Interpretable Machine Learning with Brain Image and Survival Data
Abstract
:1. Introduction
1.1. Background on Methodical Approaches to Survival Prediction
1.2. Background on MRI Regression/Classification on CNNs
1.3. Explainable AI in MRI Imaging
2. Concept and Implementation
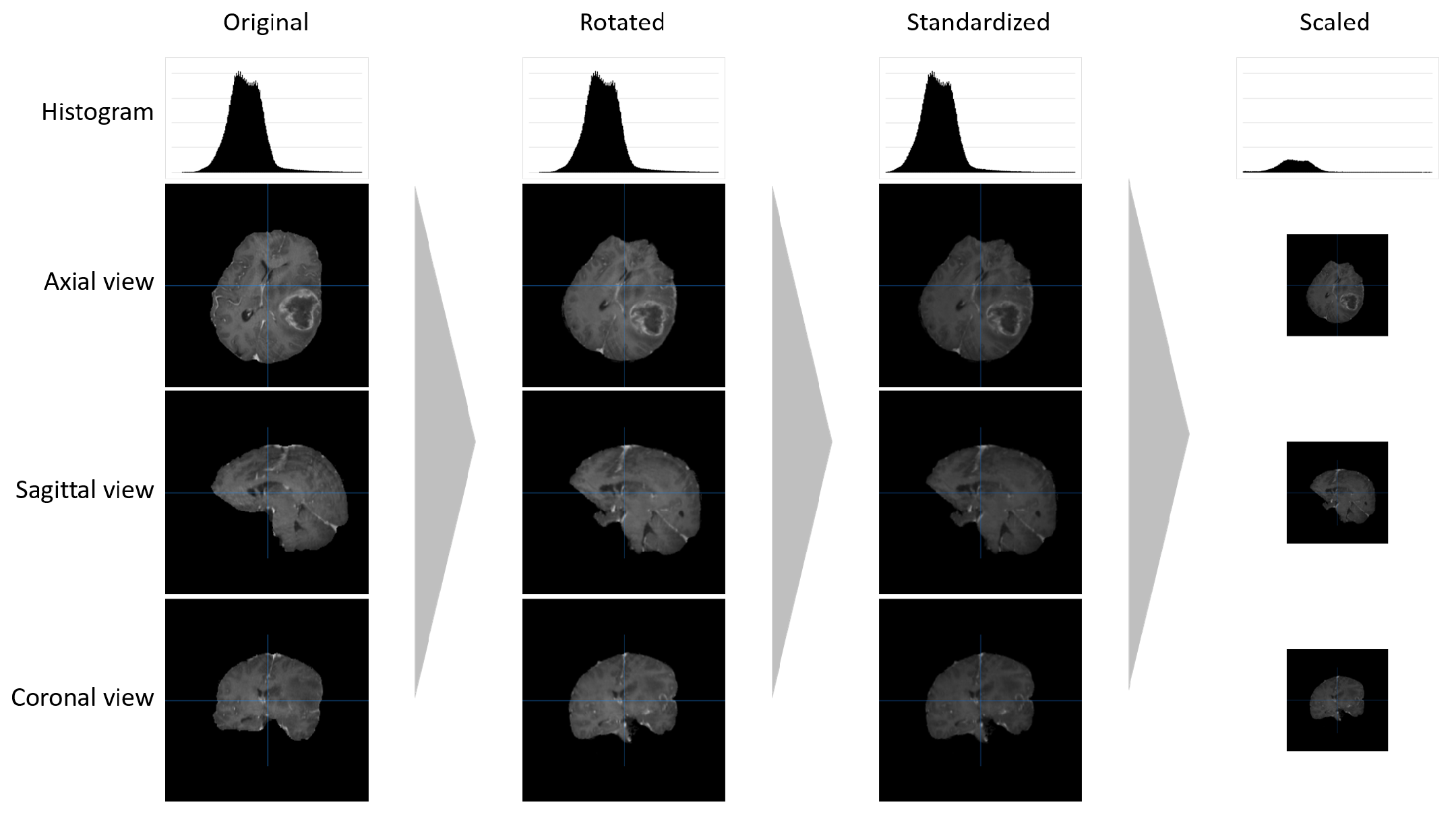
2.1. Data Pre-Processing
2.2. CNN Structure and Preparation
2.3. Explainability
3. Evaluation and Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chaddad, A.; Kucharczyk, M.J.; Daniel, P.; Sabri, S.; Jean-Claude, B.J.; Niazi, T.; Abdulkarim, B. Radiomics in glioblastoma: Current status and challenges facing clinical implementation. Front. Oncol. 2019, 9, 374. [Google Scholar] [CrossRef] [PubMed]
- Bakas, S.; Akbari, H.; Sotiras, A.; Bilello, M.; Rozycki, M.; Kirby, J.; Freymann, J.; Farahani, K.; Davatzikos, C. Advancing the Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci. Data 2017, 4, 170117. [Google Scholar] [CrossRef] [PubMed]
- Gusev, Y.; Bhuvaneshwar, K.; Song, L.; Zenklusen, J.C.; Fine, H.; Madhavan, S. The REMBRANDT study, a large collection of genomic data from brain cancer patients. Sci. Data 2018, 5, 180158. [Google Scholar] [CrossRef] [PubMed]
- Menze, B.; Isensee, F.; Wiest, R.; Wiestler, B.; Maier-Hein, K.; Reyes, M.; Bakas, S. Analyzing magnetic resonance imaging data from glioma patients using deep learning. Comput. Med. Imaging Graph. 2021, 88, 101828. [Google Scholar] [CrossRef]
- Singh, G.; Manjila, S.; Sakla, N.; True, A.; Wardeh, A.H.; Beig, N.; Vaysberg, A.; Matthews, J.; Prasanna, P.; Spektor, V. Radiomics and radiogenomics in gliomas: A contemporary update. Br. J. Cancer 2021, 125, 641–657. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO classification of tumors of the central nervous system: A summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Alderton, G.K. The origins of glioma. Nat. Rev. Cancer 2011, 11, 627. [Google Scholar] [CrossRef]
- Miller, K.D.; Fidler-Benaoudia, M.; Keegan, T.H.; Hipp, H.S.; Jemal, A.; Siegel, R.L. Cancer statistics for adolescents and young adults, 2020. CA Cancer J. Clin. 2020, 70, 443–459. [Google Scholar] [CrossRef]
- Masui, K.; Mischel, P.S.; Reifenberger, G. Molecular classification of gliomas. Handb. Clin. Neurol. 2016, 134, 97–120. [Google Scholar] [CrossRef]
- Jean-Quartier, C.; Jeanquartier, F.; Ridvan, A.; Kargl, M.; Mirza, T.; Stangl, T.; Markaĉ, R.; Jurada, M.; Holzinger, A. Mutation-based clustering and classification analysis reveals distinctive age groups and age-related biomarkers for glioma. BMC Med. Inform. Decis. Mak. 2021, 21, 77. [Google Scholar] [CrossRef]
- Baid, U.; Rane, S.U.; Talbar, S.; Gupta, S.; Thakur, M.H.; Moiyadi, A.; Mahajan, A. Overall survival prediction in glioblastoma with radiomic features using machine learning. Front. Comput. Neurosci. 2020, 14, 61. [Google Scholar] [CrossRef] [PubMed]
- Charlton, C.E.; Poon, M.T.C.; Brennan, P.M.; Fleuriot, J.D. Interpretable Machine Learning Classifiers for Brain Tumour Survival Prediction. arXiv 2021, arXiv:2106.09424. [Google Scholar] [CrossRef]
- Shen, D.; Wu, G.; Suk, H.I. Deep Learning in Medical Image Analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221–248. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Cheplygina, V.; de Bruijne, M.; Pluim, J.P.W. Not-so-supervised: A survey of semi-supervised, multi-instance, and transfer learning in medical image analysis. Med. Image Anal. 2019, 54, 280–296. [Google Scholar] [CrossRef] [PubMed]
- Bernal, J.; Kushibar, K.; Asfaw, D.S.; Valverde, S.; Oliver, A.; Martí, R.; Lladó, X. Deep convolutional neural networks for brain image analysis on magnetic resonance imaging: A review. Artif. Intell. Med. 2019, 95, 64–81. [Google Scholar] [CrossRef]
- Zhu, G.; Jiang, B.; Tong, L.; Xie, Y.; Zaharchuk, G.; Wintermark, M. Applications of Deep Learning to Neuro-Imaging Techniques. Front. Neurol. 2019, 10, 869. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, S.; Chen, H.; Luo, L. Brain Tumor Segmentation and Survival Prediction Using Multimodal MRI Scans with Deep Learning. Front. Neurosci. 2019, 13, 810. [Google Scholar] [CrossRef]
- Van Tulder, G.; de Bruijne, M. Combining Generative and Discriminative Representation Learning for Lung CT Analysis with Convolutional Restricted Boltzmann Machines. IEEE Trans. Med. Imaging 2016, 35, 1262–1272. [Google Scholar] [CrossRef]
- Fakhry, A.; Peng, H.; Ji, S. Deep models for brain EM image segmentation: Novel insights and improved performance. Bioinformatics 2016, 32, 2352–2358. [Google Scholar] [CrossRef] [Green Version]
- Suk, H.I.; Wee, C.Y.; Lee, S.W.; Shen, D. State-space model with deep learning for functional dynamics estimation in resting-state fMRI. NeuroImage 2016, 129, 292–307. [Google Scholar] [CrossRef] [PubMed]
- Zadeh Shirazi, A.; Fornaciari, E.; Bagherian, N.S.; Ebert, L.M.; Koszyca, B.; Gomez, G.A. DeepSurvNet: Deep survival convolutional network for brain cancer survival rate classification based on histopathological images. Med. Biol. Eng. Comput. 2020, 58, 1031–1045. [Google Scholar] [CrossRef] [PubMed]
- Tjoa, E.; Guan, C. A Survey on Explainable Artificial Intelligence (XAI): Toward Medical XAI. IEEE Trans. Neural Netw. Learn. Syst. 2020, 32, 4793–4813. [Google Scholar] [CrossRef]
- Holzinger, A.; Langs, G.; Denk, H.; Zatloukal, K.; Müller, H. Causability and explainability of artificial intelligence in medicine. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2019, 9, e1312. [Google Scholar] [CrossRef]
- Lötsch, J.; Kringel, D.; Ultsch, A. Explainable artificial intelligence (XAI) in biomedicine: Making AI decisions trustworthy for physicians and patients. BioMedInformatics 2021, 2, 1–17. [Google Scholar] [CrossRef]
- Yang, G.; Ye, Q.; Xia, J. Unbox the black-box for the medical explainable AI via multi-modal and multi-centre data fusion: A mini-review, two showcases and beyond. Inf. Fusion 2022, 77, 29–52. [Google Scholar] [CrossRef] [PubMed]
- Wijethilake, N.; Meedeniya, D.; Chitraranjan, C.; Perera, I.; Islam, M.; Ren, H. Glioma Survival Analysis Empowered with Data Engineering—A Survey. IEEE Access 2021, 9, 43168–43191. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S.I. A Unified Approach to Interpreting Model Predictions. In Proceedings of the Advances in Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; Volume 31. [Google Scholar]
- Kan, L.K.; Drummond, K.; Hunn, M.; Williams, D.; O’Brien, T.J.; Monif, M. Potential biomarkers and challenges in glioma diagnosis, therapy and prognosis. BMJ Neurol. Open 2020, 2, e000069. [Google Scholar] [CrossRef]
- Komori, T. Grading of adult diffuse gliomas according to the 2021 WHO Classification of Tumors of the Central Nervous System. Lab. Investig. 2021, 102, 126–133. [Google Scholar] [CrossRef]
- Upadhyay, N.; Waldman, A. Conventional MRI evaluation of gliomas. Br. J. Radiol. 2011, 84, S107–S111. [Google Scholar] [CrossRef] [Green Version]
- Li, W.B.; Tang, K.; Chen, Q.; Li, S.; Qiu, X.G.; Li, S.W.; Jiang, T. MRI manifestions correlate with survival of glioblastoma multiforme patients. Cancer Biol. Med. 2012, 9, 120. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Vallières, M.; Bai, H.X.; Su, C.; Tang, H.; Oldridge, D.; Zhang, Z.; Xiao, B.; Liao, W.; Tao, Y.; et al. MRI features predict survival and molecular markers in diffuse lower-grade gliomas. Neuro-Oncology 2017, 19, 862–870. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Ruiz, A.; Naval-Baudin, P.; Ligero, M.; Pons-Escoda, A.; Bruna, J.; Plans, G.; Calvo, N.; Cos, M.; Majós, C.; Perez-Lopez, R. Precise enhancement quantification in post-operative MRI as an indicator of residual tumor impact is associated with survival in patients with glioblastoma. Sci. Rep. 2021, 11, 695. [Google Scholar] [CrossRef] [PubMed]
- Pope, W.B.; Sayre, J.; Perlina, A.; Villablanca, J.P.; Mischel, P.S.; Cloughesy, T.F. MR imaging correlates of survival in patients with high-grade gliomas. AJNR Am. J. Neuroradiol. 2005, 26, 2466–2474. [Google Scholar]
- Wu, C.C.; Jain, R.; Radmanesh, A.; Poisson, L.M.; Guo, W.Y.; Zagzag, D.; Snuderl, M.; Placantonakis, D.G.; Golfinos, J.; Chi, A.S. Predicting Genotype and Survival in Glioma Using Standard Clinical MR Imaging Apparent Diffusion Coefficient Images: A Pilot Study from The Cancer Genome Atlas. AJNR Am. J. Neuroradiol. 2018, 39, 1814–1820. [Google Scholar] [CrossRef]
- Gates, E.; Pauloski, J.G.; Schellingerhout, D.; Fuentes, D. Glioma Segmentation and a Simple Accurate Model for Overall Survival Prediction. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries; Crimi, A., Bakas, S., Kuijf, H., Keyvan, F., Reyes, M., van Walsum, T., Eds.; Springer International Publishing: Cham, Switzerland, 2019; Volume 11384, pp. 476–484. [Google Scholar] [CrossRef]
- Islam, M.; Jose, V.J.M.; Ren, H. Glioma Prognosis: Segmentation of the Tumor and Survival Prediction Using Shape, Geometric and Clinical Information. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries; Crimi, A., Bakas, S., Kuijf, H., Keyvan, F., Reyes, M., van Walsum, T., Eds.; Springer International Publishing: Cham, Switzerland, 2019; Volume 11384, pp. 142–153. [Google Scholar] [CrossRef]
- Mazurowski, M.A.; Zhang, J.; Peters, K.B.; Hobbs, H. Computer-extracted MR imaging features are associated with survival in glioblastoma patients. J. Neuro-Oncol. 2014, 120, 483–488. [Google Scholar] [CrossRef]
- Zacharaki, E.I.; Morita, N.; Bhatt, P.; O’Rourke, D.M.; Melhem, E.R.; Davatzikos, C. Survival analysis of patients with high-grade gliomas based on data mining of imaging variables. AJNR Am. J. Neuroradiol. 2012, 33, 1065–1071. [Google Scholar] [CrossRef]
- Kang, J.; Ullah, Z.; Gwak, J. MRI-Based Brain Tumor Classification Using Ensemble of Deep Features and Machine Learning Classifiers. Sensors 2021, 21, 2222. [Google Scholar] [CrossRef] [PubMed]
- Aswathy, A.; Vinod Chandra, S. Detection of Brain Tumor Abnormality from MRI FLAIR Images using Machine Learning Techniques. J. Inst. Eng. (India) Ser. B 2022, 103, 1097–1104. [Google Scholar] [CrossRef]
- Badža, M.M.; Barjaktarović, M.Č. Classification of Brain Tumors from MRI Images Using a Convolutional Neural Network. Appl. Sci. 2020, 10, 1999. [Google Scholar] [CrossRef]
- Choi, K.S.; Choi, S.H.; Jeong, B. Prediction of IDH genotype in gliomas with dynamic susceptibility contrast perfusion MR imaging using an explainable recurrent neural network. Neuro-Oncology 2019, 21, 1197–1209. [Google Scholar] [CrossRef] [PubMed]
- Reddy, B.V.; Reddy, P.; Kumar, P.S.; Reddy, S. Developing An Approach to Brain MRI Image Preprocessing for Tumor Detection. Int. J. Res. 2014, 1, 725–731. [Google Scholar]
- Baraiya, N.; Modi, H. Comparative Study of Different Methods for Brain Tumor Extraction from MRI Images using Image Processing. Indian J. Sci. Technol. 2016, 9, 85624. [Google Scholar] [CrossRef]
- Kleesiek, J.; Urban, G.; Hubert, A.; Schwarz, D.; Maier-Hein, K.; Bendszus, M.; Biller, A. Deep MRI brain extraction: A 3D convolutional neural network for skull stripping. NeuroImage 2016, 129, 460–469. [Google Scholar] [CrossRef]
- Hashemzehi, R.; Mahdavi, S.J.S.; Kheirabadi, M.; Kamel, S.R. Detection of brain tumors from MRI images base on deep learning using hybrid model CNN and NADE. Biocybern. Biomed. Eng. 2020, 40, 1225–1232. [Google Scholar] [CrossRef]
- Hoseini, F.; Shahbahrami, A.; Bayat, P. AdaptAhead Optimization Algorithm for Learning Deep CNN Applied to MRI Segmentation. J. Digit. Imaging 2019, 32, 105–115. [Google Scholar] [CrossRef]
- Kamnitsas, K.; Ferrante, E.; Parisot, S.; Ledig, C.; Nori, A.V.; Criminisi, A.; Rueckert, D.; Glocker, B. DeepMedic for Brain Tumor Segmentation. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries; Springer: Cham, Switzerland, 2016; pp. 138–149. [Google Scholar] [CrossRef]
- Mzoughi, H.; Njeh, I.; Wali, A.; Slima, M.B.; BenHamida, A.; Mhiri, C.; Mahfoudhe, K.B. Deep Multi-Scale 3D Convolutional Neural Network (CNN) for MRI Gliomas Brain Tumor Classification. J. Digit. Imaging 2020, 33, 903–915. [Google Scholar] [CrossRef]
- Lundervold, A.S.; Lundervold, A. An overview of deep learning in medical imaging focusing on MRI. Zeitschrift fur medizinische Physik 2019, 29, 102–127. [Google Scholar] [CrossRef]
- Kirimtat, A.; Krejcar, O.; Selamat, A. Brain MRI modality understanding: A guide for image processing and segmentation. In Proceedings of the International Work-Conference on Bioinformatics and Biomedical Engineering, Granada, Spain, 6–8 May 2020; pp. 705–715. [Google Scholar]
- Möllenhoff, K.; Oros-Peusquens, A.M.; Shah, N.J. Introduction to the basics of magnetic resonance imaging. In Molecular Imaging in the Clinical Neurosciences; Springer: Cham, Switzerland, 2012; pp. 75–98. [Google Scholar]
- Lee, D. Mechanisms of contrast enhancement in magnetic resonance imaging. Can. Assoc. Radiol. J. (J. L’Association Can. Des Radiol.) 1991, 42, 6–12. [Google Scholar]
- Holzinger, A.; Malle, B.; Saranti, A.; Pfeifer, B. Towards Multi-Modal Causability with Graph Neural Networks enabling Information Fusion for explainable AI. Inf. Fusion 2021, 71, 28–37. [Google Scholar] [CrossRef]
- Holzinger, A.; Searle, G.; Kleinberger, T.; Seffah, A.; Javahery, H. Investigating Usability Metrics for the Design and Development of Applications for the Elderly. In Lecture Notes in Computer Science LNCS 5105; Springer: Berlin/Heidelberg, Germany; New York, NY, USA, 2008; pp. 98–105. [Google Scholar] [CrossRef]
- Holzinger, A.; Carrington, A.; Mueller, H. Measuring the Quality of Explanations: The System Causability Scale (SCS). Comparing Human and Machine Explanations. KI—KüNstliche Intell. 2020, 34, 193–198. [Google Scholar] [CrossRef] [PubMed]
- ISO 9241-11 (2018); Ergonomics of Human-System Interaction—Part 11: Usability: Definitions and Concepts. International Organization for Standardization: Geneva, Switzerland, 2018. [CrossRef]
- Pearl, J. Causality: Models, Reasoning and Inference, 2nd ed.; Cambridge University Press: Cambridge, UK, 2009. [Google Scholar]
- O’Sullivan, S.; Jeanquartier, F.; Jean-Quartier, C.; Holzinger, A.; Shiebler, D.; Moon, P.; Angione, C. Developments in AI and Machine Learning for Neuroimaging. In Artificial Intelligence and Machine Learning for Digital Pathology; Springer: Cham, Switzerland, 2020; pp. 307–320. [Google Scholar] [CrossRef]
- Manikis, G.C.; Ioannidis, G.S.; Siakallis, L.; Nikiforaki, K.; Iv, M.; Vozlic, D.; Surlan-Popovic, K.; Wintermark, M.; Bisdas, S.; Marias, K. Multicenter dsc–mri-based radiomics predict idh mutation in gliomas. Cancers 2021, 13, 3965. [Google Scholar] [CrossRef] [PubMed]
- Amann, J.; Blasimme, A.; Vayena, E.; Frey, D.; Madai, V.I. Explainability for artificial intelligence in healthcare: A multidisciplinary perspective. BMC Med. Inform. Decis. Mak. 2020, 20, 310. [Google Scholar] [CrossRef] [PubMed]
- Hwang, E.J.; Park, S.; Jin, K.N.; Im Kim, J.; Choi, S.Y.; Lee, J.H.; Goo, J.M.; Aum, J.; Yim, J.J.; Cohen, J.G.; et al. Development and validation of a deep learning–based automated detection algorithm for major thoracic diseases on chest radiographs. JAMA Netw. Open 2019, 2, e191095. [Google Scholar] [CrossRef]
- Papanastasopoulos, Z.; Samala, R.K.; Chan, H.P.; Hadjiiski, L.; Paramagul, C.; Helvie, M.A.; Neal, C.H. Explainable AI for medical imaging: Deep-learning CNN ensemble for classification of estrogen receptor status from breast MRI. In Proceedings of the Medical Imaging 2020: Computer-Aided Diagnosis, Houston, TX, USA, 15–20 February 2020; Volume 11314, pp. 228–235. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Zhou, B.; Khosla, A.; Lapedriza, A.; Oliva, A.; Torralba, A. Learning deep features for discriminative localization. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2921–2929. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. Int. J. Comput. Vis. 2020, 128, 336–359. [Google Scholar] [CrossRef] [Green Version]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why should i trust you?” Explaining the predictions of any classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 1135–1144. [Google Scholar]
- Shrikumar, A.; Greenside, P.; Kundaje, A. Learning important features through propagating activation differences. In Proceedings of the International Conference on Machine Learning, Sydney, Australia, 6–11 August 2017; pp. 3145–3153. [Google Scholar]
- Schwab, P.; Karlen, W. Cxplain: Causal explanations for model interpretation under uncertainty. Adv. Neural Inf. Process. Syst. 2019, 32, 917. [Google Scholar]
- Pintelas, E.; Liaskos, M.; Livieris, I.E.; Kotsiantis, S.; Pintelas, P. Explainable Machine Learning Framework for Image Classification Problems: Case Study on Glioma Cancer Prediction. J. Imaging 2020, 6, 37. [Google Scholar] [CrossRef]
- Gashi, M.; Vuković, M.; Jekic, N.; Thalmann, S.; Holzinger, A.; Jean-Quartier, C.; Jeanquartier, F. State-of-the-Art Explainability Methods with Focus on Visual Analytics Showcased by Glioma Classification. BioMedInformatics 2022, 2, 139–158. [Google Scholar] [CrossRef]
- Singh, A.; Sengupta, S.; Lakshminarayanan, V. Explainable deep learning models in medical image analysis. J. Imaging 2020, 6, 52. [Google Scholar] [CrossRef]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.R.; et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans. Med. Imaging 2015, 34, 1993–2024. [Google Scholar] [CrossRef] [PubMed]
- Bakas, S.; Reyes, M.; Jakab, A.; Bauer, S.; Rempfler, M.; Crimi, A.; Shinohara, R.T.; Berger, C.; Ha, S.M.; Rozycki, M.; et al. Identifying the Best Machine Learning Algorithms for Brain Tumor Segmentation, Progression Assessment, and Overall Survival Prediction in the BRATS Challenge. arXiv 2018, arXiv:1811.02629. [Google Scholar] [CrossRef]
- Gupta, S.; Jindal, V. Brain Tumor Segmentation and Survival Prediction Using Deep Neural Networks. 2020. Available online: https://github.com/shalabh147/Brain-Tumor-Segmentation-and-Survival-Prediction-using-Deep-Neural-Networks (accessed on 23 November 2021).
- Li, Y.; Shen, L. Deep Learning Based Multimodal Brain Tumor Diagnosis. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries; Crimi, A., Bakas, S., Kuijf, H., Menze, B., Reyes, M., Eds.; Springer International Publishing: Cham, Switzerland, 2018; Volume 10670, pp. 149–158. [Google Scholar] [CrossRef]
- Spearman, C. The Proof and Measurement of Association Between Two Things. In Studies in Individual Differences: The Search for Intelligence; Jenkins, J.J., Paterson, D.G., Eds.; Appleton-Century-Crofts: East Norwalk, CT, USA, 1961; pp. 45–58. [Google Scholar] [CrossRef]
- McKinley, R.; Rebsamen, M.; Daetwyler, K.; Meier, R.; Radojewski, P.; Wiest, R. Uncertainty-driven refinement of tumor-core segmentation using 3D-to-2D networks with label uncertainty. In Proceedings of the International MICCAI Brainlesion Workshop, Lima, Peru, 4–8 October 2020; pp. 401–411. [Google Scholar]
- Marti Asenjo, J.; Martinez-Larraz Solís, A. MRI Brain Tumor Segmentation Using a 2D-3D U-Net Ensemble. In Proceedings of the International MICCAI Brainlesion Workshop, Lima, Peru, 4–8 October 2020; pp. 354–366. [Google Scholar] [CrossRef]
- Verduin, M.; Primakov, S.; Compter, I.; Woodruff, H.C.; van Kuijk, S.M.; Ramaekers, B.L.; te Dorsthorst, M.; Revenich, E.G.; ter Laan, M.; Pegge, S.A.; et al. Prognostic and predictive value of integrated qualitative and quantitative magnetic resonance imaging analysis in glioblastoma. Cancers 2021, 13, 722. [Google Scholar] [CrossRef] [PubMed]
- Dequidt, P.; Bourdon, P.; Tremblais, B.; Guillevin, C.; Gianelli, B.; Boutet, C.; Cottier, J.P.; Vallée, J.N.; Fernandez-Maloigne, C.; Guillevin, R. Exploring Radiologic Criteria for Glioma Grade Classification on the BraTS Dataset. IRBM 2021, 42, 407–414. [Google Scholar] [CrossRef]









| Dataset Type | Evaluation Metrics (Test Set) | ||||
|---|---|---|---|---|---|
| Acc. (%) | MSE (d) | Median SE (d) | stdSE (d) | SpearmanR () | |
| original | 57.1 | 127,576.69 | 38,449.52 | 248,209.82 | 0.252 |
| pre-processed | 94.0 | 19,370.85 | 2310.53 | 68,774.36 | 0.934 |
| CNN Definition | Input Size () | FD | Batch Size | Epoch Size | Samples Per Iteration | Total Iterations |
|---|---|---|---|---|---|---|
| 2D full size | 2D | 32 | 100 | 50 | 13,000 | |
| 2D scaled | 2D | 64 | 400 | 400 | 23,000 | |
| 3D scaled | 3D | 16 | 100 | 50 | 6000 |
| CNN Type | MRI Sequence | Time (min) | Evaluation Metrics (Test Set) | ||||
|---|---|---|---|---|---|---|---|
| Acc. (%) | MSE (d) | Median SE (d) | stdSE (d) | SpearmanR () | |||
| 2D full size | T1 | 537 | 42.0 | 152,284.23 | 61,174.52 | 225,719.97 | 0.329 |
| 2D full size | T1CE | 499 | 43.5 | 183,690.40 | 30,002.06 | 389,383.37 | 0.383 |
| 2D full size | T2 | 523 | 41.3 | 183,086 | 53,753.30 | 322,327.76 | 0.260 |
| 2D full size | FLAIR | 518 | 44.7 | 171,695.34 | 29,713.50 | 322,903.01 | 0.337 |
| 2D scaled | T1 | 297 | 94.0 | 19,370.85 | 2310.53 | 68,774.36 | 0.934 |
| 2D scaled | T1CE | 260 | 84.8 | 15,181.36 | 2323.54 | 57,120.51 | 0.948 |
| 2D scaled | T2 | 252 | 86.2 | 13,362.36 | 1435.69 | 49,180.29 | 0.951 |
| 2D scaled | FLAIR | 266 | 87.8 | 12,619.52 | 1722.33 | 46,454.14 | 0.974 |
| 3D scaled | T1 | 832 | 73.7 | 22,853.60 | 3955.39 | 83,608.18 | 0.874 |
| 3D scaled | T1CE | 829 | 79.8 | 35,815.96 | 2771.14 | 131,752.81 | 0.957 |
| 3D scaled | T2 | 825 | 89.4 | 19,423.55 | 1672.75 | 74,759.27 | 0.950 |
| 3D scaled | FLAIR | 823 | 81.2 | 16,603.59 | 2620.99 | 60,487.03 | 0.942 |
| CNN Type | MRI Sequence | Evaluation Metrics (Test Set) | ||||
|---|---|---|---|---|---|---|
| Acc. (%) | MSE (d) | Median SE (d) | stdSE (d) | SpearmanR () | ||
| 2D full size | T1 | 44.8% * | 113,420.55 | 65,536.00 | 146,473.46 | 0.267 |
| 2D full size | T1CE | 44.8% * | 127,564.79 | 20,449.00 | 197,625.03 | 0.324 |
| 2D full size | T2 | 37.9% | 147,032.52 | 69,169.00 | 211,555.71 | 0.132 |
| 2D full size | FLAIR | 55.2% | 69,941.35 | 12,769.00 | 116,749.59 | 0.435 |
| 2D scaled | T1 | 48.3% * | 135,167.90 | 33,856.00 | 194,580.94 | 0.024 * |
| 2D scaled | T1CE | 48.3% * | 94,662.03 | 41,616.00 | 153,731.94 | 0.352 |
| 2D scaled | T2 | 48.8% * | 94,288.35 | 18,225.00 | 147,035.02 | 0.218 |
| 2D scaled | FLAIR | 31.0% | 119,759.07 | 44,944.00 | 176,681.82 | 0.184 |
| 3D scaled | T1 | 44.8% * | 90,073.24 | 44,100.00 | 154,947.23 | 0.249 |
| 3D scaled | T1CE | 48.3% * | 105,020.93 | 25,600.00 | 184,385.83 | 0.270 |
| 3D scaled | T2 | 44.8% * | 130,643.76 | 33,856.00 | 170,256.56 | 0.134 |
| 3D scaled | FLAIR | 31.0% | 111,939.31 | 45,796.00 | 164,669.90 | −0.020 * |
| CNN Type | MRI Sequence | Evaluation Metrics (Test Set) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| (%) | (%) | (%) | ||||||||
| 2D full size | T1 | 46 | 27 | 63.0 | 36 | 95 | 27.5 | 153 | 202 | 43.1 |
| 2D full size | T1CE | 158 | 205 | 43.5 | 42 | 87 | 32.5 | 43 | 24 | 64.2 |
| 2D full size | T2 | 82 | 71 | 53.4 | 38 | 102 | 27.1 | 111 | 155 | 41.7 |
| 2D full size | FLAIR | 112 | 112 | 50.0 | 179 | 119 | 33.2 | 78 | 78 | 50.0 |
| 2D scaled | T1 | 179 | 32 | 84.8 | 129 | 42 | 75.4 | 170 | 7 | 96.0 |
| 2D scaled | T1CE | 179 | 33 | 84.4 | 128 | 45 | 74.0 | 167 | 7 | 96.0 |
| 2D scaled | T2 | 177 | 24 | 88.1 | 140 | 47 | 74.9 | 165 | 6 | 96.5 |
| 2D scaled | FLAIR | 192 | 35 | 84.6 | 130 | 31 | 80.7 | 169 | 2 | 98.8 |
| 3D scaled | T1 | 156 | 19 | 89.1 | 79 | 49 | 61.7 | 177 | 79 | 69.1 |
| 3D scaled | T1CE | 194 | 51 | 79.2 | 117 | 62 | 65.4 | 135 | 0 | 100.0 |
| 3D scaled | T2 | 177 | 12 | 93.7 | 139 | 32 | 81.3 | 184 | 15 | 92.5 |
| 3D scaled | FLAIR | 194 | 63 | 75.5 | 107 | 39 | 73.3 | 153 | 3 | 98.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eder, M.; Moser, E.; Holzinger, A.; Jean-Quartier, C.; Jeanquartier, F. Interpretable Machine Learning with Brain Image and Survival Data. BioMedInformatics 2022, 2, 492-510. https://doi.org/10.3390/biomedinformatics2030031
Eder M, Moser E, Holzinger A, Jean-Quartier C, Jeanquartier F. Interpretable Machine Learning with Brain Image and Survival Data. BioMedInformatics. 2022; 2(3):492-510. https://doi.org/10.3390/biomedinformatics2030031
Chicago/Turabian StyleEder, Matthias, Emanuel Moser, Andreas Holzinger, Claire Jean-Quartier, and Fleur Jeanquartier. 2022. "Interpretable Machine Learning with Brain Image and Survival Data" BioMedInformatics 2, no. 3: 492-510. https://doi.org/10.3390/biomedinformatics2030031
APA StyleEder, M., Moser, E., Holzinger, A., Jean-Quartier, C., & Jeanquartier, F. (2022). Interpretable Machine Learning with Brain Image and Survival Data. BioMedInformatics, 2(3), 492-510. https://doi.org/10.3390/biomedinformatics2030031







