Explainable Artificial Intelligence (XAI) in Biomedicine: Making AI Decisions Trustworthy for Physicians and Patients
Abstract
:1. Introduction
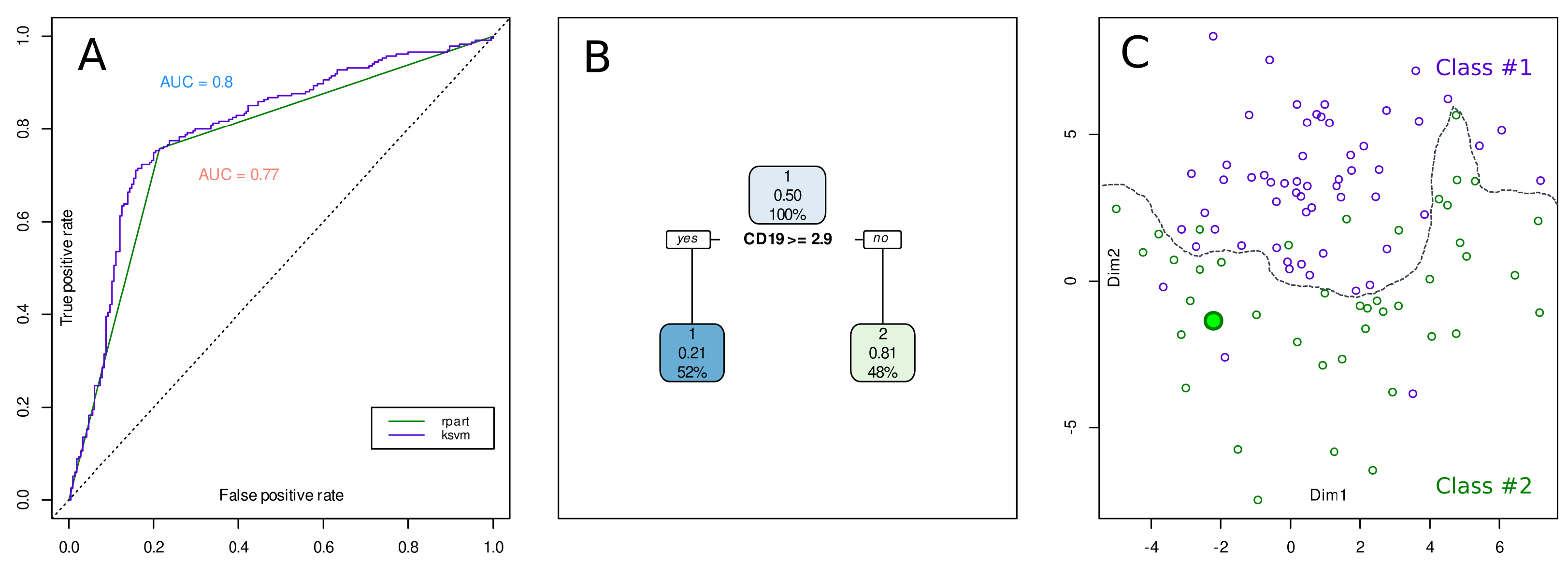
2. An Example Case of XAI versus Standard AI
3. Historical Origins of the Need for Explainable AI
3.1. Knowledge Representation in Expert Systems
3.2. Knowledge-Based Systems
3.3. Skill-Based Systems
4. Transition from AI to XAI in Biomedical Data Science
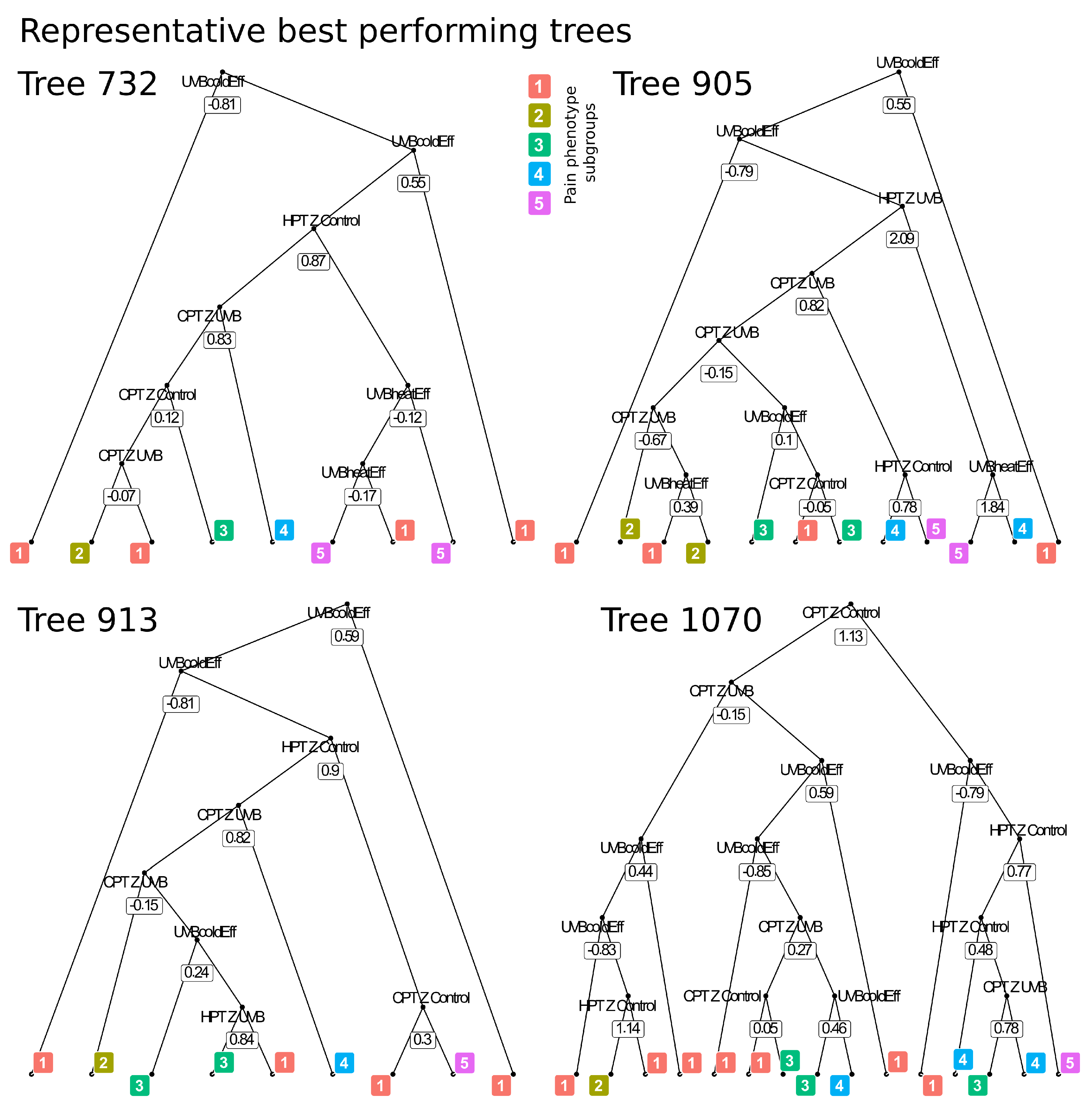
4.1. Methods to Identify the Decision Processes of Subsymbolic “Black-Box” Algorithms
4.2. XAI Designed for Non-Developers
5. Main Biomedical Goals of XAI
5.1. Trustworthiness
5.2. Further and Related Goals of AI in Biomedicine
5.2.1. Transparency
5.2.2. Comprehensibility
5.2.3. Informativeness
5.2.4. Accessibility
5.2.5. Transferability
6. Concerted AI Interpretation between Informatics and Biomedical Domain Experts
7. XAI in Biomedical Publishing
8. Discussion
9. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Luger, G. Artificial Intelligence: Structures and Strategies for Complex Problem Solving, 5th ed.; Pearson Addison Wesley: San Francisco, CA, USA, 2004. [Google Scholar]
- Lötsch, J.; Ultsch, A. Machine learning in pain research. Pain 2017, 159, 623–630. [Google Scholar] [CrossRef]
- Murphy, K.P. Machine Learning: A Probabilistic Perspective; The MIT Press: Cambridge, MA, USA, 2012; p. 1096. [Google Scholar]
- Dhar, V. Data science and prediction. Commun. ACM 2013, 56, 64–73. [Google Scholar] [CrossRef]
- Hamon, R.; Junklewitz, H.; Sanchez, I. Robustness and Explainability of Artificial Intelligence—From Technical to Policy Solutions; Publications Office of the European Union: Luxembourg, 2020. [Google Scholar] [CrossRef]
- Arrieta, A.B.; Díaz-Rodríguez, N.; Ser, J.D.; Bennetot, A.; Tabik, S.; Barbado, A.; García, S.; Gil-López, S.; Molina, D.; Benjamins, R.; et al. Explainable Artificial Intelligence (XAI): Concepts, Taxonomies, Opportunities and Challenges toward Responsible AI. Inf. Fusion 2020, 58, 82–115. [Google Scholar] [CrossRef] [Green Version]
- Turek, M. Explainable Artificial Intelligence (XAI); Defense Advanced Research Projects Agency: Arlington County, VA, USA, 2016.
- Hutson, M. Has artificial intelligence become alchemy? Science 2018, 360, 478. [Google Scholar] [CrossRef]
- Brasko, C.; Smith, K.; Molnar, C.; Farago, N.; Hegedus, L.; Balind, A.; Balassa, T.; Szkalisity, A.; Sukosd, F.; Kocsis, K.; et al. Intelligent image-based in situ single-cell isolation. Nat. Commun. 2018, 9, 226. [Google Scholar] [CrossRef] [PubMed]
- Lötsch, J.; Malkusch, S.; Ultsch, A. Optimal distribution-preserving downsampling of large biomedical data sets (opdisDownsampling). PLoS ONE 2021, 16, e0255838. [Google Scholar] [CrossRef]
- Williams, G.J. Data Mining with Rattle and R: The Art of Excavating Data for Knowledge Discovery; Use R; Springer: Berlin/Heidelberg, Germany, 2011. [Google Scholar]
- Williams, G.J. Rattle: A Data Mining GUI for R. R J. 2009, 1, 45–55. [Google Scholar] [CrossRef] [Green Version]
- Breimann, L.; Friedman, J.; Olshen, R.; Stone, C. Classification and Regression Trees; Chapman and Hall: Boca Raton, FL, USA, 1993. [Google Scholar]
- Cortes, C.; Vapnik, V. Support-Vector Networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Therneau, T.; Atkinson, B. Rpart: Recursive Partitioning and Regression Trees; R Package Version 4.1-15; 2019; Available online: https://cran.r-project.org/package=rpart (accessed on 15 December 2021).
- Karatzoglou, A.; Smola, A.; Hornik, K.; Zeileis, A. kernlab—An S4 Package for Kernel Methods in R. J. Stat. Softw. 2004, 11, 1–20. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Inkscape Project. Inkscape, Version 0.92.5. 2020. Available online: https://inkscape.org (accessed on 15 December 2021).
- De Rie, M.A.; Schumacher, T.N.M.; van Schijndel, G.M.W.; van Lier, R.W.; Miedema, F. Regulatory role of CD19 molecules in B-cell activation and differentiation. Cell. Immunol. 1989, 118, 368–381. [Google Scholar] [CrossRef]
- Hastie, T.; Rosset, S.; Tibshirani, R.; Zhu, J. The entire regularization path for the support vector machine. J. Mach. Learn. Res. 2004, 5, 1391–1415. [Google Scholar]
- Goebel, R.; Chander, A.; Holzinger, K.; Lecue, F.; Akata, Z.; Stumpf, S.; Kieseberg, P.; Holzinger, A. Explainable AI: The New 42? In Machine Learning and Knowledge Extraction. CD-MAKE 2018; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2018; Volume 11015. [Google Scholar] [CrossRef] [Green Version]
- Bayes, M.; Price, M. An Essay towards Solving a Problem in the Doctrine of Chances. By the Late Rev. Mr. Bayes, FRS Communicated by Mr. Price, in a Letter to John Canton, AMFRS. Philos. Trans. 1763, 53, 370–418. [Google Scholar] [CrossRef]
- Kyburg, H.E.T.C.M. Uncertain Inference; Cambridge University Press: Cambridge, UK, 2001. [Google Scholar]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Murray, C.D.; Dermott, S.F. Solar System Dynamics; Cambridge University Press: Cambridge, UK, 2000. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and Regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Hryniewska, W.; Bombiński, P.; Szatkowski, P.; Tomaszewska, P.; Przelaskowski, A.; Biecek, P. Checklist for responsible deep learning modeling of medical images based on COVID-19 detection studies. Pattern Recognit. 2021, 118, 108035. [Google Scholar] [CrossRef]
- Murschel, A. The Structure and Function of Ptolemy’s Physical Hypotheses of Planetary Motion. J. Hist. Astron. 1995, 26, 33–61. [Google Scholar] [CrossRef]
- Hanson, N.R. The Mathematical Power of Epicyclical Astronomy. Isis 1960, 51, 150–158. [Google Scholar] [CrossRef]
- Cover, T.; Hart, P. Nearest neighbor pattern classification. IEEE Trans. Inf. Theor. 1967, 13, 21–27. [Google Scholar] [CrossRef]
- Newell, A.; Simon, H.A. Computer science as empirical inquiry: Symbols and search. Commun. ACM 1976, 19, 113–126. [Google Scholar] [CrossRef] [Green Version]
- Smolensky, P. On the proper treatment of connectionism. Behav. Brain Sci. 2010, 11, 1–23. [Google Scholar] [CrossRef]
- Ho, T.K. Random Decision Forests. In ICDAR ’95: Proceedings of the Third International Conference on Document Analysis and Recognition; IEEE Computer Society: Washington, DC, USA, August 1995; Volume 1, p. 278. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, M.; Ding, Y.; Noone, A.M. Identifying representative trees from ensembles. Stat. Med. 2012, 31, 1601–1616. [Google Scholar] [CrossRef] [Green Version]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why Should I Trust You?”: Explaining the Predictions of Any Classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 1135–1144. [Google Scholar] [CrossRef]
- Lötsch, J.; Malkusch, S. Interpretation of cluster structures in pain-related phenotype data using explainable artificial intelligence (XAI). Eur. J. Pain 2021, 25, 442–465. [Google Scholar] [CrossRef]
- Dasgupta, A. Reprtree: Representative Trees from Ensembles. 2014. Available online: https://github.com/araastat/reprtree/blob/master/R/ReprTree.R (accessed on 15 December 2021).
- Pedersen, T.L. Ggraph: An Implementation of Grammar of Graphics for Graphs and Networks; R package version 2.0.5; 2021. Available online: https://cran.r-project.org/package=ggraph (accessed on 15 December 2021).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Knutson, M.L. Credit Scoring Approaches Guidelines-Final-Web; The World Bank Group: Washington, DC, USA, 2020; Available online: https://thedocs.worldbank.org/en/doc/935891585869698451-0130022020/original/CREDITSCORINGAPPROACHESGUIDELINESFINALWEB.pdf (accessed on 15 December 2021).
- Rumelhart, D.E.; McClelland, J.L. Parallel Distributed Processing: Explorations in the Microstructure of Cognition; MIT Press: Cambridge, MA, USA, 1986; Volume 1. [Google Scholar]
- Huang, X.; Baker, J.; Reddy, R. A Historical Perspective of Speech Recognition. Commun. ACM 2014, 57, 94–103. [Google Scholar] [CrossRef]
- Li, J.; Lavrukhin, V.; Ginsburg, B.; Leary, R.; Kuchaiev, O.; Cohen, J.M.; Nguyen, H.; Gadde, R.T. Jasper: An End-to-End Convolutional Neural Acoustic Model. arXiv 2019, arXiv:1904.03288. [Google Scholar]
- Michalski, R.S. A theory and methodology of inductive learning. In Machine Learning; Michalski, R.S., Carbonell, J.G., Mitchell, T.M., Eds.; Morgan Kaufmann: San Francisco, CA, USA, 1983; pp. 83–134. [Google Scholar] [CrossRef]
- Craven, M.W.; Shavlik, J.W. Extracting Comprehensible Models from Trained Neural Networks; Computer Sciences Department, University of Wisconsin-Madison: Madison, WI, USA, 1996. [Google Scholar]
- Yanase, J.; Triantaphyllou, E. The seven key challenges for the future of computer-aided diagnosis in medicine. Int. J. Med. Inf. 2019, 129, 413–422. [Google Scholar] [CrossRef] [PubMed]
- Ultsch, A.; Kleine, T.; Korus, D.; Farsch, S.; Guimaraes, G.; Pietzuch, W.; Simon, J. Evaluation of Automatic and Manual Knowledge Acquisition for Cerebrospinal Fluid (CSF) Diagnosis. In Artificial Intelligence in Medicine; Keravnou, E., Garbay, C., Baud, R., Wyatt, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; Volume 1211. [Google Scholar] [CrossRef] [Green Version]
- Hodges, W. Classical Logic I: First Order Logic. In The Blackwell Guide to Philosophical Logic; Wiley-Blackwell: Hoboken, NJ, USA, 2001. [Google Scholar]
- Quinlan, J.R. Induction of Decision Trees. Mach. Learn. 1986, 1, 81–106. [Google Scholar] [CrossRef] [Green Version]
- Loh, W.Y.; Vanichsetakul, N. Tree-Structured Classification via Generalized Discriminant Analysis. J. Am. Stat. Assoc. 1988, 83, 715–725. [Google Scholar] [CrossRef]
- Loh, W.Y. Classification and regression trees. WIREs Data Min. Knowl. Discov. 2011, 1, 14–23. [Google Scholar] [CrossRef]
- Loh, W.Y. Fifty Years of Classification and Regression Trees. Int. Stat. Rev. 2014, 82, 329–348. [Google Scholar] [CrossRef] [Green Version]
- Cohen, W.W. Fast Effective Rule Induction. In Proceedings of the 12th International Conference on Machine Learning, Tahoe City, CA, USA, 9–12 July 1995; Morgan Kaufmann: Burlington, MA, USA, 1995; pp. 115–123. [Google Scholar]
- Gigerenzer, G.; Todd, P.M. Fast and frugal heuristics: The adaptive toolbox. In Simple Heuristics That Make Us Smart; Evolution and Cognition; Oxford University Press: New York, NY, USA, 1999; pp. 3–34. [Google Scholar]
- Martignon, L.; Katsikopoulos, K.V.; Woike, J.K. Categorization with limited resources: A family of simple heuristics. J. Math. Psychol. 2008, 52, 352–361. [Google Scholar] [CrossRef]
- Marewski, J.N.; Gigerenzer, G. Heuristic decision making in medicine. Dialogues Clin. Neurosci. 2012, 14, 77–89. [Google Scholar] [PubMed]
- Miller, G.A. The magical number seven plus or minus two: Some limits on our capacity for processing information. Psychol. Rev. 1956, 63, 81–97. [Google Scholar] [CrossRef] [Green Version]
- Holzinger, A. Explainable AI and Multi-Modal Causability in Medicine. i-com 2020, 19, 171–179. [Google Scholar] [CrossRef]
- Bach, S.; Binder, A.; Müller, K.R.; Samek, W. Controlling Explanatory Heatmap Resolution and Semantics via Decomposition Depth. In Proceedings of the 2016 IEEE International Conference on Image Processing (ICIP), Phoenix, AZ, USA, 25–28 September 2016. [Google Scholar]
- Montavon, G. Gradient-Based vs. Propagation-Based Explanations: An Axiomatic Comparison; Springer: Cham, Switzerland, 2019; pp. 253–265. [Google Scholar] [CrossRef]
- Linardatos, P.; Papastefanopoulos, V.; Kotsiantis, S. Explainable AI: A Review of Machine Learning Interpretability Methods. Entropy 2020, 23, 18. [Google Scholar] [CrossRef]
- Thiebes, S.; Lins, S.; Sunyaev, A. Trustworthy artificial intelligence. Electron. Mark. 2021, 31, 447–464. [Google Scholar] [CrossRef]
- Skantzos, N.; Castelein, N. Credit Scoring—Case Study in Data Analytics; Deloitte Touche Tohmatsu Limited: London, UK, 2016. [Google Scholar]
- Rosenblatt, F. The perceptron: A probabilist@articleic model for information storage and organization in the brain. Psychol. Rev. 1958, 65, 386–408. [Google Scholar] [CrossRef] [Green Version]
- Ultsch, A. Emergence in Self-Organizing Feature Maps. In Proceedings of the International Workshop on Self-Organizing Maps (WSOM ’07), Bielefield, Germany, 3–6 September 2007; Ritter, H., Haschke, R., Eds.; Neuroinformatics Group, Bielefeld University: Bielefeld, Germany, 2007. [Google Scholar]
- Kringel, D.; Ultsch, A.; Zimmermann, M.; Jansen, J.P.; Ilias, W.; Freynhagen, R.; Griessinger, N.; Kopf, A.; Stein, C.; Doehring, A.; et al. Emergent biomarker derived from next-generation sequencing to identify pain patients requiring uncommonly high opioid doses. Pharmacogenomics J. 2017, 17, 419–426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stephan, A. Emergenz: Von der Unvorhersagbarkeit zur Selbstorganisation. 4. Auflage; Brill | Mentis: Leiden, The Netherlands, 2020. [Google Scholar] [CrossRef]
- Gilpin, L.H.; Bau, D.; Yuan, B.Z.; Bajwa, A.; Specter, M.; Kagal, L. Explaining Explanations: An Overview of Interpretability of Machine Learnin. In Proceedings of the 2018 IEEE 5th International Conference on Data Science and Advanced Analytics (DSAA), Turin, Italy, 1–4 October 2018. [Google Scholar]
- Lee, E.; Choi, J.S.; Kim, M.; Suk, H.I. Toward an interpretable Alzheimer’s disease diagnostic model with regional abnormality representation via deep learning. Neuroimage 2019, 202, 116113. [Google Scholar] [CrossRef]
- Papadimitroulas, P.; Brocki, L.; Christopher Chung, N.; Marchadour, W.; Vermet, F.; Gaubert, L.; Eleftheriadis, V.; Plachouris, D.; Visvikis, D.; Kagadis, G.C.; et al. Artificial intelligence: Deep learning in oncological radiomics and challenges of interpretability and data harmonization. Phys. Med. 2021, 83, 108–121. [Google Scholar] [CrossRef]
- Ultsch, A.; Thrun, M.C.; Hansen-Goos, O.; Lötsch, J. Identification of Molecular Fingerprints in Human Heat Pain Thresholds by Use of an Interactive Mixture Model R Toolbox (AdaptGauss). Int. J. Mol. Sci. 2015, 16, 25897–25911. [Google Scholar] [CrossRef] [PubMed]
- Malkusch, S.; Hahnefeld, L.; Gurke, R.; Lötsch, J. Visually guided preprocessing of bioanalytical laboratory data using an interactive R notebook (pguIMP). CPT Pharmacometrics Syst. Pharmacol. 2021, 10, 1371–1381. [Google Scholar] [CrossRef] [PubMed]
- Lötsch, J.; Sipilä, R.; Tasmuth, T.; Kringel, D.; Estlander, A.M.; Meretoja, T.; Kalso, E.; Ultsch, A. Machine-learning-derived classifier predicts absence of persistent pain after breast cancer surgery with high accuracy. Breast Cancer Res Treat. 2018, 171, 399–411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Fujiwara, T.; Choi, Y.K.; Kim, K.K.; Ma, K.L. A visual analytics system for multi-model comparison on clinical data predictions. Vis. Inform. 2020, 4, 122–131. [Google Scholar] [CrossRef]
- Liao, Q.V.; Gruen, D.; Miller, S. Questioning the AI: Informing Design Practices for Explainable AI User Experiences. In Proceedings of the 2020 CHI Conference on Human Factors in Computing Systems, New York, NY, USA, 25–30 April 2020. [Google Scholar]
- Lötsch, J.; Sipilä, R.; Dimova, V.; Kalso, E. Machine-learned selection of psychological questionnaire items relevant to the development of persistent pain after breast cancer surgery. Br. J. Anaesth. 2018, 121, 1123–1132. [Google Scholar] [CrossRef]
- Benioff, M.R.; Lazowska, E.D.; Bajcsy, R.; Beese, J.C.; Celis, P.; Evans, P.T.; Yang, G. Report to the President: Computational Science: Ensuring America’s Competitiveness; President’s Information Technology Advisory Committee: Washington, DC, USA, 2005.
- Datta, A.; Matlock, M.K.; Le Dang, N.; Moulin, T.; Woeltje, K.F.; Yanik, E.L.; Joshua Swamidass, S. ‘Black Box’ to ‘Conversational’ Machine Learning: Ondansetron Reduces Risk of Hospital-Acquired Venous Thromboembolism. IEEE J. Biomed. Health Inf. 2021, 25, 2204–2214. [Google Scholar] [CrossRef]
- Bhattacharya, J.; Vogt, W.B. Do Instrumental Variables Belong in Propensity Scores? Int. J. Stat. Econ. 2012, 9, A12. [Google Scholar] [CrossRef]
- VanderWeele, T.J. Principles of confounder selection. Eur. J. Epidemiol. 2019, 34, 211–219. [Google Scholar] [CrossRef] [Green Version]
- Datta, A.; Flynn, N.R.; Barnette, D.A.; Woeltje, K.F.; Miller, G.P.; Swamidass, S.J. Machine learning liver-injuring drug interactions with non-steroidal anti-inflammatory drugs (NSAIDs) from a retrospective electronic health record (EHR) cohort. PLoS Comput. Biol. 2021, 17, e1009053. [Google Scholar] [CrossRef] [PubMed]
- Holzinger, A. Interactive machine learning for health informatics: When do we need the human-in-the-loop? Brain Inf. 2016, 3, 119–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lötsch, J.; Daiker, H.; Hähner, A.; Ultsch, A.; Hummel, T. Drug-target based cross-sectional analysis of olfactory drug effects. Eur. J. Clin. Pharmacol. 2015, 71, 461–471. [Google Scholar] [CrossRef] [PubMed]
- Kovalchik, S. RISmed: Download Content from NCBI Databases; R Package Version 2.3.0; 2021; Available online: https://cran.r-project.org/package=RISmed (accessed on 15 December 2021).
- Fletcher, K.H. Matter with a mind; a neurological research robot. Research 1951, 4, 305–307. [Google Scholar] [PubMed]
- Lanzola, G.; Stefanelli, M.; Barosi, G.; Magnani, L. NEOANEMIA: A knowledge-based system emulating diagnostic reasoning. Comput. Biomed. Res. 1990, 23, 560–582. [Google Scholar] [CrossRef]
- Ching, T.; Himmelstein, D.S.; Beaulieu-Jones, B.K.; Kalinin, A.A.; Do, B.T.; Way, G.P.; Ferrero, E.; Agapow, P.M.; Zietz, M.; Hoffman, M.M.; et al. Opportunities and obstacles for deep learning in biology and medicine. J. R. Soc. Interface 2018, 15, 20170387. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lötsch, J.; Kringel, D.; Ultsch, A. Explainable Artificial Intelligence (XAI) in Biomedicine: Making AI Decisions Trustworthy for Physicians and Patients. BioMedInformatics 2022, 2, 1-17. https://doi.org/10.3390/biomedinformatics2010001
Lötsch J, Kringel D, Ultsch A. Explainable Artificial Intelligence (XAI) in Biomedicine: Making AI Decisions Trustworthy for Physicians and Patients. BioMedInformatics. 2022; 2(1):1-17. https://doi.org/10.3390/biomedinformatics2010001
Chicago/Turabian StyleLötsch, Jörn, Dario Kringel, and Alfred Ultsch. 2022. "Explainable Artificial Intelligence (XAI) in Biomedicine: Making AI Decisions Trustworthy for Physicians and Patients" BioMedInformatics 2, no. 1: 1-17. https://doi.org/10.3390/biomedinformatics2010001
APA StyleLötsch, J., Kringel, D., & Ultsch, A. (2022). Explainable Artificial Intelligence (XAI) in Biomedicine: Making AI Decisions Trustworthy for Physicians and Patients. BioMedInformatics, 2(1), 1-17. https://doi.org/10.3390/biomedinformatics2010001







