Abstract
Protein three-dimensional structural analysis using artificial intelligence is attracting attention in various fields, such as the estimation of vaccine structure and stability. In particular, when using the spike protein in vaccines, the major issues in the construction of SARS-CoV-2 vaccines are their weak abilities to attack the virus and elicit immunity for a short period. Structural information about new viruses is essential for understanding their properties and creating effective vaccines. However, determining the structure of a protein through experiments is a lengthy and laborious process. Therefore, a new computational approach accelerated the elucidation process and made predictions more accurate. Using advanced machine learning technology called deep neural networks, it has become possible to predict protein structures directly from protein and gene sequences. We summarize the advances in antiviral therapy with the SARS-CoV-2 vaccine and extracellular vesicles via computational analysis.
1. Introduction
Vaccines for the new coronavirus disease (COVID-19) are on track around the world, but it is still difficult to predict when this pandemic will end. Furthermore, the possibility of achieving “herd immunity” that if a sufficient proportion of people develop immunity to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is beginning to be considered unlikely. This thinking reflects the complexity and difficulty of responding to a pandemic and does not deny the fact that vaccination is beneficial. As more people in the population acquire immunity, another problem arises. A higher percentage of people who acquire immunity creates selective pressure, favoring mutant strains that can infect those who acquire immunity. Furthermore, new SARS-CoV-2 variants emerge that are highly contagious and are resistant to vaccine, and once acquired immunity is attenuated. Thus, antibodies induced by current vaccines are ‘strain-specific’ and cannot respond to antigenic mutation of virus strains, and it is necessary to activate antibodies that match the latest epidemic strains. By vaccinating as many as possible as soon as possible, it is possible to prevent new variants from gaining footholds. However, it is almost inevitable that vaccines will create new selective pressure and lead to the emergence of mutant strains, so it is necessary to develop infrastructure and processes to monitor this. In this way, vaccines are a double-edged sword that can immunize many people and create many new patients. Furthermore, the persistence of induced antibodies is not as good as that of live vaccines, such as the measles vaccine [1,2]. It will also be important to clarify how long immunity from vaccines lasts and whether booster vaccinations are necessary after vaccination.
Additionally, considerable attention has been focused on antibodies that acquire ‘cross-reactivity’ by targeting epitopes that are difficult to mutate to improve the strain specificity of vaccines [3,4,5,6,7]. Because this cross-reactive antibody is a rare antibody that is difficult to induce with current vaccines, structural analysis has clarified the binding sites and B cell epitopes of monoclonal cross-reactive antibodies, and it has become possible to produce vaccines with artificially increased antigenicity to facilitate the induction of these antibodies through structural biology approaches, such as epitope-focused vaccines [8,9]. Although vaccine formulations based on this strategy have shown steady efficacy in animal models, clinical studies have suggested that the persistence of induced cross-reactive antibodies may be even lower than that of normal antibodies. Therefore, in the future, it will be necessary to devise ways to increase the amount and persistence of antibodies induced. To develop vaccines that are both safe and effective, it is important to understand the in vivo infection mechanisms of the virus. The amount and persistence of antibodies induced by influenza vaccines are largely dependent on the amount and quality of helper signals supplied by activated T cells to B cells [10,11]. Therefore, vaccine antigens must bind to T cell antigen receptors in addition to binding to antibodies, which are B cell antigen receptors that elicit helper signals from T cells [12,13,14,15,16,17,18,19,20]. Because T-cell epitopes consist of peptides of 20 amino acids or less, antigenicity is mainly determined by the primary amino acid sequence [21,22]. On the other hand, by mutating the part that binds to the antibody made by the vaccine, the virus can escape from the antibody while maintaining the ability to invade cells. At the time, a new vaccine containing the mutated part will be needed. In such a case, although there is a protein property prediction that predicts a change in stability for a single amino acid mutation from the amino acid sequence of the protein, not only the static structure but also the dynamic structure greatly contributes to the expression of protein function. Therefore, the molecular dynamics (MD) method has come to be used frequently as a means of analyzing the dynamic structure of proteins by simulation, but the amount of trajectory, molecular motion, obtained as a result of MD simulation is enormous. Moreover, since it is time-series data, in silico technology, including machine learning or deep learning is actively applied. Then, using the learning results, pseudo-MD is performed for single amino acid mutants of the protein without performing MD simulation calculation, which takes a long time, similar results, such as trajectory etc. can be obtained. Therefore, it is relatively easy to predict antigenicity using the bioinformatics tools. In this review, we summarize the applications of in silico analysis including deep learning for SARS-CoV-2 vaccine.
2. Anti-Virus Therapy via Vaccine
Two strategies are available for the development of antiviral drugs: (1) suppress the life cycle of the virus in the host cell and (2) control the runaway of the host immune system [23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44]. Three-dimensional (3D) protein structure information is extremely useful in searching for drug candidates that inhibit the functions of viral proteins based on strategy (1) [45,46]. Therefore, it is necessary to develop therapeutic drugs and vaccines as soon as possible; therapeutic drugs and vaccines against COVID-19 are underway. SARS-CoV-2 is classified as a single-stranded positive-strand RNA virus, and its genome size is approximately 30,000 bases, encoding 11 open reading frames and genes (Figure 1A) [47,48,49,50,51,52,53,54,55,56,57]. Each gene contains one non-structural protein (orf1ab) and four structural proteins (spike (S) protein, envelope (E), membrane (M), and nucleocapsid (N) protein) and encodes six accessory proteins (ORF3a, ORF6, ORF7a, ORF7b, ORF8, and ORF10) (Figure 1B) [58,59,60,61,62,63,64]. After translation, orf1ab is cleaved by the papain-like protease (nsp3, PL-pro) and the main protease (M-pro) that it encodes and is divided into 16 proteins (Nsp1 to Nsp16) [65,66,67]. SARS-CoV-2 is similar to SARS coronavirus (SARS-CoV), the pathogen of the severe acute respiratory syndrome (SARS), with approximately 80% genome sequence identity, and many encoded proteins are highly conserved [68,69]. Homology of the amino acid sequence of the SARS-CoV-2 protein revealed that 17 of the 26 proteins had structurally known proteins with significant sequence similarity, of which 16, excluding nsp4, had SARS-CoV protein conformations. Many SARS-CoV-2 proteins have postulated conformational models in the form of homo- or hetero- multimers [70,71]. For example, M-pro is a homodimer, nsp10 is a heterodimer with exonuclease (ExoN), respectively, and 2-O’-ribose methyltransferase (2oMT), and a model of inhibitor complex is assumed [72,73,74,75]. In addition, the S protein forms a homotrimer, and a complex model of the receptor-binding domain (RBD) and human angiotensin-converting enzyme 2 (ACE2) is assumed [76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95]. Virtual screening is an in silico analysis method for identifying drug candidates, which are compounds that bind to specific sites on viral proteins, based on 3D structural models. Typically, this method defines the site at which the drug molecule is bound to the 3D structure of the target protein. Compound library molecules are comprehensively docked on a computer, and candidate compounds are extracted by evaluating bond stability using evaluation functions, such as the energy function [96,97]. Although significant seed-up has been achieved using parallel computing and machine learning, it is not easy to apply in situations where the target protein or compound library has not been narrowed down. A ligand bound to a target protein homologue in a known complex structure is highly likely to contain a pharmacophore, where a structural feature is specifically recognized by the site where the compound is bound, such as the ligand-binding site. If there is an approved drug with a structure similar to that of the ligand that can be reasonably docked to the structural model, the molecule is expected to become a therapeutic drug candidate. Three of the SARS-CoV-2 protein models, the M-pro homodimer, S protein-ACE2 complex, and 2oMT-nsp10 heterodimer, have ligand molecules bound to the template structure [98,99,100,101]. M-pro is an essential enzyme for viral protein production and is considered a promising drug target for SARS-CoV-2. Therefore, complex structures with many peptidomimetic inhibitors have been analyzed; however, no existing drug molecules showing high similarity to these known ligands have been found. This suggests that the M-pro of SARS-CoV-2 is a cysteine protease, whereas many of the targets of existing antiviral protease inhibitors, such as the HIV protease, are aspartate or zinc proteases [23,102,103,104]. Moreover, carfilzomib, which showed the highest similarity among known ligands, is an irreversible inhibitor of proteasome and approved for the clinical treatment of multiple myeloma or Walden Strom’s macroglobulinemia [105,106,107]. The target of carfilzomib is a threonine protease with a nucleophilic attacking group: Thr; however, it also reacts with the nucleophilic attacking group Cys of M-pro. Because the S protein on the virus surface uses human angiotensin-converting enzyme 2 (ACE2) as a receptor when infecting host cells, the S protein-ACE2 binding site is an important target. ACE2 is a homologue, with 44% amino acid sequence identity, of ACE, which is a major target of anti-hypertensive ACE-inhibitor complex structures [108]. Approved drugs analogous to these inhibitors were found to be lisinopril, enalaprilat, and captopril, all of which are antihypertensive drugs.
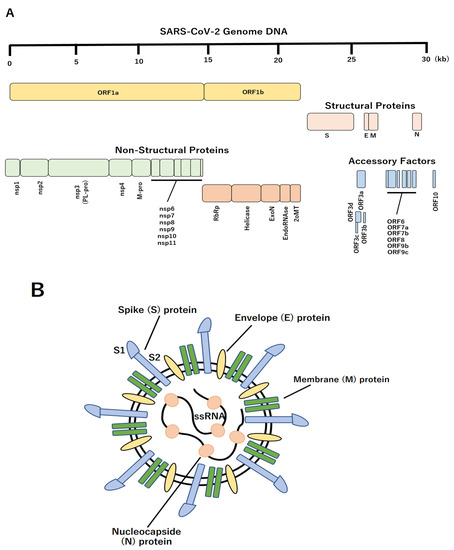
Figure 1.
Schematic structure of SARS-CoV-2. (A) The genomic organization of SARS-CoV-2. Upper line indicates genomic scale. Sixteen non-structural proteins, four structural proteins, and eleven accessory factors were represented. (B) Schematic diagram of the SARS-CoV-2 virus. The four structural proteins, including S, M, N and E proteins are shown.
However, these molecules were bound at a position different from the S protein-ACE2 interaction site; therefore, they could not directly inhibit the interaction with the S protein. Clinical trials of antibody drugs targeting the receptor-binding domain (RGD) as antigens are currently being conducted for drugs that target the S protein [109,110]. As a low-molecular-weight drug targeting the site, catharanthine, a component derived from Tamasaki Tsutsurugi, which has been approved as a treatment for alopecia areata and leukopenia, inhibits S protein-ACE2 interaction and suppresses SARS-CoV-2 infection [111,112]. This finding suggests the possibility of developing a drug to prevent COVID-19 infection by expanding catharanthine. The 2oMT-nsp10 complex is an enzyme that modifies the methyl group on the 5′-terminal cap structure of viral RNA. The cap structure protects viral RNA from degradation by the host and is essential for synthesizing its proteins using the host’s translational machinery [113]. Several existing drugs were discovered from the 2oMT ligand complex structure of SARS-CoV, which was used as the template for the model, and antiviral activity was reported in in vitro experiments. Additionally, the adenosine A1 receptor agonists tecadenoson, serodenosone, and travodenosone are expected to target 2oMT, of which tecadenoson and travodenosone have passed phase I clinical trials, and their safety has been confirmed [114]. In addition, there should be a guanylyl transferase that adds a cap structure to the 5′ end of the viral RNA molecule, but the protein that plays that role is currently unknown; if it is identified in the future, it can become a drug discovery target [115].
Since protein interactions between humans and viruses play a crucial role in viral infections, their identification will lead to elucidation of viral infection mechanisms and discovery of targets for antiviral drugs. However, since biological experiments for this identification require a huge amount of time and cost, the prediction of the interaction by in silico analysis is expected. Conventional computer prediction method of the protein interaction is docking simulation using molecular dynamics method based on protein 3D-structure information, which examines the shape of the key and the keyhole of the protein and uses computer simulation to find the conditions that the key fits into the keyhole. However, it is difficult to elucidate the 3D-structure information, and the application of the molecular dynamics method for mutant viruses is limited. On the other hand, high-throughput experimental methods make it easy to obtain amino acid sequence information of viral proteins. By applying a deep learning model that predicts the future from time series data and taking the amino acid sequence of a protein as a flow of context, it is possible to extract 3D features of keys and keyholes from the order patterns of long-chain amino acid sequences. Thus, COVID-19 runaway of the host immune system is investigated by AI-based analytical approaches [116,117].
3. SARS-CoV-2 Vaccine with Extracellular Vesicles
Furthermore, modified extracellular vesicles (EV), i.e., vesicles with a heterogeneous lipid bilayer structure that are secreted from almost all living cells, are roughly divided into three types: exosomes, macrovesicles, and apoptotic bodies, based on differences in intracellular production mechanisms, loaded with an antibody consisting only of a heavy chain, which is a type of low-molecular-weight antibody against the spike protein of SARS-CoV-2, and IFN-b, a cytokine with antiviral effect, which inhibits the SARS-CoV-2 pseudo-virus derived from infecting cells and can induce the cells into an antiviral state [118]. In particular, exosomes are expected as new preventive and therapeutic strategies that exhibit antiviral activity. As the new coronavirus establishes infection by binding the SARS-CoV-2 spike protein to ACE2 on cells, blocking the spike protein with antibodies to render it incapable of binding to ACE2 is an important strategy for preventing SARS-CoV-2 infection and aggravation. Anti-spike neutralizing antibodies are expected to be therapeutic agents for COVID-19. Although the SARS-CoV-2 vaccine also promotes antibody production against the spike protein, among mutant strains, such as the Omicron strain, some strains that reduce the infection prevention effect of the SARS-CoV-2 vaccine have appeared [119,120,121,122,123,124]. Therefore, it is difficult to completely prevent SARS-CoV-2 infection and aggravation using the anti-spike neutralizing antibody alone. A large number of modified EV-mounted fusion proteins consisting of IFN-b, which induces an antiviral state in cells, an antibody comprising only heavy chains, which is a type of low-molecular-weight anti-spike antibody, and MFG-E8 protein, which can bind to EVs, showed significant anti-inhibitory effects on SARS-CoV-2 pseudo virus infectivity [118].
In addition, two mRNA vaccines have been developed against SARS-CoV-2, designed to induce systemic immunity via intramuscular injection [125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146]. However, it is necessary to develop a cold chain for real-world inoculation. Therefore, it has been reported that the vaccine is administered directly to the lungs, not via intramuscular injection, and EVs secreted from lung spheroid cells (LSC) are used as carriers [147,148]. The receptor binding domain (RBD) is more tightly retained in both muscle-lined respiratory airways and lung parenchyma than in liposome-based vaccines by inhaling LSC-EV virus-like particles (VLPs) modified with the RBD of the recombinant SARS-CoV-2 spike protein. In mice, this vaccine induces lung CD4+/CD8+ T cells with RBD-specific IgG antibodies, mucosal IgA responses, and a Th1-like cytokine expression profile, leading to the removal of the challenged SARS-CoV-2 pseudo virus [149]. In hamsters, two doses of this vaccine attenuated severe pneumonia and reduced inflammatory infiltrates after the SARS-CoV-2 challenge. RBD-modified LSC-EV vaccines (RBD-EVs) induce mucosal and systemic immunity in the lungs.
4. Vaccination Process and Nuclei Acids
Plasmid DNA (pDNA) is a safe and highly productive vector for DNA vaccines and gene therapies [150,151]. Antigen-presenting cells, such as macrophages and dendritic cells, which play an important role in the immune response and defense against foreign substances, recognize pDNA administered to the body as a ‘foreign substance’, and have a significant effect on its pharmacokinetics and gene expression. Therefore, it is important to optimize the gene expression profile obtained by pDNA administration for each target disease. DNA derived from bacteria, including pDNA, has a high frequency of unmethylated CpG sequences, known as CpG motifs. When mammalian macrophages and dendritic cells take them up, they are recognized as danger signals via intracellular toll-like receptor 9 (TLR9), and immune activation reactions, such as the production of various inflammatory cytokines, are induced [152,153,154,155]. Inflammatory cytokines are responsible for reducing gene expression in target cells owing to their cytotoxic effects. However, in the case of cancer treatment, in addition to the effects of transgenes, immune activation by inflammatory cytokine production can be expected, and the immune response to pDNA is thought to have complex effects on therapeutic efficacy. However, the mechanism of cellular uptake and activity of pDNA in macrophages and dendritic cells has not been fully elucidated. In particular, in the case of complexes with cationic carriers, which are commonly used to increase gene expression, immune activation by a mechanism different from cell activation by CpG motifs has been suggested, but the details are unknown. Non-parenchymal cells in the liver are significantly involved in the pharmacokinetics of pDNA, and this cellular uptake involves a mechanism similar to that of scavenger receptors, which specifically recognize the conformation of polyanions. In addition, a similar uptake mechanism exists in dendritic cells [156]. In contrast, by complexing DNA with cationic liposomes, cytokines are produced from macrophages
Regardless of the presence or absence of CpG motifs [157]. TLR9 is not involved in this CpG motif-independent phenomenon in mouse-derived macrophages, and a similar CpG motif-independent activation occurs in mouse dendritic cells as well as in human-derived cells. Furthermore, cell activation is highly dependent on the type of liposomes used for complex formation. In contrast, mouse peritoneal macrophages and RAW264.7, a cultured macrophage cell line, differ significantly in DNA uptake and cytokine production between the two cell groups. Because peritoneal macrophages efficiently take up naked pDNA but produce few cytokines, inhibition of TLR9 recognition by DNA binding factors is envisioned. Th1-type cytokine production induced by CpG DNA administration exhibits effective therapeutic effects against cancer and allergic diseases [158]. Y-shaped DNA is constructed by combining three short DNA strands with partially complementary sequences, and this unique structure induces cytokine production more efficiently than identical double-stranded DNA.
Chemokines are secretory proteins that promote cell migration and contribute to inflammatory reactions by attracting leukocytes. In addition, CXCL14, a chemokine, binds to CpG DNA and significantly enhances the induction of innate immunity and inflammatory responses through its uptake by dendritic cells (Figure 2) [159]. Furthermore, CXCL4, the CXC-type chemokine CXCL14, has functions similar to those of CXCL14 and enhances CpG DNA-induced dendritic cell activation [160]. CXCL14 has both CpG DNA and cell surface receptor-binding domains, and uptake of the CXCL14/CpG DNA complex into dendritic cells via the clathrin-dependent endocytosis pathway is required for the enhancement of CpG DNA activity [161]. In addition, by simulating the binding of CXCL14/CpG DNA, multiple amino acids on the N-terminal and C-terminal sides of CXCL14 act cooperatively to stabilize binding. Thus, the activation of dendritic cells by CXCL14 and CpG DNA is expected to function as a vaccine adjuvant to enhance vaccine efficacy [162]. Further elucidation of the cooperative action of CXCL14 and CpG DNA may lead to the development of more efficient cancer immunopotentiators and vaccine adjuvants. However, the immunological mechanisms of action of DNA vaccines, which are next-generation vaccines under development against infectious diseases, such as influenza, cancer, and allergies, are still not well understood. In contrast, the right-handed double-helical structure of DNA acts as an endogenous adjuvant for vaccines by activating the innate immune system via tank-binding kinase 1 (TBK1) in cells, and signals for activating the innate immune system are essential for the efficacy in DNA vaccines [163]. Among the effects of DNA vaccines, activation of TBK1-dependent innate immunity in immune cells, such as dendritic cells, is important for antibody production. Activation of TBK1 in non-immune cells, such as muscle cells, that take up DNA is important for the activation of cell-mediated immunity by T cells. In other words, the effects of DNA vaccines involve a pathway that induces type I interferon without being mediated by TLRs. Although the innate immunostimulatory action of nucleic acids is due to a special base sequence, CpG motif, often found in pathogens, such as bacteria and viruses, mediated by TLR9, it was shown that the right-handed structure of double-stranded DNA found in both viruses and host cells has a strong TLR-independent ability to produce interferon. Furthermore, innate immune activations in both immune and nonimmune cells interact with each other.
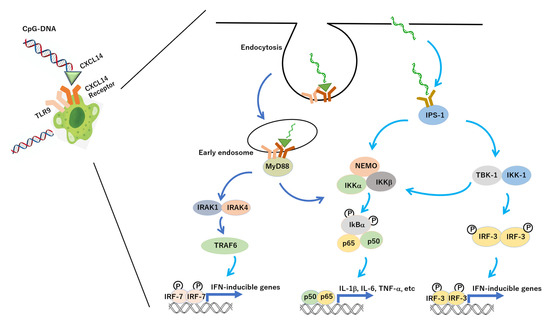
Figure 2.
Molecular pathways of inflammation induced by CXCL14 and CpG-DNA. CXCL14: chemokine (C-X-C motif) ligand 14, TLR9: toll-like receptor 9, TRAF6:TNF receptor-associated factor 6, MyD88: myeloid differentiation primary response gene 88, NEMO: NF-κB essential modulator, IκB: inhibitor kappa B, IKK: IκB kinase, p50: NF-κB p50, p65: NF-κB p65, IPS-1: IFN-inducing β promoter stimulator-1, TBK1: TANK-binding kinase 1, IRF: interferon regulatory factor, IFN: interferon, IRAK: IL-1 receptor associated kinase, IL-6: interleukin-6, IL-1b: interleukin-1b, TNF-α: tumor necrosis factor α.
5. Construction of Vaccine and Protein Structure in Silico Analysis
The methodology of analyzing the results of experiments using the information science method is the same as that of bioinformatics and computational biology and is a pioneering study that utilizes bioinformatics in virology (Figure 3) [164,165,166,167,168,169,170]. There are ethical issues with artificial intelligence (AI), but it has the potential to revolutionize science and solve some of the most complex problems facing modern biology. In particular, it is expected to predict the structure of unknown proteins, solve the mysteries of cells, and quickly elucidate diseases that affect cells. However, determining the structure of a protein through experiments is a lengthy and laborious process. Structural information about new viruses is essential for understanding their properties and for creating effective vaccines. Thus, researchers have accelerated the unravelling process and made predictions more accurate with a new computational approach. With the remarkable development of AI, it is now possible to predict the 3D structure of complex proteins with a high degree of accuracy. The AI system AlphaFold2 has accomplished a feat of identified several protein structures that make up the previously little-known novel SARS-CoV-2 within a fairly short time [171]. Thus, the tireless efforts of scientists and international collaboration, combined with cutting-edge AI technologies, such as AlphaFold2, have enabled a rapid response to the pandemic. AlphaFold2 uses advanced machine learning techniques, called deep learning neural networks, to predict protein structures directly from protein gene sequences [172,173,174,175,176,177]. In addition, AI must first learn the sequences and structures of approximately 100,000 known proteins from the experimental data published in the scientific community. This has made it possible to predict the 3D models of any protein with high accuracy. Because protein structure is related to protein function, it is important to clarify protein function and is essential and even more important information. There are several methods for experimentally determining protein structures, such as NMR and X-ray crystallography, but they are both time-consuming and expensive [178]. Therefore, researchers have been actively researching to predict 3D structures for some time, and many modelling methods have been devised [173,179,180,181,182]. There are various modelling techniques, and with regard to comparative modelling, different proteins used as templates yield different results; thus, a variety of predicted 3D structures can be obtained. However, it is necessary to choose the most natural structure among the predicted 3D structures. Herein, ‘natural structure-like’ implies that the structure is highly similar to the natural structure, and this is called the model quality assessment program (MQAP) [183]. Many MQAPs comprise single or multiple statistical potential functions that express natural structure-likeness, and prediction models with machine learning based on explicitly created feature values have also been proposed [184]. This statistical potential function is a statistically constructed potential function based on the distribution of structural features from the natural structures known in the Protein Data Bank and has been devised many times. Many of these statistical potential functions mainly capture the interactions between two bodies, such as the original pairs and residue pairs. However, because proteins have a 3D structure, it is difficult to capture their features. Therefore, although many-body potential functions have been devised, they are not as accurate as existing two-body functions. This is because the problem becomes more complicated, and the number of parameters increases in the case of many bodies. Therefore, to capture the interactions between many bodies, a new method that differs from the conventional method of creating a statistical potential function is required.
Figure 3.
Schematic procedure of vaccine design by bioinformatics of virus characterizations. Structural information and biological activity of viruses can automatically extract the molecular futures by AI.
Convolutional neural networks (CNN), which are neural networks with convolutional layers, have been successfully applied in many fields [185]. A 3D CNN, which is an extension of this to 3D, has been used for motion recognition and object recognition in the past, but it is also beginning to be used for the analysis of the 3D structures of proteins. Among them, 3D-CNN achieved better accuracy than existing methods that used machine learning with explicit feature values, suggesting the effectiveness of 3D-CNN in analyzing 3D structures of proteins. Based on this, it was expected that 3D-CNN would be effective in the MQAP field. Therefore, to develop a method for evaluating the predicted 3D structure that captures the interaction between many bodies, a method for evaluating the predicted conformation that analyses the local environment of a protein using 3D-CNN and outputs the overall score of the protein as the average of the evaluations of the local environment was developed. Consequently, the validity of evaluating the local structure of proteins using a 3D-CNN was suggested [184,185,186,187,188,189].
In addition, many studies have been conducted to predict the local and secondary structures of proteins from amino acid sequence information using machine learning [190,191,192,193,194]. The secondary structure can be classified into two types: α-helix and β-sheet (Table 1). The alpha-helix is a right-handed helical structure with an average of 3.6 residues per cycle. In this helical structure, all the amino acids form hydrogen bounds with amino acids residues to maintain an energetically stable structure. In contrast, the beta-strand contains a series of amino acids in a straight line. This secondary structure prediction is defined as a classification problem called sequence labelling, which predicts secondary structures from information, such as amino acid sequences. Furthermore, a secondary structure prediction model using a deep neural network (DNN) has been proposed, and it has been reported that highly accurate predictions can be made [195,196,197]. Conversely, a DNN is a nonlinear function involving a large number of parameters ranging from thousands to millions [198,199]. As the inside is a black box, it is unclear whether the prediction is based on biologically plausible features, and the prediction results for unknown proteins cannot be guaranteed. DNN can be input from both ends of the amino acid sequence using bidirectional LSTM with a convolution layer and bidirectional LSTM layer [200,201,202]. The output layer of the DNN had the same number of neurons as the number of classes to be discriminated. Given an input vector x0 ∈ R c, we find the largest output value SI of each neuron l = {L, B, E, G, I, H, S, T, NoSeq} in the output layer. Then, the label argmaxlSl corresponding to that neuron was selected as the prediction result. At this time, saliency, which is a characteristic of the spatial arrangement of visual stimuli that induces bottom-up attention, is defined as the value of the partial differential with respect to the input x, as shown in Equation (1):
(Saliency) = maxc|∂Sl /∂x| x0|

Table 1.
Secondary structure of amino acids.
Saliency represents the result of a type of sensitivity analysis [203,204,205,206]. For example, consider the case of obtaining saliency for neurons in the output layer corresponding to an α-helix, where saliency indicates the part of the input that should be changed locally to fire the neuron in the output layer corresponding to the α-helix. For example, a large value at a certain position in the amino acid sequence on saliency indicates that changing the input at that position has a large effect on the output. By using saliency, when predicting the secondary structure label Lx of a certain position x, it is possible to determine which amino acid, feature value, at the surrounding position contributes greatly. For example, it is expected that the effect tends to approach zero at positions that are not related to the prediction, such as positions far enough away. If these results are consistent with what is known biologically, a trained DNN can be considered to capture biologically plausible features. In particular, for the α-helix and β-strand, which have high prediction accuracy, visualization with saliency is important to determine what type of amino acid exists at a position three or four residues away when predicting whether the secondary structure at a certain position in an amino acid sequence is an α-helix, because the α-helix has a right-handed helical structure with an average of 3.6 residues. Conversely, the β-strand has a structure in which amino acids are linked in a straight chain, and when making predictions, the relationship with amino acids that are close to each other is important. When the DNN acquires the correct prediction model, the saliency values at positions three or four residues away are higher when predicting the α-helix than when predicting the β-strand. This saliency is a method to obtain the value corresponding to each feature quantity of each input for each output neuron. In β-strand prediction, the saliency value gradually decreases as the distance between the sequences increases. Conversely, regarding α-helix prediction, the saliency value did not decrease from the first residue, that is, from the next amino acid to the third residue, which is consistent with the α-helix cycle length of 3.6 residues. However, when a DNN that predicts the secondary structure is visualized using saliency, a large amount of saliency is created. For human interpretation, it is necessary to obtain statistics from that saliency, and design the types of statistics to obtain. Therefore, activation maximization has been proposed in addition to saliency as a visualization method for DNN. By using these alternative visualization methods, we may extract insights without explicitly designing the statistics. Moreover, it is reported some AI-based prediction systems of protein structure with high-performance [207,208,209,210,211,212,213,214,215,216,217,218,219,220]. However, there are issues about time-consumption, high-throughput, or versatility, etc.
Research on the structures of such proteins and their associated functions has been applied to vaccine development. In particular, simulating the ‘spike protein’ present on the surface of SARS-CoV-2 and clarifying the molecular mechanism that causes the structural change of the spike protein necessary for viral infection will lead to the establishment of infection prevention and treatment methods.
6. Conclusions
By more accurately predicting the distance between the beta carbon of each amino acid residue and the beta carbon of another amino acid residue, it is possible to more accurately predict the formation of 3D structures from the amino acid sequences of proteins. Running computer simulations related to SARS-CoV-2 vaccine development can dramatically accelerate the design process and may further aid drug discovery to improve diagnostic and therapeutic outcomes.
Author Contributions
Writing, review, and editing, Y.M.; supervision, R.Y.; funding acquisition, Y.M. All authors have read and agreed to the published version of the manuscript.
Funding
This review was funded by the Fukuda Foundation for Medical Technology, and APC was funded by the Fukuda Foundation for Medical Technology.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Winkler, N.E.; Dey, A.; Quinn, H.E.; Pourmarzi, D.; Lambert, S.; McIntyre, P.; Beard, F. Australian vaccine preventable disease epidemiological review series: Measles, 2012–2019. Commun. Dis. Intell. 2022, 46. in press. [Google Scholar] [CrossRef] [PubMed]
- Paret, M.; Trillo, R.; Lighter, J.; Youngster, I.; Ratner, A.J.; Pellett Madan, R. Poor Uptake of MMR Vaccine 1-year Post-Measles Outbreak: New York City and Israel. J. Pediatric Infect. Dis. Soc. 2022, 11, 322–328. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Kelsoe, G. Role of germinal centers for the induction of broadly-reactive memory B cells. Curr. Opin. Immunol. 2017, 45, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Ura, T.; Takeuchi, M.; Kawagoe, T.; Mizuki, N.; Okuda, K.; Shimada, M. Current Vaccine Platforms in Enhancing T-Cell Response. Vaccines 2022, 10, 1367. [Google Scholar] [CrossRef] [PubMed]
- Antoñanzas, J.; Rodríguez-Garijo, N.; Estenaga, Á.; Morelló-Vicente, A.; España, A.; Aguado, L. Generalized morphea following the COVID vaccine: A series of two patients and a bibliographic review. Dermatol. Ther. 2022, 35, e15709. [Google Scholar] [CrossRef]
- Bostan, H.; Ucan, B.; Kizilgul, M.; Calapkulu, M.; Hepsen, S.; Gul, U.; Ozturk Unsal, I.; Cakal, E. Relapsed and newly diagnosed Graves’ disease due to immunization against COVID-19: A case series and review of the literature. J. Autoimmun. 2022, 128, 102809. [Google Scholar] [CrossRef]
- Pereira, D.F.S.; Ribeiro, H.S.; Gonçalves, A.A.M.; da Silva, A.V.; Lair, D.F.; de Oliveira, D.S.; Boas, D.F.V.; Conrado, I.D.S.S.; Leite, J.C.; Barata, L.M.; et al. Rhipicephalus microplus: An overview of vaccine antigens against the cattle tick. Ticks. Tick. Borne Dis. 2022, 13, 101828. [Google Scholar] [CrossRef]
- Gan, S.K.; Phua, S.X.; Yeo, J.Y. Sagacious epitope selection for vaccines, and both antibody-based therapeutics and diagnostics: Tips from virology and oncology. Antib. Ther. 2022, 5, 63–72. [Google Scholar] [CrossRef]
- Stepanova, E.; Matyushenko, V.; Rudenko, L.; Isakova-Sivak, I. Prospects of and Barriers to the Development of Epitope-Based Vaccines against Human Metapneumovirus. Pathogens 2020, 9, 481. [Google Scholar] [CrossRef]
- Moritzky, S.A.; Richards, K.A.; Glover, M.A.; Krammer, F.; Chaves, F.A.; Topham, D.J.; Branche, A.; Nayak, J.L.; Sant, A.J. The negative effect of pre-existing immunity on influenza vaccine responses transcends the impact of vaccine formulation type and vaccination history. J. Infect. Dis. 2022, in press. [Google Scholar] [CrossRef]
- Devarajan, P.; Vong, A.M.; Castonguay, C.H.; Kugler-Umana, O.; Bautista, B.L.; Jones, M.C.; Kelly, K.A.; Xia, J.; Swain, S.L. Strong influenza-induced TFH generation requires CD4 effectors to recognize antigen locally and receive signals from continuing infection. Proc. Natl. Acad. Sci. USA 2022, 119, e2111064119. [Google Scholar] [CrossRef]
- Wylie, B.; Ong, F.; Belhoul-Fakir, H.; Priebatsch, K.; Bogdawa, H.; Stirnweiss, A.; Watt, P.; Cunningham, P.; Stone, S.R.; Waithman, J. Targeting Cross-Presentation as a Route to Improve the Efficiency of Peptide-Based Cancer Vaccines. Cancers 2021, 13, 6189. [Google Scholar] [CrossRef]
- Tagliamonte, M.; Mauriello, A.; Cavalluzzo, B.; Ragone, C.; Manolio, C.; Luciano, A.; Barbieri, A.; Palma, G.; Scognamiglio, G.; Di Mauro, A.; et al. MHC-Optimized Peptide Scaffold for Improved Antigen Presentation and Anti-Tumor Response. Front. Immunol. 2021, 12, 769799. [Google Scholar] [CrossRef]
- Li, W.H.; Su, J.Y.; Li, Y.M. Rational Design of T-Cell- and B-Cell-Based Therapeutic Cancer Vaccines. Acc. Chem. Res. 2022, 55, 2660–2671. [Google Scholar] [CrossRef]
- He, B.; Liu, S.; Xu, M.; Hu, Y.; Lv, K.; Wang, Y.; Ma, Y.; Zhai, Y.; Yue, X.; Liu, L.; et al. Comparative global B cell receptor repertoire difference induced by SARS-CoV-2 infection or vaccination via single-cell V(D)J sequencing. Emerg. Microbes Infect. 2022, 11, 2007–2020. [Google Scholar] [CrossRef]
- Kumar, P.; Shiraz, M.; Akif, M. Multiepitope-based vaccine design by exploring antigenic potential among leptospiral lipoproteins using comprehensive immunoinformatics and structure-based approaches. Biotechnol. Appl. Biochem. 2022, in press. [Google Scholar] [CrossRef]
- Gupta, S.L.; Khan, N.; Basu, S.; Soni, V. B-Cell-Based Immunotherapy: A Promising New Alternative. Vaccines 2022, 10, 879. [Google Scholar] [CrossRef]
- Lee, A.; Wimmers, F.; Pulendran, B. Epigenetic adjuvants: Durable reprogramming of the innate immune system with adjuvants. Curr. Opin. Immunol. 2022, 77, 102189. [Google Scholar] [CrossRef]
- Jarisch, A.; Wiercinska, E.; Huenecke, S.; Bremm, M.; Cappel, C.; Hauler, J.; Rettinger, E.; Soerensen, J.; Hellstern, H.; Klusmann, J.H.; et al. Immune Responses to SARS-CoV-2 Vaccination in Young Patients with Anti-CD19 Chimeric Antigen Receptor T Cell-Induced B Cell Aplasia. Transplant Cell Ther. 2022, 28, 366.e1–366.e7. [Google Scholar] [CrossRef]
- Srinivasan, S.; Selvaraj, G.F.; Gopalan, V.; Padmanabhan, P.; Ramesh, K.; Govindan, K.; Chandran, A.; Dhandapani, P.; Krishnasamy, K.; Kitambi, S.S. Epitope Identification and Designing a Potent Multi-epitope Vaccine Construct against SARS-CoV-2 Including the Emerging Variants. J. Glob. Infect. Dis. 2022, 14, 24–30. [Google Scholar] [CrossRef]
- Khanum, S.; Carbone, V.; Gupta, S.K.; Yeung, J.; Shu, D.; Wilson, T.; Parlane, N.A.; Altermann, E.; Estein, S.M.; Janssen, P.H.; et al. Mapping immunogenic epitopes of an adhesin-like protein from Methanobrevibacter ruminantium M1 and comparison of empirical data with in silico prediction methods. Sci. Rep. 2022, 12, 10394. [Google Scholar] [CrossRef] [PubMed]
- Dhiman, G.; Lohia, N.; Jain, S.; Baranwal, M. Metadherin peptides containing CD4(+) and CD8(+) T cell epitopes as a therapeutic vaccine candidate against cancer. Microbiol. Immunol. 2016, 60, 646–652. [Google Scholar] [CrossRef] [PubMed]
- La Monica, G.; Bono, A.; Lauria, A.; Martorana, A. Targeting SARS-CoV-2 Main Protease for Treatment of COVID-19: Covalent Inhibitors Structure-Activity Relationship Insights and Evolution Perspectives. J. Med. Chem. 2022, 65, 12500–12534. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Jia, C.; Wu, J.; Zhang, J.; Jiang, Z.; Ma, K. Towards the Antiviral Agents and Nanotechnology-Enabled Approaches Against Parvovirus B19. Front. Cell Infect. Microbiol. 2022, 12, 916012. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Park, J.; Cho, I.H. Target-Specific Drug Discovery of Natural Products against SARS-CoV-2 Life Cycle and Cytokine Storm in COVID-19. Am. J. Chin. Med. 2022, 50, 927–959. [Google Scholar] [CrossRef]
- Leowattana, W.; Leowattana, T. Chronic hepatitis B: New potential therapeutic drugs target. World J. Virol. 2022, 11, 57–72. [Google Scholar] [CrossRef]
- Gorai, S.; Junghare, V.; Kundu, K.; Gharui, S.; Kumar, M.; Patro, B.S.; Nayak, S.K.; Hazra, S.; Mula, S. Synthesis of Dihydrobenzofuro[3,2-b]chromenes as Potential 3CLpro Inhibitors of SARS-CoV-2: A Molecular Docking and Molecular Dynamics Study. Chem. Med. Chem. 2022, 17, e202100782. [Google Scholar] [CrossRef]
- Zhai, J.; He, X.; Man, V.H.; Sun, Y.; Ji, B.; Cai, L.; Wang, J. A multiple-step in silico screening protocol to identify allosteric inhibitors of Spike-hACE2 binding. Phys. Chem. Chem. Phys. 2022, 24, 4305–4316. [Google Scholar] [CrossRef]
- Rashid, F.; Xie, Z.; Suleman, M.; Shah, A.; Khan, S.; Luo, S. Roles and functions of SARS-CoV-2 proteins in host immune evasion. Front. Immunol. 2022, 13, 940756. [Google Scholar] [CrossRef]
- Jiang, J.; Li, J.; Zhang, Y.; Zhou, C.; Guo, C.; Zhou, Z.; Ming, Y. The Protective Effect of the Soluble Egg Antigen of Schistosoma japonicum in A Mouse Skin Transplantation Model. Front. Immunol. 2022, 13, 884006. [Google Scholar] [CrossRef]
- Wang, M.; Tan, W.; Li, J.; Fang, L.; Yue, M. The Endless Wars: Severe Fever with Thrombocytopenia Syndrome Virus, Host Immune and Genetic Factors. Front. Cell Infect. Microbiol. 2022, 12, 808098. [Google Scholar] [CrossRef]
- Gori Savellini, G.; Anichini, G.; Gandolfo, C.; Cusi, M.G. Nucleopore Traffic Is Hindered by SARS-CoV-2 ORF6 Protein to Efficiently Suppress IFN-β and IL-6 Secretion. Viruses 2022, 14, 1273. [Google Scholar] [CrossRef]
- Naman, Z.T.; Kadhim, S.; Al-Isawi, Z.J.K.; Butch, C.J.; Muhseen, Z.T. Computational Investigations of Traditional Chinese Medicinal Compounds against the Omicron Variant of SARS-CoV-2 to Rescue the Host Immune System. Pharmaceuticals 2022, 15, 741. [Google Scholar] [CrossRef]
- Dong, S.; Kong, N.; Wang, C.; Li, Y.; Sun, D.; Qin, W.; Zhai, H.; Zhai, X.; Yang, X.; Ye, C.; et al. FUBP3 Degrades the Porcine Epidemic Diarrhea Virus Nucleocapsid Protein and Induces the Production of Type I Interferon. J. Virol. 2022, 96, e0061822. [Google Scholar] [CrossRef]
- Zhang, H.; Sha, H.; Qin, L.; Wang, N.; Kong, W.; Huang, L.; Zhao, M. Research Progress in Porcine Reproductive and Respiratory Syndrome Virus-Host Protein Interactions. Animals 2022, 12, 1381. [Google Scholar] [CrossRef]
- Wu, S.; Yi, W.; Gao, Y.; Deng, W.; Bi, X.; Lin, Y.; Yang, L.; Lu, Y.; Liu, R.; Chang, M.; et al. Immune Mechanisms Underlying Hepatitis B Surface Antigen Seroclearance in Chronic Hepatitis B Patients with Viral Coinfection. Front. Immunol. 2022, 13, 893512. [Google Scholar] [CrossRef]
- Chakraborty, A.; Diwan, A.; Arora, V.; Thakur, Y.; Chiniga, V.; Tatake, J.; Pandey, R.; Holkar, P.; Holkar, N.; Pond, B. Mechanism of Antiviral Activities of Nanoviricide’s Platform Technology based Biopolymer (NV-CoV-2). AIMS Public Health 2022, 9, 415–422. [Google Scholar] [CrossRef]
- Poirson, J.; Suarez, I.P.; Straub, M.L.; Cousido-Siah, A.; Peixoto, P.; Hervouet, E.; Foster, A.; Mitschler, A.; Mukobo, N.; Chebaro, Y.; et al. High-Risk Mucosal Human Papillomavirus 16 (HPV16) E6 Protein and Cutaneous HPV5 and HPV8 E6 Proteins Employ Distinct Strategies To Interfere with Interferon Regulatory Factor 3-Mediated Beta Interferon Expression. J. Virol. 2022, 96, e0187521. [Google Scholar] [CrossRef]
- Farzana, M.; Shahriar, S.; Jeba, F.R.; Tabassum, T.; Araf, Y.; Ullah, M.A.; Tasnim, J.; Chakraborty, A.; Naima, T.A.; Marma, K.K.S.; et al. Functional food: Complementary to fight against COVID-19. Beni. Suef. Univ. J. Basic Appl. Sci. 2022, 11, 33. [Google Scholar] [CrossRef]
- Ramdhan, P.; Li, C. Targeting Viral Methyltransferases: An Approach to Antiviral Treatment for ssRNA Viruses. Viruses 2022, 14, 379. [Google Scholar] [CrossRef]
- Gong, L.; Ou, X.; Hu, L.; Zhong, J.; Li, J.; Deng, S.; Li, B.; Pan, L.; Wang, L.; Hong, X.; et al. The Molecular Mechanism of Herpes Simplex Virus 1 UL31 in Antagonizing the Activity of IFN-β. Microbiol. Spectr. 2022, 10, e0188321. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Truong, A.D.; Vu, T.H.; Lee, S.; Heo, J.; Kang, S.; Lillehoj, H.S.; Hong, Y.H. Exosomes from H5N1 avian influenza virus-infected chickens regulate antiviral immune responses of chicken immune cells. Dev. Comp. Immunol. 2022, 130, 104368. [Google Scholar] [CrossRef] [PubMed]
- Sencanski, M.; Perovic, V.; Milicevic, J.; Todorovic, T.; Prodanovic, R.; Veljkovic, V.; Paessler, S.; Glisic, S. Identification of SARS-CoV-2 Papain-like Protease (PLpro) Inhibitors Using Combined Computational Approach. Chem. Open 2022, 11, e202100248. [Google Scholar] [CrossRef] [PubMed]
- Qian, Z.; Yang, C.; Xu, L.; Mickael, H.K.; Chen, S.; Zhang, Y.; Xia, Y.; Li, T.; Yu, W.; Huang, F. Hepatitis E virus-encoded microRNA promotes viral replication by inhibiting type I interferon. FASEB J. 2022, 36, e22104. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, R.l.; Ghosh, M.; Sahoo, S.; Padhi, S.; Misra, N.; Raina, V.; Suar, M.; Son, Y.O. Next-Generation Bioinformatics Approaches and Resources for Coronavirus Vaccine Discovery and Development-A Perspective Review. Vaccines 2021, 9, 812. [Google Scholar] [CrossRef]
- Chen, J.; Gao, K.; Wang, R.; Nguyen, D.D.; Wei, G.W. Review of COVID-19 Antibody Therapies. Annu. Rev. Biophys. 2021, 50, 1–30. [Google Scholar] [CrossRef]
- Park, C.; Kim, K.W.; Park, D.; Hassan, Z.U.; Park, E.C.; Lee, C.S.; Rahman, M.T.; Yi, H.; Kim, S. Rapid and sensitive amplicon-based genome sequencing of SARS-CoV-2. Front. Microbiol. 2022, 13, 876085. [Google Scholar] [CrossRef]
- Asif, M.; Amir, M.; Hussain, A.; Achakzai, N.M.; Natesan Pushparaj, P.; Rasool, M. Role of tyrosine kinase inhibitor in chronic myeloid leukemia patients with SARS-CoV-2 infection: A narrative Review. Medicine 2022, 101, e29660. [Google Scholar] [CrossRef]
- Srivastava, K.; Singh, M.K. Drug repurposing in COVID-19: A review with past, present and future. Metabol. Open 2021, 12, 100121. [Google Scholar] [CrossRef]
- Zhu, M.; Shen, J.; Zeng, Q.; Tan, J.W.; Kleepbua, J.; Chew, I.; Law, J.X.; Chew, S.P.; Tangathajinda, A.; Latthitha, N.; et al. Molecular Phylogenesis and Spatiotemporal Spread of SARS-CoV-2 in Southeast Asia. Front. Public Health 2021, 9, 685315. [Google Scholar] [CrossRef]
- Amarilla, A.A.; Sng, J.D.J.; Parry, R.; Deerain, J.M.; Potter, J.R.; Setoh, Y.X.; Rawle, D.J.; Le, T.T.; Modhiran, N.; Wang, X.; et al. A versatile reverse genetics platform for SARS-CoV-2 and other positive-strand RNA viruses. Nat. Commun. 2021, 12, 3431. [Google Scholar] [CrossRef]
- Islam, M.A.; Rahman, M.A.; Jakariya, M.; Bahadur, N.M.; Hossen, F.; Mukharjee, S.K.; Hossain, M.S.; Tasneem, A.; Haque, M.A.; Sera, F.; et al. A 30-day follow-up study on the prevalence of SARS-COV-2 genetic markers in wastewater from the residence of COVID-19 patient and comparison with clinical positivity. Sci. Total Environ. 2023, 858, 159350. [Google Scholar] [CrossRef]
- Azzarà, A.; Cassano, I.; Paccagnella, E.; Tirindelli, M.C.; Nobile, C.; Schittone, V.; Lintas, C.; Sacco, R.; Gurrieri, F. Genetic variants determine intrafamilial variability of SARS-CoV-2 clinical outcomes in 19 Italian families. PLoS ONE 2022, 17, e0275988. [Google Scholar] [CrossRef]
- Reno, U.; Regaldo, L.; Ojeda, G.; Schmuck, J.; Romero, N.; Polla, W.; Kergaravat, S.V.; Gagneten, A.M. Wastewater-Based Epidemiology: Detection of SARS-CoV-2 RNA in Different Stages of Domestic Wastewater Treatment in Santa Fe, Argentina. Water Air Soil. Pollut. 2022, 233, 372. [Google Scholar] [CrossRef]
- Iqbal, N.; Rafiq, M.; Tareen, S.; Ahmad, M.; Nawaz, F.; Khan, S.; Riaz, R.; Yang, T.; Fatima, A.; Jamal, M.; et al. The SARS-CoV-2 differential genomic adaptation in response to varying UVindex reveals potential genomic resources for better COVID-19 diagnosis and prevention. Front. Microbiol. 2022, 13, 922393. [Google Scholar] [CrossRef]
- Kim, H.S.; Lee, H.; Park, J.; Abbas, N.; Kang, S.; Hyun, H.; Seong, H.; Yoon, J.G.; Noh, J.Y.; Kim, W.J.; et al. Collection and detection of SARS-CoV-2 in exhaled breath using face mask. PLoS ONE 2022, 17, e0270765. [Google Scholar] [CrossRef]
- Huang, J.; Zhang, Z.; Hao, C.; Qiu, Y.; Tan, R.; Liu, J.; Wang, X.; Yang, W.; Qu, H. Identifying Drug-Induced Liver Injury Associated With Inflammation-Drug and Drug-Drug Interactions in Pharmacologic Treatments for COVID-19 by Bioinformatics and System Biology Analyses: The Role of Pregnane X Receptor. Front. Pharmacol. 2022, 13, 804189. [Google Scholar] [CrossRef]
- Chen, Z.; Ng, R.W.Y.; Lui, G.; Ling, L.; Chow, C.; Yeung, A.C.M.; Boon, S.S.; Wang, M.H.; Chan, K.C.C.; Chan, R.W.Y.; et al. Profiling of SARS-CoV-2 Subgenomic RNAs in Clinical Specimens. Microbiol. Spectr. 2022, 10, e0018222. [Google Scholar] [CrossRef]
- Hassan, S.S.; Choudhury, P.P.; Dayhoff, G.W., II; Aljabali, A.A.A.; Uhal, B.D.; Lundstrom, K.; Rezaei, N.; Pizzol, D.; Adadi, P.; Lal, A.; et al. The importance of accessory protein variants in the pathogenicity of SARS-CoV-2. Arch. Biochem. Biophys. 2022, 717, 109124. [Google Scholar] [CrossRef]
- Mohammed, M.E.A. SARS-CoV-2 Proteins: Are They Useful as Targets for COVID-19 Drugs and Vaccines? Curr. Mol. Med. 2022, 22, 50–66. [Google Scholar] [CrossRef]
- Dolan, K.A.; Dutta, M.; Kern, D.M.; Kotecha, A.; Voth, G.A.; Brohawn, S.G. Structure of SARS-CoV-2 M protein in lipid nanodiscs. Elife 2022, 11, e81702. [Google Scholar] [CrossRef] [PubMed]
- Araújo, L.P.; Dias, M.E.C.; Scodeler, G.C.; Santos, A.S.; Soares, L.M.; Corsetti, P.P.; Padovan, A.C.B.; Silveira, N.J.F.; de Almeida, L.A. Epitope identification of SARS-CoV-2 structural proteins using in silico approaches to obtain a conserved rational immunogenic peptide. Immunoinformatics 2022, 7, 100015. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Enríquez, A.; Herrera-Camacho, I.; Millán-Pérez-Peña, L.; Reyes-Leyva, J.; Santos-López, G.; Rivera-Benítez, J.F.; Rosas-Murrieta, N.H. Predicted 3D model of the M protein of Porcine Epidemic Diarrhea Virus and analysis of its immunogenic potential. PLoS ONE 2022, 17, e0263582. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S. Towards Determining the Epitopes of the Structural Proteins of SARS-CoV-2. Methods Mol. Biol. 2022, 2410, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Emam, M.; Oweda, M.; Antunes, A.; El-Hadidi, M. Positive selection as a key player for SARS-CoV-2 pathogenicity: Insights into ORF1ab, S and E genes. Virus Res. 2021, 302, 198472. [Google Scholar] [CrossRef]
- Boccia, A.; Tufano, R.; Ferrucci, V.; Sepe, L.; Bianchi, M.; Pascarella, S.; Zollo, M.; Paolella, G. SARS-CoV-2 Pandemic Tracing in Italy Highlights Lineages with Mutational Burden in Growing Subsets. Int. J. Mol. Sci. 2022, 23, 4155. [Google Scholar] [CrossRef]
- Urrutia-Cabrera, D.; Liou, R.H.; Wang, J.H.; Chan, J.; Hung, S.S.; Hewitt, A.W.; Martin, K.R.; Edwards, T.L.; Kwan, P.; Wong, R.C. Comparative analysis of loop-mediated isothermal amplification (LAMP)-based assays for rapid detection of SARS-CoV-2 genes. Sci. Rep. 2021, 11, 22493. [Google Scholar] [CrossRef]
- Li, L.; Zhang, L.; Zhou, J.; He, X.; Yu, Y.; Liu, P.; Huang, W.; Xiang, Z.; Chen, J. Epidemiology and Genomic Characterization of Two Novel SARS-Related Coronaviruses in Horseshoe Bats from Guangdong, China. mBio 2022, 13, e0046322. [Google Scholar] [CrossRef]
- Portakal, S.H.; Kanat, B.; Sayan, M.; Berber, B.; Doluca, O. A novel method for conserved sequence extraction with prospective mutation prediction for SARS-CoV-2 PCR primer design. J. Virol. Methods 2021, 293, 114146. [Google Scholar] [CrossRef]
- Yan, W.; Zheng, Y.; Zeng, X.; He, B.; Cheng, W. Structural biology of SARS-CoV-2: Open the door for novel therapies. Signal Transduct. Target Ther. 2022, 7, 26. [Google Scholar] [CrossRef]
- Selvaraj, C.; Dinesh, D.C.; Krafcikova, P.; Boura, E.; Aarthy, M.; Pravin, M.A.; Singh, S.K. Structural Understanding of SARS-CoV-2 Drug Targets, Active Site Contour Map Analysis and COVID-19 Therapeutics. Curr. Mol. Pharmacol. 2022, 15, 418–433. [Google Scholar] [CrossRef]
- Ebrahim, A.; Riley, B.T.; Kumaran, D.; Andi, B.; Fuchs, M.R.; McSweeney, S.; Keedy, D.A. The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (Mpro). IUCrJ. 2022, 9, 682–694. [Google Scholar] [CrossRef]
- Siddiqa, M.A.; Rao, D.S.; Suvarna, G.; Chennamachetty, V.K.; Verma, M.K.; Rao, M.V.R. In-Silico Drug Designing of Spike Receptor with Its ACE2 Receptor and Nsp10/Nsp16 MTase Complex Against SARS-CoV-2. Int. J. Pept. Res. Ther. 2021, 27, 1633–1640. [Google Scholar] [CrossRef]
- Sharma, K.; Morla, S.; Goyal, A.; Kumar, S. Computational guided drug repurposing for targeting 2’-O-ribose methyltransferase of SARS-CoV-2. Life Sci. 2020, 259, 118169. [Google Scholar] [CrossRef]
- El Khoury, L.; Jing, Z.; Cuzzolin, A.; Deplano, A.; Loco, D.; Sattarov, B.; Hédin, F.; Wendeborn, S.; Ho, C.; El Ahdab, D.; et al. Computationally driven discovery of SARS-CoV-2 Mpro inhibitors: From design to experimental validation. Chem. Sci. 2022, 13, 3674–3687. [Google Scholar] [CrossRef]
- Kim, Y.S.; Kim, B.; Kwon, E.B.; Chung, H.S.; Choi, J.G. Mulberrofuran G, a Mulberry Component, Prevents SARS-CoV-2 Infection by Blocking the Interaction between SARS-CoV-2 Spike Protein S1 Receptor-Binding Domain and Human Angiotensin-Converting Enzyme 2 Receptor. Nutrients 2022, 14, 4170. [Google Scholar] [CrossRef]
- Verkhivker, G.; Agajanian, S.; Kassab, R.; Krishnan, K. Probing Mechanisms of Binding and Allostery in the SARS-CoV-2 Spike Omicron Variant Complexes with the Host Receptor: Revealing Functional Roles of the Binding Hotspots in Mediating Epistatic Effects and Communication with Allosteric Pockets. Int. J. Mol. Sci. 2022, 23, 11542. [Google Scholar] [CrossRef]
- Sarma, S.; Herrera, S.M.; Xiao, X.; Hudalla, G.A.; Hall, C.K. Computational Design and Experimental Validation of ACE2-Derived Peptides as SARS-CoV-2 Receptor Binding Domain Inhibitors. J. Phys. Chem. B. 2022, 126, 8129–8139. [Google Scholar] [CrossRef]
- Eka Saputri, M.; Aisyah Rahmalia Effendi, S.; Nadila, R.; Azzam Fajar, S.; Damajanti Soejoedono, R.; Handharyani, E.; Nadia Poetri, O. Immunoglobulin yolk targeting spike 1, receptor binding domain of spike glycoprotein and nucleocapsid of SARS-CoV-2 blocking RBD-ACE2 binding interaction. Int. Immunopharmacol. 2022, 112, 109280. [Google Scholar] [CrossRef]
- Lv, N.; Cao, Z. RBD spatial orientation of the spike protein and its binding to ACE2: Insight into the high infectivity of the SARS-CoV-2 Delta variant from MD simulations. Phys. Chem. Chem. Phys. 2022, 24, 24155–24165. [Google Scholar] [CrossRef]
- Singh, J.; Vashishtha, S.; Rahman, S.A.; Ehtesham, N.Z.; Alam, A.; Kundu, B.; Dobrindt, U. Energetics of Spike Protein Opening of SARS-CoV-1 and SARS-CoV-2 and Its Variants of Concern: Implications in Host Receptor Scanning and Transmission. Biochemistry 2022, 61, 2188–2197. [Google Scholar] [CrossRef] [PubMed]
- Taft, J.M.; Weber, C.R.; Gao, B.; Ehling, R.A.; Han, J.; Frei, L.; Metcalfe, S.W.; Overath, M.D.; Yermanos, A.; Kelton, W.; et al. Deep mutational learning predicts ACE2 binding and antibody escape to combinatorial mutations in the SARS-CoV-2 receptor-binding domain. Cell 2022, 185, 4008–4022.e14. [Google Scholar] [CrossRef] [PubMed]
- Seifert, S.N.; Bai, S.; Fawcett, S.; Norton, E.B.; Zwezdaryk, K.J.; Robinson, J.; Gunn, B.; Letko, M. An ACE2-dependent Sarbecovirus in Russian bats is resistant to SARS-CoV-2 vaccines. PLoS Pathog. 2022, 18, e1010828. [Google Scholar] [CrossRef] [PubMed]
- Huhn, G.; Poorbaugh, J.; Zhang, L.; Beasley, S.; Nirula, A.; Brothers, J.; Welbel, S.; Wilson, J.; Gillani, S.; Weber, K.M.; et al. COVID-19 symptom relationship to antibody response and ACE2 neutralization in recovered health systems employees before and after mRNA BNT162b2 COVID-19 vaccine. PLoS ONE 2022, 17, e0273323. [Google Scholar] [CrossRef] [PubMed]
- Ching, W.Y.; Adhikari, P.; Jawad, B.; Podgornik, R. Effect of Delta and Omicron Mutations on the RBD-SD1 Domain of the Spike Protein in SARS-CoV-2 and the Omicron Mutations on RBD-ACE2 Interface Complex. Int. J. Mol. Sci. 2022, 23, 10091. [Google Scholar] [CrossRef] [PubMed]
- Lai, H.T.T.; Nguyen, L.H.; Phan, A.D.; Kranjc, A.; Nguyen, T.T.; Nguyen-Manh, D. A comparative study of receptor interactions between SARS-CoV and SARS-CoV-2 from molecular modeling. J. Mol. Model. 2022, 28, 305. [Google Scholar] [CrossRef]
- Thébault, S.; Lejal, N.; Dogliani, A.; Donchet, A.; Urvoas, A.; Valerio-Lepiniec, M.; Lavie, M.; Baronti, C.; Touret, F.; Da Costa, B.; et al. Biosynthetic proteins targeting the SARS-CoV-2 spike as anti-virals. PLoS Pathog. 2022, 18, e1010799. [Google Scholar] [CrossRef]
- Barroso da Silva, F.L.; Giron, C.C.; Laaksonen, A. Electrostatic Features for the Receptor Binding Domain of SARS-COV-2 Wildtype and Its Variants. Compass to the Severity of the Future Variants with the Charge-Rule. J. Phys. Chem. B. 2022, 126, 6835–6852. [Google Scholar] [CrossRef]
- Pitsillou, E.; Liang, J.J.; Beh, R.C.; Hung, A.; Karagiannis, T.C. Molecular dynamics simulations highlight the altered binding landscape at the spike-ACE2 interface between the Delta and Omicron variants compared to the SARS-CoV-2 original strain. Comput. Biol. Med. 2022, 149, 106035. [Google Scholar] [CrossRef]
- Erausquin, E.; Glaser, F.; Fernández-Recio, J.; López-Sagaseta, J. Structural bases for the higher adherence to ACE2 conferred by the SARS-CoV-2 spike Q498Y substitution. Acta. Crystallogr. D Struct. Biol. 2022, 78, 1156–1170. [Google Scholar] [CrossRef]
- Verma, S.; Patil, V.M.; Gupta, M.K. Mutation informatics: SARS-CoV-2 receptor-binding domain of the spike protein. Drug Discov. Today 2022, 27, 103312. [Google Scholar] [CrossRef]
- Singh, D.D.; Sharma, A.; Lee, H.J.; Yadav, D.K. SARS-CoV-2: Recent Variants and Clinical Efficacy of Antibody-Based Therapy. Front. Cell Infect. Microbiol. 2022, 12, 839170. [Google Scholar] [CrossRef]
- Liu, H.; Wei, P.; Kappler, J.W.; Marrack, P.; Zhang, G. SARS-CoV-2 Variants of Concern and Variants of Interest Receptor Binding Domain Mutations and Virus Infectivity. Front. Immunol. 2022, 13, 825256. [Google Scholar] [CrossRef]
- Ghosh, N.; Nandi, S.; Saha, I. A review on evolution of emerging SARS-CoV-2 variants based on spike glycoprotein. Int. Immunopharmacol. 2022, 105, 108565. [Google Scholar] [CrossRef]
- Kumar, A.; Parashar, R.; Kumar, S.; Faiq, M.A.; Kumari, C.; Kulandhasamy, M.; Narayan, R.K.; Jha, R.K.; Singh, H.N.; Prasoon, P.; et al. Emerging SARS-CoV-2 variants can potentially break set epidemiological barriers in COVID-19. J. Med. Virol. 2022, 94, 1300–1314. [Google Scholar] [CrossRef]
- Afolabi, R.; Chinedu, S.; Ajamma, Y.; Adam, Y.; Koenig, R.; Adebiyi, E. Computational identification of Plasmodium falciparum RNA pseudouridylate synthase as a viable drug target, its physicochemical properties, 3D structure prediction and prediction of potential inhibitors. Infect. Genet. Evol. 2022, 97, 105194. [Google Scholar] [CrossRef]
- Shimizu, Y.; Yonezawa, T.; Sakamoto, J.; Furuya, T.; Osawa, M.; Ikeda, K. Identification of novel inhibitors of Keap1/Nrf2 by a promising method combining protein-protein interaction-oriented library and machine learning. Sci. Rep. 2021, 11, 7420. [Google Scholar] [CrossRef]
- Santiago-Silva, K.M.; Camargo, P.; Felix da Silva Gomes, G.; Sotero, A.P.; Orsato, A.; Perez, C.C.; Nakazato, G.; da Silva Lima, C.H.; Bispo, M. In silico approach identified benzoylguanidines as SARS-CoV-2 main protease (Mpro) potential inhibitors. J. Biomol. Struct. Dyn. 2022, in press. [Google Scholar] [CrossRef]
- Macip, G.; Garcia-Segura, P.; Mestres-Truyol, J.; Saldivar-Espinoza, B.; Ojeda-Montes, M.J.; Gimeno, A.; Cereto-Massagué, A.; Garcia-Vallvé, S.; Pujadas, G. Haste makes waste: A critical review of docking-based virtual screening in drug repurposing for SARS-CoV-2 main protease (M-pro) inhibition. Med. Res. Rev. 2022, 42, 744–769. [Google Scholar] [CrossRef]
- de Azevedo Junior, W.F.; Bitencourt-Ferreira, G.; Godoy, J.R.; Adriano, H.M.A.; Dos Santos Bezerra, W.A.; Dos Santos Soares, A.M. Protein-Ligand Docking Simulations with AutoDock4 Focused on the Main Protease of SARS-CoV-2. Curr. Med. Chem. 2021, 28, 7614–7673. [Google Scholar] [CrossRef]
- Zhang, Y.; Yan, R.; Zhou, Q. ACE2, B0AT1, and SARS-CoV-2 spike protein: Structural and functional implications. Curr. Opin. Struct. Biol. 2022, 74, 102388. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Xiong, Y.; Zhu, G.H.; Zhang, Y.N.; Zhang, Y.W.; Huang, P.; Ge, G.B. The SARS-CoV-2 main protease (Mpro): Structure, function, and emerging therapies for COVID-19. MedComm 2022, 3, e151. [Google Scholar] [CrossRef] [PubMed]
- Nocentini, A.; Capasso, C.; Supuran, C.T. Perspectives on the design and discovery of α-ketoamide inhibitors for the treatment of novel coronavirus: Where do we stand and where do we go? Expert. Opin. Drug Discov. 2022, 17, 547–557. [Google Scholar] [CrossRef] [PubMed]
- Mahato, S. Recent Development in Small Molecules for SARS-CoV-2 and the Opportunity for Fragment-Based Drug Discovery. Med. Chem. 2022, 18, 847–858. [Google Scholar] [CrossRef] [PubMed]
- Georgoulis, V.; Haidich, A.B.; Bougioukas, K.I.; Hatzimichael, E. Efficacy and safety of carfilzomib for the treatment of multiple myeloma: An overview of systematic reviews. Crit. Rev. Oncol. Hematol. 2022, 180, 103842. [Google Scholar] [CrossRef]
- Terao, T.; Tsushima, T.; Miura, D.; Ikeda, D.; Fukumoto, A.; Kuzume, A.; Tabata, R.; Narita, K.; Takeuchi, M.; Matsue, K. Carfilzomib-induced thrombotic microangiopathy is underestimated in clinical practice: A report of five patients and literature review. Leuk. Lymphoma 2022, 63, 1102–1110. [Google Scholar] [CrossRef]
- Chaudhry, M.; Steiner, R.; Claussen, C.; Patel, K.; Lee, H.; Weber, D.; Thomas, S.; Feng, C.; Amini, B.; Orlowski, R.; et al. Carfilzomib-based combination regimens are highly effective frontline therapies for multiple myeloma and Waldenström’s macroglobulinemia. Leuk. Lymphoma 2019, 60, 964–970. [Google Scholar] [CrossRef]
- Carlos-Escalante, J.A.; de Jesús-Sánchez, M.; Rivas-Castro, A.; Pichardo-Rojas, P.S.; Arce, C.; Wegman-Ostrosky, T. The Use of Antihypertensive Drugs as Coadjuvant Therapy in Cancer. Front. Oncol. 2021, 11, 660943. [Google Scholar] [CrossRef]
- Zhan, Y.; Zhu, Y.; Wang, S.; Jia, S.; Gao, Y.; Lu, Y.; Zhou, C.; Liang, R.; Sun, D.; Wang, X.; et al. SARS-CoV-2 immunity and functional recovery of COVID-19 patients 1-year after infection. Signal Transduct. Target Ther. 2021, 6, 368. [Google Scholar] [CrossRef]
- Zhang, P.; Li, B.; Wang, Y.; Min, W.; Wang, X.; Zhou, Y.; Li, Z.; Zhao, Y.; Zhang, H.; Jiang, M.; et al. Development and multi-center clinical trials of an up-converting phosphor technology-based point-of-care (UPT-POCT) assay for rapid COVID-19 diagnosis and prediction of protective effects. BMC Microbiol. 2022, 22, 42. [Google Scholar] [CrossRef]
- Grau-Expósito, J.; Perea, D.; Suppi, M.; Massana, N.; Vergara, A.; Soler, M.J.; Trinite, B.; Blanco, J.; García-Pérez, J.; Alcamí, J.; et al. Evaluation of SARS-CoV-2 entry, inflammation and new therapeutics in human lung tissue cells. PLoS Pathog. 2022, 18, e1010171. [Google Scholar] [CrossRef]
- Ohashi, H.; Watashi, K.; Saso, W.; Shionoya, K.; Iwanami, S.; Hirokawa, T.; Shirai, T.; Kanaya, S.; Ito, Y.; Kim, K.S.; et al. Potential anti-COVID-19 agents, cepharanthine and nelfinavir, and their usage for combination treatment. iScience 2021, 24, 102367. [Google Scholar] [CrossRef]
- Kasprzyk, R.; Jemielity, J. Enzymatic Assays to Explore Viral mRNA Capping Machinery. Chembiochem 2021, 22, 3236–3253. [Google Scholar] [CrossRef]
- Jiang, Y.; Liu, L.; Manning, M.; Bonahoom, M.; Lotvola, A.; Yang, Z.-Q. Repurposing Therapeutics to Identify Novel Inhibitors Targeting 2′-O-Ribose Methyltransferase Nsp16 of SARS-CoV-2. ChemRxiv 2020, 25, 2965. [Google Scholar] [CrossRef]
- Walker, A.P.; Fan, H.; Keown, J.R.; Knight, M.L.; Grimes, J.M.; Fodor, E. The SARS-CoV-2 RNA polymerase is a viral RNA capping enzyme. Nucleic Acids Res. 2021, 49, 13019–13030. [Google Scholar] [CrossRef]
- Zhang, H.; Saravanan, K.M.; Yang, Y.; Hossain, M.T.; Li, J.; Ren, X.; Pan, Y.; Wei, Y. Deep Learning Based Drug Screening for Novel Coronavirus 2019-nCov. Interdiscip. Sci. 2020, 12, 368–376. [Google Scholar] [CrossRef]
- Gao, K.; Wang, R.; Chen, J.; Cheng, L.; Frishcosy, J.; Huzumi, Y.; Qiu, Y.; Schluckbier, T.; Wei, X.; Wei, G.W. Methodology-Centered Review of Molecular Modeling, Simulation, and Prediction of SARS-CoV-2. Chem. Rev. 2022, 122, 11287–11368. [Google Scholar] [CrossRef]
- Lyu, X.; Imai, S.; Yamano, T.; Hanayama, R. Preventing SARS-CoV-2 Infection Using Anti-spike Nanobody-IFN-β Conjugated Exosomes. Pharm. Res. 2022, in press. [Google Scholar] [CrossRef]
- Hielscher, F.; Schmidt, T.; Klemis, V.; Wilhelm, A.; Marx, S.; Abu-Omar, A.; Ziegler, L.; Guckelmus, C.; Urschel, R.; Sester, U.; et al. NVX-CoV2373-induced cellular and humoral immunity towards parental SARS-CoV-2 and VOCs compared to BNT162b2 and mRNA-1273-regimens. J. Clin. Virol. 2022, 157, 105321. [Google Scholar] [CrossRef]
- Wiedemann, A.; Pellaton, C.; Dekeyser, M.; Guillaumat, L.; Déchenaud, M.; Krief, C.; Lacabaratz, C.; Grimbert, P.; Pantaleo, G.; Lévy, Y.; et al. Longitudinal evaluation of the impact of immunosuppressive regimen on immune responses to COVID-19 vaccination in kidney transplant recipients. Front. Med. 2022, 9, 978764. [Google Scholar] [CrossRef]
- Grikscheit, K.; Rabenau, H.F.; Ghodratian, Z.; Widera, M.; Wilhelm, A.; Toptan Grabmair, T.; Hoehl, S.; Layer, E.; Helfritz, F.; Ciesek, S. Characterization of the Antibody and Interferon-Gamma Release Response after a Second COVID-19 Booster Vaccination. Vaccines 2022, 10, 1163. [Google Scholar] [CrossRef] [PubMed]
- Seki, Y.; Yoshihara, Y.; Nojima, K.; Momose, H.; Fukushi, S.; Moriyama, S.; Wagatsuma, A.; Numata, N.; Sasaki, K.; Kuzuoka, T.; et al. Safety and immunogenicity of the Pfizer/BioNTech SARS-CoV-2 mRNA third booster vaccine dose against the BA.1 and BA.2 Omicron variants. Med 2022, 3, 406–421.e4. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.K.; Go, J.; Sung, H.; Kim, S.W.; Walter, M.; Knabl, L.; Furth, P.A.; Hennighausen, L.; Huh, J.W. Heterologous ChAdOx1-BNT162b2 vaccination in Korean cohort induces robust immune and antibody responses that includes Omicron. iScience 2022, 25, 104473. [Google Scholar] [CrossRef] [PubMed]
- Karaba, A.H.; Johnston, T.S.; Aytenfisu, T.Y.; Akinde, O.; Eby, Y.; Ruff, J.E.; Abedon, A.T.; Alejo, J.L.; Blankson, J.N.; Cox, A.L.; et al. A Fourth Dose of COVID-19 Vaccine Does Not Induce Neutralization of the Omicron Variant Among Solid Organ Transplant Recipients with Suboptimal Vaccine Response. Transplantation 2022, 106, 1440–1444. [Google Scholar] [CrossRef] [PubMed]
- Benfield, T.L.; Iversen, K.K.; Mustafa, A.B.; Juhl, M.R.; Petersen, K.T.; Ostrowski, S.R.; Lindvig, S.O.; Rasmussen, L.D.; Schleimann, M.H.; Andersen, S.D.; et al. Comparison of vaccine-induced antibody neutralization against SARS-CoV-2 variants of concern following primary and booster doses of COVID-19 vaccines. Front. Med. 2022, 9, 994160. [Google Scholar] [CrossRef]
- Zafar, U.; Zafar, H.; Ahmed, M.S.; Khattak, M. Link between COVID-19 vaccines and myocardial infarction. World J. Clin. Cases 2022, 10, 10109–10119. [Google Scholar] [CrossRef]
- Morgan, M.C.; Atri, L.; Harrell, S.; Al-Jaroudi, W.; Berman, A. COVID-19 vaccine-associated myocarditis. World J. Cardiol. 2022, 14, 382–391. [Google Scholar] [CrossRef]
- Ho, J.Y.K.; Siu, I.C.H.; Ng, K.H.L.; Tam, M.; Chow, S.C.Y.; Lim, K.; Kwok, M.W.T.; Wan, S.; Fujikawa, T.; Wong, R.H.L. Retrospective record review on timing of COVID-19 vaccination and cardiac surgery. J. Card. Surg. 2022, 37, 3634–3638. [Google Scholar] [CrossRef]
- Risk, M.; Hayek, S.S.; Schiopu, E.; Yuan, L.; Shen, C.; Shi, X.; Freed, G.; Zhao, L. COVID-19 vaccine effectiveness against omicron (B.1.1.529) variant infection and hospitalisation in patients taking immunosuppressive medications: A retrospective cohort study. Lancet Rheumatol. 2022, 4, E775–E784. [Google Scholar] [CrossRef]
- Tan, E.; Salman, S. Unusual Case of Painful Glossitis and Xerostomia Following Vaccination with Pfizer-BioNTech SARS-CoV-2 (BNT162b2). Am. J. Case Rep. 2022, 23, e937212. [Google Scholar] [CrossRef]
- Numakura, T.; Murakami, K.; Tamada, T.; Yamaguchi, C.; Inoue, C.; Ohkouchi, S.; Tode, N.; Sano, H.; Aizawa, H.; Sato, K.; et al. A Novel Development of Sarcoidosis Following COVID-19 Vaccination and a Literature Review. Intern. Med. 2022, 61, 3101–3106. [Google Scholar] [CrossRef]
- Patel, S.; Wu, E.; Mundae, M.; Lim, K. Myocarditis and pericarditis following mRNA vaccination in autoimmune inflammatory rheumatic disease patients: A single-center experience. Rheumatol. Autoimmun. 2022, 2, 92–97. [Google Scholar] [CrossRef]
- Chandra, P.; Roldao, M.; Drachenberg, C.; Santos, P.; Washida, N.; Clark, A.; Bista, B.; Mitsuna, R.; Yango, A. Minimal change disease and COVID-19 vaccination: Four cases and review of literature. Clin. Nephrol. Case Stud. 2022, 10, 54–63. [Google Scholar] [CrossRef]
- Yong, S.J.; Halim, A.; Halim, M.; Al Mutair, A.; Alhumaid, S.; Al-Sihati, J.; Albayat, H.; Alsaeed, M.; Garout, M.; Al Azmi, R.; et al. Rare Adverse Events Associated with BNT162b2 mRNA Vaccine (Pfizer-BioNTech): A Review of Large-Scale, Controlled Surveillance Studies. Vaccines 2022, 10, 1067. [Google Scholar] [CrossRef]
- Ritskes-Hoitinga, M.; Barella, Y.; Kleinhout-Vliek, T. The Promises of Speeding Up: Changes in Requirements for Animal Studies and Alternatives during COVID-19 Vaccine Approval-A Case Study. Animals 2022, 12, 1735. [Google Scholar] [CrossRef]
- Zou, Y.; Huang, D.; Jiang, Q.; Guo, Y.; Chen, C. The Vaccine Efficacy Against the SARS-CoV-2 Omicron: A Systemic Review and Meta-Analysis. Front. Public Health 2022, 10, 940956. [Google Scholar] [CrossRef]
- Jawalagatti, V.; Kirthika, P.; Lee, J.H. Oral mRNA Vaccines Against Infectious Diseases- A Bacterial Perspective. Front. Immunol. 2022, 13, 884862. [Google Scholar] [CrossRef]
- Shi, J.; Huang, M.W.; Lu, Z.D.; Du, X.J.; Shen, S.; Xu, C.F.; Wang, J. Delivery of mRNA for regulating functions of immune cells. J. Control. Release 2022, 345, 494–511. [Google Scholar] [CrossRef]
- Banerjee, S.; Banerjee, D.; Singh, A.; Saharan, V.A. A Comprehensive Investigation Regarding the Differentiation of the Procurable COVID-19 Vaccines. AAPS PharmSciTech 2022, 23, 95. [Google Scholar] [CrossRef]
- Gasmi, A.; Srinath, S.; Dadar, M.; Pivina, L.; Menzel, A.; Benahmed, A.G.; Chirumbolo, S.; Bjørklund, G. A global survey in the developmental landscape of possible vaccination strategies for COVID-19. Clin. Immunol. 2022, 237, 108958. [Google Scholar] [CrossRef]
- Feikin, D.R.; Higdon, M.M.; Abu-Raddad, L.J.; Andrews, N.; Araos, R.; Goldberg, Y.; Groome, M.J.; Huppert, A.; O’Brien, K.L.; Smith, P.G.; et al. Duration of effectiveness of vaccines against SARS-CoV-2 infection and COVID-19 disease: Results of a systematic review and meta-regression. Lancet 2022, 399, 924–944. [Google Scholar] [CrossRef] [PubMed]
- Buckley, J.E.; Landis, L.N.; Rapini, R.P. Pityriasis rosea-like rash after messenger RNA COVID-19 vaccination: A case report and review of the literature. JAAD Int. 2022, 7, 164–168. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.; Kaki, M.; Potluri, V.S.; Kahar, P.; Khanna, D. A comprehensive review of SARS-CoV-2 vaccines: Pfizer, Moderna & Johnson & Johnson. Hum. Vaccin. Immunother. 2022, 18, 2002083. [Google Scholar] [CrossRef] [PubMed]
- Pratama, N.R.; Wafa, I.A.; Budi, D.S.; Putra, M.; Wardhana, M.P.; Wungu, C.D.K. mRNA Covid-19 vaccines in pregnancy: A systematic review. PLoS ONE 2022, 17, e0261350. [Google Scholar] [CrossRef] [PubMed]
- Simnani, F.Z.; Singh, D.; Kaur, R. COVID-19 phase 4 vaccine candidates, effectiveness on SARS-CoV-2 variants, neutralizing antibody, rare side effects, traditional and nano-based vaccine platforms: A review. 3 Biotech. 2022, 12, 15. [Google Scholar] [CrossRef]
- Sapkota, B.; Saud, B.; Shrestha, R.; Al-Fahad, D.; Sah, R.; Shrestha, S.; Rodriguez-Morales, A.J. Heterologous prime-boost strategies for COVID-19 vaccines. J. Travel. Med. 2022, 29, taab191. [Google Scholar] [CrossRef]
- Wang, Z.; Popowski, K.D.; Zhu, D.; de Juan Abad, B.L.; Wang, X.; Liu, M.; Lutz, H.; De Naeyer, N.; DeMarco, C.T.; Denny, T.N.; et al. Exosomes decorated with a recombinant SARS-CoV-2 receptor-binding domain as an inhalable COVID-19 vaccine. Nat. Biomed. Eng. 2022, 6, 791–805. [Google Scholar] [CrossRef]
- Mustajab, T.; Kwamboka, M.S.; Choi, D.A.; Kang, D.W.; Kim, J.; Han, K.R.; Han, Y.; Lee, S.; Song, D.; Chwae, Y.J. Update on Extracellular Vesicle-Based Vaccines and Therapeutics to Combat COVID-19. Int. J. Mol. Sci. 2022, 23, 11247. [Google Scholar] [CrossRef]
- Stewart, E.L.; Counoupas, C.; Johansen, M.D.; Nguyen, D.H.; Miemczyk, S.; Hansbro, N.G.; Ferrell, K.C.; Ashhurst, A.; Alca, S.; Ashley, C.; et al. Mucosal immunization with a delta-inulin adjuvanted recombinant spike vaccine elicits lung-resident immune memory and protects mice against SARS-CoV-2. Mucosal. Immunol. 2022, 15, 1405–1415. [Google Scholar] [CrossRef]
- Martínez-Puente, D.H.; Pérez-Trujillo, J.J.; Zavala-Flores, L.M.; García-García, A.; Villanueva-Olivo, A.; Rodríguez-Rocha, H.; Valdés, J.; Saucedo-Cárdenas, O.; Montes de Oca-Luna, R.; Loera-Arias, M.J. Plasmid DNA for Therapeutic Applications in Cancer. Pharmaceutics 2022, 14, 1861. [Google Scholar] [CrossRef]
- Fomsgaard, A.; Liu, M.A. The Key Role of Nucleic Acid Vaccines for One Health. Viruses 2021, 13, 258. [Google Scholar] [CrossRef]
- Kayraklioglu, N.; Horuluoglu, B.; Klinman, D.M. CpG Oligonucleotides as Vaccine Adjuvants. Methods Mol. Biol. 2021, 2197, 51–85. [Google Scholar] [CrossRef]
- Zhang, Z.; Kuo, J.C.; Yao, S.; Zhang, C.; Khan, H.; Lee, R.J. CpG Oligodeoxynucleotides for Anticancer Monotherapy from Preclinical Stages to Clinical Trials. Pharmaceutics 2021, 14, 73. [Google Scholar] [CrossRef]
- Chen, W.; Jiang, M.; Yu, W.; Xu, Z.; Liu, X.; Jia, Q.; Guan, X.; Zhang, W. CpG-Based Nanovaccines for Cancer Immunotherapy. Int. J. Nanomedicine. 2021, 16, 5281–5299. [Google Scholar] [CrossRef]
- Jin, Y.; Zhuang, Y.; Dong, X.; Liu, M. Development of CpG oligodeoxynucleotide TLR9 agonists in anti-cancer therapy. Expert Rev. Anticancer Ther. 2021, 21, 841–851. [Google Scholar] [CrossRef]
- Putzke, S.; Feldhues, E.; Heep, I.; Ilg, T.; Lamprecht, A. Cationic lipid/pDNA complex formation as potential generic method to generate specific IRF pathway stimulators. Eur. J. Pharm. Biopharm. 2020, 155, 112–121. [Google Scholar] [CrossRef]
- Yasuda, S.; Yoshida, H.; Nishikawa, M.; Takakura, Y. Comparison of the type of liposome involving cytokine production induced by non-CpG Lipoplex in macrophages. Mol. Pharm. 2010, 7, 533–542. [Google Scholar] [CrossRef]
- Gupta, G.K.; Agrawal, D.K. CpG oligodeoxynucleotides as TLR9 agonists: Therapeutic application in allergy and asthma. BioDrugs 2010, 24, 225–235. [Google Scholar] [CrossRef]
- Tsujihana, K.; Tanegashima, K.; Santo, Y.; Yamada, H.; Akazawa, S.; Nakao, R.; Tominaga, K.; Saito, R.; Nishito, Y.; Hata, R.I.; et al. Circadian protection against bacterial skin infection by epidermal CXCL14-mediated innate immunity. Proc. Natl. Acad. Sci. USA 2022, 119, e2116027119. [Google Scholar] [CrossRef]
- Bi, X.; Liu, W.; Ding, X.; Liang, S.; Zheng, Y.; Zhu, X.; Quan, S.; Yi, X.; Xiang, N.; Du, J.; et al. Proteomic and metabolomic profiling of urine uncovers immune responses in patients with COVID-19. Cell Rep. 2022, 38, 110271. [Google Scholar] [CrossRef]
- Iwase, R.; Naruse, N.; Nakagawa, M.; Saito, R.; Shigenaga, A.; Otaka, A.; Hara, T.; Tanegashima, K. Identification of Functional Domains of CXCL14 Involved in High-Affinity Binding and Intracellular Transport of CpG DNA. J. Immunol. 2021, 207, 459–469. [Google Scholar] [CrossRef] [PubMed]
- Tanegashima, K.; Takahashi, R.; Nuriya, H.; Iwase, R.; Naruse, N.; Tsuji, K.; Shigenaga, A.; Otaka, A.; Hara, T. CXCL14 Acts as a Specific Carrier of CpG DNA into Dendritic Cells and Activates Toll-like Receptor 9-mediated Adaptive Immunity. EBioMedicine 2017, 24, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Larsen, K.C.; Spencer, A.J.; Goodman, A.L.; Gilchrist, A.; Furze, J.; Rollier, C.S.; Kiss-Toth, E.; Gilbert, S.C.; Bregu, M.; Soilleux, E.J.; et al. Expression of tak1 and tram induces synergistic pro-inflammatory signalling and adjuvants DNA vaccines. Vaccine 2009, 27, 5589–5598. [Google Scholar] [CrossRef] [PubMed]
- Hoque, M.N.; Sarkar, M.M.H.; Khan, M.A.; Hossain, M.A.; Hasan, M.I.; Rahman, M.H.; Habib, M.A.; Akter, S.; Banu, T.A.; Goswami, B.; et al. Differential gene expression profiling reveals potential biomarkers and pharmacological compounds against SARS-CoV-2: Insights from machine learning and bioinformatics approaches. Front. Immunol. 2022, 13, 918692. [Google Scholar] [CrossRef] [PubMed]
- Maghsoudi, S.; Taghavi Shahraki, B.; Rameh, F.; Nazarabi, M.; Fatahi, Y.; Akhavan, O.; Rabiee, M.; Mostafavi, E.; Lima, E.C.; Saeb, M.R.; et al. A review on computer-aided chemogenomics and drug repositioning for rational COVID-19 drug discovery. Chem. Biol. Drug Des. 2022, 100, 699–721. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Kumar, G.S.; Maitra, S.S.; Malý, P.; Bharadwaj, S.; Sharma, P.; Dwivedi, V.D. Viral informatics: Bioinformatics-based solution for managing viral infections. Brief Bioinform. 2022, 23, bbac326. [Google Scholar] [CrossRef]
- Marques-Pereira, C.; Pires, M.; Moreira, I.S. Discovery of Virus-Host interactions using bioinformatic tools. Methods Cell Biol. 2022, 169, 169–198. [Google Scholar] [CrossRef]
- Swain, S.S.; Singh, S.R.; Sahoo, A.; Panda, P.K.; Hussain, T.; Pati, S. Integrated bioinformatics-cheminformatics approach toward locating pseudo-potential antiviral marine alkaloids against SARS-CoV-2-Mpro. Proteins 2022, 90, 1617–1633. [Google Scholar] [CrossRef]
- Ghaznavi, H.; Shirvaliloo, M.; Sargazi, S.; Mohammadghasemipour, Z.; Shams, Z.; Hesari, Z.; Shahraki, O.; Nazarlou, Z.; Sheervalilou, R.; Shirvalilou, S. SARS-CoV-2 and influenza viruses: Strategies to cope with coinfection and bioinformatics perspective. Cell Biol. Int. 2022, 46, 1009–1020. [Google Scholar] [CrossRef]
- Gorbalenya, A.E.; Anisimova, M. Editorial overview: Virus bioinformatics—Empowering genomics of pathogens, viromes, and the virosphere across divergence scales. Curr. Opin. Virol. 2022, 52, 161–165. [Google Scholar] [CrossRef]
- Robertson, A.J.; Courtney, J.M.; Shen, Y.; Ying, J.; Bax, A. Concordance of X-ray and AlphaFold2 Models of SARS-CoV-2 Main Protease with Residual Dipolar Couplings Measured in Solution. J. Am. Chem. Soc. 2021, 143, 19306–19310. [Google Scholar] [CrossRef]
- Beuming, T.; Martín, H.; Díaz-Rovira, A.M.; Díaz, L.; Guallar, V.; Ray, S.S. Are Deep Learning Structural Models Sufficiently Accurate for Free-Energy Calculations? Application of FEP+ to AlphaFold2-Predicted Structures. J. Chem. Inf. Model. 2022, 62, 4351–4360. [Google Scholar] [CrossRef]
- Lee, D.; Xiong, D.; Wierbowski, S.; Li, L.; Liang, S.; Yu, H. Deep learning methods for 3D structural proteome and interactome modeling. Curr. Opin. Struct. Biol. 2022, 73, 102329. [Google Scholar] [CrossRef]
- Tsaban, T.; Varga, J.K.; Avraham, O.; Ben-Aharon, Z.; Khramushin, A.; Schueler-Furman, O. Harnessing protein folding neural networks for peptide-protein docking. Nat. Commun. 2022, 13, 176. [Google Scholar] [CrossRef]
- McCoy, A.J.; Sammito, M.D.; Read, R.J. Implications of AlphaFold2 for crystallographic phasing by molecular replacement. Acta. Crystallogr. D Struct. Biol. 2022, 78, 1–13. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Applying and improving AlphaFold at CASP14. Proteins 2021, 89, 1711–1721. [Google Scholar] [CrossRef]
- Cramer, P. AlphaFold2 and the future of structural biology. Nat. Struct. Mol. Biol. 2021, 28, 704–705. [Google Scholar] [CrossRef]
- Marzolf, D.R.; Seffernick, J.T.; Lindert, S. Protein Structure Prediction from NMR Hydrogen-Deuterium Exchange Data. J. Chem. Theory Comput. 2021, 17, 2619–2629. [Google Scholar] [CrossRef]
- Andreini, C.; Rosato, A. Structural Bioinformatics and Deep Learning of Metalloproteins: Recent Advances and Applications. Int. J. Mol. Sci. 2022, 23, 7684. [Google Scholar] [CrossRef]
- Park, S.; Seok, C. GalaxyWater-CNN: Prediction of Water Positions on the Protein Structure by a 3D-Convolutional Neural Network. J. Chem. Inf. Model. 2022, 62, 3157–3168. [Google Scholar] [CrossRef]
- Perez, M.A.S.; Cuendet, M.A.; Röhrig, U.F.; Michielin, O.; Zoete, V. Structural Prediction of Peptide-MHC Binding Modes. Methods Mol. Biol. 2022, 2405, 245–282. [Google Scholar] [CrossRef] [PubMed]
- Yalcin-Ozkat, G. Molecular Modeling Strategies of Cancer Multidrug Resistance. Drug Resist. Updat. 2021, 59, 100789. [Google Scholar] [CrossRef] [PubMed]
- Jing, X.; Dong, Q. MQAPRank: Improved global protein model quality assessment by learning-to-rank. BMC Bioinform. 2017, 18, 275. [Google Scholar] [CrossRef] [PubMed]
- Sato, R.; Ishida, T. Protein model accuracy estimation based on local structure quality assessment using 3D convolutional neural network. PLoS ONE 2019, 14, e0221347. [Google Scholar] [CrossRef] [PubMed]
- Dankelman, L.H.M.; Schilstra, S.; IJpma, F.F.A.; Doornberg, J.N.; Colaris, J.W.; Verhofstad, M.H.J.; Wijffels, M.M.E.; Prijs, J. Artificial intelligence fracture recognition on computed tomography: Review of literature and recommendations. Eur. J. Trauma Emerg. Surg. 2022, in press. [Google Scholar] [CrossRef]
- Islam, M.M.; Nooruddin, S.; Karray, F.; Muhammad, G. Human activity recognition using tools of convolutional neural networks: A state of the art review, data sets, challenges, and future prospects. Comput. Biol. Med. 2022, 149, 106060. [Google Scholar] [CrossRef]
- Baur, D.; Kroboth, K.; Heyde, C.E.; Voelker, A. Convolutional Neural Networks in Spinal Magnetic Resonance Imaging: A Systematic Review. World Neurosurg. 2022, 166, 60–70. [Google Scholar] [CrossRef]
- Lin, Y.; Xu, J.; Zhang, Y. Identification Method of Citrus Aurantium Diseases and Pests Based on Deep Convolutional Neural Network. Comput. Intell. Neurosci. 2022, 2022, 7012399. [Google Scholar] [CrossRef]
- Loddo, A.; Fadda, C.; Di Ruberto, C. An Empirical Evaluation of Convolutional Networks for Malaria Diagnosis. J. Imaging. 2022, 8, 66. [Google Scholar] [CrossRef]
- Ren, H.; Zhang, Q.; Wang, Z.; Zhang, G.; Liu, H.; Guo, W.; Mukamel, S.; Jiang, J. Machine learning recognition of protein secondary structures based on two-dimensional spectroscopic descriptors. Proc. Natl. Acad. Sci. USA 2022, 119, e2202713119. [Google Scholar] [CrossRef]
- Yu, C.H.; Chen, W.; Chiang, Y.H.; Guo, K.; Martin Moldes, Z.; Kaplan, D.L.; Buehler, M.J. End-to-End Deep Learning Model to Predict and Design Secondary Structure Content of Structural Proteins. ACS Biomater. Sci. Eng. 2022, 8, 1156–1165. [Google Scholar] [CrossRef]
- Robson, B. Testing machine learning techniques for general application by using protein secondary structure prediction. A brief survey with studies of pitfalls and benefits using a simple progressive learning approach. Comput. Biol. Med. 2021, 138, 104883. [Google Scholar] [CrossRef]
- Goodswen, S.J.; Kennedy, P.J.; Ellis, J.T. Predicting Protein Therapeutic Candidates for Bovine Babesiosis Using Secondary Structure Properties and Machine Learning. Front. Genet. 2021, 12, 716132. [Google Scholar] [CrossRef]
- Bouvier, B. Protein-Protein Interface Topology as a Predictor of Secondary Structure and Molecular Function Using Convolutional Deep Learning. J. Chem. Inf. Model. 2021, 61, 3292–3303. [Google Scholar] [CrossRef]
- Chelur, V.R.; Priyakumar, U.D. BiRDS—Binding Residue Detection from Protein Sequences Using Deep ResNets. J. Chem. Inf. Model. 2022, 62, 1809–1818. [Google Scholar] [CrossRef]
- Du, Z.; Su, H.; Wang, W.; Ye, L.; Wei, H.; Peng, Z.; Anishchenko, I.; Baker, D.; Yang, J. The trRosetta server for fast and accurate protein structure prediction. Nat. Protoc. 2021, 16, 5634–5651. [Google Scholar] [CrossRef]
- Liu, Z.; Gong, Y.; Bao, Y.; Guo, Y.; Wang, H.; Lin, G.N. TMPSS: A Deep Learning-Based Predictor for Secondary Structure and Topology Structure Prediction of Alpha-Helical Transmembrane Proteins. Front. Bioeng. Biotechnol. 2021, 8, 629937. [Google Scholar] [CrossRef]
- Wu, B.; Zheng, C. Pattern Recognition of Holographic Image Library Based on Deep Learning. J. Healthc. Eng. 2022, 2022, 2129168. [Google Scholar] [CrossRef]
- Yu, B.; Zhou, L.; Wang, L.; Yang, W.; Yang, M.; Bourgeat, P.; Fripp, J. SA-LuT-Nets: Learning Sample-Adaptive Intensity Lookup Tables for Brain Tumor Segmentation. IEEE Trans. Med. Imaging 2021, 40, 1417–1427. [Google Scholar] [CrossRef]
- Jeong, S.; Cheon, W.; Cho, S.; Han, Y. Clinical applicability of deep learning-based respiratory signal prediction models for four-dimensional radiation therapy. PLoS ONE 2022, 17, e0275719. [Google Scholar] [CrossRef]
- Wang, C.; Garlick, S.; Zloh, M. Deep Learning for Novel Antimicrobial Peptide Design. Biomolecules 2021, 11, 471. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Shrivastava, S.; Kumar Singh, S.; Kumar, A.; Saxena, S.; Kumar Singh, R. Deep-ABPpred: Identifying antibacterial peptides in protein sequences using bidirectional LSTM with word2vec. Brief Bioinform. 2021, 22, bbab065. [Google Scholar] [CrossRef] [PubMed]
- Ayhan, M.S.; Kümmerle, L.B.; Kühlewein, L.; Inhoffen, W.; Aliyeva, G.; Ziemssen, F.; Berens, P. Clinical validation of saliency maps for understanding deep neural networks in ophthalmology. Med. Image Anal. 2022, 77, 102364. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Yao, R.; Liao, G.; Liu, B.; Qiu, G. Visual Saliency via Embedding Hierarchical Knowledge in a Deep Neural Network. IEEE Trans. Image Process. 2020, 29, 8490–8505. [Google Scholar] [CrossRef] [PubMed]
- Yan, K.; Wang, X.; Kim, J.; Feng, D. A New Aggregation of DNN Sparse and Dense Labeling for Saliency Detection. IEEE Trans. Cybern. 2021, 51, 5907–5920. [Google Scholar] [CrossRef]
- Lu, Y.; Liu, A.A.; Chen, M.; Nie, W.Z.; Su, Y.T. Sequential Saliency Guided Deep Neural Network for Joint Mitosis Identification and Localization in Time-Lapse Phase Contrast Microscopy Images. IEEE J. Biomed. Health Inform. 2020, 24, 1367–1378. [Google Scholar] [CrossRef]
- Jumper, J.; Hassabis, D. Protein structure predictions to atomic accuracy with AlphaFold. Nat. Methods 2022, 19, 11–12. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Peng, Z.; Wang, W.; Han, R.; Zhang, F.; Yang, J. Protein structure prediction in the deep learning era. Curr. Opin. Struct. Biol. 2022, 77, 102495. [Google Scholar] [CrossRef]
- Nikam, R.; Yugandhar, K.; Gromiha, M.M. DeepBSRPred: Deep learning-based binding site residue prediction for proteins. Amino Acids 2022, in press. [Google Scholar] [CrossRef]
- Ferruz, N.; Heinzinger, M.; Akdel, M.; Goncearenco, A.; Naef, L.; Dallago, C. From sequence to function through structure: Deep learning for protein design. Comput. Struct. Biotechnol. J. 2022, 21, 238–250. [Google Scholar] [CrossRef]
- Lee, S.; Kim, S.; Lee, G.R.; Kwon, S.; Woo, H.; Seok, C.; Park, H. Evaluating GPCR modeling and docking strategies in the era of deep learning-based protein structure prediction. Comput. Struct. Biotechnol. J. 2022, 21, 158–167. [Google Scholar] [CrossRef]
- Wang, P.H.; Zhu, Y.H.; Yang, X.; Yu, D.J. GCmapCrys: Integrating graph attention network with predicted contact map for multi-stage protein crystallization propensity prediction. Anal. Biochem. 2022, 663, 115020. [Google Scholar] [CrossRef]
- Derry, A.; Altman, R.B. COLLAPSE: A representation learning framework for identification and characterization of protein structural sites. Protein Sci. 2022, 15, e4541. [Google Scholar] [CrossRef]
- Yuan, L.; Hu, X.; Ma, Y.; Liu, Y. DLBLS_SS: Protein secondary structure prediction using deep learning and broad learning system. RSC Adv. 2022, 12, 33479–33487. [Google Scholar] [CrossRef]
- Lin, P.; Yan, Y.; Huang, S.Y. DeepHomo2.0: Improved protein-protein contact prediction of homodimers by transformer-enhanced deep learning. Brief Bioinform. 2022, in press. [Google Scholar] [CrossRef]
- Kang, Y.; Xu, Y.; Wang, X.; Pu, B.; Yang, X.; Rao, Y.; Chen, J. HN-PPISP: A hybrid network based on MLP-Mixer for protein-protein interaction site prediction. Brief Bioinform. 2022, 19, bbac480. [Google Scholar] [CrossRef]
- Aybey, E.; Gümüş, Ö. SENSDeep: An Ensemble Deep Learning Method for Protein-Protein Interaction Sites Prediction. Interdiscip. Sci. 2022, in press. [Google Scholar] [CrossRef]
- Soleymani, F.; Paquet, E.; Viktor, H.; Michalowski, W.; Spinello, D. Protein-protein interaction prediction with deep learning: A comprehensive review. Comput. Struct. Biotechnol. J. 2022, 20, 5316–5341. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, C.; Yu, D.J.; Zhang, Y. Deep learning geometrical potential for high-accuracy ab initio protein structure prediction. iScience 2022, 25, 104425. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).