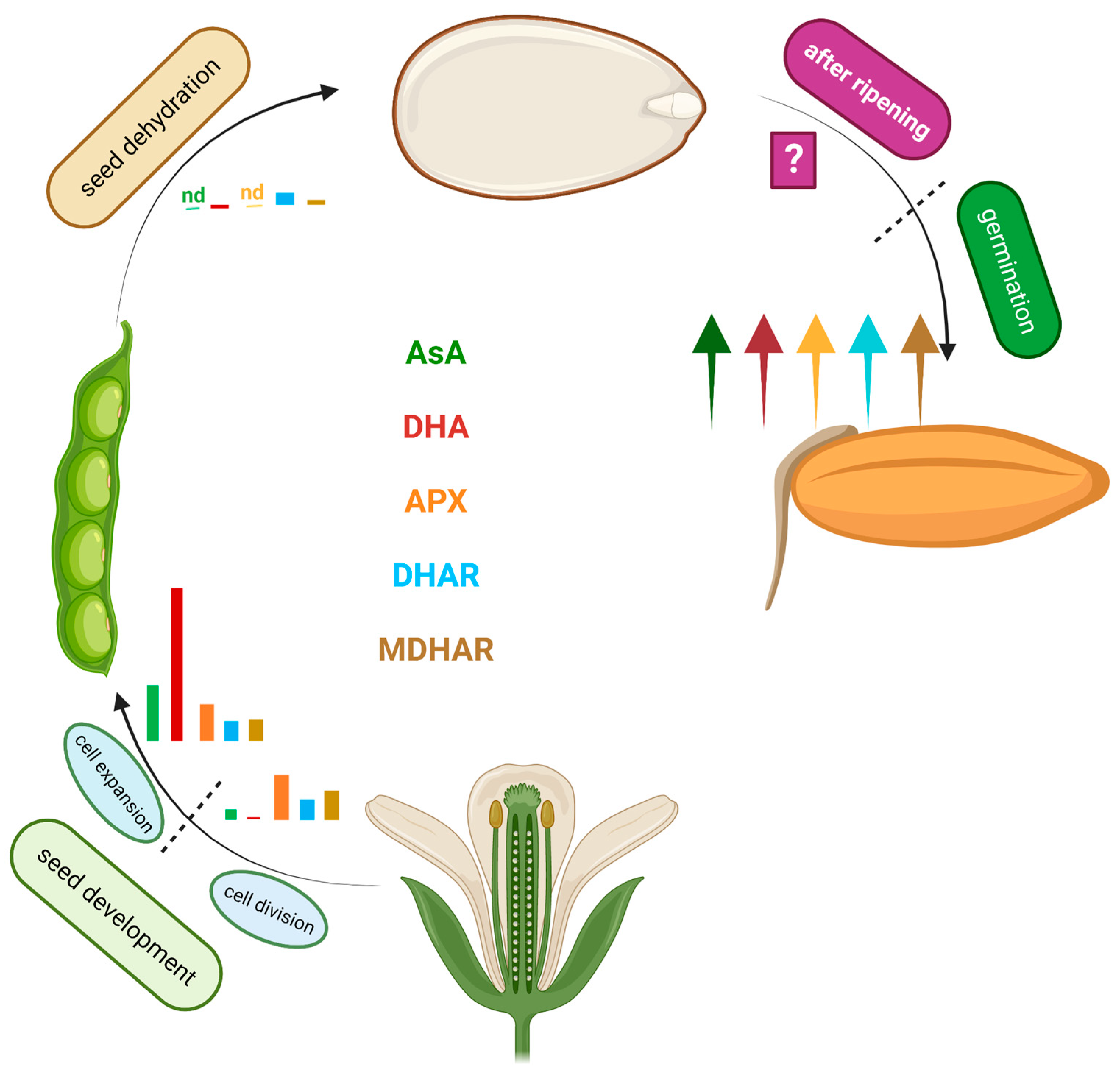
3. Dynamic Regulation of the AsA System during Seed Development
4. AsA in Seed Dehydration, Dormancy, and Germination: To Be or Not to Be (There)?
5. Priming with AsA
6. What’s Next?
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Linkies, A.; Graeber, K.; Knight, C.; Leubner-Metzger, G. The evolution of seeds. New Phytol. 2010, 186, 817–831. [Google Scholar] [CrossRef] [PubMed]
- Finch-Savage, W.E.; Leubner-Metzger, G. Seed dormancy and the control of germination. New Phytol. 2006, 171, 501–523. [Google Scholar] [CrossRef] [PubMed]
- Kravets, O.P.; Sokolova, D.O. Epigenetic mechanisms regulating seed germination rate. Cytol. Genet. 2017, 51, 346–351. [Google Scholar] [CrossRef]
- Smolikova, G.; Strygina, K.; Krylova, E.; Leonova, T.; Frolov, A.; Khlestkina, E.; Medvedev, S. Transition from Seeds to Seedlings: Hormonal and Epigenetic Aspects. Plants 2021, 10, 1884. [Google Scholar] [CrossRef]
- De Tullio, M.C.; Arrigoni, O. The ascorbic acid system in seeds: To protect and to serve. Seed Sci. Res. 2003, 13, 249–260. [Google Scholar] [CrossRef]
- Wheeler, G.L.; Jones, M.A.; Smirnoff, N. The biosynthetic pathway of vitamin C in higher plants. Nature 1998, 393, 365–369. [Google Scholar] [CrossRef]
- Lorence, A.; Chevone, B.I.; Mendes, P.; Nessler, C.L. myo-inositol oxygenase offers a possible entry point into plant ascorbate biosynthesis. Plant Physiol. 2004, 134, 1200–1205. [Google Scholar] [CrossRef]
- Agius, F.; González-Lamothe, R.; Caballero, J.L.; Muñoz-Blanco, J.; Botella, M.A.; Valpuesta, V. Engineering increased vitamin C levels in plants by overexpression of a D-galacturonic acid reductase. Nat. Biotechnol. 2003, 21, 177–181. [Google Scholar] [CrossRef]
- Terzaghi, M.; De Tullio, M.C. The perils of planning strategies to increase vitamin C content in plants: Beyond the hype. Front. Plant Sci. 2022, 13, 1096549. [Google Scholar] [CrossRef]
- Fenech, M.; Amorim-Silva, V.; Esteban Del Valle, A.; Arnaud, D.; Ruiz-Lopez, N.; Castillo, A.G.; Smirnoff, N.; Botella, M.A. The role of GDP-l-galactose phosphorylase in the control of ascorbate biosynthesis. Plant Physiol. 2021, 185, 1574–1594. [Google Scholar] [CrossRef]
- Bulley, S.; Laing, W. The regulation of ascorbate biosynthesis. Curr. Opin. Plant Biol. 2016, 33, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Leferink, N.G.; van den Berg, W.A.; van Berkel, W.J. l-Galactono-gamma-lactone dehydrogenase from Arabidopsis thaliana, a flavoprotein involved in vitamin C biosynthesis. Febs. J. 2008, 275, 713–726. [Google Scholar] [CrossRef] [PubMed]
- Duque, P.; Vieira, C.P.; Vieira, J. Advances in Novel Animal Vitamin C Biosynthesis Pathways and the Role of Prokaryote-Based Inferences to Understand Their Origin. Genes 2022, 13, 1917. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, G.; Ishikawa, T.; Pornsaksit, V.; Smirnoff, N. Evolution of alternative biosynthetic pathways for vitamin C following plastid acquisition in photosynthetic eukaryotes. eLife 2015, 4, e06369. [Google Scholar] [CrossRef] [PubMed]
- Zechmann, B.; Stumpe, M.; Mauch, F. Immunocytochemical determination of the subcellular distribution of ascorbate in plants. Planta 2011, 233, 1–12. [Google Scholar] [CrossRef]
- Horemans, N.; Foyer, C.H.; Asard, H. Transport and action of ascorbate at the plant plasma membrane. Trends Plant Sci. 2000, 5, 263–267. [Google Scholar] [CrossRef]
- Łukawski, M.; Dałek, P.; Witkiewicz, W.; Przybyło, M.; Langner, M. Experimental evidence and physiological significance of the ascorbate passive diffusion through the lipid bilayer. Chem. Phys. Lipids 2020, 232, 104950. [Google Scholar] [CrossRef]
- Corti, A.; Casini, A.F.; Pompella, A. Cellular pathways for transport and efflux of ascorbate and dehydroascorbate. Arch Biochem. Biophys. 2010, 500, 107–115. [Google Scholar] [CrossRef]
- Scalera, V.; Giangregorio, N.; De Leonardis, S.; Console, L.; Carulli, E.S.; Tonazzi, A. Characterization of a Novel Mitochondrial Ascorbate Transporter From Rat Liver and Potato Mitochondria. Front. Mol. Biosci. 2018, 5, 58. [Google Scholar] [CrossRef]
- Miyaji, T.; Kuromori, T.; Takeuchi, Y.; Yamaji, N.; Yokosho, K.; Shimazawa, A.; Sugimoto, E.; Omote, H.; Ma, J.F.; Shinozaki, K.; et al. AtPHT4;4 is a chloroplast-localized ascorbate transporter in Arabidopsis. Nat. Commun. 2015, 6, 5928. [Google Scholar] [CrossRef]
- Maruta, T.; Inoue, T.; Noshi, M.; Tamoi, M.; Yabuta, Y.; Yoshimura, K.; Ishikawa, T.; Shigeoka, S. Cytosolic ascorbate peroxidase 1 protects organelles against oxidative stress by wounding- and jasmonate-induced H(2)O(2) in Arabidopsis plants. Biochim. Biophys. Acta. 2012, 1820, 1901–1907. [Google Scholar] [CrossRef]
- Yin, B.; Zhang, J.; Liu, Y.; Pan, X.; Zhao, Z.; Li, H.; Zhang, C.; Li, C.; Du, X.; Li, Y.; et al. PtomtAPX, a mitochondrial ascorbate peroxidase, plays an important role in maintaining the redox balance of Populus tomentosa Carr. Sci. Rep. 2019, 9, 19541. [Google Scholar] [CrossRef]
- Narendra, S.; Venkataramani, S.; Shen, G.; Wang, J.; Pasapula, V.; Lin, Y.; Kornyeyev, D.; Holaday, A.S.; Zhang, H. The Arabidopsis ascorbate peroxidase 3 is a peroxisomal membrane-bound antioxidant enzyme and is dispensable for Arabidopsis growth and development. J. Exp. Bot. 2006, 57, 3033–3042. [Google Scholar] [CrossRef]
- Maruta, T.; Sawa, Y.; Shigeoka, S.; Ishikawa, T. Diversity and Evolution of Ascorbate Peroxidase Functions in Chloroplasts: More Than Just a Classical Antioxidant Enzyme? Plant Cell Physiol. 2016, 57, 1377–1386. [Google Scholar] [CrossRef]
- Smirnoff, N.; Arnaud, D. Hydrogen peroxide metabolism and functions in plants. New Phytol. 2019, 221, 1197–1214. [Google Scholar] [CrossRef]
- Chin, D.-C.; Senthil Kumar, R.; Suen, C.-S.; Chien, C.-Y.; Hwang, M.-J.; Hsu, C.-H.; Xuhan, X.; Lai, Z.X.; Yeh, K.-W. Plant Cytosolic Ascorbate Peroxidase with Dual Catalytic Activity Modulates Abiotic Stress Tolerances. iScience 2019, 16, 31–49. [Google Scholar] [CrossRef]
- Stevens, R.; Truffault, V.; Baldet, P.; Gautier, H. Ascorbate Oxidase in Plant Growth, Development, and Stress Tolerance. In Ascorbic Acid in Plant Growth, Development and Stress Tolerance; Hossain, M.A., Munné-Bosch, S., Burritt, D.J., Diaz-Vivancos, P., Fujita, M., Lorence, A., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 273–295. [Google Scholar] [CrossRef]
- Balestrini, R.; Ott, T.; Güther, M.; Bonfante, P.; Udvardi, M.K.; De Tullio, M.C. Ascorbate oxidase: The unexpected involvement of a ‘wasteful enzyme’ in the symbioses with nitrogen-fixing bacteria and arbuscular mycorrhizal fungi. Plant Physiol. Biochem. 2012, 59, 71–79. [Google Scholar] [CrossRef]
- Hagel, J.M.; Facchini, P.J. Expanding the roles for 2-oxoglutarate-dependent oxygenases in plant metabolism. Nat. Prod. Rep. 2018, 35, 721–734. [Google Scholar] [CrossRef]
- Kawai, Y.; Ono, E.; Mizutani, M. Evolution and diversity of the 2–oxoglutarate-dependent dioxygenase superfamily in plants. Plant J. 2014, 78, 328–343. [Google Scholar] [CrossRef]
- Lucibelli, F.; Valoroso, M.C.; Aceto, S. Plant DNA Methylation: An Epigenetic Mark in Development, Environmental Interactions, and Evolution. Int. J. Mol. Sci. 2022, 23, 8299. [Google Scholar] [CrossRef]
- Chen, X.; Hu, Y.; Zhou, D.X. Epigenetic gene regulation by plant Jumonji group of histone demethylase. Biochim. Biophys. Acta. 2011, 1809, 421–426. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Ali, S.; Zhang, X.; Zhang, Y.; Wang, M.; Zhang, Q.; Xie, L. Genome-wide identification, classification, and expression analysis of the JmjC domain-containing histone demethylase gene family in birch. BMC Genom. 2021, 22, 772. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, A.M.; Dunwell, J.M. 2-oxoglutarate-dependent dioxygenases: A renaissance in attention for ascorbic acid in plants. PLoS ONE 2020, 15, e0242833. [Google Scholar] [CrossRef]
- Shen, J.; Griffiths, P.T.; Campbell, S.J.; Utinger, B.; Kalberer, M.; Paulson, S.E. Ascorbate oxidation by iron, copper and reactive oxygen species: Review, model development, and derivation of key rate constants. Sci. Rep. 2021, 11, 7417. [Google Scholar] [CrossRef]
- Möller, M.N.; Cuevasanta, E.; Orrico, F.; Lopez, A.C.; Thomson, L.; Denicola, A. Diffusion and Transport of Reactive Species Across Cell Membranes. In Bioactive Lipids in Health and Disease; Trostchansky, A., Rubbo, H., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 3–19. [Google Scholar] [CrossRef]
- Arrigoni, O. Ascorbate system in plant development. J. Bioenerg. Biomembr. 1994, 26, 407–419. [Google Scholar] [CrossRef]
- Leterrier, M.; Cagnac, O. Function of the Various MDAR Isoforms in Higher Plants. In Antioxidants and Antioxidant Enzymes in Higher Plants; Gupta, D.K., Palma, J.M., Corpas, F.J., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 83–94. [Google Scholar] [CrossRef]
- Ding, H.; Wang, B.; Han, Y.; Li, S. The pivotal function of dehydroascorbate reductase in glutathione homeostasis in plants. J. Exp. Bot. 2020, 71, 3405–3416. [Google Scholar] [CrossRef]
- De Tullio, M.C.; De Gara, L.; Paciolla, C.; Arrigoni, O. Dehydroascorbate-reducing proteins in maize are induced by the ascorbate biosynthesis inhibitor lycorine. Plant Physiol. Biochem. 1998, 36, 433–440. [Google Scholar] [CrossRef]
- Dowdle, J.; Ishikawa, T.; Gatzek, S.; Rolinski, S.; Smirnoff, N. Two genes in Arabidopsis thaliana encoding GDP-L-galactose phosphorylase are required for ascorbate biosynthesis and seedling viability. Plant J. 2007, 52, 673–689. [Google Scholar] [CrossRef]
- De Tullio, M.C.; Paciolla, C.; Arrigoni, O. Identification and Analysis of Proteins Sharing Dehydroascorbate Reductase Activity. Biol. Plant. 2002, 45, 145–147. [Google Scholar] [CrossRef]
- Njus, D.; Kelley, P.M.; Tu, Y.J.; Schlegel, H.B. Ascorbic acid: The chemistry underlying its antioxidant properties. Free Radic. Biol. Med. 2020, 159, 37–43. [Google Scholar] [CrossRef]
- Fotopoulos, V.; De Tullio, M.C.; Barnes, J.; Kanellis, A.K. Altered stomatal dynamics in ascorbate oxidase over-expressing tobacco plants suggest a role for dehydroascorbate signalling. J. Exp. Bot. 2008, 59, 729–737. [Google Scholar] [CrossRef] [PubMed]
- Chavan, S.N.; De Kesel, J.; Desmedt, W.; Degroote, E.; Singh, R.R.; Nguyen, G.T.; Demeestere, K.; De Meyer, T.; Kyndt, T. Dehydroascorbate induces plant resistance in rice against root-knot nematode Meloidogyne graminicola. Mol. Plant Pathol. 2022, 23, 1303–1319. [Google Scholar] [CrossRef] [PubMed]
- Fiorani, M.; Azzolini, C.; Guidarelli, A.; Cerioni, L.; Cantoni, O. A novel biological role of dehydroascorbic acid: Inhibition of Na+-dependent transport of ascorbic acid. Pharmacol. Res. 2014, 84, 12–17. [Google Scholar] [CrossRef]
- Saaranen, M.J.; Karala, A.R.; Lappi, A.K.; Ruddock, L.W. The role of dehydroascorbate in disulfide bond formation. Antioxid. Redox Signal. 2010, 12, 15–25. [Google Scholar] [CrossRef]
- Pignocchi, C.; Foyer, C.H. Apoplastic ascorbate metabolism and its role in the regulation of cell signalling. Curr. Opin. Plant Biol. 2003, 6, 379–389. [Google Scholar] [CrossRef]
- Green, M.A.; Fry, S.C. Vitamin C degradation in plant cells via enzymatic hydrolysis of 4-O-oxalyl-L-threonate. Nature 2005, 433, 83–87. [Google Scholar] [CrossRef]
- Parsons, H.T.; Yasmin, T.; Fry, S.C. Alternative pathways of dehydroascorbic acid degradation in vitro and in plant cell cultures: Novel insights into vitamin C catabolism. Biochem. J. 2011, 440, 375–383. [Google Scholar] [CrossRef]
- Parsons, H.T.; Fry, S.C. Oxidation of dehydroascorbic acid and 2,3-diketogulonate under plant apoplastic conditions. Phytochemistry 2012, 75, 41–49. [Google Scholar] [CrossRef]
- Truffault, V.; Fry, S.C.; Stevens, R.G.; Gautier, H. Ascorbate degradation in tomato leads to accumulation of oxalate, threonate and oxalyl threonate. Plant J. 2017, 89, 996–1008. [Google Scholar] [CrossRef]
- Dewhirst, R.A.; Fry, S.C. The oxidation of dehydroascorbic acid and 2,3-diketogulonate by distinct reactive oxygen species. Biochem. J. 2018, 475, 3451–3470. [Google Scholar] [CrossRef]
- Gerna, D.; Arc, E.; Holzknecht, M.; Roach, T.; Jansen-Dürr, P.; Weiss, A.K.H.; Kranner, I. AtFAHD1a: A New Player Influencing Seed Longevity and Dormancy in Arabidopsis? Int. J. Mol. Sci. 2021, 22, 2997. [Google Scholar] [CrossRef]
- Stack, T.M.M.; Morrison, K.N.; Dettmer, T.M.; Wille, B.; Kim, C.; Joyce, R.; Jermain, M.; Naing, Y.T.; Bhatti, K.; Francisco, B.S.; et al. Characterization of an l-Ascorbate Catabolic Pathway with Unprecedented Enzymatic Transformations. J. Am. Chem. Soc. 2020, 142, 1657–1661. [Google Scholar] [CrossRef] [PubMed]
- Dewhirst, R.A.; Fry, S.C. Oxalyltransferase, a plant cell-wall acyltransferase activity, transfers oxalate groups from ascorbate metabolites to carbohydrates. Plant J. 2018, 95, 743–757. [Google Scholar] [CrossRef] [PubMed]
- Arrigoni, O.; De Gara, L.; Tommasi, F.; Liso, R. Changes in the Ascorbate System during Seed Development of Vicia faba L. Plant Physiol. 1992, 99, 235–238. [Google Scholar] [CrossRef] [PubMed]
- De Gara, L.; de Pinto, M.C.; Moliterni, V.M.C.; D’Egidio, M.G. Redox regulation and storage processes during maturation in kernels of Triticum durum. J. Exp. Bot. 2003, 54, 249–258. [Google Scholar] [CrossRef]
- Tommasi, F.; Paciolla, C.; Arrigoni, O. The ascorbate system in recalcitrant and orthodox seeds. Physiol. Plant. 1999, 105, 193–198. [Google Scholar] [CrossRef]
- Klepikova, A.V.; Kasianov, A.S.; Gerasimov, E.S.; Logacheva, M.D.; Penin, A.A. A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J. 2016, 88, 1058–1070. [Google Scholar] [CrossRef]
- Barba-Espin, G.; Diaz-Vivancos, P.; Clemente-Moreno, M.J.; Albacete, A.; Faize, L.; Faize, M.; Pérez-Alfocea, F.; Hernández, J.A. Interaction between hydrogen peroxide and plant hormones during germination and the early growth of pea seedlings. Plant Cell Environ. 2010, 33, 981–994. [Google Scholar] [CrossRef]
- Barba-Espin, G.; Nicolas, E.; Almansa, M.S.; Cantero-Navarro, E.; Albacete, A.; Hernández, J.A.; Díaz-Vivancos, P. Role of thioproline on seed germination: Interaction ROS-ABA and effects on antioxidative metabolism. Plant Physiol. Biochem. 2012, 59, 30–36. [Google Scholar] [CrossRef]
- Kozaki, A.; Aoyanagi, T. Molecular Aspects of Seed Development Controlled by Gibberellins and Abscisic Acids. Int. J. Mol. Sci. 2022, 23, 1876. [Google Scholar] [CrossRef]
- Hedden, P. The Current Status of Research on Gibberellin Biosynthesis. Plant Cell Physiol. 2020, 61, 1832–1849. [Google Scholar] [CrossRef]
- Frey, A.; Effroy, D.; Lefebvre, V.; Seo, M.; Perreau, F.; Berger, A.; Sechet, J.; To, A.; North, H.M.; Marion-Poll, A. Epoxycarotenoid cleavage by NCED5 fine-tunes ABA accumulation and affects seed dormancy and drought tolerance with other NCED family members. Plant J. 2012, 70, 501–512. [Google Scholar] [CrossRef] [PubMed]
- Bulley, S.M.; Cooney, J.M.; Laing, W. Elevating Ascorbate in Arabidopsis Stimulates the Production of Abscisic Acid, Phaseic Acid, and to a Lesser Extent Auxin (IAA) and Jasmonates, Resulting in Increased Expression of DHAR1 and Multiple Transcription Factors Associated with Abiotic Stress Tolerance. Int. J. Mol. Sci. 2021, 22, 6743. [Google Scholar] [PubMed]
- Kobrehel, K.; Wong, J.H.; Balogh, A.; Kiss, F.; Yee, B.C.; Buchanan, B.B. Specific reduction of wheat storage proteins by thioredoxin h. Plant Physiol. 1992, 99, 919–924. [Google Scholar] [CrossRef] [PubMed]
- Oliver, M.J.; Farrant, J.M.; Hilhorst, H.W.M.; Mundree, S.; Williams, B.; Bewley, J.D. Desiccation Tolerance: Avoiding Cellular Damage During Drying and Rehydration. Annu. Rev. Plant. Biol. 2020, 71, 435–460. [Google Scholar] [CrossRef]
- Sergeant, M.J.; Li, J.J.; Fox, C.; Brookbank, N.; Rea, D.; Bugg, T.D.; Thompson, A.J. Selective inhibition of carotenoid cleavage dioxygenases: Phenotypic effects on shoot branching. J. Biol. Chem. 2009, 284, 5257–5264. [Google Scholar] [CrossRef] [PubMed]
- Nakata, P.A. Influence of calcium oxalate crystal accumulation on the calcium content of seeds from Medicago truncatula. Plant Sci. 2012, 185, 246–249. [Google Scholar] [CrossRef] [PubMed]
- Minor, E.A.; Court, B.L.; Young, J.I.; Wang, G. Ascorbate induces ten-eleven translocation (Tet) methylcytosine dioxygenase-mediated generation of 5-hydroxymethylcytosine. J. Biol. Chem. 2013, 288, 13669–13674. [Google Scholar] [CrossRef] [PubMed]
- Kawakatsu, T.; Nery, J.R.; Castanon, R.; Ecker, J.R. Dynamic DNA methylation reconfiguration during seed development and germination. Genome. Biol. 2017, 18, 171. [Google Scholar] [CrossRef]
- Chung, T.L.; Brena, R.M.; Kolle, G.; Grimmond, S.M.; Berman, B.P.; Laird, P.W.; Pera, M.F.; Wolvetang, E.J. Vitamin C promotes widespread yet specific DNA demethylation of the epigenome in human embryonic stem cells. Stem. Cells 2010, 28, 1848–1855. [Google Scholar] [CrossRef]
- Jeevan Kumar, S.P.; Rajendra Prasad, S.; Banerjee, R.; Thammineni, C. Seed birth to death: Dual functions of reactive oxygen species in seed physiology. Ann. Bot. 2015, 116, 663–668. [Google Scholar] [CrossRef] [PubMed]
- Farooq, M.A.; Zhang, X.; Zafar, M.M.; Ma, W.; Zhao, J. Roles of Reactive Oxygen Species and Mitochondria in Seed Germination. Front. Plant Sci. 2021, 12, 781734. [Google Scholar] [CrossRef] [PubMed]
- Katsuya-Gaviria, K.; Caro, E.; Carrillo-Barral, N.; Iglesias-Fernández, R. Reactive Oxygen Species (ROS) and Nucleic Acid Modifications during Seed Dormancy. Plants 2020, 9, 679. [Google Scholar] [CrossRef] [PubMed]
- Jurdak, R.; Rodrigues, G.A.G.; Chaumont, N.; Schivre, G.; Bourbousse, C.; Barneche, F.; Bou Dagher Kharrat, M.; Bailly, C. Intracellular reactive oxygen species trafficking participates in seed dormancy alleviation in Arabidopsis seeds. New Phytol. 2022, 234, 850–866. [Google Scholar] [CrossRef]
- Iwasaki, M.; Penfield, S.; Lopez-Molina, L. Parental and Environmental Control of Seed Dormancy in Arabidopsis thaliana. Annu. Rev. Plant Biol. 2022, 73, 355–378. [Google Scholar] [CrossRef]
- Carrillo-Barral, N.; Rodríguez-Gacio, M.d.C.; Matilla, A.J. Delay of Germination-1 (DOG1): A Key to Understanding Seed Dormancy. Plants 2020, 9, 480. [Google Scholar] [CrossRef]
- Chen, F.; Li, Y.; Li, X.; Li, W.; Xu, J.; Cao, H.; Wang, Z.; Li, Y.; Soppe, W.J.J.; Liu, Y. Ectopic expression of the Arabidopsis florigen gene FLOWERING LOCUS T in seeds enhances seed dormancy via the GA and DOG1 pathways. Plant J. 2021, 107, 909–924. [Google Scholar] [CrossRef]
- Chen, M.; Penfield, S. Feedback regulation of COOLAIR expression controls seed dormancy and flowering time. Science 2018, 360, 1014–1017. [Google Scholar] [CrossRef]
- Müller, K.; Bouyer, D.; Schnittger, A.; Kermode, A.R. Evolutionarily conserved histone methylation dynamics during seed life-cycle transitions. PLoS ONE 2012, 7, e51532. [Google Scholar] [CrossRef]
- Barth, C.; De Tullio, M.; Conklin, P.L. The role of ascorbic acid in the control of flowering time and the onset of senescence. J. Exp. Bot. 2006, 57, 1657–1665. [Google Scholar] [CrossRef]
- Jurdak, R.; Launay-Avon, A.; Paysant-Le Roux, C.; Bailly, C. Retrograde signalling from the mitochondria to the nucleus translates the positive effect of ethylene on dormancy breaking of Arabidopsis thaliana seeds. New Phytol. 2021, 229, 2192–2205. [Google Scholar] [CrossRef] [PubMed]
- Shanmugabalaji, V.; Chahtane, H.; Accossato, S.; Rahire, M.; Gouzerh, G.; Lopez-Molina, L.; Kessler, F. Chloroplast Biogenesis Controlled by DELLA-TOC159 Interaction in Early Plant Development. Curr. Biol. 2018, 28, 2616–2623.e2615. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Dai, Y.; Zheng, C.; Yang, Y.; Chen, W.; Wang, Q.; Chandrasekaran, U.; Du, J.; Liu, W.; Shu, K. The ABI4-RbohD/VTC2 regulatory module promotes reactive oxygen species (ROS) accumulation to decrease seed germination under salinity stress. New Phytol. 2021, 229, 950–962. [Google Scholar] [CrossRef] [PubMed]
- Ye, N.; Zhu, G.; Liu, Y.; Zhang, A.; Li, Y.; Liu, R.; Shi, L.; Jia, L.; Zhang, J. Ascorbic acid and reactive oxygen species are involved in the inhibition of seed germination by abscisic acid in rice seeds. J. Exp. Bot. 2012, 63, 1809–1822. [Google Scholar] [CrossRef] [PubMed]
- Stanley, L.; Paolo, B.; Lukasz, W.; Szymon Kubala, S.; Roberta, P.; Katzarina, L.; Muriel, Q.; Malgorzata, G. Seed Priming: New Comprehensive Approaches for an Old Empirical Technique. In New Challenges in Seed Biology; Susana, A., Alma, B., Eds.; IntechOpen: Rijeka, Croatian, 2016; Chapter 1. [Google Scholar] [CrossRef]
- Paparella, S.; Araújo, S.S.; Rossi, G.; Wijayasinghe, M.; Carbonera, D.; Balestrazzi, A. Seed priming: State of the art and new perspectives. Plant Cell. Rep. 2015, 34, 1281–1293. [Google Scholar] [CrossRef]
- Zulfiqar, F.; Nafees, M.; Chen, J.; Darras, A.; Ferrante, A.; Hancock, J.T.; Ashraf, M.; Zaid, A.; Latif, N.; Corpas, F.J.; et al. Chemical priming enhances plant tolerance to salt stress. Front. Plant Sci. 2022, 13, 946922. [Google Scholar] [CrossRef]
- Ibrahim, E.A. Seed priming to alleviate salinity stress in germinating seeds. J. Plant Physiol. 2016, 192, 38–46. [Google Scholar] [CrossRef]
- Farooq, M.; Tabassum, R.; Afzal, I. Enhancing the performance of direct seeded fine rice by seed priming. Plant Prod. Sci. 2006, 9, 446–456. [Google Scholar] [CrossRef]
- Jafar, M.Z.; Farooq, M.; Cheema, M.A.; Afzal, I.; Basra, S.M.A.; Wahid, M.A.; Aziz, T.; Shahid, M. Improving the Performance of Wheat by Seed Priming Under Saline Conditions. J. Agron. Crop Sci. 2012, 198, 38–45. [Google Scholar] [CrossRef]
- Shah, T.; Latif, S.; Khan, H.; Munsif, F.; Nie, L.X. Ascorbic Acid Priming Enhances Seed Germination and Seedling Growth of Winter Wheat under Low Temperature Due to Late Sowing in Pakistan. Agronomy 2019, 9, 757. [Google Scholar] [CrossRef]
- Ahmad, I.; Khaliq, T.; Ahmad, A.; Basra, S.M.; Hasnain, Z.; Ali, A. Effect of seed priming with ascorbic acid, salicylic acid and hydrogen peroxide on emergence, vigor and antioxidant activities of maize. Afr. J. Biotechnol. 2012, 11, 1127–1132. [Google Scholar]
- Farooq, M.; Irfan, M.; Aziz, T.; Ahmad, I.; Cheema, S.A. Seed Priming with Ascorbic Acid Improves Drought Resistance of Wheat. J. Agron. Crop Sci. 2013, 199, 12–22. [Google Scholar] [CrossRef]
- Alcantara, B.K.; Rizzi, V.; Gaziola, S.A.; Azevedo, R.A. Soluble amino acid profile, mineral nutrient and carbohydrate content of maize kernels harvested from plants submitted to ascorbic acid seed priming. An. Acad. Bras. Cienc. 2017, 89, 695–704. [Google Scholar] [CrossRef] [PubMed]
- Baig, Z.; Khan, N.; Sahar, S.; Sattar, S.; Zehra, R. Effects of seed priming with ascorbic acid to mitigate salinity stress on three wheat (Triticum aestivum L.) cultivars. Acta Ecologica Sinica 2021, 41, 491–498. [Google Scholar] [CrossRef]
- Alves, R.D.; Rossatto, D.R.; da Silva, J.D.; Checchio, M.V.; de Oliveira, K.R.; Oliveira, F.D.; de Queiroz, S.F.; da Cruz, M.C.P.; Gratao, P.L. Seed priming with ascorbic acid enhances salt tolerance in micro-tom tomato plants by modifying the antioxidant defense system components. Biocatal. Agric. Biotechnol. 2021, 31, 101927. [Google Scholar] [CrossRef]
- Kasim, W.A.; Nessem, A.A.; Gaber, A. Alleviation of Drought Stress in Vicia faba by Seed Priming with Ascorbic Acid or Extracts of Garlic and Carrot. Egypt. J. Bot. 2017, 57, 45–59. [Google Scholar] [CrossRef]
- Kasim, W.A.; Nessem, A.A.; Gaber, A. Effect of seed priming with aqueous extracts of carrot roots, garlic cloves or ascorbic acid on the yield of Vicia faba grown under drought stress. Pak. J. Bot. 2019, 51, 1979–1985. [Google Scholar] [CrossRef]
- Shahnawaz, M.; Sanadhya, D. Aluminium induced oxidative stress and antioxidants system in two barley varieties and its alleviation through ascorbic acid and salicylic acid seed priming approach. Int. J. Life Sci. Pharma Res. 2017, 7, L26–L37. [Google Scholar]
- Xia, F.; Cheng, H.; Chen, L.; Zhu, H.; Mao, P.; Wang, M. Influence of exogenous ascorbic acid and glutathione priming on mitochondrial structural and functional systems to alleviate aging damage in oat seeds. BMC Plant Biol. 2020, 20, 104. [Google Scholar] [CrossRef]
- Fercha, A.; Capriotti, A.L.; Caruso, G.; Cavaliere, C.; Samperi, R.; Stampachiacchiere, S.; Laganà, A. Comparative analysis of metabolic proteome variation in ascorbate-primed and unprimed wheat seeds during germination under salt stress. J. Proteom. 2014, 108, 238–257. [Google Scholar] [CrossRef]
- Munns, R.; Gilliham, M. Salinity tolerance of crops—What is the cost? New Phytol. 2015, 208, 668–673. [Google Scholar] [CrossRef] [PubMed]
- Gulyas, L.; Powell, J.R. Predicting the Future: Parental Progeny Investment in Response to Environmental Stress Cues. Front. Cell Dev. Biol. 2019, 7, 115. [Google Scholar] [CrossRef] [PubMed]
- Segerstrom, S.C. Resources, stress, and immunity: An ecological perspective on human psychoneuroimmunology. Ann. Behav. Med. 2010, 40, 114–125. [Google Scholar] [CrossRef] [PubMed]
- Richards, C.L.; Alonso, C.; Becker, C.; Bossdorf, O.; Bucher, E.; Colomé-Tatché, M.; Durka, W.; Engelhardt, J.; Gaspar, B.; Gogol-Döring, A.; et al. Ecological plant epigenetics: Evidence from model and non-model species, and the way forward. Ecol. Lett. 2017, 20, 1576–1590. [Google Scholar] [CrossRef] [PubMed]
| AsA Concentration | MET (Days Ahead of Controls) | Species | Ref. |
|---|---|---|---|
| 10 mg/L | 0.36 (ns) | Oryza sativa | [92] |
| 50 mg/L | 2 | Triticum aestivum | [93] |
| 50 mg/L | 1.1 | Triticum aestivum | [94] |
| 40 mg/L | 0.68 | Zea mays | [95] |
| 2 mM | 0.92 | Triticum aestivum | [96] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Terzaghi, M.; De Tullio, M.C. Ascorbic Acid in Seeds, Priming and Beyond. Seeds 2023, 2, 421-435. https://doi.org/10.3390/seeds2040032
Terzaghi M, De Tullio MC. Ascorbic Acid in Seeds, Priming and Beyond. Seeds. 2023; 2(4):421-435. https://doi.org/10.3390/seeds2040032
Chicago/Turabian StyleTerzaghi, Mattia, and Mario C. De Tullio. 2023. "Ascorbic Acid in Seeds, Priming and Beyond" Seeds 2, no. 4: 421-435. https://doi.org/10.3390/seeds2040032
APA StyleTerzaghi, M., & De Tullio, M. C. (2023). Ascorbic Acid in Seeds, Priming and Beyond. Seeds, 2(4), 421-435. https://doi.org/10.3390/seeds2040032









