Abstract
Background: Pulmonary hypertension (PH) is a complex disease caused by a wide range of underlying conditions, Tanshinone IIA (Tan IIA) has been widely used in PH patients. The study aimed to explore the possible molecular mechanism of Tan IIA against PH by network pharmacology and molecular docking. Methods: Tan IIA and PH-related targets were retrieved from public databases. Gene ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis, and protein–protein interaction (PPI) network were used to investigate the protein targets and mechanism. The binding activity of core targets and Tan IIA were verified by molecular docking. Results: A total of 26 overlapping target proteins between Tan IIA and PH were screened. PPI network identified HSP90AA1, PTPN11, ATM, CA2, TERT, PRKDC, and APEX1 as key pharmacological targets. The results of GO function enrichment analysis included regulation of smooth muscle cell proliferation and migration, regulation of mitotic cell cycle, and regulation of G1/S transition of mitotic cell cycle. KEGG pathway analysis showed that nitrogen metabolism, NF-kappa B signaling pathway, cell cycle, necroptosis, apoptosis, and JAK-STAT signaling pathway were associated with Tan IIA in PH. The molecular docking results showed that Tan IIA can closely bind three core targets (HSP90AA1, PTPN11, and CA2). Conclusions: The present work initially clarified the effective therapeutic targets, biological processes, and signaling pathways of Tan IIA treatment of PH, which lay a foundation for further research on the pharmacological effects of Tan IIA.
1. Introduction
Pulmonary hypertension (PH) is characterized by elevated blood pressure in the lungs at rest, which links to high mortality due to right heart failure [1]. PH arises from a wide range of underlying diseases, including pulmonary arterial hypertension (PAH), PH secondary to left heart, and lung diseases [2]. The treatment of PH has improved significantly over the last decades with targeted drug therapies. Currently, commonly used clinical drugs included PDE-5 Inhibitors, Soluble Guanylate Cyclase Stimulators, Endothelin Receptor Antagonists, Prostacyclin Analogues, and Prostacyclin Receptor Agonists [3]. However, significant worsening symptom over time in PH patients irrespective of the treatment procedure because of the complexity and heterogeneity of PH. Therefore, developing a new effective therapy for the treatment of PH and to reveal their underlying mechanisms is of utmost importance.
Tanshinone IIA (Tan IIA, C19H18O3, PubChem CID:164676) isolated from Salvia miltiorrhiza Bunge is used widely in the prevention and treatment of coronary artery disease, diabetes, obesity, ischemic stroke and liver fibrosis with few toxic side effects in China for many years [4]. Clinical research also proved that PH patients received beneficial effects from Tan IIA treatment [5]. However, the mechanism of action of Tan IIA against PH remain elusive.
Network pharmacology is a powerful tool in illuminating the drug-target-disease network and generates new ideas and methods for research on medicinal plants [6]. Therefore, we investigate the therapeutic effects of Tan IIA in PH using network pharmacology, then preliminarily verified therapeutic targets by using molecular docking. Figure 1 shows the flowchart of this study.
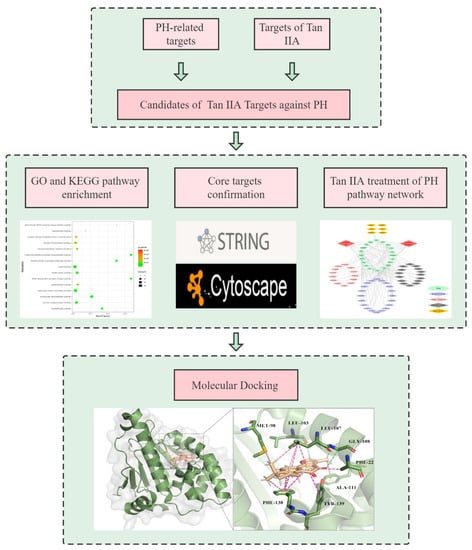
Figure 1.
The flowchart of this study.
2. Methods
2.1. Data Mining
The two-dimensional (2D) structure of TIIA was obtained from PubChem (http://pubchem.ncbi.nlm.nlh.gov/) (accessed on 14 April 2022). According to this structure, several databases including BindingDB (http://bindingdb.org/) (accessed on 15 April 2022), ChEMBL (http://ebi.ac.ek/chembl/) (accessed on 15 April 2022), SwissTarget prediction (http://www.swisstargetprediction.ch) (accessed on 15 April 2022), and STITCH database (http://www.stitch.embl.de/) (accessed on 16 April 2022) were used to obtain as many potential targets as possible.
The PH-related protein targets were screened using the GeneCards database (https://www.genecards.org/) (accessed on 15 April 2022), DisGeNET platform (https://www.disgenet.org/) (accessed on 16 April 2022), and CTD database (https://ctdbase.org/) (accessed on 16 April 2022), and the keywords “pulmonary hypertension” and “pulmonary arterial hypertension”. Finally, all proteins were standardized by the Uniprot database (https://www.uniprot.org/) (accessed on 20 April 2022) with the species “homo sapiens”.
2.2. Protein–Protein Interaction (PPI) Analysis and Core Gene Screening
The intersecting protein targets of Tan IIA and PH were identified by STRING platform (https://cn.string-db.org/) (accessed on 21 April 2022). Cytoscape software v.3.7.1 (https://cytoscape.org/) (accessed on 1 August 2022) and plug-in Cytohubba were used to analyze the interaction network and the hub targets in the PPI network [7].
2.3. Enrichment Analysis
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were carried out to examine the potential therapeutic targets of Tan IIA against PH. GO terms and KEGG pathway analysis with p value < 0.05 were considered statistically significant.
2.4. Molecular Docking
To further understand the mechanism of Tan IIA with PH disease targets, the receptor-ligand molecular docking method was applied to verify the activity of binding between Tan IIA and hub targets. Tan IIA was used as the ligand molecule and the crystal structure of the relevant target protein was used as the receptor molecule for molecular docking. The three-dimensional (3D) structure of Tan IIA was downloaded from PubChem database (https://pubchem.ncbi.nlm.nih.gov/) (accessed on 14 April 2022) and protein crystal structures of some relevant targets were obtained from the UniProt database (https://www.uniprot.org/) (accessed on 22 April 2022) and RCSB PDB database (https://www.rcsb.org/) (accessed on 22 April 2022).
Molecular docking analysis was conducted by Discovery Studio 2019 software (BIOVIA, San Diego, CA, USA); the Docking results of three-dimension images drawn by Pymol 2.5.Dock ligands (CDOCKER) uses a CHARMm-based molecular dynamics scheme to dock ligands into a receptor binding site. The CDOCKER method was performed to identify the binding affinity of Tan IIA receptors. The smaller the binding energy (docking score), the more stable the binding of Tan IIA to the target site. A docking score <−5.0 kcal/mol was considered a good binding activity, while a docking score < −7.0 kcal/mol indicated a high-affinity association between Tan IIA and the identified core targets [8].
2.5. Construction of a Compound-Target-Pathway Network
To obtain a comprehensive understanding of the associations between Tan IIA and multiple targets and pathways, Cytoscape 3.7.1 software was used to build Tan IIA-therapy target pathway networks. The Closeness centrality is calculated by CentiScaPe of Cytoscape [9].
3. Results
3.1. Identification of Potential Targets for Tan IIA for the Treatment of Pulmonary Hypertension
Based on the data structure and algorithm, we obtained 31 potential targets of TIIA and 12,144 genes related to PH after deleting the redundant data. There were 26 overlapping targets that were filtered out as candidate genes of Tan IIA for the treatment of PH by using the online tool VEENY2.1 (https://bioinfop.cnb.csic.es/tools/venny/) (accessed on 20 April 2022), and they were thoroughly analyzed, as is shown in Figure 2.
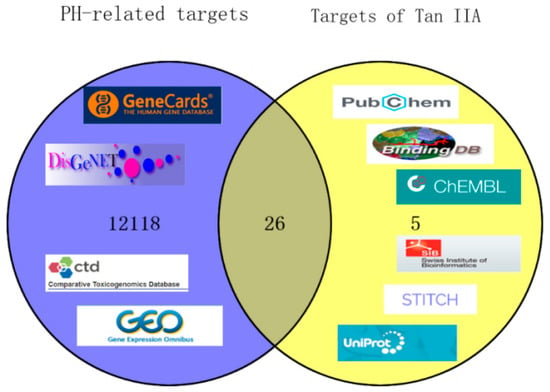
Figure 2.
Matching of target genes of pulmonary hypertension and Tan IIA. The purple circle represents the PH-related targets and the yellow circle represents the drug targets of Tan IIA.
3.2. The PPI of Potential Targets for Tan IIA in the Treatment of Pulmonary Hypertension
The 26 obtained targets were imported into the STRING database to build a whole PPI network containing 26 nodes and 25 edges (Figure 3A). Based on network topological analysis, only the genes with higher levels of “degree” and “closeness” were collected as the key targets of Tan IIA against PH. Ultimately, seven key targets were collected, with heat shock protein HSP 90-alpha (HSP90AA1), tyrosine protein phosphatase nonreceptor type 11 (PTPN11), ATM serine/threonine kinase (ATM), carbonic Anhydrase II (CA2), telomerase reverse transcriptase (TERT), protein kinase, DNA- activated, catalytic polypeptide (PRKDC), and apurinic/apyrimidinic endonuclease 1 (APEX1) as the hub target genes (Table 1, Figure 3B).
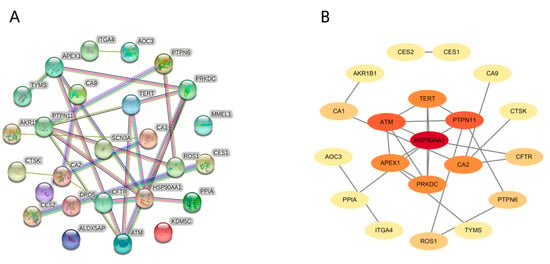
Figure 3.
Protein–protein interaction (PPI) network construction and core targets. (A) A complete PPI network; (B) A PPI network indicating hub target genes, with darker colors indicating a higher degree.

Table 1.
Topological analysis of the Tan IIA target network.
3.3. Enrichment Analysis of Tan IIA in the Treatment of Pulmonary Hypertension
GO and KEGG enrichment analyses were conducted to understand the functions and related pathways of Tan IIA in the treatment PH. Go enrichment analysis revealed that the top 20 items of biological processes annotations included regulation of smooth muscle cell (SMC) proliferation, regulation of SMC migration, regulation of mitotic cell cycle, and regulation of G1/S transition of mitotic cell cycle. The majority of molecular functions were involved in lyase activity, hydro-lyase activity, carbonate dehydratase activity, protein N-terminus binding, carboxylic ester hydrolase activity, etc. For cellular components, the targets were enriched in microvillus, chromosome, telomeric region, actin-based cell projection, chromosomal region, and DNA repair complex, etc. (Figure 4A–C). KEGG pathway analysis showed that these targets were related to nitrogen metabolism, NF-kappa B signaling pathway, cell cycle, necroptosis, apoptosis, and JAK-STAT signaling pathway (Figure 4D).
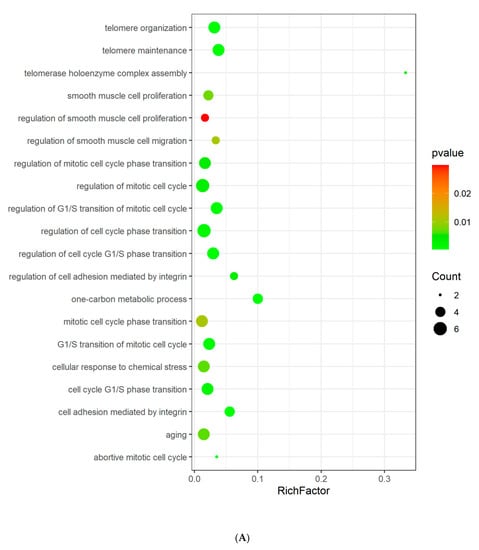
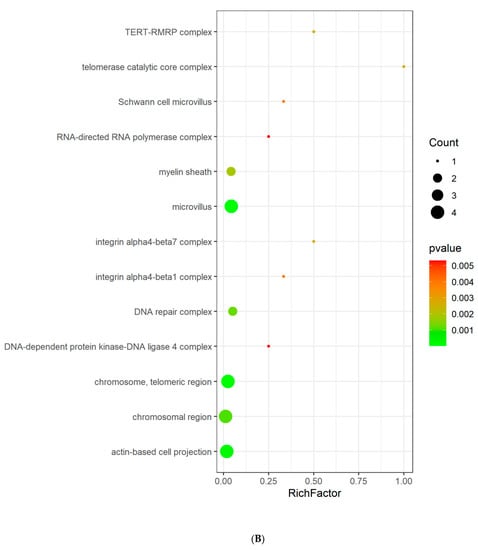
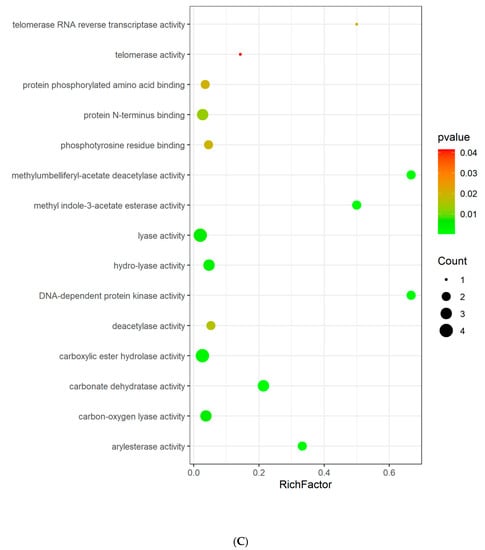
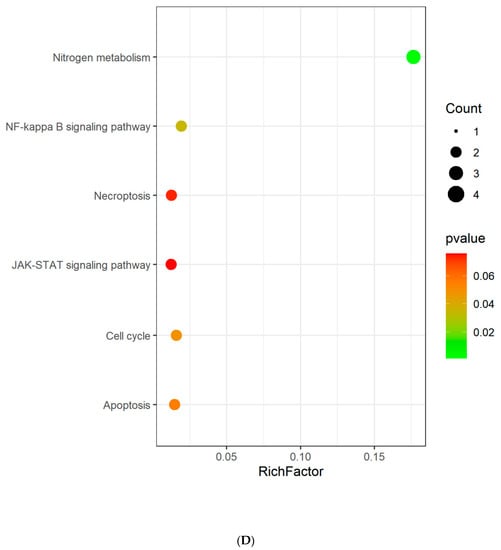
Figure 4.
GO analysis and KEGG pathway analysis of the Tan IIA target. (A) Biological process; (B) cellular components; (C) molecular function; (D) KEGG pathway. The color and size of each bubble represent the p value and gene count, respectively. The X-axis is a rich factor representing the ratio of differentially expressed proteins (DEPs) over the total number of proteins in each category.
3.4. Molecular Docking
Molecular docking was employed to validate the binding interactions of Tan IIA with PH-related key targets according to the network analysis. The binding energies for receptor-ligand interactions were calculated using the -CDOCKER energy. Table 2 shows the binding energies (kcal/mol) of the top three key targets and Tan IIA. The molecular docking results showed that the docking scores of Tan IIA to the HSP90AA1, PTPN11, and CA2 proteins were all less than −5.0 kcal/mol, showing a good overlapping effect.

Table 2.
Tan IIA molecular docking energy scoring results (kcal/mol).
As shown in Figure 5A, the carbon hydrogen bond could combine Tan IIA with GLY108 and LEU103 in HSP90AA1. In addition, there were Pi-alkyl interactions with PHE22, ALA111, LEU107, and PHE138, respectively. Tan IIA could form Pi-Pi stacked with PHE138 and TYR139 in HSP90AA1 and interact with HSP90AA1 via Alkyl with LEU107 and MET98.
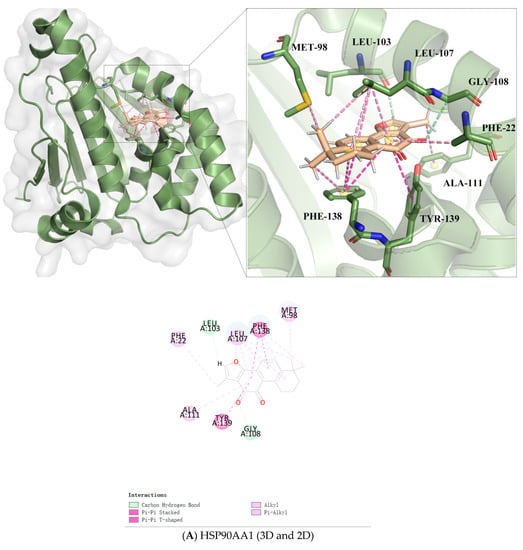
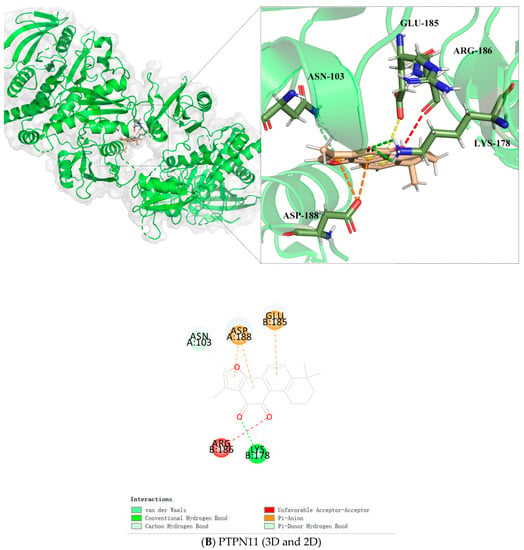
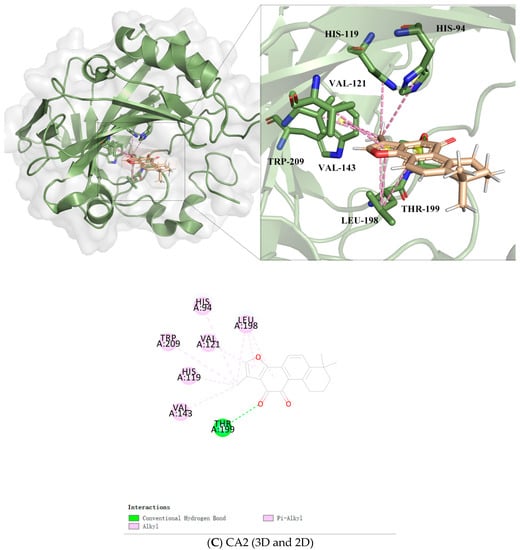
Figure 5.
Docking patterns of Tan IIA interacting with HSP90AA1 (A), PTPN11 (B), and CA2 (C) as produced by CDOCKER, with three-dimensional plots and two-dimensional plots. The 3D plots include complete docking and binding site details, while the different colors of the 2D plots represent different intermolecular forces.
Tan IIA interacts with PTPN11 through conventional hydrogen bond with residues LYS178; Pi-donor hydrogen bond with ASN103; Pi-anion with ASP188 and GLU185, respectively; and unfavorable acceptor-acceptor with ARG186 (Figure 5B).
Similarly, the Tan IIA-CA2 was stabilized by a conventional hydrogen bond with residues THR199; Alkyl with VAL143 and LEU198. The structure of Tan IIA was linked to HIS199, HIS94, TRP209, VAL121, and LEU198 through Pi-Alkyl, respectively (Figure 5C).
3.5. Component-Target-Pathway Network of Pulmonary Hypertension Treatment
To obtain a comprehensive understanding of the potential mechanisms of Tan IIA on PH, we constructed a “component-target-pathway” network (Figure 6). The network revealed that Tan IIA treated PH through multiple targets and pathways.
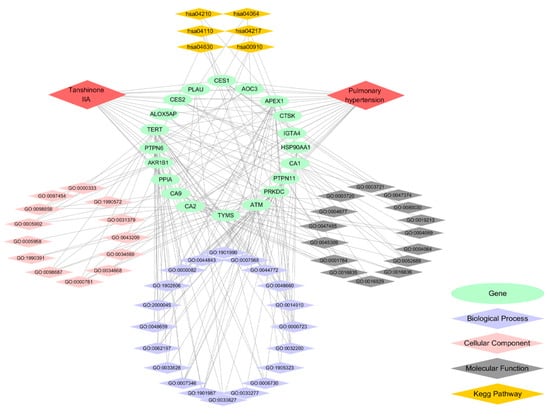
Figure 6.
Component-target-pathway network of treating pulmonary hypertension. The green ovals represent genes. The purple diamonds represent the biological processes. The pink diamonds represent the cellular components. The gray diamonds represent the molecular functions. The orange diamonds represent the KEGG pathway. The gray lines represent the connections.
4. Discussion
PH is a progressive disease, and the pathogenesis of pulmonary vascular remodeling is associated with SMC proliferation, chronic inflammation, apoptotic, and necroptotic signaling pathways [10,11]. Network pharmacology allows the systematic study of drugs, proteins or genes and diseases at the molecular level, improving our understanding of some diseases [12].
In this study, we initially screened out 26 potential targets of Tan IIA in the treatment of PH through data mining. The PPI analysis offers deep knowledge of the drug–target interaction network [13]. The results of network pharmacology and PPI analysis showed that the Tan IIA-related core drug targets involved HSP90AA1, PTPN11, ATM, CA2, TERT, PRKDC, and APEX1. Previous studies showed that HSP90AA1 was highly expressed in the lung tissues of PAH patients and associated with inflammation activation and SMC phenotype change in pulmonary arteries [14]. Meanwhile, the human PTPN11 gene encodes the protein tyrosine phosphatase Shp2, which play critical roles in the development of PAH [15]. It has long been considered that ATM protein kinase mediates DNA damage-induced cell death through inducing apoptosis in human disease [16]. The proteomics discovery of CA2 is a serum biomarker for PAH and participate in the pathogenesis of PAH [17]. TERT was increased in pulmonary vasculature of PH patients, while experimental studies showed that TERT promotes proliferation and migration of pulmonary artery SMC and regulates cell cycle transition from the G0/G1 phase to the S phase [18]. PRKDC is involved in SMC proliferation, innate immune response, and cell apoptosis, which is associated with the pathobiology of PH [19,20]. Studies have also shown that APEX1 affects DNA repair and redox regulation of transcriptional factors, thus increasing the risk of developing PH [21]. Based on the above analysis, we can conclude that these seven core targets are related to Tan IIA-based therapy in PH.
To elucidate the signaling pathways of Tan IIA in treating PH, KEGG pathway analysis was conducted to demonstrate that Tan IIA targeted proteins were involved in nitrogen metabolism, NF-kappa B signaling pathway, cell cycle, necroptosis, apoptosis, and JAK-STAT signaling pathway. Previous studies have demonstrated that nitrogen metabolism influences redox homeostasis, and impairs vascular tone, leading to cell proliferation and obliteration of the pulmonary vasculature [22]. The role of the NF-kappa B pathway in chronic inflammation is well understood, and it has a key role in pulmonary arterial SMC proliferation, migration, and apoptosis, thereby promoting PH development [23]. JAK-STAT pathways have been found to induce cell proliferation, survival, migration, inflammation, and vasoconstriction, which are associated with PH pathology [24]. Besides, increasing evidence supports that the cell cycle signaling pathway is closely related to the pathogenesis of PH. Qin Y et al. found that lncRNA AC068039.4 negatively regulates pulmonary artery SMC proliferation by interrupting each cell cycle through the G0/G1 to the S phase [25]. Necroptosis and apoptosis are the two major modes of cell death, which regulate cell proliferation, survival, and cell homeostasis [26]. Animal models demonstrated that these two cell death forms also promote pulmonary vascular remodeling and inflammation [27].
5. Conclusions
Taken together, our study utilized a network pharmacology and molecular docking technology to demonstrate that Tan IIA may achieve a role in the treatment of PH by intervening in a series of targets (such as HSP90AA1, PTPN11, and CA2), biological processes (such as regulation of SMC proliferation and migration, regulation of mitotic cell cycle, and regulation of G1/S transition of mitotic cell cycle), and signaling pathways (such as nitrogen metabolism, NF-kappa B pathway, cell cycle, necroptosis, apoptosis, and JAK-STAT pathway). However, limitations still exist. This study used only a network for prediction and analysis to discover potential therapeutic targets and disease treatment pathways, and further clinical and experimental validation is needed.
Author Contributions
B.Z. conceived of the idea for this study; K.Z., H.S., K.H. and Z.S. performed the research and analyzed the data; B.Z. and K.Z. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by research grants from the AnHui Provincial Health Commission (No: AHWJ 2021b082) and the Natural Science Foundation of Anhui Province (No: 2208085MH196).
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.
Acknowledgments
We would like to express our gratitude to all those who helped us during the writing of this manuscript. Thanks to all the peer reviewers for their opinions and suggestions.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Morell, E.; Gaies, M.; Fineman, J.R.; Charpie, J.; Rao, R.; Sasaki, J.; Zhang, W.; Reichle, G.; Banerjee, M.; Tabbutt, S. Mortality from Pulmonary Hypertension in the Pediatric Cardiac ICU. Am. J. Respir. Crit. Care Med. 2021, 204, 454–461. [Google Scholar] [CrossRef] [PubMed]
- Beshay, S.; Sahay, S.; Humbert, M. Evaluation and management of pulmonary arterial hypertension. Respir. Med. 2020, 171, 106099. [Google Scholar] [CrossRef] [PubMed]
- Coons, J.C.; Pogue, K.; Kolodziej, A.R.; Hirsch, G.A.; George, M.P. Pulmonary Arterial Hypertension: A Pharmacotherapeutic Update. Curr. Cardiol. Rep. 2019, 21, 141. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Li, L.; Su, J.; Li, S.; Duncan, S.E.; Liu, Z.; Fan, G. Pharmacological Activity and Mechanism of Tanshinone IIA in Related Diseases. Drug Des. Dev. Ther. 2020, 14, 4735–4748. [Google Scholar] [CrossRef]
- Wang, J.; Lu, W.; Wang, W.; Zhang, N.; Wu, H.; Liu, C.; Chen, X.; Chen, Y.; Chen, Y.; Jiang, Q.; et al. Promising therapeutic effects of sodium tanshinone IIA sulfonate towards pulmonary arterial hypertension in patients. J. Thorac Dis. 2013, 5, 169–172. [Google Scholar] [CrossRef] [PubMed]
- Leem, J.; Jung, W.; Park, H.-J.; Kim, K. A network pharmacology-based approach to explore mechanism of action of medicinal herbs for alopecia treatment. Sci. Rep. 2022, 12, 2852. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.-H.; Chen, S.-H.; Wu, H.-H.; Ho, C.-W.; Ko, M.-T.; Lin, C.-Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8 (Suppl. S4), S11. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.-M.; Wang, D.; Lu, F.; Zhao, R.; Ye, X.; He, L.; Ai, L.; Wu, C.-J. Identification of the active substances and mechanisms of ginger for the treatment of colon cancer based on network pharmacology and molecular docking. BioData Min. 2021, 14, 1. [Google Scholar] [CrossRef]
- Scardoni, G.; Petterlini, M.; Laudanna, C. Analyzing biological network parameters with CentiScaPe. Bioinformatics 2009, 25, 2857–2859. [Google Scholar] [CrossRef]
- Poch, D.; Mandel, J. Pulmonary Hypertension. Ann. Intern. Med. 2021, 174, ITC49–ITC64. [Google Scholar] [CrossRef]
- Jarabicová, I.; Horváth, C.; Veľasová, E.; Piváčková, L.B.; Vetešková, J.; Klimas, J.; Křenek, P.; Adameová, A. Analysis of necroptosis and its association with pyroptosis in organ damage in experimental pulmonary arterial hypertension. J. Cell. Mol. Med. 2022, 26, 2633–2645. [Google Scholar] [CrossRef] [PubMed]
- Kibble, M.; Saarinen, N.; Tang, J.; Wennerberg, K.; Mäkelä, S.; Aittokallio, T. Network pharmacology applications to map the unexplored target space and therapeutic potential of natural products. Nat. Prod. Rep. 2015, 32, 1249–1266. [Google Scholar] [CrossRef]
- Kotlyar, M.; Fortney, K.; Jurisica, I. Network-based characterization of drug-regulated genes, drug targets, and toxicity. Methods 2012, 57, 499–507. [Google Scholar] [CrossRef]
- Deng, Y.-X.; Zhong, J.; Liu, Z.-J.; Wang, X.-Q.; Zhang, B. Active ingredients targeting Nrf2 in the Mongolian medicine Qiwei Putao powder: Systematic pharmacological prediction and validation for chronic obstructive pulmonary disease treatment. J. Ethnopharmacol. 2021, 265, 113385. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Yu, M.; Xu, J.; He, M.; Wang, H.; Kong, H.; Xie, W. Inhibition of Shp2 ameliorates monocrotaline-induced pulmonary arterial hypertension in rats. BMC Pulm. Med. 2018, 18, 130. [Google Scholar] [CrossRef]
- Lee, J.-H.; Paull, T.T. Cellular functions of the protein kinase ATM and their relevance to human disease. Nat. Rev. Mol. Cell Biol. 2021, 22, 796–814. [Google Scholar] [CrossRef] [PubMed]
- Nies, M.K.; Yang, J.; Griffiths, M.; Damico, R.; Zhu, J.; Vaydia, D.; Fu, Z.; Brandal, S.; Austin, E.D.; Ivy, D.D.; et al. Proteomics discovery of pulmonary hypertension biomarkers: Insulin-like growth factor binding proteins are associated with disease severity. Pulm. Circ. 2022, 12, e12039. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.; Ma, J.; Zhang, L.; Yu, X.; Liu, M.; Hou, Y.; Wang, Y.; Ma, C.; Li, S.; Zhu, D. Positive Feedback-Loop of Telomerase Reverse Transcriptase and 15-Lipoxygenase-2 Promotes Pulmonary Hypertension. PLoS ONE 2013, 8, e83132. [Google Scholar] [CrossRef]
- Tan, K.T.; Yeh, C.-N.; Chang, Y.-C.; Cheng, J.-H.; Fang, W.-L.; Yeh, Y.-C.; Wang, Y.-C.; Hsu, D.S.-S.; Wu, C.-E.; Lai, J.-I.; et al. PRKDC: New biomarker and drug target for checkpoint blockade immunotherapy. J. Immunother. Cancer 2020, 8, e000485. [Google Scholar] [CrossRef]
- Tajsic, T.; Morrell, N.W. Smooth muscle cell hypertrophy, proliferation, migration and apoptosis in pulmonary hypertension. Compr. Physiol. 2011, 1, 295–317. [Google Scholar] [CrossRef]
- Pei, D.-S.; Jia, P.-P.; Luo, J.-J.; Liu, W.; Strauss, P.R. AP endonuclease 1 (Apex1) influences brain development linking oxidative stress and DNA repair. Cell Death Dis. 2019, 10, 348. [Google Scholar] [CrossRef] [PubMed]
- Fulton, D.J.; Li, X.; Bordan, Z.; Haigh, S.; Bentley, A.; Chen, F.; Barman, S.A. Reactive Oxygen and Nitrogen Species in the Development of Pulmonary Hypertension. Antioxidants 2017, 6, 54. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, S.; Zhang, Y.; Gao, Q.; Sun, W.; Fu, L.; Cao, J. Histone demethylase JARID1B regulates proliferation and migration of pulmonary arterial smooth muscle cells in mice with chronic hypoxia-induced pulmonary hypertension via nuclear factor-kappa B (NFkB). Cardiovasc. Pathol. 2018, 37, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Yerabolu, D.; Weiss, A.; Kojonazarov, B.; Boehm, M.; Schlueter, B.C.; Ruppert, C.; Günther, A.; Jonigk, D.; Grimminger, F.; Ghofrani, H.-A.; et al. Targeting Jak–Stat Signaling in Experimental Pulmonary Hypertension. Am. J. Respir. Cell Mol. Biol. 2021, 64, 100–114. [Google Scholar] [CrossRef]
- Qin, Y.; Zhu, B.; Li, L.; Wang, D.; Qiao, Y.; Liu, B.; Luo, E.; Hou, J.; Yan, G.; Tang, C. Overexpressed lncRNA AC068039.4 Contributes to Proliferation and Cell Cycle Progression of Pulmonary Artery Smooth Muscle Cells Via Sponging miR-26a-5p/TRPC6 in Hypoxic Pulmonary Arterial Hypertension. Shock 2021, 55, 244–255. [Google Scholar] [CrossRef]
- Dannappel, M.; Vlantis, K.; Kumari, S.; Polykratis, A.; Kim, C.; Wachsmuth, L.; Eftychi, C.; Lin, J.; Corona, T.; Hermance, N.; et al. RIPK1 maintains epithelial homeostasis by inhibiting apoptosis and necroptosis. Nature 2014, 513, 90–94. [Google Scholar] [CrossRef]
- Zemskova, M.; McClain, N.; Niihori, M.; Varghese, M.V.; James, J.; Rafikov, R.; Rafikova, O. Necrosis-Released HMGB1 (High Mobility Group Box 1) in the Progressive Pulmonary Arterial Hypertension Associated with Male Sex. Hypertension 2020, 76, 1787–1799. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).