Analysis of Differentially Expressed Genes, MMP3 and TESC, and Their Potential Value in Molecular Pathways in Colon Adenocarcinoma: A Bioinformatics Approach
Abstract
1. Introduction
2. Materials and Methods
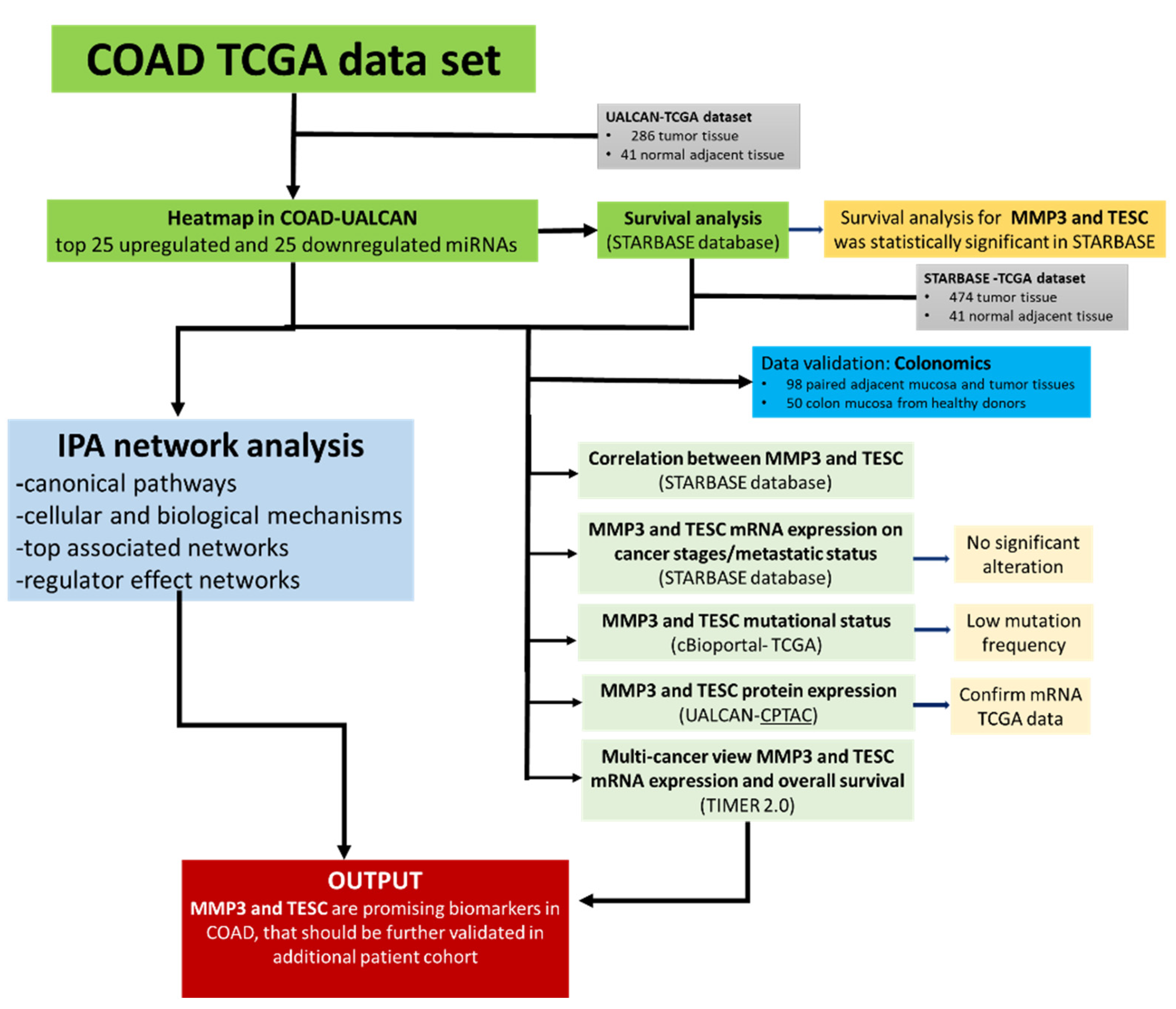
2.1. Study Design
2.2. Data Mining of TCGA Data Set in COAD
2.3. Inclusion and Exclusion Criteria
2.4. Survival Analysis STARBASE Database
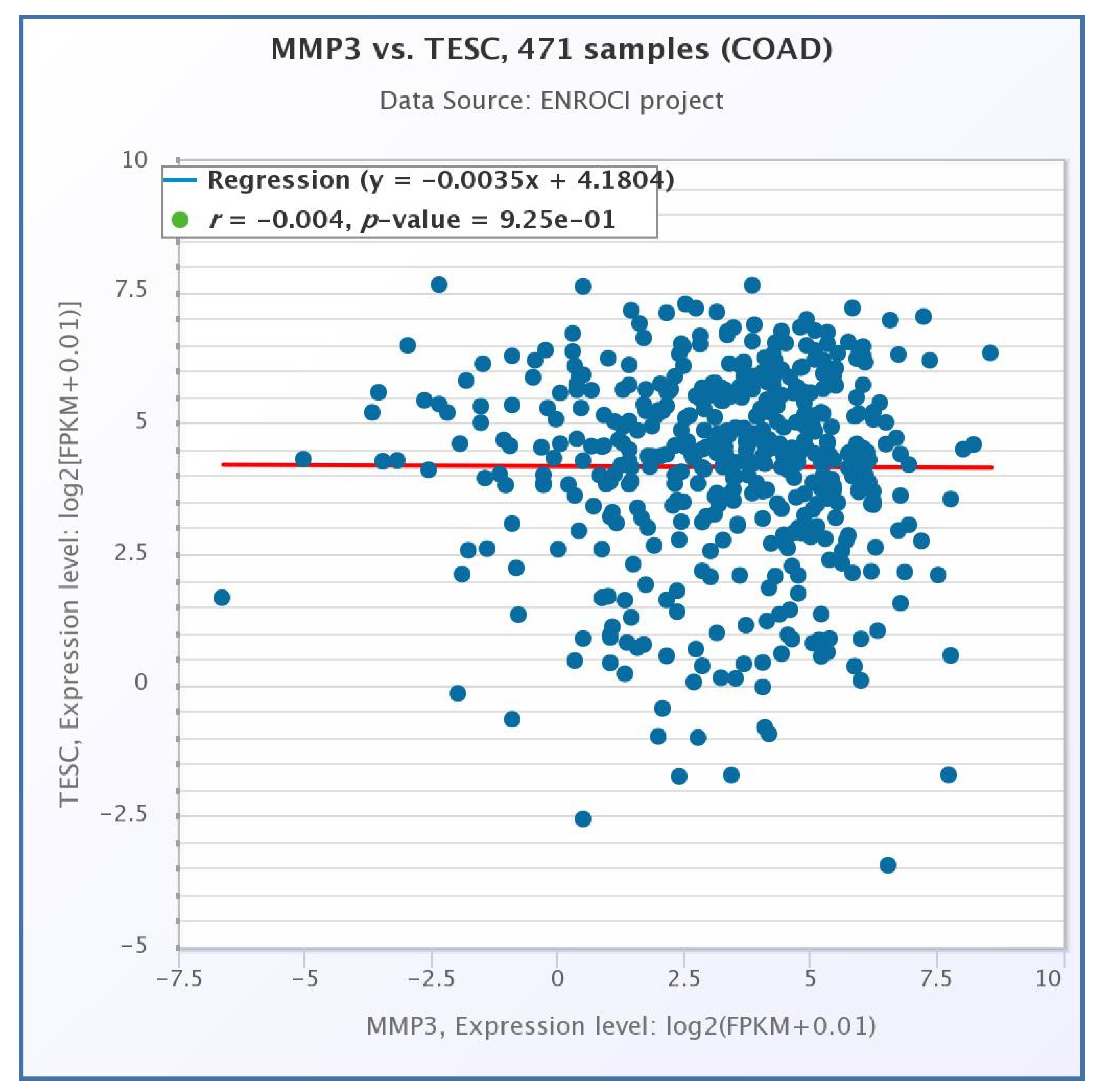
2.5. Pearson Correlation Analysis for Gene Expression Data
2.6. Genetic Alterations Using cBioPortal
2.7. MMP3 with TESC in COAD Protein Expression Levels
2.8. Validation of MMP3 and TESC Expression Level with the Colonomics Database
2.9. Pathway Analysis in COAD
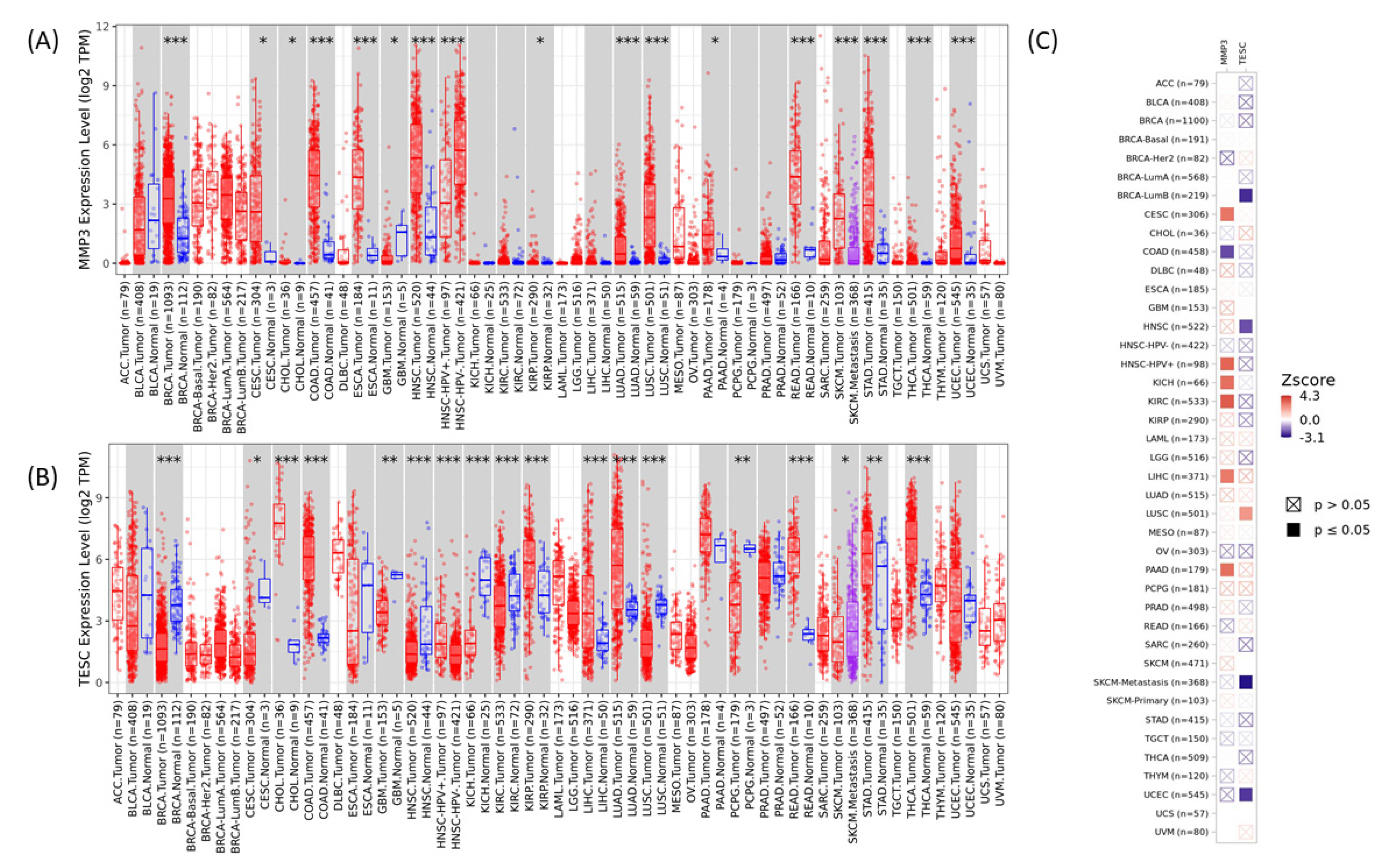
2.10. Multi-Cancer View of MMP3 and TESC in Cancer
3. Results
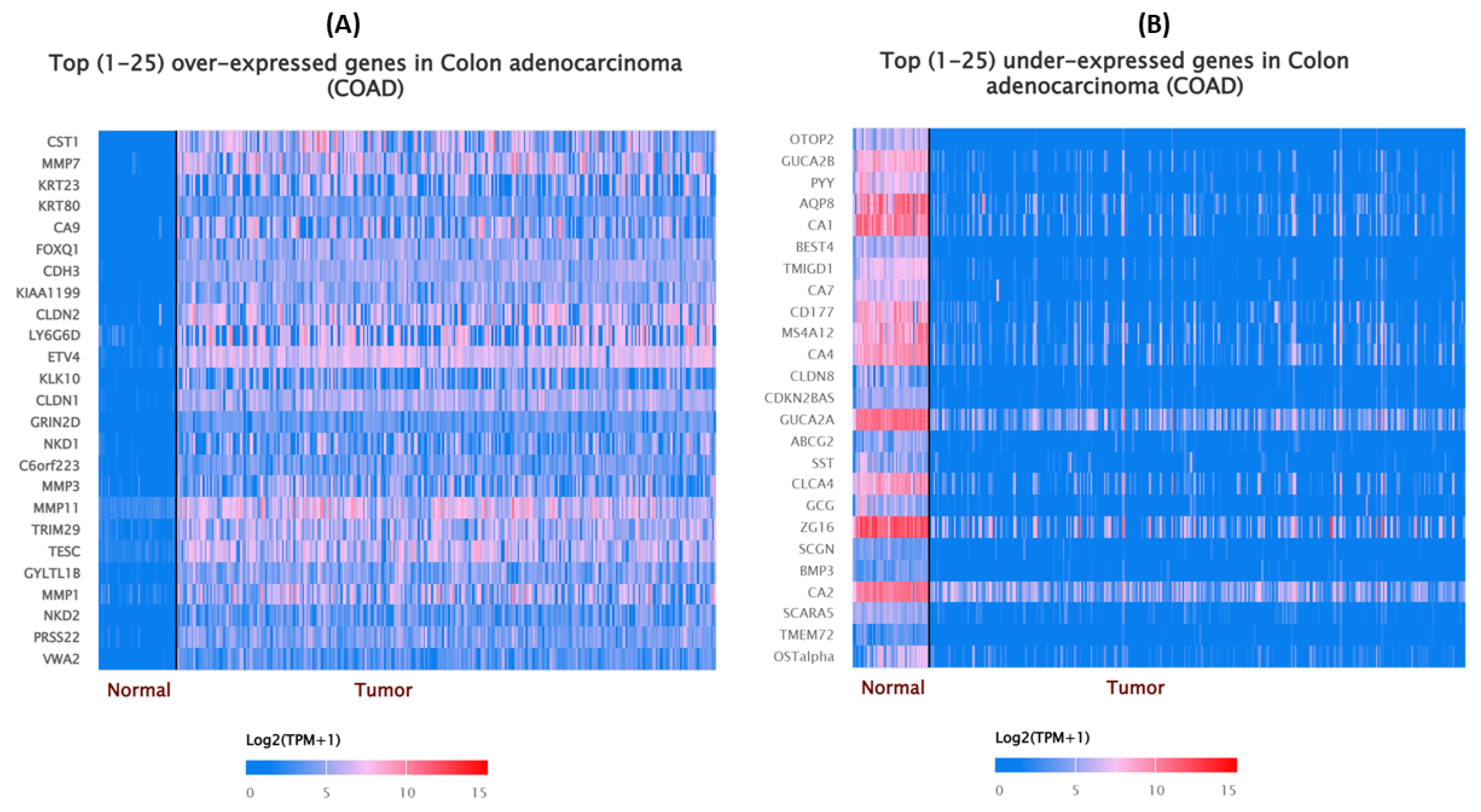
3.1. Altered Gene Expression Pattern in COAD Based on TCGA Dataset
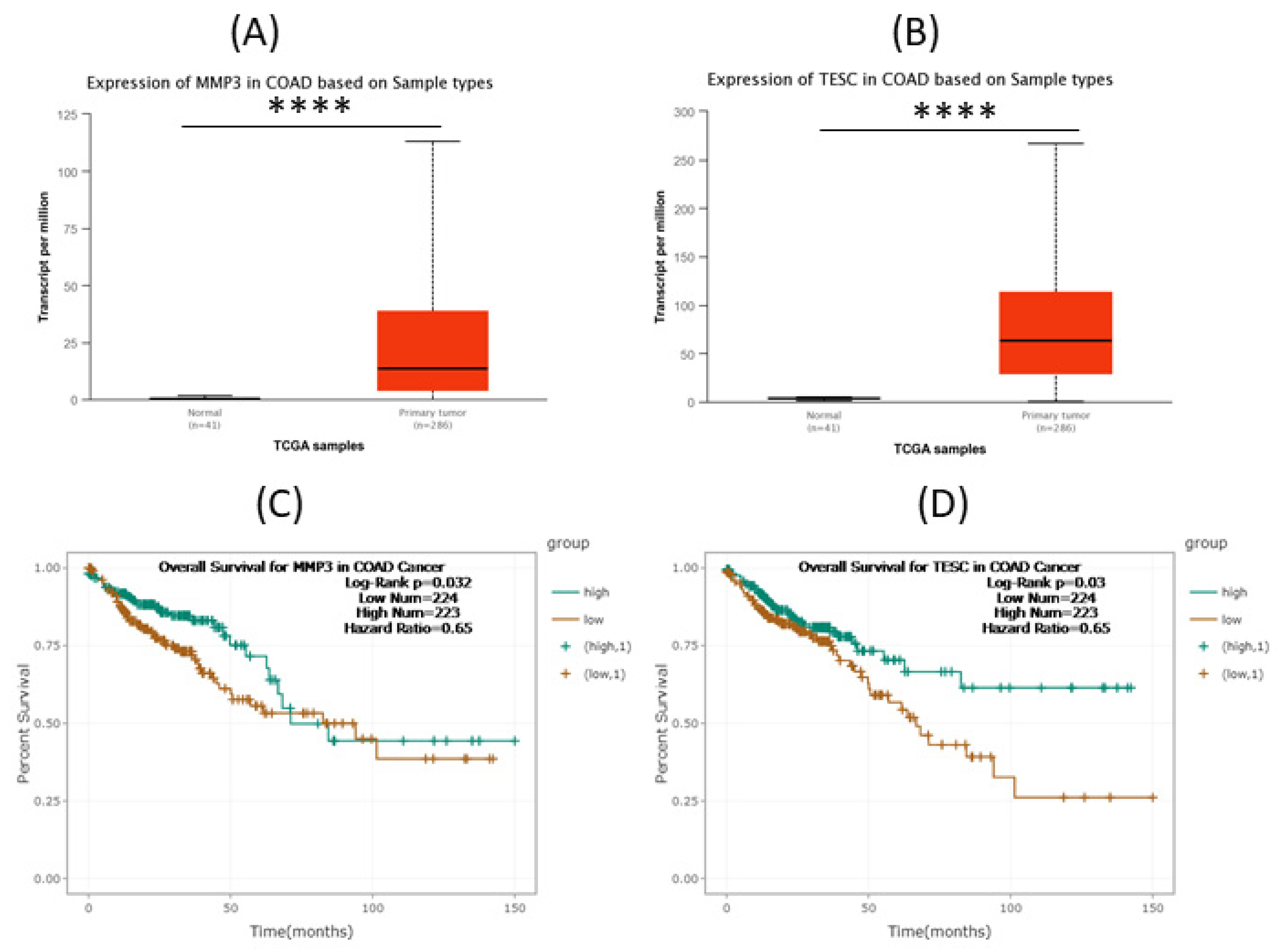
3.2. Significance of the MMP3 and TESC in COAD
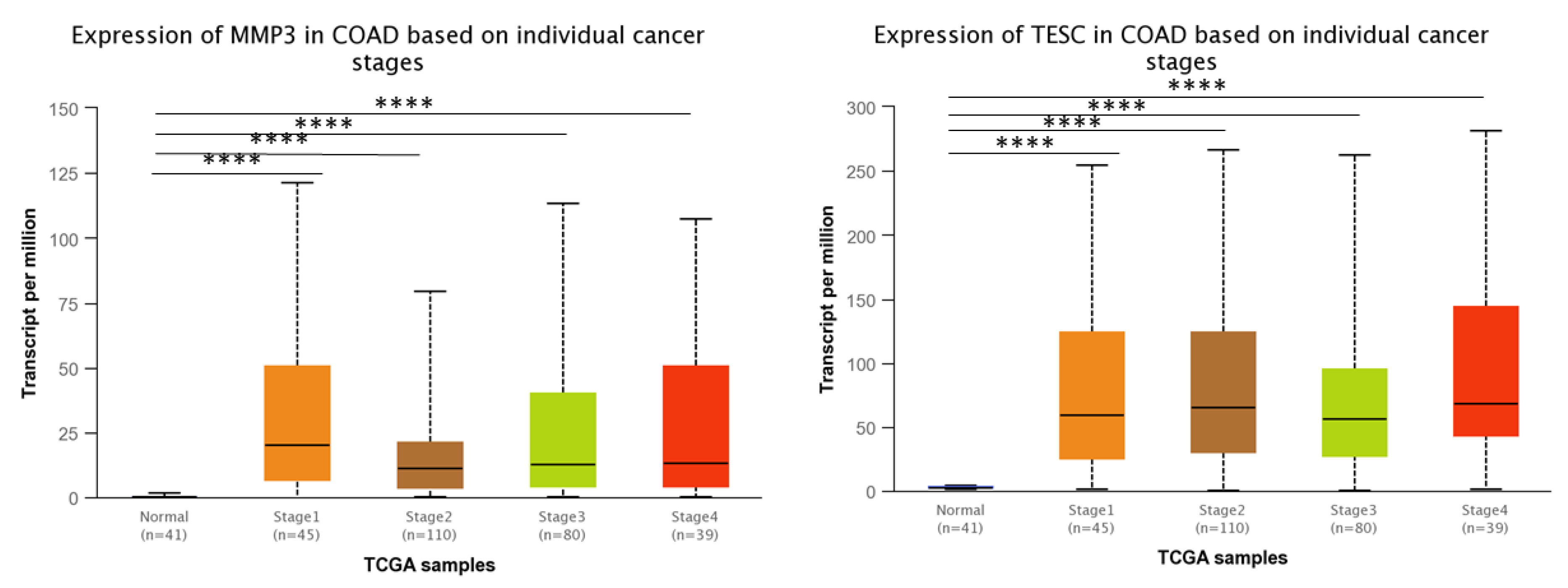
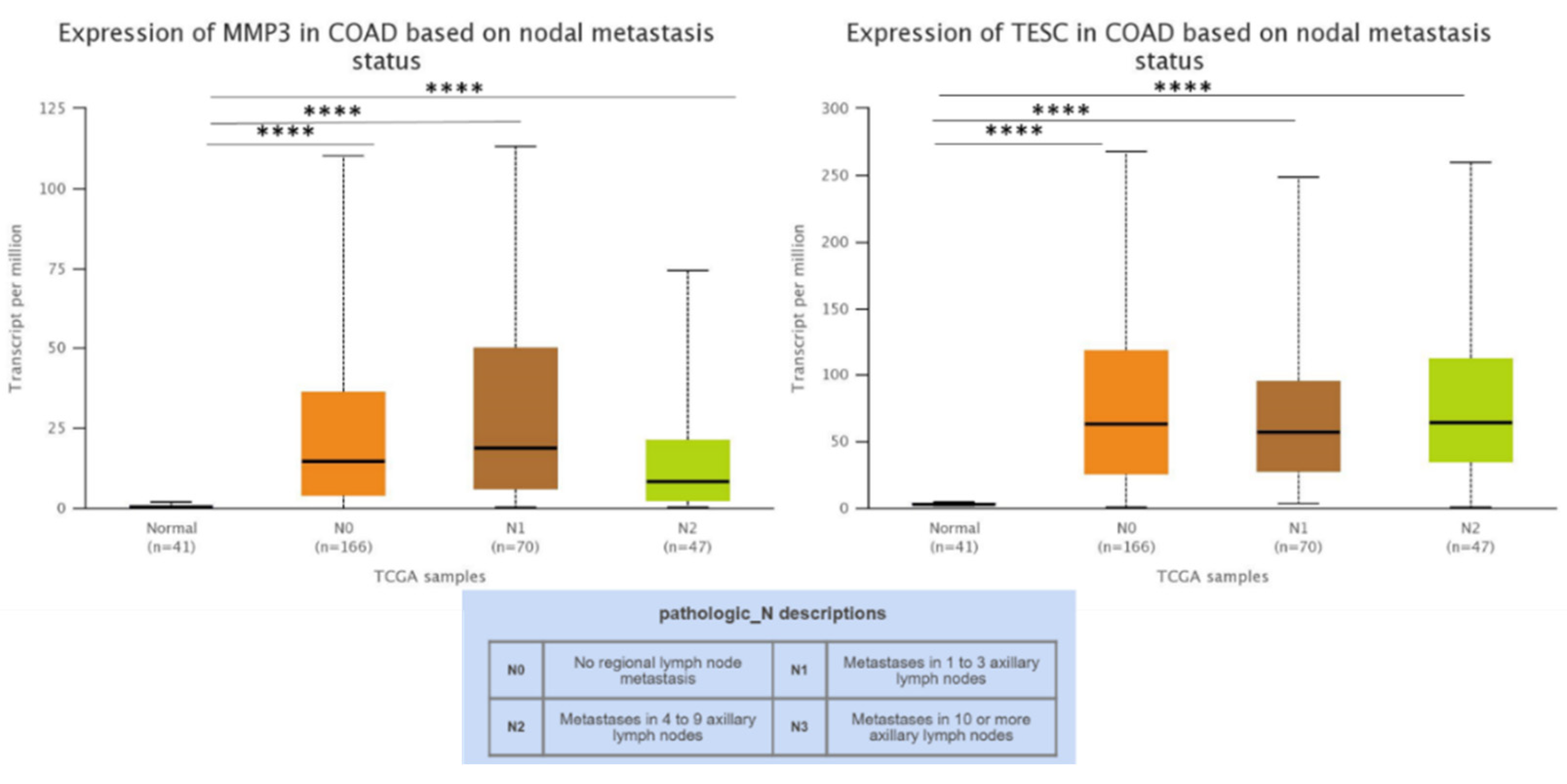
3.3. MMP3 and TESC mRNA Expression and Cancer Stages and Lymph Node-Positive/Metastatic Status in COAD
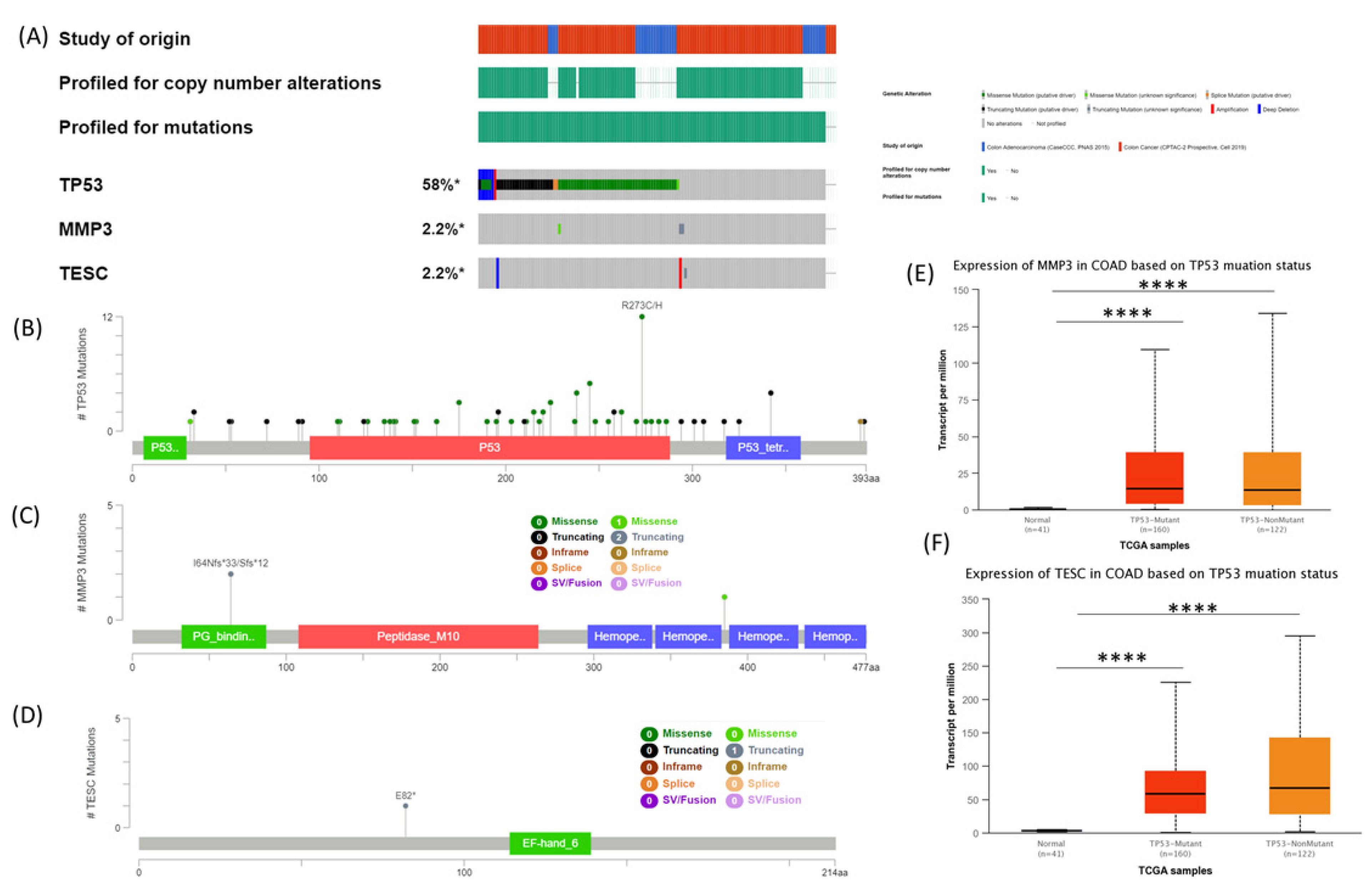
3.4. MMP3 and TESC Mutational Signature in COAD Evaluated Using cBioPortal
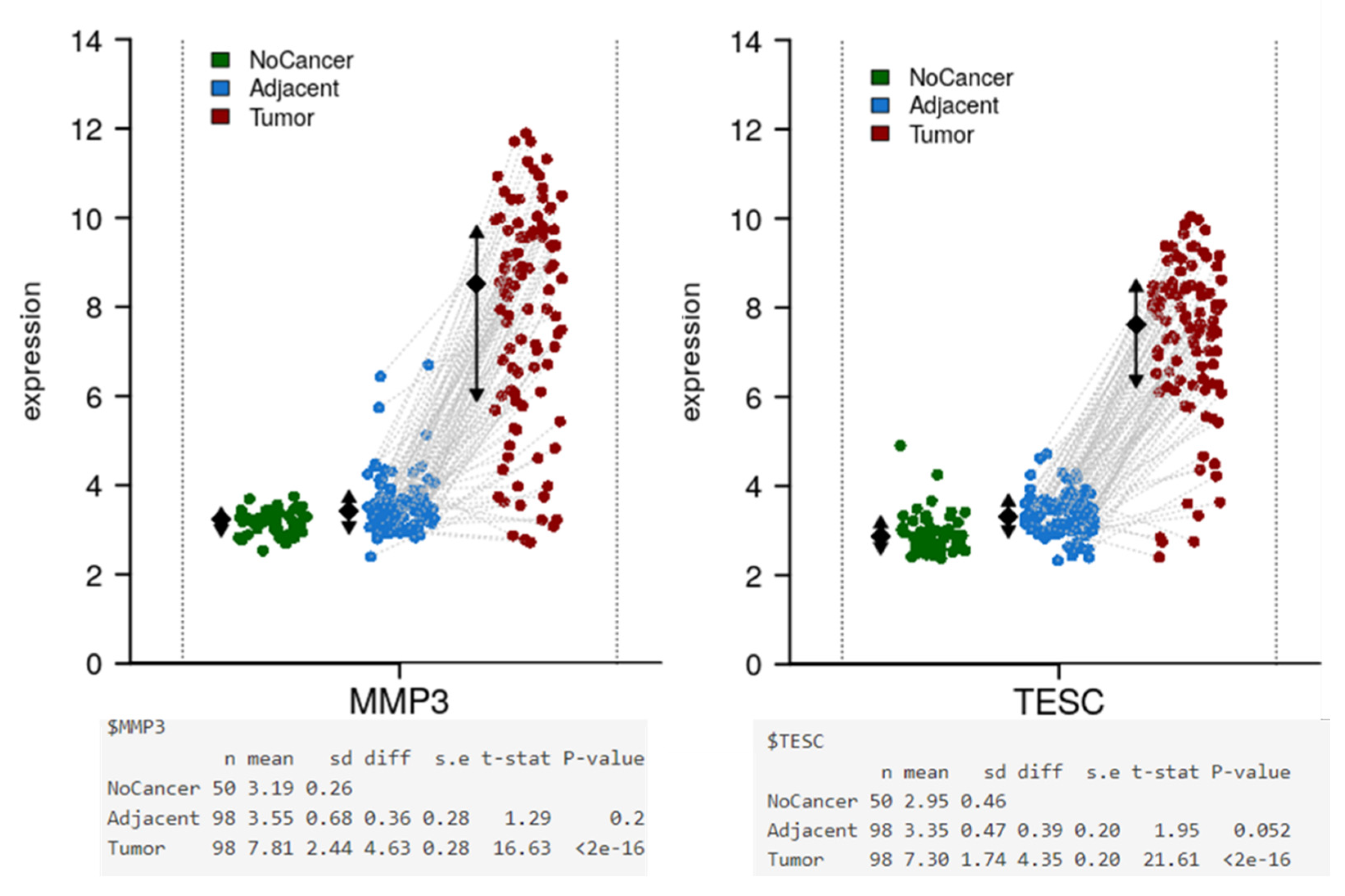
3.5. Validation of MMP3 and TESC with the Colonomics Patient Cohort
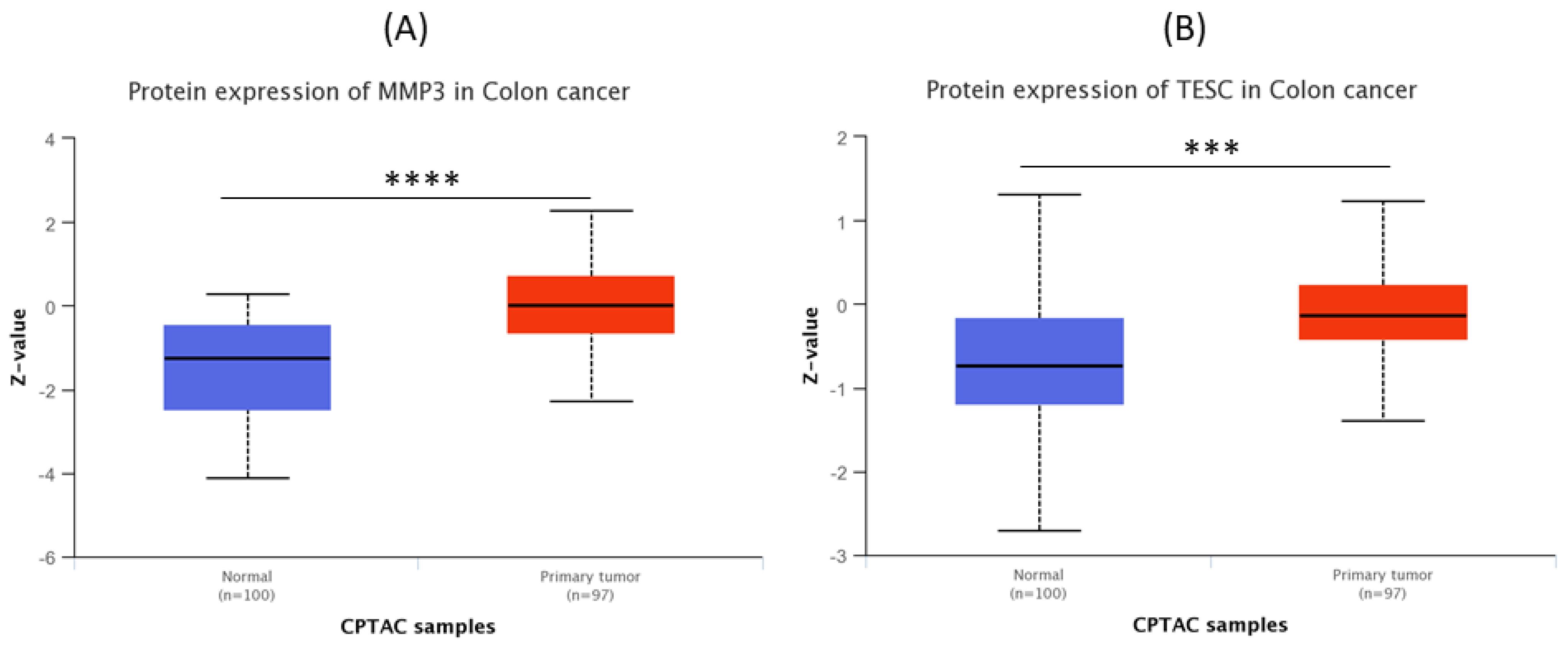
3.6. MMP3 and TESC Protein Expression Levels
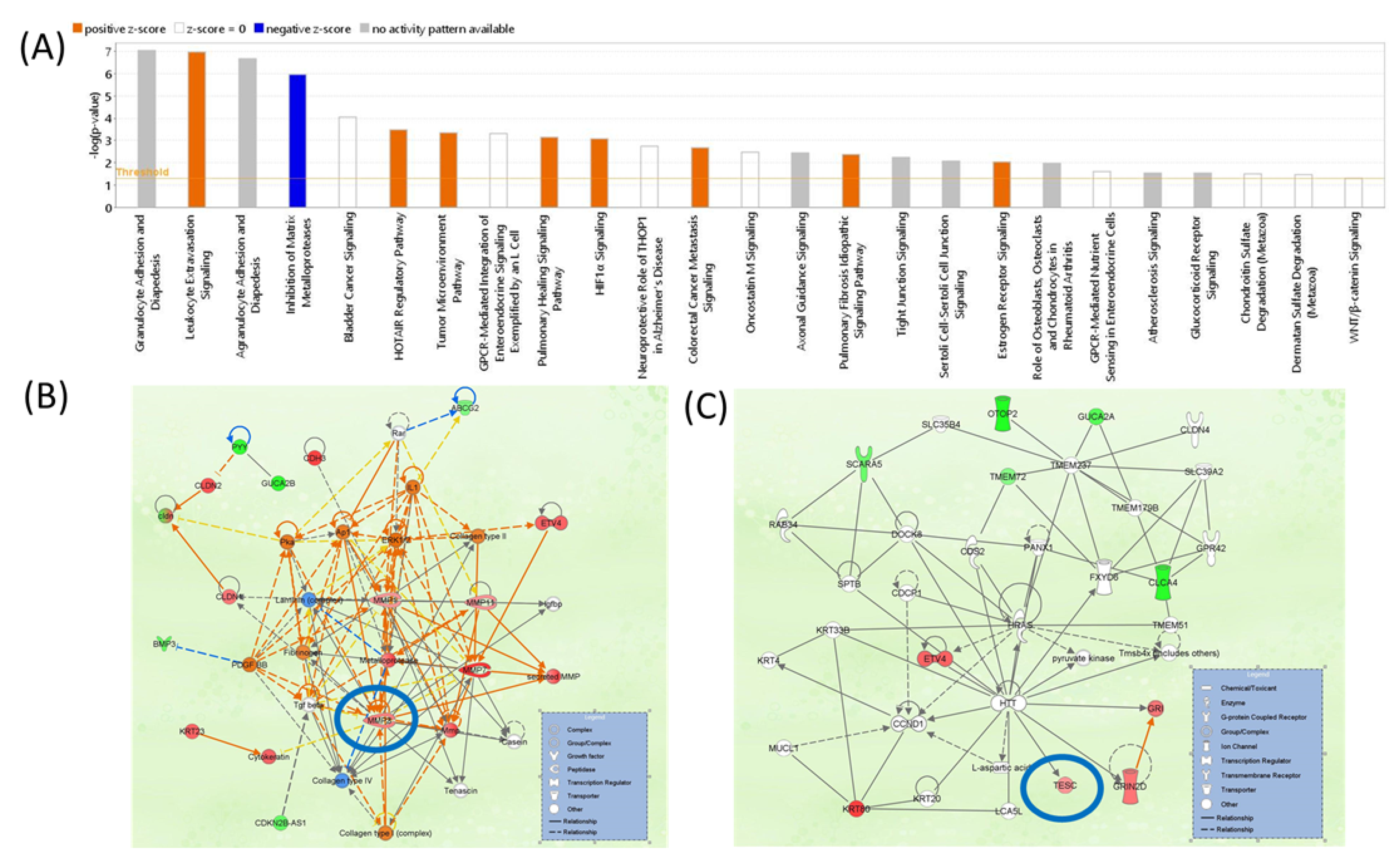
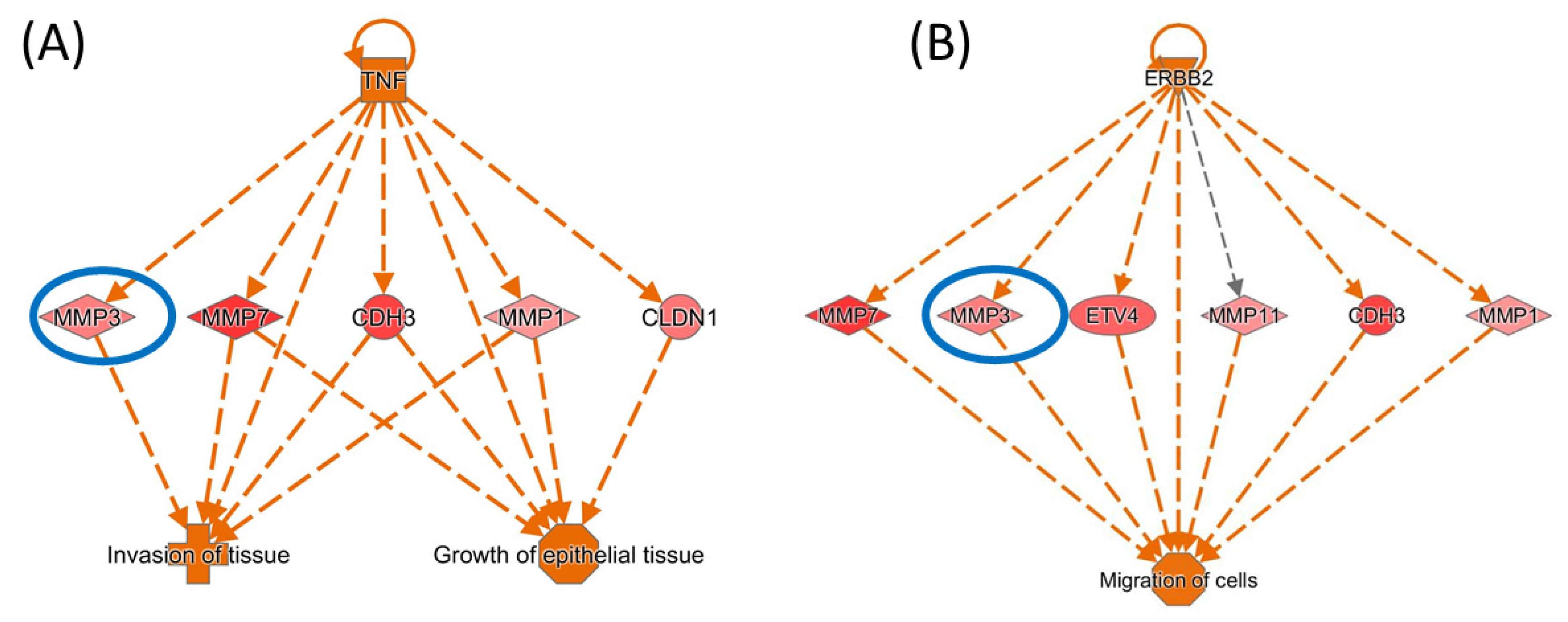
3.7. IPA Network Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yue, F.R.; Wei, Z.B.; Yan, R.Z.; Guo, Q.H.; Liu, B.; Zhang, J.H.; Li, Z. SMYD3 promotes colon adenocarcinoma (COAD) progression by mediating cell proliferation and apoptosis. Exp. Ther. Med. 2020, 20, 11. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Schliemann, D.; Ramanathan, K.; Matovu, N.; O’Neill, C.; Kee, F.; Su, T.T.; Donnelly, M. The implementation of colorectal cancer screening interventions in low-and middle-income countries: A scoping review. BMC Cancer 2021, 21, 1125. [Google Scholar] [CrossRef]
- Gurzu, S.; Silveanu, C.; Fetyko, A.; Butiurca, V.; Kovacs, Z.; Jung, I. Systematic review of the old and new concepts in the epithelial-mesenchymal transition of colorectal cancer. World J. Gastroenterol. 2016, 22, 6764–6775. [Google Scholar] [CrossRef]
- Busuioc, C.; Ciocan-Cartita, C.A.; Braicu, C.; Zanoaga, O.; Raduly, L.; Trif, M.; Muresan, M.S.; Ionescu, C.; Stefan, C.; Crivii, C.; et al. Epithelial-Mesenchymal Transition Gene Signature Related to Prognostic in Colon Adenocarcinoma. J. Pers. Med. 2021, 11, 476. [Google Scholar] [CrossRef]
- Nguyen, H.T.; Duong, H.Q. The molecular characteristics of colorectal cancer: Implications for diagnosis and therapy. Oncol. Lett. 2018, 16, 9–18. [Google Scholar] [CrossRef]
- Gurzu, S.; Banias, L.; Kovacs, Z.; Jung, I. Epithelial-mesenchymal transition of tumor budding in colorectal cancer: The mystery of CD44-positive stromal cells. Hum. Pathol. 2018, 71, 168–169. [Google Scholar] [CrossRef]
- Cojocneanu, R.; Braicu, C.; Raduly, L.; Jurj, A.; Zanoaga, O.; Magdo, L.; Irimie, A.; Muresan, M.S.; Ionescu, C.; Grigorescu, M.; et al. Plasma and Tissue Specific miRNA Expression Pattern and Functional Analysis Associated to Colorectal Cancer Patients. Cancers 2020, 12, 843. [Google Scholar] [CrossRef]
- Ionescu, C.; Braicu, C.; Chiorean, R.; Cojocneanu Petric, R.; Neagoe, E.; Pop, L.; Chira, S.; Berindan-Neagoe, I. TIMP-1 expression in human colorectal cancer is associated with SMAD3 gene expression levels: A pilot study. J. Gastrointestin. Liver Dis. 2014, 23, 413–418. [Google Scholar] [CrossRef]
- Tomuleasa, C.; Braicu, C.; Irimie, A.; Craciun, L.; Berindan-Neagoe, I. Nanopharmacology in translational hematology and oncology. Int. J. Nanomed. 2014, 9, 3465–3479. [Google Scholar] [CrossRef]
- Jurj, A.; Zanoaga, O.; Braicu, C.; Lazar, V.; Tomuleasa, C.; Irimie, A.; Berindan-Neagoe, I. A Comprehensive Picture of Extracellular Vesicles and Their Contents. Molecular Transfer to Cancer Cells. Cancers 2020, 12, 298. [Google Scholar] [CrossRef]
- Alajez, N.M. Large-Scale Analysis of Gene Expression Data Reveals a Novel Gene Expression Signature Associated with Colorectal Cancer Distant Recurrence. PLoS ONE 2016, 11, e0167455. [Google Scholar] [CrossRef]
- Bochis, O.V.; Irimie, A.; Pichler, M.; Berindan-Neagoe, I. The role of Skp2 and its substrate CDKN1B (p27) in colorectal cancer. J. Gastrointestin. Liver Dis. 2015, 24, 225–234. [Google Scholar] [CrossRef]
- Zeng, C.; Chen, Y. HTR1D, TIMP1, SERPINE1, MMP3 and CNR2 affect the survival of patients with colon adenocarcinoma. Oncol. Lett. 2019, 18, 2448–2454. [Google Scholar] [CrossRef]
- Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- Wang, Y.; Gao, X.; Ru, X.; Sun, P.; Wang, J. Identification of gene signatures for COAD using feature selection and Bayesian network approaches. Sci. Rep. 2022, 12, 8761. [Google Scholar] [CrossRef]
- Lin, J.; Cao, Z.; Yu, D.; Cai, W. Identification of Transcription Factor-Related Gene Signature and Risk Score Model for Colon Adenocarcinoma. Front. Genet. 2021, 12, 709133. [Google Scholar] [CrossRef]
- Sanz-Pamplona, R.; Berenguer, A.; Cordero, D.; Molleví, D.G.; Crous-Bou, M.; Sole, X.; Paré-Brunet, L.; Guino, E.; Salazar, R.; Santos, C.; et al. Aberrant gene expression in mucosa adjacent to tumor reveals a molecular crosstalk in colon cancer. Mol. Cancer 2014, 13, 46. [Google Scholar] [CrossRef]
- Buttacavoli, M.; Di Cara, G.; Roz, E.; Pucci-Minafra, I.; Feo, S.; Cancemi, P. Integrated Multi-Omics Investigations of Metalloproteinases in Colon Cancer: Focus on MMP2 and MMP9. Int. J. Mol. Sci. 2021, 22, 12389. [Google Scholar] [CrossRef]
- Available online: https://www.colonomics.org (accessed on 28 August 2022).
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Karthikeyan, S.K.; Korla, P.K.; Patel, H.; Shovon, A.R.; Athar, M.; Netto, G.J.; Qin, Z.S.; Kumar, S.; Manne, U.; et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022, 25, 18–27. [Google Scholar] [CrossRef]
- Sun, Q.; Li, M.; Wang, X. The Cancer Omics Atlas: An integrative resource for cancer omics annotations. BMC Med. Genom. 2018, 11, 63. [Google Scholar] [CrossRef]
- Li, J.-H.; Liu, S.; Zhou, H.; Qu, L.-H.; Yang, J.-H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.R.; Liu, S.; Cai, L.; Bin, L. ENCORI: The Encyclopedia of RNA Interactomes. Available online: http://starbase.sysu.edu.cn/index.php (accessed on 20 August 2022).
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Chen, F.; Chandrashekar, D.S.; Varambally, S.; Creighton, C.J. Pan-cancer molecular subtypes revealed by mass-spectrometry-based proteomic characterization of more than 500 human cancers. Nature Commun. 2019, 10, 5679. [Google Scholar] [CrossRef] [PubMed]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Li, B.; Severson, E.; Pignon, J.C.; Zhao, H.; Li, T.; Novak, J.; Jiang, P.; Shen, H.; Aster, J.C.; Rodig, S.; et al. Comprehensive analyses of tumor immunity: Implications for cancer immunotherapy. Genome Biol. 2016, 17, 174. [Google Scholar] [CrossRef]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef]
- Sharma, A.; Yadav, D.; Rao, P.; Sinha, S.; Goswami, D.; Rawal, R.M.; Shrivastava, N. Identification of potential therapeutic targets associated with diagnosis and prognosis of colorectal cancer patients based on integrated bioinformatics analysis. Comput. Biol. Med. 2022, 146, 105688. [Google Scholar] [CrossRef]
- Kang, Y.H.; Han, S.R.; Kim, J.T.; Lee, S.J.; Yeom, Y.I.; Min, J.K.; Lee, C.H.; Kim, J.W.; Yoon, S.R.; Yoon, D.Y.; et al. The EF-hand calcium-binding protein tescalcin is a potential oncotarget in colorectal cancer. Oncotarget 2014, 5, 2149–2160. [Google Scholar] [CrossRef]
- Liang, M.; Wang, J.; Wu, C.; Wu, M.; Hu, J.; Dai, J.; Ruan, H.; Xiong, S.; Dong, C. Targeting matrix metalloproteinase MMP3 greatly enhances oncolytic virus mediated tumor therapy. Transl. Oncol. 2021, 14, 101221. [Google Scholar] [CrossRef] [PubMed]
- Curran, S.; Murray, G.I. Matrix metalloproteinases: Molecular aspects of their roles in tumour invasion and metastasis. Eur. J. Cancer 2000, 36, 1621–1630. [Google Scholar] [CrossRef]
- Zinzindohoué, F.; Lecomte, T.; Ferraz, J.M.; Houllier, A.M.; Cugnenc, P.H.; Berger, A.; Blons, H.; Laurent-Puig, P. Prognostic significance of MMP-1 and MMP-3 functional promoter polymorphisms in colorectal cancer. Clin. Cancer Res. 2005, 11, 594–599. [Google Scholar] [CrossRef]
- Chen, H.; Ye, Y.; Yang, Y.; Zhong, M.; Gu, L.; Han, Z.; Qiu, J.; Liu, Z.; Qiu, X.; Zhuang, G. TIPE-mediated up-regulation of MMP-9 promotes colorectal cancer invasion and metastasis through MKK-3/p38/NF-κB pro-oncogenic signaling pathway. Signal Transduct. Target. Ther. 2020, 5, 163. [Google Scholar] [CrossRef]
- Pezeshkian, Z.; Nobili, S.; Peyravian, N.; Shojaee, B.; Nazari, H.; Soleimani, H.; Asadzadeh-Aghdaei, H.; Ashrafian Bonab, M.; Nazemalhosseini-Mojarad, E.; Mini, E. Insights into the Role of Matrix Metalloproteinases in Precancerous Conditions and in Colorectal Cancer. Cancers 2021, 13, 6226. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; He, Z.; He, X.; Luo, Z.; Lian, L.; Wu, B.; Lan, P.; Chen, H. Comprehensive Analysis of the Expression and Prognosis for MMPs in Human Colorectal Cancer. Front. Oncol. 2021, 11. [Google Scholar] [CrossRef]
- Baker, E.A.; Bergin, F.G.; Leaper, D.J. Matrix metalloproteinases, their tissue inhibitors and colorectal cancer staging. Br. J. Surg. 2000, 87, 1215–1221. [Google Scholar] [CrossRef]
- Coussens, L.M.; Fingleton, B.; Matrisian, L.M. Matrix metalloproteinase inhibitors and cancer: Trials and tribulations. Science 2002, 295, 2387–2392. [Google Scholar] [CrossRef]
- Honda, T.; Yamamoto, I.; Inagawa, H. Angiogenesis-, Metastasis- and Signaling Pathway-related Factor Dynamics in Human Colon Cancer Cells Following Interaction with Monocytes. Anticancer Res. 2013, 33, 2895–2900. [Google Scholar]
- Gobin, E.; Bagwell, K.; Wagner, J.; Mysona, D.; Sandirasegarane, S.; Smith, N.; Bai, S.; Sharma, A.; Schleifer, R.; She, J.-X. A pan-cancer perspective of matrix metalloproteases (MMP) gene expression profile and their diagnostic/prognostic potential. BMC Cancer 2019, 19, 581. [Google Scholar] [CrossRef]
- Sipos, F.; Germann, T.M.; Wichmann, B.; Galamb, O.; Spisák, S.; Krenács, T.; Tulassay, Z.; Molnár, B.; Műzes, G. MMP3 and CXCL1 are potent stromal protein markers of dysplasia-carcinoma transition in sporadic colorectal cancer. Eur. J. Cancer Prev. 2014, 23, 336–343. [Google Scholar] [CrossRef] [PubMed]
- Calu, V.; Ionescu, A.; Stanca, L.; Geicu, O.I.; Iordache, F.; Pisoschi, A.M.; Serban, A.I.; Bilteanu, L. Key biomarkers within the colorectal cancer related inflammatory microenvironment. Sci. Rep. 2021, 11, 7940. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Zhang, X.; Li, X.; Tian, L.; Shen, S.; Ma, J.; Ai, F. Histone deacetylase (HDAC) 11 inhibits matrix metalloproteinase (MMP) 3 expression to suppress colorectal cancer metastasis. J. Cancer 2022, 13, 1923–1932. [Google Scholar] [CrossRef]
- Mehner, C.; Miller, E.; Khauv, D.; Nassar, A.; Oberg, A.L.; Bamlet, W.R.; Zhang, L.; Waldmann, J.; Radisky, E.S.; Crawford, H.C.; et al. Tumor cell-derived MMP3 orchestrates Rac1b and tissue alterations that promote pancreatic adenocarcinoma. Mol. Cancer Res. 2014, 12, 1430–1439. [Google Scholar] [CrossRef] [PubMed]
- El-Sharkawi, F.; El Sabah, M.; Hassan, Z.; Khaled, H. The biochemical value of urinary metalloproteinases 3 and 9 in diagnosis and prognosis of bladder cancer in Egypt. J. Biomed. Sci. 2014, 21, 72. [Google Scholar] [CrossRef]
- Ligi, D.; Mannello, F. Do matrix metalloproteinases represent reliable circulating biomarkers in colorectal cancer? Br. J. Cancer 2016, 115, 633–634. [Google Scholar] [CrossRef][Green Version]
- Ji, Y.; Li, J.; Li, P.; Wang, L.; Yang, H.; Jiang, G. C/EBPβ Promotion of MMP3-Dependent Tumor Cell Invasion and Association with Metastasis in Colorectal Cancer. Genet. Test Mol. Biomark. 2018, 22, 5–10. [Google Scholar] [CrossRef]
- Piskór, B.M.; Przylipiak, A.; Dąbrowska, E.; Niczyporuk, M.; Ławicki, S. Matrilysins and Stromelysins in Pathogenesis and Diagnostics of Cancers. Cancer Manag. Res. 2020, 12, 10949–10964. [Google Scholar] [CrossRef]
- Kolobynina, K.G.; Solovyova, V.V.; Levay, K.; Rizvanov, A.A.; Slepak, V.Z. Emerging roles of the single EF-hand Ca2+ sensor tescalcin in the regulation of gene expression, cell growth and differentiation. J. Cell Sci. 2016, 129, 3533–3540. [Google Scholar] [CrossRef]
- Kang, J.; Kang, Y.H.; Oh, B.M.; Uhm, T.G.; Park, S.Y.; Kim, T.W.; Han, S.R.; Lee, S.J.; Lee, Y.; Lee, H.G. Tescalcin expression contributes to invasive and metastatic activity in colorectal cancer. Tumour. Biol. 2016, 37, 13843–13853. [Google Scholar] [CrossRef]
- Zhou, Z.-G.; Chen, J.-B.; Zhang, R.-X.; Ye, L.; Wang, J.-C.; Pan, Y.-X.; Wang, X.-H.; Li, W.-X.; Zhang, Y.-J.; Xu, L.; et al. Tescalcin is an unfavorable prognosis factor that regulats cell proliferation and survival in hepatocellular carcinoma patients. Cancer Commun. 2020, 40, 355–369. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.W.; Han, S.R.; Kim, J.T.; Yoo, S.M.; Lee, M.S.; Lee, S.H.; Kang, Y.H.; Lee, H.G. Differential expression of tescalcin by modification of promoter methylation controls cell survival in gastric cancer cells. Oncol. Rep. 2019, 41, 3464–3474. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Choi, S.I.; Kim, R.K.; Cho, E.W.; Kim, I.G. Tescalcin/c-Src/IGF1Rβ-mediated STAT3 activation enhances cancer stemness and radioresistant properties through ALDH1. Sci. Rep. 2018, 8, 10711. [Google Scholar] [CrossRef]
- Karolak, A.; Levatić, J.; Supek, F. A framework for mutational signature analysis based on DNA shape parameters. PLoS ONE 2022, 17, e0262495. [Google Scholar] [CrossRef] [PubMed]
- Biondi, M.L.; Turri, O.; Leviti, S.; Seminati, R.; Cecchini, F.; Bernini, M.; Ghilardi, G.; Guagnellini, E. MMP1 and MMP3 Polymorphisms in Promoter Regions and Cancer. Clin. Chem. 2000, 46, 2023–2024. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, F.; Zhang, X.; Lin, H.-K.; Xu, C. Insights into the post-translational modification and its emerging role in shaping the tumor microenvironment. Signal Transduct. Target. Ther. 2021, 6, 422. [Google Scholar] [CrossRef]
| Demographics | COAD Tumor (n = 286) Normal (n = 41) | |
|---|---|---|
| Age—Range (years) | 31–90 | |
| Gender | F | 127 |
| M | 156 | |
| Unknown | 3 | |
| Histological subtype | Adenocarcinoma | 243 |
| Mucinous adenocarcinoma | 37 | |
| Unknown | 3 | |
| Tumor stage | I | 45 |
| II | 110 | |
| III | 80 | |
| IV | 39 | |
| Unknown | 2 | |
| Nodal metastasis status | N0 | 166 |
| N1 | 70 | |
| N2 | 47 | |
| Unknown | 3 | |
| ID | Associated Network | Score |
|---|---|---|
| N1 | Post-Translational Modification, Protein Degradation, Protein Synthesis | 32 |
| N2 | Developmental Disorder, Ophthalmic Disease, Organismal Injury and Abnormalities | 29 |
| N3 | Hereditary Disorder, Ophthalmic Disease, Organismal Injury and Abnormalities | 29 |
| N4 | Cell Cycle, Cancer, Neurological Disease | 18 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Busuioc, C.; Nutu, A.; Braicu, C.; Zanoaga, O.; Trif, M.; Berindan-Neagoe, I. Analysis of Differentially Expressed Genes, MMP3 and TESC, and Their Potential Value in Molecular Pathways in Colon Adenocarcinoma: A Bioinformatics Approach. BioMedInformatics 2022, 2, 474-491. https://doi.org/10.3390/biomedinformatics2030030
Busuioc C, Nutu A, Braicu C, Zanoaga O, Trif M, Berindan-Neagoe I. Analysis of Differentially Expressed Genes, MMP3 and TESC, and Their Potential Value in Molecular Pathways in Colon Adenocarcinoma: A Bioinformatics Approach. BioMedInformatics. 2022; 2(3):474-491. https://doi.org/10.3390/biomedinformatics2030030
Chicago/Turabian StyleBusuioc, Constantin, Andreea Nutu, Cornelia Braicu, Oana Zanoaga, Monica Trif, and Ioana Berindan-Neagoe. 2022. "Analysis of Differentially Expressed Genes, MMP3 and TESC, and Their Potential Value in Molecular Pathways in Colon Adenocarcinoma: A Bioinformatics Approach" BioMedInformatics 2, no. 3: 474-491. https://doi.org/10.3390/biomedinformatics2030030
APA StyleBusuioc, C., Nutu, A., Braicu, C., Zanoaga, O., Trif, M., & Berindan-Neagoe, I. (2022). Analysis of Differentially Expressed Genes, MMP3 and TESC, and Their Potential Value in Molecular Pathways in Colon Adenocarcinoma: A Bioinformatics Approach. BioMedInformatics, 2(3), 474-491. https://doi.org/10.3390/biomedinformatics2030030










