Abstract
High expression of the anti-apoptotic TNFAIP8 is associated with poor survival of the patients with diffuse large B-cell lymphoma (DLBCL), and one of the functions of TNFAIP8 is to inhibit the pro-apoptosis Caspase-8. We aimed to analyze the immunohistochemical expression of Caspase-8 (active subunit p18; CASP8) in a series of 97 cases of DLBCL from Tokai University Hospital, and to correlate with other Caspase-8 pathway-related markers, including cleaved Caspase-3, cleaved PARP, BCL2, TP53, MDM2, MYC, Ki67, E2F1, CDK6, MYB and LMO2. After digital image quantification, the correlation with several clinicopathological characteristics of the patients showed that high protein expression of Caspase-8 was associated with a favorable overall and progression-free survival (Hazard Risks = 0.3; p = 0.005 and 0.03, respectively). Caspase-8 also positively correlated with cCASP3, MDM2, E2F1, TNFAIP8, BCL2 and Ki67. Next, the Caspase-8 protein expression was modeled using predictive analytics, and a high overall predictive accuracy (>80%) was obtained with CHAID decision tree, Bayesian network, discriminant analysis, C5 tree, logistic regression, and Artificial Intelligence Neural Network methods (both Multilayer perceptron and Radial basis function); the most relevant markers were cCASP3, E2F1, TP53, cPARP, MDM2, BCL2 and TNFAIP8. Finally, the CASP8 gene expression was also successfully modeled in an independent DLBCL series of 414 cases from the Lymphoma/Leukemia Molecular Profiling Project (LLMPP). In conclusion, high protein expression of Caspase-8 is associated with a favorable prognosis of DLBCL. Predictive modeling is a feasible analytic strategy that results in a solution that can be understood (i.e., explainable artificial intelligence, “white-box” algorithms).
1. Introduction
Diffuse Large B-cell Lymphoma (DLBCL) is one of the most frequent non-Hodgkin lymphomas (NHLs) in western countries. DLBCL accounts for an approximate 25% of NHLs and is characterized by being heterogeneous from a clinicopathological point of view, including histological morphological features, genetic changes and biological characteristics [1,2,3]. Within the category of DLBCL there are several distinct subtypes that are separated, such as the T cell histiocyte rich large B cell lymphoma, the primary DLBCL of mediastinum, the intravascular lymphoma and the lymphomatoid granulomatosis [2]. The prognosis of DLBCL is variable, and with current treatment the disease is curable in 50% of the cases [2,4]. As DLBCL is heterogeneous, it is necessary to identify biomarkers with prognostic value.
The prognosis of DLBCL correlates with the International Prognostic Index (IPI) score, which includes the factors of the age, the serum lactate dehydrogenase, Eastern Cooperative Oncology Group (ECOG) performance status, the clinical stage and the number of extranodal disease sites [5,6,7,8]. A variation of the original IPI that incorporates more detailed information about these used clinical variables is the National Comprehensive Cancer Network (NCCN)-IPI [9]. In this research both IPIs will be used.
The molecular genetics has also managed to stratify the patients according to their prognosis. The gene expression profiling identified three groups according to the postulated cell-of-origin: the germinal center B-cell type (GCB), the activated B-cell type (ABC), and the unclassified. The Hans’ algorithm also identifies the GCB and the non-GCB (ABC) groups, but is based on a stepwise progression of 3 immunohistochemical markers of CD10, BCL6 and MUM1 (IRF4) [10]. Other prognostic markers are the cytogenetic abnormalities of the MYC, BCL2 and BCL6 oncogenes [11,12,13,14,15,16,17,18,19], M2-like tumor-associated macrophages (M2-like TAMs) [20,21] and RGS1 (among others) [22].
In comparison to the GCB, the ABC subtype is characterized by a more aggressive clinical evolution and constitutive activation of the anti-apoptotic nuclear factor kappa B (NF-kB) pathway [23,24,25,26]. We have recently described the prognostic value of a negative mediator of apoptosis in DLBCL, the tumor necrosis factor alpha-induced protein 8 (TNFAIP8) [27,28]. In this research, we had used artificial intelligence—the multilayer perceptron neural network—to analyze the gene expression of the DLBCL series of the Lymphoma/Leukemia Molecular Profiling Project (LLMPP) and to identify the genes that were associated with the overall survival of the patients. The TNFAIP8 was identified within the top 20 most relevant genes of the LLMPP series. Then, we validated the importance of TNFAIP8 by immunohistochemistry and by digital quantification using a machine-learning Weka-based segmentation method in a series of DLBCL from Tokai University Hospital, and we confirmed that high TNFAIP8 was associated with a poor overall survival of the patients [28]. TNFAIP8 acts as a negative mediator of apoptosis and may play a role in tumor progression. TNFAIP8 suppresses the TNF-mediated apoptosis by inhibiting Caspase-8 activity but not the processing of procaspase-8, subsequently resulting in inhibition of BID cleavage and Caspase-3 activation [29,30,31].
In our previous research, we quantified the protein expression of TNFAIP8 in a series from Tokai University Hospital and we also correlated with two markers related to the proliferation cycle, the Ki67 and MYC. We found that through immunohistochemistry, the expression of TNFAIP8 was associated with a poor survival of the patients and also positively correlated with Ki67 and MYC in a moderate manner. Nevertheless, in our previous work we had the limitation of not knowing how in DLBCL the TNFAIP8 expression correlated with the apoptosis pathway (Caspase-8, Caspase-3, PARP), which is the main function of TNFAIP8. In Figure 1 the protein–protein interactions of TNFAIP8 are shown. These interactions highlight the apoptosis (including Caspase-8), cell cycle and the p53 signaling pathways. In addition, in our previous research our correlations included only a linear analysis, and more complex nonlinear analyses (that may fit better in the biological processes) had not been performed. Statistics and machine learning differ in their aim: statistical models infer relationship between variables. Conversely, machine learning is designed to make the most accurate predictions.
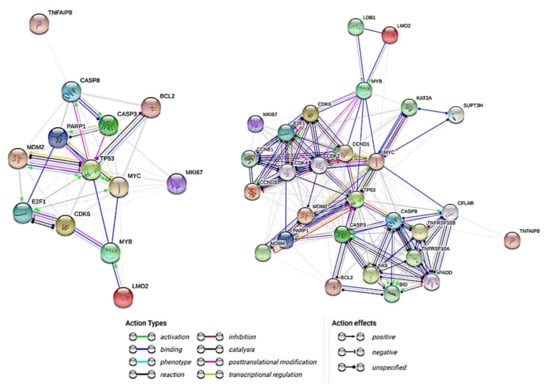
Figure 1.
Interactions between the Caspase-8 and the Caspase-8-related proteins. The aim of this research is to analyze the role of Caspase-8 in Diffuse large B-cell lymphoma, focusing in the investigation of the possible pathological mechanism, the correlations with Caspase-8-related markers and the clinicopathological correlations. This network summarizes the predicted associations of Caspase-8 with the group of pathway-related proteins. The nodes are the proteins and the edges represent the predicted functional associations: action types (activation, binding, inhibition, etc.) and effects types (positive, negative, and unspecified). The basic network only has the markers (nodes) of this project (left), the extended network (right) includes additional nodes for better action types and action effects information.
The purpose of this research was to analyze the expression of Caspase-8 (CASP8) in DLBCL. A series of DLBCL from Tokai University was immunostained with Caspase-8 and the protein expression was quantified by digital image analysis, and other markers of the Caspase-8 pathway including BCL2, cCASP3, CDK6, E2F1, LMO2, MDM2, MKI67, MYB, MYC, cPARP and TP53 were analyzed as well. We performed statistics and machine learning analyses to investigate the correlations between them and with the clinicopathological characteristics of the samples. Then, we also used the multilayer perceptron neural network analysis to identify other genes related to CASP8 using the LLMPP dataset. We found that high expression of Caspase-8 was associated with a good prognosis of the patients.
2. Materials and Methods
2.1. Patients and Samples
2.1.1. Series of DLBCL from Tokai University Hospital
The DLBCL series of Tokai University Hospital is comprised of 97 cases, collected from the years 2006 to 2011. The clinicopathological characteristics are shown in Table 1. In summary, the male/female ratio was 54/43 (1.3) and the age ranged from 14 to 97 years, with a median of 67 and a mean of 64.2 ± 14.5. According to the International Prognostic Index (IPI), 38.3% of the patients were low, 30.9% were low-intermediate, 17.3% were high-intermediate and 13.6% were high. Serum IL2R was high in 77% of the cases and B symptoms were present in 24% of the cases. The location was nodal (including the spleen) in 55% of the cases. The treatment was RCHOP or RCHOP-like in 93.4% of the cases. Clinical response was achieved in 74% of the patients. The pathological characteristics showed that the cell-of-origin was non-GCB in 67% of the cases, and the immune phenotype was CD5+ in 15%, CD10 in 30%, MUM1+ in 79%, BCL2+ in 79% and BCL6 in 67% of the cases. The immunohistochemical expression of Regulator of G-protein signaling 1 (RGS1), which is a marker associated with the chemotaxis of B-lymphocytes, with the germinal centers formation and with a poor prognosis of DLBCL [22,32], was high in 54% of the cases. The clinicopathological variables associated with the overall survival of the patients are shown in Table 1. Relevant variables were the IPI, sIL2R, Epstein-Barr virus infection and the cell-of-origin molecular classification according to the Hans’ classifier [10].

Table 1.
Clinicopathological characteristics of the DLBCL series of Tokai University Hospital.
2.1.2. Series of DLBCL from the Lymphoma/Leukemia Molecular Profiling Project (LLMPP)
We used the series of the LLMPP for gene expression analysis [33,34]. This series, the GSE10846, is a robust and well annotated series of 414 cases of DLBCL from Western countries that is publicly archived and available for downloading at the webpage https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE10846 (accessed on 16 April 2021). This series was last updated on 25 March 2019 (contact: Prof. Louis M. Staudt, Center for Cancer Research, National Cancer Institute, Building 10, Room 5A02, Bethesda, MD 20892, USA).
The clinicopathological features of this series are shown in detail in the Table 2. In summary, the male/female ratio was 224/172 (1.3) and the age ranged from 14 to 92 years, with a median of 62.5 and a mean of 61 ± 15.5. The 5 and 10-years overall survival of the patients was 57% and 47%, respectively. The variables with prognostic value for the overall survival included, among others, the National Comprehensive Cancer Network International Prognostic Index (the enhanced NCCN IPI) and the cell-of-origin molecular subtypes of germinal center B-cell (GCB), activated B-cell (ABC) and unclassified types (Table 2). This series is comparable to the one from Tokai University Hospital.

Table 2.
Clinicopathological characteristics of the DLBCL series of the LLMPP.
2.2. Immunohistochemistry and Digital Image Quantification
The immunohistochemical procedures were performed using formalin-fixed paraffin-embedded tissue sections of the lymphoma samples. The immunostaining was performed in a fully automated stainer for immunohistochemistry and in-situ hybridization (Leica Biosystems Bond-Max, Leica K.K., Tokyo, Japan), including the manufacturer’s ancillary reagents and consumables such as the Dewax solution (AR9222), Wash solution (AR9590), Bond epitope retrieval solution 1 and 2 (AR9961 and AR9640) and Polymer refine detection (DS9800). The staining process included the following steps: dewaxing, antigen retrieval, peroxide block, post-primary, polymer, DAB and hematoxylin. The mounting was performed in a Leica CV5030 coverslipper. The slides were visualized in an Olympus BX53 upright microscope, with a DP74 digital camera and cellSens imaging software (Olympus LifeScience, Olympus K.K., Tokyo, Japan). The whole slides were also digitalized using a Hamamatsu digital slide scanner, the NanoZoomer S360, and visualized with the NDP.view2 Viewing software (Hamamatsu Photonics K.K., Hamamatsu, Japan). The representative areas of each marker were stored as a jpeg image for futher digital image quantification using the Fiji (ImageJ) image processing package, in a RGB and threshold strategy as we have recently described [28].
The primary antibodies that were used in the immunophenotype were the following: CD3e [1:200, clone LN10, Novocastra (NV), Leica K.K., Tokyo, Japan], CD5 (1:400, 4C7, NV), CD20 (1:200, L26, NV), CD10 (1:100, 56C6, NV), MUM1 (1:100, IRF4, EAU32, NV), BCL2 (1:400, bcl2/100/D5, NV), BCL6 (1:100, LN22, NV) and RGS1 (1:100, Rabbit polyclonal, Thermo Fisher Scientific K.K., Yokohama, Japan). More than 30% expression of the tumoral B-lymphocytes of the DLBCL was assessed as positive. The presence of Epstein–Barr virus was also tested (EBER in-situ hybridization #PB0589, Leica K.K.) and the molecular characterization included the gene translocation status of BCL2 and MYC by FISH (split probes, #Y5407 and #Y5410, Dako/Agilent), and the MYD88 (L265P) mutation assessment [22,35,36].
The target markers of this research were Caspase-8, BCL2, cleaved Caspase-3, CDK6, E2F1, LMO2, MDM2, Ki67, A-B-C MYB, MYC, cleaved PARP, TP53 and TNFAIP8.
The primary antibodies and the staining conditions for the target markers were the following: Caspase-8 (active subunit p18, found in caspases 8a, 8b and 8h)(1:30, 11B6, NCL-CASP-8, NV), BCL2 (1:400, mouse monoclonal, bcl2/100/D5, NV), cleaved Caspase-3 (Asp175) [1:300, rabbit polyclonal, #9661, Cell Signalling (CST)], CDK6 [1:5, mouse monoclonal, 98D, Monoclonal Antibodies Unit, Spanish National Cancer Research Center (CNIO), Madrid, Spain], E2F1 (1:14, rat monoclonal, Agro368V, CNIO), LMO2 (1:10, mouse monoclonal, 299B, CNIO), MDM2 (1:50, mouse monoclonal, IF2, Invitrogen K.K., Tokyo, Japan), Ki67 (RTU, mouse monoclonal, MM1, NV), A-B-C MYB (1:10, rat monoclonal, DANI51, CNIO), MYC (1:50, rabbit monoclonal, Y69, Abcam K.K., Tokyo, Japan), cleaved PARP (Asp214) (1:100, rabbit monoclonal, D64E10, CST), TP53 (1:100, DO-7, NV) and TNFAIP8 (1:30,000, mouse monoclonal, #14559-MM01, Sino Biological, Beijing, China).
2.3. Bioinformatics and Statistical Analyses
2.3.1. Comparison between Groups
Comparisons between groups was performed when needed using non-parametric tests, with the Mann–Whitney U test or the Kruskal–Wallis test, and with crosstabulations that included the Pearson Chi-Square, The Fisher’s Exact test, and the Likelihood Ratio test. Correlations between two quantitative variables were performed with Pearson and Spearman correlations. Binary logistic regression was performed to calculate Odds Ratios and to correlate the expression of Caspase-8 (as dichotomic variable) and the rest of clinicopathological variables (also as dichotomic variables).
2.3.2. Survival Analysis
The definition of overall and progression-free survivals were the standards as described by Cheson BD et al. [37,38]. The overall survival was calculated from the time of diagnosis to the time of the death or the last follow-up. The Kaplan–Meier analysis with the Log rank test was used to calculate survival times, as well as for group comparisons; and the analysis included the Breslow and Tarone–Ware tests when necessary. Survival analysis was also performed with the Cox regression (enter method). The significance threshold was set a priori at p < 0.05.
2.3.3. Software and Artificial Neural Network Analysis
Several software were used in this research according to the manufacturer’s instructions: R software for statistical computing version 3.6.3 (https://www.r-project.org/ (accessed on 29 February 2020)) and the integrated development environment R Studio (version 1.3.959; https://www.rstudio.com/products/rstudio/#rstudio-desktop (accessed on 16 April 2021)), the Gene set enrichment analysis software (GSEA 4.1.0, build: 27, Broad Institute, Cambridge, MA, USA; https://www.gsea-msigdb.org/gsea/index.jsp (accessed on 16 April 2021)), IBM SPSS statistics (IBM Corp. Released 2019. IBM SPSS Statistics for Windows, Version 26.0. Armonk, NY, USA: IBM Corp; https://www.ibm.com/jp-ja/analytics/spss-statistics-software (accessed on 16 April 2021)), IBM data mining and predictive analytics (Modeler version 18), Xlstat (version 2018.1, Addinsoft, USA; https://www.xlstat.com/ja/solutions/premium (accessed on 16 April 2021)), Excel (version 16.0.13127.21062, Microsoft, Redmond, WA, USA; https://www.microsoft.com/ja-jp/microsoft-365/excel (accessed on 16 April 2021)) and EditPad Lite (version 8.1.2 x64, Just Great Software Co. Ltd., Rawai Phuket, Thailand; https://www.editpadlite.com/ (accessed on 16 April 2021)). The IBM SPSS Statistics documentation can be found in the following link: https://www.ibm.com/support/knowledgecenter/en/SSLVMB_26.0.0/statistics_mainhelp_ddita/spss/base/overvw_auto_0.html (accessed on 16 April 2021). The statistics algorithms are found at https://www.ibm.com/support/pages/node/874712#en (accessed on 16 April 2021) and ftp://public.dhe.ibm.com/software/analytics/spss/documentation/statistics/26.0/en/client/Manuals/IBM_SPSS_Statistics_Algorithms.pdf (accessed on 16 April 2021). The IBM Modeler can be accessed in the following link: http://127.0.0.1:57379/help/index.jsp?topic=/com.ibm.spss.modeler.help/clementine/clem_intro.htm (accessed on 16 April 2021). A package for survival analysis in R can be accessed at https://cran.r-project.org/web/packages/survival/vignettes/survival.pdf (accessed on 16 April 2021). The Multilayer Perceptron (Figure 2a–d) and Radial Basis Function analysis using the immunohistochemical data was performed following the manufacturer’s instructions, and as we have thoroughly described in our previous publications [39,40]. In this neural network analysis, the prediction of Caspase-8 by the other related markers of the pathway was performed using the immunohistochemical data of Caspase-8 as a dichotomic variable (high vs. low, with the same cut-off of the overall survival).
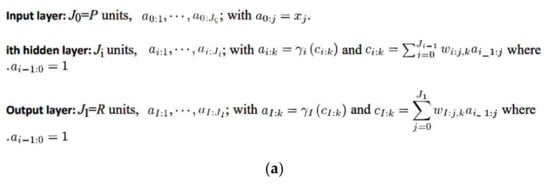

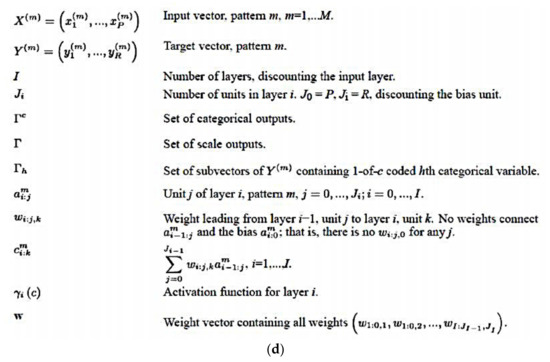
Figure 2.
(a) General architecture for the multilayer perceptron artificial neural network. (b) Activation functions for the multilayer perceptron artificial neural network. (c) Error functions for the multilayer perceptron artificial neural network. (d) Notation for the multilayer perceptron artificial neural network.
The LLMPP dataset was downloaded from the Gene Expression Omnibus (GEO) repository located on the National Center for Biotechnology Information (NCBI) webpage. The gene expression data of the GSE10846 was normalized and log2 transformed. The probes were collapsed according to the maximum probe values. Therefore, each gene had one expression value and the final series was comprised of a total 20,684 genes and 414 cases. Using an Artificial Intelligence approach, we aimed to predict the gene expression of Caspase-8 (CASP8) by the rest of the genes of the array (20,683 genes), using the series of 414 cases of DLBCL from the LLMPP. We used the multilayer perceptron (MLP) procedure, which produced a predictive model for CASP8 (dependent, target variable) based on the values of the predictor variables. Therefore, the dependent variable was the CASP8 and the covariates were the 20,683 genes. In this analysis, the dependent variable was treated as a scale (continuous) because the values represent ordered categories with a meaningful metric, so that distance comparisons between values are appropriate. Of note, this differs from our previous publications in which the dependent (target) variables were dichotomic (high vs. low, or dead vs. alive) [27,28]. Another difference from our previous publications [27,28] is that we are using the values of the collapsed probes. In the setup, CASP8 was the dependent variable, while for the rest of the genes the covariates and the rescaling of the covariates were standardized. As partitions, 70% of the cases corresponded with the training set, while 30% corresponded with the testing set (the holdout was 0%). In the partition dataset the cases were randomly assigned based on the relative number of cases. The architecture had a series of parameters. The hidden layers setup included the number of hidden layers (one or two), the activation function (hyperbolic tangent or sigmoid), and the number of units (automatically computed or custom). The output layer setup included the activation function (identity, softmax, hyperbolic tangent or sigmoid), and the rescaling of scale dependent variables (standardized, normalized, adjusted normalized or none). The type of training could be batch, online or mini-batch; and the optimization algorithm included the scaled conjugate gradient or gradient descent. In the training options the initial lambda value was 0.0000005, the initial sigma 0.00005, the interval center 0, and the interval offset ±0.5. The output displayed the network structure (description, diagram, and the synaptic weights), and the network performance (model summary, predicted by observed chart and residual by predicted chart). In addition, the output also showed the case processing summary and the independent variable importance analysis. The predicted value or category for the dependent variable was saved as a new variable. The synaptic weight estimates were also exported as an xml file. The setup also included the user-missing values and the stopping rules.
3. Results
3.1. Immunohistochemical Protein Expression of Caspase-8 in DLBCL (Tokai Series)
The immunohistochemical protein expression of Caspase-8 in the series of 97 cases of DLBCL from Tokai University Hospital showed a histological localization in the cytoplasm of the cells (compatible with B-lymphocytes), that had a morphology of middle or large sized centroblasts, or immunoblasts in some cases. In some cases, with high Caspase-8 expression the localization was perinuclear including some extension into the nucleus. After digital image quantification, the Caspase-8 expression ranged from 0.0% to 40.2%, with a median of 3.1% and a mean of 6.7% ± 8.3. In Figure 3, the immunohistochemical expression of Caspase-8 is shown, with a characteristic low and high expression pictures. In addition, the immunohistochemistry of the other markers is also shown in the Figure 3, Figure 4 and Figure 5.
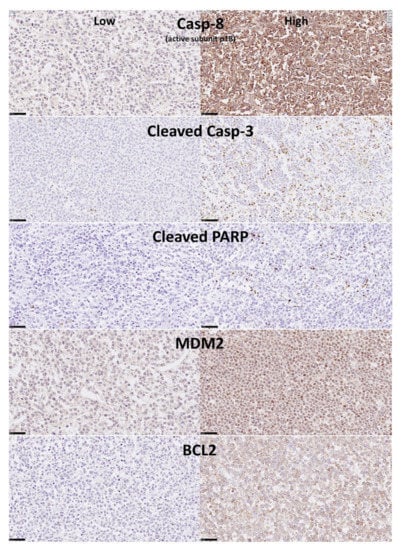
Figure 3.
Immunohistochemical expression in the DLBCL samples of Caspase-8, cleaved Caspase-3, cleaved PARP, MDM2 and BCL2 (Tokai series). Caspase-8 protein is a protease with a key role in the programmed cell death (extrinsic apoptosis). Once activated, Caspase-8 cleaves and activates other effector caspases including Caspase-3 and PARP1. It also regulates necroptosis and innate immunity. MDM2 is a ligase that inhibits the p53 and p73-mediated cell cycle arrest and apoptosis. BCL2 is an apoptosis inhibitor, controlling the mitochondrial membrane activity and inhibiting caspase activity [31]. By immunohistochemistry, Caspase-8 protein expression was cytoplasmic and perinuclear, with some staining in the nucleus when the protein expression was high. Cleaved Caspase-3, cleaved PARP and MDM2 staining was nuclear. BCL2 expression was mainly cytoplasmic and perinuclear.
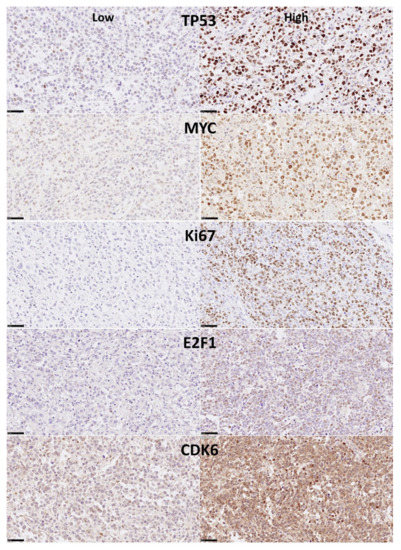
Figure 4.
Immunohistochemical expression in the DLBCL samples of TP53, MYC, Ki67, E2F1 and CDK6 (Tokai series). P53 is a tumor suppressor that controls the cell cycle and induces apoptosis. MYC proto-oncogene is a transcription factor that binds the DNA and activates the transcription of growth-related genes, promotes angiogenesis and regulates somatic reprogramming. Ki67 plays a key role in cell proliferation, with a role in chromatin organization maintaining the mitotic chromosomes dispersed. E2F1 is a transcription factor involved in cell cycle regulation (progression from G1 to S phase) and DNA replication. E2F1 binds RB1 and can mediate both cell proliferation and p53 apoptosis. CDK6 is a kinase involved in the control of cell cycle (G1/S transition) and cell differentiation [31]. By immunohistochemistry all the markers show nuclear staining. CDK6 also shown cytoplasmic localization.
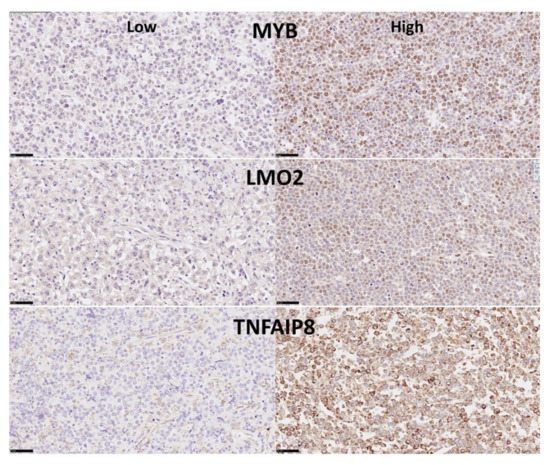
Figure 5.
Immunohistochemical expression of MYB, LMO2 and TNFAIP8 (Tokai series). MYB is a transcriptional activator that binds the DNA and plays a role in the control of cell proliferation and differentiation. LMO2 is a nuclear marker expressed by normal B lymphocytes in the germinal centers. It also regulates hematopoietic stem cell differentiation. TNFAIP8 is a negative regulator of apoptosis and play a role in tumor progression. It inhibits Caspase-8, subsequently resulting in inhibiting the activation of Caspase-3 [31]. We have recently described that high expression of TNFAIP8 correlates with poor survival of DLBCL patients [28]. MYB and LMO2 protein expression is nuclear, TNFAIP8 is in the cytoplasm and perinuclear.
3.2. Correlation between Caspase-8 and the Clinicopathological Variables in DLBCL (Tokai Series)
The protein expression of Caspase-8 as a quantitative variable correlated with the overall survival of the patients (Figure 6). The Cox regression analysis showed a trend of correlation with the overall survival, with high values associated with better survival, Beta = −0.045, p value = 0.071, Hazard risk = 0.956 (95% CI 0.911–1.004). A cut-off was searched and at 8.7% two groups of patients were identified, with different overall survival. The group of high Caspase-8 expression (>8.8%, n = 27/97, 27.8%) was characterized by a more favorable overall survival than the group of low expression (<8.8%, n = 70/97, 72.2%): Beta = −1.3, p value = 0.009, Hazard risk = 0.3 (95% CI, 0.1–0.7). This means, on average, a 70% lower risk of death, and a 233% increase in survival time. When the overall survival was compared using the Kaplan–Meier and the Log rank test, the group of high Caspase-8 expression was characterized by a favorable prognosis, with a 3, 5 and 10-year overall survival of 85%, 85% and 75%. Conversely, the group of low expression had an unfavorable prognosis, with a 3, 5 and 10-year survival of 56%, 52% and 40% (p value = 0.005).
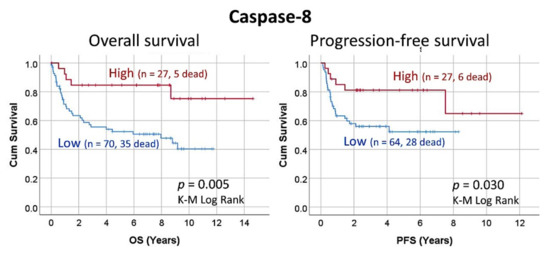
Figure 6.
Overall and progression-free survival according to the Caspase-8 expression by immunohistochemistry (Tokai series, immunohistochemical data). High percentages of Caspase-8 associated with a favorable prognosis of the patients with DLBCL, including both the overall survival and the progression-free survival.
The protein expression of Caspase-8 was also correlated with the progression-free survival of the patients. As a quantitative variable, Caspase-8 did not correlate with the progression-free survival (p value = 0.251). Using the same cut-off as the overall survival (8.8%), high Caspase-8 expression was associated with a more favorable progression-free survival of the patients (Beta = −0.952, p value = 0.036, Hazard risk = 0.386 (95% CI, 0.2–0.9).
Using the same cut-off of the survival analysis (8.8%), a correlation was performed with several clinicopathological characteristics of the series. Nevertheless, no significant correlations were found (Table 3). Therefore, other factors that are not the conventionally tested in DLBCL may be related to the Caspase-8 expression.

Table 3.
Correlation between the clinicopathological characteristics of the DLBCL cases and high immunohistochemical expression of Caspase-8 (Tokai series).
3.3. Correlation between Caspase-8 and the Related Markers in DLBCL (Tokai Series)
The immunohistochemical protein expression of Caspase-8-related markers was also analyzed in the series of 97 cases of DLBCL from Tokai University Hospital. In the Table 4 the distribution of the markers, including cleaved Caspase-3, cleaved PARP, MDM2, BCL2, TP53, MYC, Ki67, E2F1, CDK6, MYB, LMO2 and TNFAIP8 is shown in detail. The expression of Caspase-8 correlated with these markers, and positive correlation was found for cleaved Caspase-3 (correlation coefficient 0.435), MDM2 (0.389), E2F1 (0.324), TNFAIP8 (0.248), BCL2 (0.217), and Ki67 (0.204) (Table 5). Of note, cleaved Caspase-3 positively correlated with cleaved PARP (correlation coefficient = 0.679, p = 0.003).

Table 4.
Immunohistochemical expression of Caspase-8-related markers in DLBCL (Tokai series).

Table 5.
Correlation between Caspase-8 and the Caspase-8-related markers (Tokai series).
The correlation with overall survival and the progression-free survival of these markers is also shown in the Table 6 and Table 7.

Table 6.
Overall survival of the Caspase-8-related markers in DLBCL (Tokai series).

Table 7.
Progression-free survival (PFS) of the Caspase-8-related markers in DLBCL (Tokai series).
3.4. Predictive Modeling of Caspase-8 Protein Expression by the Rest of Caspase-8-Related Markers (Tokai Series)
Predictive analytics was performed to model the immunohistochemical expression of Caspase-8 as a dichotomic variable (high vs. low, using the same 8.7% cut-off) with all the other Caspase-8-related markers, which were used as quantitative variables.
Twelve different models were executed, including the algorithms of C5.0 node that builds a decision tree or a rule set, logistic regression, Bayesian Network, discriminant analysis, k-Nearest Neighbor (KNN), Support Vector Machine (SVM), Tree-AS decision tree, Chi-squared Automatic Interaction Detection (CHAID) decision tree, Classification and Regression (C&R) Tree and Neural Network.
Results of the analysis showed that 9 models predicted the Caspase-8 expression. When ranked according to overall accuracy, they were as follows: CHAID (92%, 4 variables), Bayesian Network (88%, 12 variables), SVM (87%, 12 variables), Discriminant (86%, 12 variables), C5 (85%, 2 variables), Logistic regression (83%, 12 variables), Neural network (80%, 12 variables), C&R Tree (72%, 12 variables), and KNN Algorithm (69%, 12 variables).
3.4.1. Chi-Squared Automatic Interaction Detection (CHAID) Decision Tree
The CHAID node graph is shown in Figure 7; this decision tree predicted the Caspase-8 expression using cCASP3, BCL2, LMO2 and cPARP. The CHAID classification method builds decision trees by using chi-square statistics to identify optimal cut-offs (splits). Unlike the C&R Tree and the QUEST nodes, the CHAID method can generate non-binary trees. Therefore, the splits can be of more than 2, and the trees are wider.
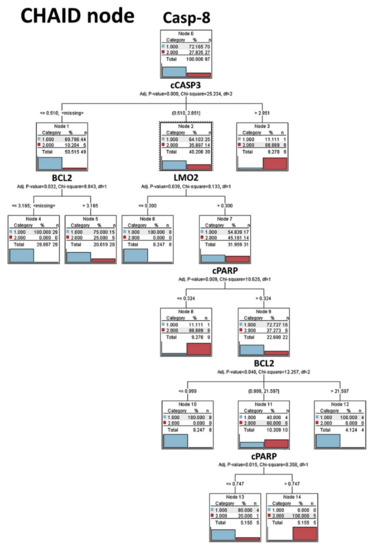
Figure 7.
CHAID node decision tree analysis (Tokai series, immunohistochemical data). The Chi-squared automatic interaction detection (CHAID) is a classification method for building decision trees that identify optimal splits by using chi-square statistics. CHAID examines the crosstabulations between each input field and the outcome, and tests for significance. CHAID can generate nonbinary trees (splits of more than two branches). In this analysis we aimed to predict the Caspase-8 expression as low (1) versus high (2), which is the same cut-off used for the survival analysis. The Caspase-8 expression could be predicted using cleaved Caspase-3, BCL2, cleaved and PARP. This decision tree is highlighting the Caspase-8, cCaspase-3, cPARP apoptosis pathway.
3.4.2. Bayesian Network
The bayesian network model is shown in Figure 8. A Bayesian network is a graphical model that shows variables (i.e., nodes) in a dataset and the probabilistic, or conditional, independencies between them. Causal relationships between nodes may be represented, but the links in the network (i.e., arcs) do not necessarily represent direct cause and effect. The basic view contains a network graph of nodes that displays the relationship between the target (dependent) variable and the predictor variables, and the relationship between the predictors. The distribution view shows the conditional probabilities for each node in the network as a mini graph, the corresponding tables for cleaved Caspase-3 and E2F1 are shown below.
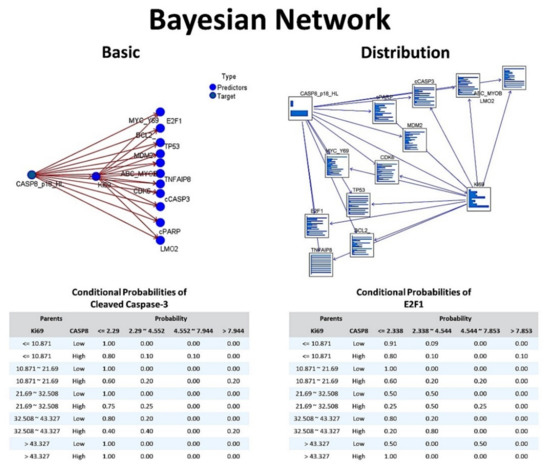
Figure 8.
Bayesian Network (Tokai series, immunohistochemical data). The Bayesian network allows to build a probabilistic model combining observed and recorded evidence with “common-sense” real-world knowledge to establish the likelihood of occurrences by using seemingly unlinked attributes. Therefore, Bayesian networks are used for making predictions. Each of the nodes is one of the markers that have been analyzed by immunohistochemistry in the Tokai series of DLBCL. In this analysis we aimed to predict the Caspase-8 expression (target) by the rest of the markers (predictors). Bayesian networks are very robust where information is missing and make the best possible prediction using whatever information is present. In this figure, the conditional probabilities of cCaspase-3 and E2F1 are also shown.
3.4.3. Discriminant Analysis
The discriminant analysis had 6 excluded cases due to having at least one missing discriminant variable, so the valid cases were 91 of 97 (93.8%). The number of discriminant functions was 1, with an eigen value (discriminating ability) of 0.612 (p = 0.83 × 10−4). The standardized canonical discriminant function coefficients for the different markers were Ki67 (−0.178), LMO2 (0.125), MYC (0.148), MDM2 (−0.329), CDK6 (0.035), E2F1 (0.569), BCL2 (0.190), MYB (0.109), TP53 (−0.328), cPARP (−0.345), cCASP3 (1.118) and TNFAIP8 (0.180).
3.4.4. C5.0 Decision Tree
The C5.0 algorithm builds a decision tree by splitting the sample based on the field that provides the maximum information gain. The C5.0 node can predict only a categorical target. In this model, the Caspase-8 expression (high vs. low) was predicted by cleaved Caspase-3 and E2F1 variables as shown in the Figure 9.
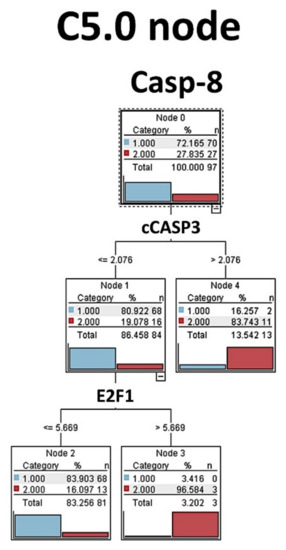
Figure 9.
C5.0 node decision tree analysis (Tokai series, immunohistochemical data). The C5.0 algorithm was used to predict the Caspase-8 expression as a categorical target (low versus high, same cut-off for the survival analysis) by the rest of the markers (predictors). C5.0 models are quite robust when missing data is present and there are large numbers of input fields. C5.0 models tend to be easier to understand. In this analysis we found that Caspase-8 expression could be predicted by cCaspase-3 and E2F1, highlighting the apoptosis pathway.
3.4.5. Logistic Regression
The logistic regression (i.e., nominal regression) classifies records based on values of input fields. It is comparable to the linear regression, but the target variable is categorical instead of a numeric one. The logistic regression equation for the High Caspase-8 expression was the following: −0.02362*Ki67 + 0.01278*LMO2 + 0.1576*MYC + −0.2012*MDM2 + 0.009816*CDK6 + 0.9908*E2F1 + 0.9499*BCL2 + 0.05347*MYB + −0.2845*TP53 + −1.631*cPARP + 3.21*cCASP3 + −0.004535*TNFAIP8 + −2.65. The predictor importance, from most to less was the following: cCASP3, E2F1, BCL2, TP53, MYC, MYOB, CDK6, LMO2, TNFAIP8, Ki69, cPARP and MDM2. As shown in Table 8, in this model the significant variables were cCASP3, cPARP, MDM2 and E2F1.

Table 8.
Logistic regression of Caspase-8 by the Caspase-8-related markers (Tokai series).
When the logistic regression was repeated using the backward method, the predictor importance rank was cCASP3 (most important), E2F1, TP53, BCL2, MYC, cPARP and MDM2 (less). In this case, the equation for High Caspase-8 expression was 0.1527*MYC + −0.1942*MDM2 + 0.8855*E2F1 + 0.09246*BCL2 + −0.3098*TP53 + −1.666*cPARP + 3.114*cCASP3 + −2.697.
3.4.6. Artificial Neural Network
The Neural Network analysis predicted the Caspase-8 expression as a dichotomic variable (high vs. low) with an overall accuracy of 80.4%, using the quantitative values of the Caspase-8-related markers as predictors. This analysis was repeated with two consecutive but independent multilayer perceptron (MLP) and radial basis function (RBF) artificial neural network (ANN) analyses. The details of the neural networks and the results are shown in Table 9 and Figure 10. In summary, the MLP was characterized by a better “performance” because of a lower percent of incorrect predictions both in the training (9.4% vs. 15.7%) and the testing (7.4% vs. 23.8), better overall % of correct classification of training (90.6% vs. 84.3%) and testing (92.6% vs. 76.2%), and a slightly better area under the curve (0.891 vs. 0.880). According to the MLP analysis, the most relevant markers for predicting the Caspase-8 expression as a dichotomic variable were cleaved Caspase-3 (100%), E2F1 (93%), CDK6 (58.8%), TP53 (46.8%), MYC (42.5%), MYB (30.2%), Ki69 (30%), cleaved PARP (12.7%), BCL2 (9%), TNFAIP8 (8.2%) and LMO2 (1.5%).

Table 9.
Artificial Neural Network analysis for Caspase-8 prediction by the Caspase-8-related markers (Tokai series).
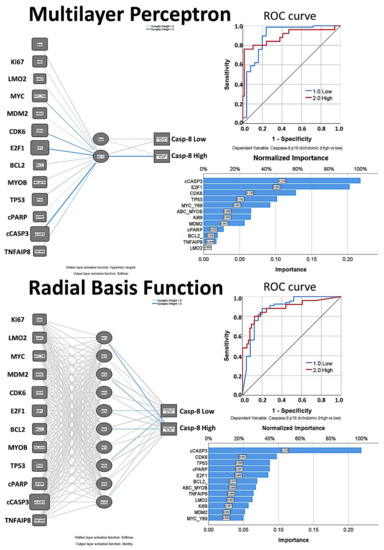
Figure 10.
Artificial Neural Network analysis for the prediction of Caspase-8 by the Caspase-8-related markers (Tokai series, immunohistochemical data). The neural network model determines how the network connects the predictors (our series of 12 markers, input layer) to the targets (the Caspase-8, output layer, as a dichotomic variable high versus low, same cutoff used for the survival analysis) through the hidden layers. The multilayer perceptron (MLP) allows for more complex relationships. Conversely, the radial basis function (RBF) is generally faster and has only one hidden layer, but at the cost of reduced predictive power. The hidden layer(s) contains unobservable units. The value of each hidden unit is some function of the predictors. In this figure, the relevance of each marker for prediction of Caspase-8 is shown by the width of the node and by the value of the normalized importance for prediction. The performance of the network can be checked by the area under the curve ROC curve, of which the higher it is, the better the prediction of Caspase-8 expression. The synaptic weights from the output of the network are available on request from the corresponding author (Carreras J).
3.4.7. Integrated Analysis
The results of several tests were integrated to calculate the percentage of importance for the association to Caspase-8. The most relevant markers were cCASP3, E2F1, TP53, MDM2, BCL2 and TNFAIP8 (Table 10).

Table 10.
Integrated analysis, ranking of markers according to relevance of Caspase-8 association.
3.5. Gene Expression Analysis Based on CASP8 Expression in DLBCL (LLMPP Series)
The LLMPP DLBCL dataset that is comprised of 20,684 genes was used to identify in an unsupervised manner which genes are associated with the CASP8 expression. A multilayer perceptron analysis was performed, with CASP8 as dependent variable (quantitative data) and the rest of 20,863 as predictors (also as quantitative variables). As a result of the artificial neutral network, the genes were ranked according to their normalized importance for prediction of the CASP8 expression. The neural network moderately managed to predict the CASP8 expression. According to their normalized importance, the top most relevant genes were: MED29 (1st), PRH1, YIPF3, PLEKHH1, PRB4, IKZF1, CYSRT1, ACTC1, FAM160B1, TBC1D10C, TMEM176B, ADAMTS10, CTSV, CEP20, AZGP1, ZNF557, SDCCAG8, CSKMT, BGLAP and SRP54 (20th).
To understand the relationship between CASP8 expression and the top 20 genes, the expression of CASP8 was modeled using the top 20 genes. The analysis included the following model types: regression, generalized liner, linear-AS, LSVM, random trees, Tree-AS, linear, CHAID, C&R tree and neural network. The most relevant models were the following: CHAID (correlation 0.806), neural network (0.712), regression (0.668), generalized linear (0.668), linear (0.667) and C&R tree (0.647).
A visualization of the CHAID and neural network is shown in Figure 11. The regression output was the following: CASP8 = MED29*0.1483 + PRH1* − 0.1032 + YIPF3* − 0.1555 + PLEKHH1*0.1117 + PRB4* − 0.001069 + IKZF1*0.1014 + CYSRT1*0.008583 + ACTC1*0.04482 + FAM160B1*0.2315 + TBC1D10C*0.2088 + TMEM176B*0.1449 + ADAMTS10*0.1131 + CTSV* − 0.0005433 + CEP20*0.1234 + AZGP1*0.06398 + ZNF5571*0.08978 + SDCCAG8* − 0.04932 + CSKMT*0.05439 + BGLAP*0.08571 + SRP54*0.3457 − 6.131.
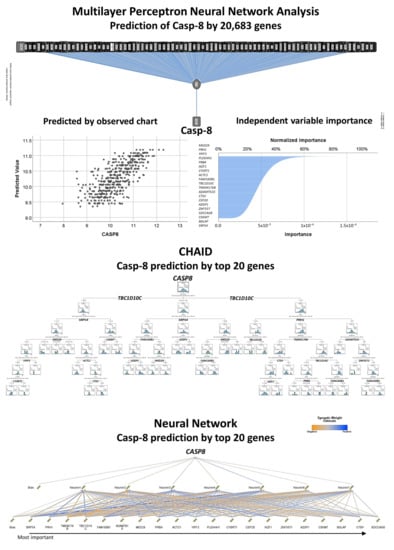
Figure 11.
Prediction of CASP8 by 20,683 genes of the LLMPP series and modeling using the top 20 most relevant genes (gene expression data). The DLBCL gene expression data of the GEO dataset GSE10846 of the Lymphoma/Leukemia Molecular Profiling Project (LLMPP) was used to predict the expression of the CASP8 as a quantitative target variable. In this analysis, the predictors were the 20,863 genes of the gene expression array. Conversely to the analysis of the Tokai cases, in the LLMPP data analyses the CASP8 is predicted as a quantitative variable, which we have not performed in our previous publications (thus the novelty). In neural networks, the predicted by observed chart is used for continuous targets and displays a binned scatterplot of the predicted values on the vertical axis by the observed values on the horizontal axis. The importance of each predictor in making the prediction is shown in the independent variable importance figure. The synaptic weights from the output of the network and the normalized importance chart are available on request from the corresponding author (Carreras J). Typically, the modelling will focus on the predictor fields that matter most and those that matter least will be dropped or ignored. Therefore, the neural network was repeated only with the top 20 genes. In addition to the neural network analysis, this figure also shown the result of the CHAID decision tree.
Further analysis was performed focusing on CASP8 as a dichotomic variable in the DLBCL GEO GSE10846. Using a ROC curve analysis, the best cut-off of CASP8 for the overall survival phenotype (dead/alive) was searched, and the value was 10.3805. Among the 414 cases of the series, CASP8 was high in 180 (48.3%) and low in 234 (69.2%). We confirmed the association of most of the previously identified 20 top genes of the neural network analysis with a high CASP8 expression. The Gene Set Enrichment Analysis (GSEA) is a biostatistical method that confirms if a defined set of genes correlates between two biological states (e.g., phenotypes). We used GSEA to correlate the phenotype CASP8 high vs. low with several set of genes (pathways). The whole collection of the MSigDB gene sets were used (23,677 genes sets in total, MSigDB database v7.3 updated March 2021), which include 9 major collections: H (hallmark genes), C1 (positional), C2 (curated), C3 (regulatory target), C4 (computational), C5 (ontology), C6 (oncogenic signature), C7 (immunologic signature), and C8 (cell type signature). From the 23,677 tested genes sets, 843 gene sets were significantly enriched at nominal p value < 5%, either towards high or low CASP8. For example, significantly enriched pathways of the oncogenic signature that associated to high CASP8 were ALK, KRAS, PGF, P53 and CYCLIND1. Other correlations included sets of the immunologic signature such as macrophages (Genes up-regulated in bone marrow-derived macrophages treated with IL4, GSE25088). The complete results are available on request from the corresponding author (Carreras J).
4. Discussion
This research focused on the analysis of Caspase-8 in DLBCL from Tokai University Hospital. The protein expression of Caspase-8 was evaluated by immunohistochemistry, followed by marker quantification by digital image analysis. We found that high Caspase-8 protein expression was associated with a favorable prognosis of the patients, including a favorable overall and progression-free survival.
Apoptosis is a term to designate programmed cell death. The mechanism of cell death has multiple roles, including a function in the pathogenesis, homeostasis, and control of several types of infection, as well as in cancer [41]. Excessive cell damage results in passive necrosis. On the other hand, the mechanism of cell death can be triggered by several molecular programs including cellular stress, oncogenic changes that involve tumor suppressor genes and oncogenes, several pathogens, and other immune mechanisms. Apoptosis is one of the most known and studied types of programmed cell death [41]; other types of programmed cell death are necroptosis, pyroptosis, ferroptosis, mitotic catastrophy and autophagic cell death, among others [41]. The pathway of apoptosis includes an extrinsic (controlled death receptors of the TNFR superfamily) and an intrinsic (mitochondrial) pathway. Interestingly, ligation of these death receptors induces both activation of extrinsic apoptosis and necroptosis, and the balance between these two pathways determines whether the cell lives. Caspase-8 has a role in initiating of extrinsic apoptosis and inhibiting necroptosis [41]. Caspase-8 activates Caspase-3 by proteolytic cleavage, and then Caspase-3 cleaves other vital cellular proteins or other caspases, which result in activation of cPARP, which eventually leads to apoptosis [42,43,44].
In DLBCL, the mechanisms of cell survival are dysregulated [45]. Dysregulation of an inhibitor of apoptosis proteins (IAPs) has been described in DLBCL [45]. For example, overexpression of XIAP (an apoptosis inhibitor) was associated with a worse outcome in DLBCL [46]. Another inhibitor, the Survivin, was also found overexpressed in DLBCL [47] and in ABC molecular type DLBCL the overexpression was also associated to a poor prognosis [47]. Besides, we recently described that high expression of another apoptosis inhibitor (TNFAIP8) was associated with a poor prognosis of DLBCL [40]. In this project the protein expression of Caspase-8 was analyzed in a series of Tokai University’s, and we found that high expression was associated with a favorable survival of the patients. Therefore, while anti-apoptosis seems to be associated to a poor prognosis of DLBCL, the pro-apoptosis Caspase-8 associates to a favorable outcome of the patients.
In DLBCL there is also dysregulation of TP53 [45], which includes not only mutations or deletions of TP53, but also alterations of TP53 pathways-related markers of BCL6, MDM2, CDKN2A, etc. In this research some of these markers were analyzed by immunohistochemistry in the Tokai series, and the relationship between them as well as with Caspase-8 was explored as shown in Figure 1. In addition, using several modeling analyses, we showed how these markers correlated with the Caspase-8 expression, either as positive or negative correlation, so a pathogenic model can be postulated. For example, the Caspase-8 expression could be calculated as 0.2*MYC + −0.2*MDM2 + 0.9*E2F1 + 0.1*BCL2 + −0.3*TP53 + −1.7*cPARP + 3.1*cCASP3 − 2.697.
This research focused on the analysis of Caspase-8 in a series of Tokai University’s and we found that high protein expression of Caspase-8 correlated with a favorable outcome of the patients, both the overall survival and the progression-free survival. As shown in the Figure 6, the 30% of the patients with high Caspase-8 expression had a favorable overall survival. At the 10-years’ time, around 80% of the patients with high Caspase-8 expression were still alive. Conversely, at that time only 40% were alive in the low expression group. This finding was important and to the best of our knowledge, to date, this association has not been reported in DLBCL. Nevertheless, the Caspase-8 did not correlate with the conventional clinicopathological variables that are usually associated with the prognosis of DLBCL such as the cell-of-origin molecular classifications (Hans’ algorithm) and the International Prognostic Index (IPI) that integrates the clinical variables of age, performance status, LDH, extranodal sites and stage. Therefore, a functional network association analysis was performed, markers associated to Caspase-8 were identified (Figure 1), and finally several types of predictive modeling were tested.
Predictive analytics was performed to model the immunohistochemical expression of Caspase-8 as a dichotomic variable (high vs. low, using the same 8.7% cut-off for the overall survival analysis) with the other Caspase-8-related markers, which were used as quantitative variables.
Twelve different models were executed, including the algorithms of C5.0 node that builds a decision tree or a rule set, logistic regression, Bayesian Network, discriminant analysis, k-Nearest Neighbor (KNN), Support Vector Machine (SVM), Tree-AS decision tree, Chi-squared Automatic Interaction Detection (CHAID) decision tree, Classification and Regression (C&R) Tree and Neural Network. All these models of data mining are tools that enable to develop predictive models using the research experimental data. This data mining process allowed better results and data interpretation, and integrated methods of machine learning, artificial intelligence, and statistics. Of note, each method had certain strengths and was best suited for particular types of problems. Among the 12 different models that were executed, 9 models predicted the Caspase-8 protein expression as a dichotomic variable (high vs. low). When ranked according to their overall accuracy for Caspase-8 prediction, the results were as follows: CHAID tree (92%, 4 variables), Bayesian Network (88%, 12 variables), C5 tree (85%, 2 variables), Logistic regression (83%, 12 variables) and Neural network (80%, 12 variables). The results of all these types of analysis were compatible between them, and each model provided insights into the relationship between Caspase-8 and the rest of the markers. Nevertheless, as previously stated, each method had strengths and weaknesses. For example, the decision trees had an overall accuracy that ranged from 92% for the CHAID tree to 85% of the C5 tree. This means that prediction of Caspase-8 was successful, although variable. Nevertheless, in these models not all the markers were used in the final model, so the relevance of some of the markers cannot be properly assessed. The Bayesian Network built a probabilistic model and made use of all the markers. Bayesian Networks are very robust where information is missing and make the best possible prediction using whatever information is present. Causal relationships between nodes may be represented but the links in the network (i.e., arcs) do not necessarily represent direct cause and effect. The logistic regression (i.e., nominal regression) classifies records based on values of input fields. It is comparable to the linear regression, but the target variable is categorical instead of numeric. This method had the strength of allowing us to know which were the most relevant markers for the prediction of Caspase-8, with information of the direction of the association (increase or decrease) and the strength of that association. Neural networks are simple models of the way the nervous system operates. The basic units are neurons, which are typically organized into layers. There are three parts in a neural network: the input, the hidden and the output layers. The network learns thorough training. Since the output is known, as the training progresses the network becomes increasingly accurate in replicating the known outcomes. Since the deep neural networks have a multilayer non-linear structure (i.e., black box model), neural networks are criticized to be non-transparent because their predictions are not traceable by humans. In our analysis we could rank the markers according to their normalized importance for Caspase-8 prediction, but the reason for this association was elusive because the synaptic weights are only sort of meaningful. In summary, we used a series of algorithms to create classification models. Each model used the values of the input fields (our markers) to predict the value of one output or target field (Caspase-8 as a dichotomic variable, high vs. low), and the integration of all the information made the results more understandable (explainable). As shown in Table 10, the most relevant markers associated with Caspase-8 were the following: cCASP3, E2F1, TP53, cPARP, MDM2, BCL2 and TNFAIP8. Caspase 3, PARP, BCL2 are known markers closely related to apoptosis. Therefore, it makes sense that they were highly associated with Caspase-8. Nevertheless, some of the markers are also associated with other pathways. MDM2 is a ligase that inhibits the p53 and p73-mediated cell cycle arrest and apoptosis [31]. The p53 protein is a tumor suppressor that also controls the cell cycle and induces apoptosis. MYC proto-oncogene is a transcription factor that activates the transcription of growth-related genes and promotes angiogenesis. Ki67 has a role in chromatin organization and it is a widely used marker of cell proliferation. E2F1 is also involved in the cell cycle. CDK6 is a kinase that also controls the G1/S cell cycle transition and the cell differentiation [31]. MYB also controls the cell cycle and cell differentiation. LMO2 is a nuclear marker of normal B lymphocytes of the germinal centers, and DLBCL is supposed to be developed from these lymphocytes. Finally, TNFAIP8 is a negative regulator of apoptosis and plays a role in tumor progression [31]. In summary, the most relevant markers that we have highlighted belonged to the apoptosis and the control of cell cycle.
Finally, the Capase-8 gene expression as a quantitative variable was also analyzed in an independent series of DLBCL of the LLMPP, as the relationship with other genes could also be successfully explored. The most relevant gene was MED29, a component of the Mediator complex that is involved in the regulation of transcription [31]. MED29 has been related to prostate cancer [48].
Future research directions should include analyzing the same markers in larger series of DLBCL to validate our findings. In addition, in-vitro or in-vivo analyses may also help to clarify the pathological function of Caspase-8 in DLBCL.
5. Conclusions
In conclusion, high immunohistochemical protein expression of Caspase-8 is associated with a favorable overall survival and progression-free survival of the patients in a series of DLBCL from Tokai University Hospital. The relationship of Caspase-8 with other related markers could also be confirmed by predictive analytics including decision trees, Bayesian network, logistic regression and artificial neural networks. Therefore, the immunohistochemical analysis of Caspase-8 could be implemented in the routine diagnosis of DLBCL as a prognostic marker.
Author Contributions
Conceptualization, J.C.; methodology, J.C.; software, J.C.; validation, R.H.; formal analysis, J.C.; investigation, Y.Y.K., M.M., S.H., S.T., H.I., Y.K., A.I., G.R., S.S., K.A.; resources, N.N., K.A., R.H., G.R.; writing—original draft preparation, J.C.; writing—review and editing, J.C.; supervision, N.N.; project administration, J.C.; funding acquisition, J.C. and R.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by THE MINISTRY OF EDUCATION, CULTURE, SPORTS, SCIENCE AND TECHNOLOGY (MEXT) and THE JAPAN SOCIETY FOR THE PROMOTION OF SCIENCE, grant number KAKEN 18K15100 to Joaquim Carreras. Rifat Hamoudi was funded by AL-JALILA FOUNDATION (grant number AJF201741), THE SHARJAH RESEARCH ACADEMY (grant number MED001) and UNIVERSITY OF SHARJAH (grant number 1901090258).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board and the Ethics Committee of Tokai University, School of Medicine (protocol code IRB14R-080 and IRB-156).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The gene expression data of DLBCL (GEO dataset GSE10846) was obtained from the publicly available database of the NCBI resources webpage, located at https://www.ncbi.nlm.nih.gov/gds (accessed on 16 April 2021). The data from Tokai University presented in this study are available on request from the corresponding author. The data are not publicly available due to data protection policy.
Acknowledgments
We would like to thank and acknowledge all the members of the Lymphoma/Leukemia Molecular Profiling Project (LLMPP) who participated in the generation of the GSE10846 dataset, including LM Staudt, E Campo, ES Jaffe, WC Chan, WH Wilson, TA Lister, RD Gascoyne, JM Conners, G Wright, SS Dave, LM Rimsza, A Ronsenwald, D Wrench, H-K Muller-Hermelink, G Ott and E Hartman (among others).
Conflicts of Interest
The authors declare no conflict of interest.
References
- WHO. Classification of Tumours of Haematopoietic and Lymphoid Tissues, 4th ed.; Swerdlow, S.H., Campo, E., Harris, N.L., Jaffe, E.S., Pileri, S.A., Thiele, J., Eds.; International Agency for Research on Cancer (IARC): Lyon, France, 2017. [Google Scholar]
- Freedman, A.S.; Aster, J.C.; Lister, A.L.; Rosmarin, A.G. Prognosis of Diffuse Large B Cell Lymphoma; UpToDate: Wellesley, MA, USA, 2017. [Google Scholar]
- Morton, L.M.; Wang, S.S.; Devesa, S.S.; Hartge, P.; Weisenburger, D.D.; Linet, M.S. Lymphoma incidence patterns by WHO subtype in the United States, 1992–2001. Blood 2006, 107, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.; Crouch, S.; Howell, D.; Burton, C.; Patmore, R.; Roman, E. Impact of age and socioeconomic status on treatment and survival from aggressive lymphoma: A UK population-based study of diffuse large B-cell lymphoma. Cancer Epidemiol. 2015, 39, 1103–1112. [Google Scholar] [CrossRef] [PubMed]
- Bari, A.; Marcheselli, L.; Sacchi, S.; Marcheselli, R.; Pozzi, S.; Ferri, P.; Balleari, E.; Musto, P.; Neri, S.; Aloe Spiriti, M.A.; et al. Prognostic models for diffuse large B-cell lymphoma in the rituximab era: A never-ending story. Ann. Oncol. 2010, 21, 1486–1491. [Google Scholar] [CrossRef] [PubMed]
- Salles, G.; de Jong, D.; Xie, W.; Rosenwald, A.; Chhanabhai, M.; Gaulard, P.; Klapper, W.; Calaminici, M.; Sander, B.; Thorns, C.; et al. Prognostic significance of immunohistochemical biomarkers in diffuse large B-cell lymphoma: A study from the Lunenburg Lymphoma Biomarker Consortium. Blood 2011, 117, 7070–7078. [Google Scholar] [CrossRef]
- Sehn, L.H.; Berry, B.; Chhanabhai, M.; Fitzgerald, C.; Gill, K.; Hoskins, P.; Klasa, R.; Savage, K.J.; Shenkier, T.; Sutherland, J.; et al. The revised International Prognostic Index (R-IPI) is a better predictor of outcome than the standard IPI for patients with diffuse large B-cell lymphoma treated with R-CHOP. Blood 2007, 109, 1857–1861. [Google Scholar] [CrossRef]
- Ziepert, M.; Hasenclever, D.; Kuhnt, E.; Glass, B.; Schmitz, N.; Pfreundschuh, M.; Loeffler, M. Standard International prognostic index remains a valid predictor of outcome for patients with aggressive CD20+ B-cell lymphoma in the rituximab era. J. Clin. Oncol. 2010, 28, 2373–2380. [Google Scholar] [CrossRef]
- Zhou, Z.; Sehn, L.H.; Rademaker, A.W.; Gordon, L.I.; Lacasce, A.S.; Crosby-Thompson, A.; Vanderplas, A.; Zelenetz, A.D.; Abel, G.A.; Rodriguez, M.A.; et al. An enhanced International Prognostic Index (NCCN-IPI) for patients with diffuse large B-cell lymphoma treated in the rituximab era. Blood 2014, 123, 837–842. [Google Scholar] [CrossRef]
- Hans, C.P.; Weisenburger, D.D.; Greiner, T.C.; Gascoyne, R.D.; Delabie, J.; Ott, G.; Muller-Hermelink, H.K.; Campo, E.; Braziel, R.M.; Jaffe, E.S.; et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood 2004, 103, 275–282. [Google Scholar] [CrossRef]
- Barrans, S.; Crouch, S.; Smith, A.; Turner, K.; Owen, R.; Patmore, R.; Roman, E.; Jack, A. Rearrangement of MYC is associated with poor prognosis in patients with diffuse large B-cell lymphoma treated in the era of rituximab. J. Clin. Oncol. 2010, 28, 3360–3365. [Google Scholar] [CrossRef]
- Copie-Bergman, C.; Cuilliere-Dartigues, P.; Baia, M.; Briere, J.; Delarue, R.; Canioni, D.; Salles, G.; Parrens, M.; Belhadj, K.; Fabiani, B.; et al. MYC-IG rearrangements are negative predictors of survival in DLBCL patients treated with immunochemotherapy: A GELA/LYSA study. Blood 2015, 126, 2466–2474. [Google Scholar] [CrossRef]
- Horn, H.; Ziepert, M.; Becher, C.; Barth, T.F.; Bernd, H.W.; Feller, A.C.; Klapper, W.; Hummel, M.; Stein, H.; Hansmann, M.L.; et al. MYC status in concert with BCL2 and BCL6 expression predicts outcome in diffuse large B-cell lymphoma. Blood 2013, 121, 2253–2263. [Google Scholar] [CrossRef]
- Iqbal, J.; Meyer, P.N.; Smith, L.M.; Johnson, N.A.; Vose, J.M.; Greiner, T.C.; Connors, J.M.; Staudt, L.M.; Rimsza, L.; Jaffe, E.; et al. BCL2 predicts survival in germinal center B-cell-like diffuse large B-cell lymphoma treated with CHOP-like therapy and rituximab. Clin. Cancer Res. 2011, 17, 7785–7795. [Google Scholar] [CrossRef]
- Papakonstantinou, G.; Verbeke, C.; Hastka, J.; Bohrer, M.; Hehlmann, R. bcl-2 expression in non-Hodgkin’s lymphomas is not associated with bcl-2 gene rearrangements. Br. J. Haematol. 2001, 113, 383–390. [Google Scholar] [CrossRef]
- Petrella, T.; Copie-Bergman, C.; Briere, J.; Delarue, R.; Jardin, F.; Ruminy, P.; Thieblemont, C.; Figeac, M.; Canioni, D.; Feugier, P.; et al. BCL2 expression but not MYC and BCL2 coexpression predicts survival in elderly patients with diffuse large B-cell lymphoma independently of cell of origin in the phase 3 LNH03-6B trial. Ann. Oncol. 2017, 28, 1042–1049. [Google Scholar] [CrossRef]
- Savage, K.J.; Johnson, N.A.; Ben-Neriah, S.; Connors, J.M.; Sehn, L.H.; Farinha, P.; Horsman, D.E.; Gascoyne, R.D. MYC gene rearrangements are associated with a poor prognosis in diffuse large B-cell lymphoma patients treated with R-CHOP chemotherapy. Blood 2009, 114, 3533–3537. [Google Scholar] [CrossRef]
- Shustik, J.; Han, G.; Farinha, P.; Johnson, N.A.; Ben Neriah, S.; Connors, J.M.; Sehn, L.H.; Horsman, D.E.; Gascoyne, R.D.; Steidl, C. Correlations between BCL6 rearrangement and outcome in patients with diffuse large B-cell lymphoma treated with CHOP or R-CHOP. Haematologica 2010, 95, 96–101. [Google Scholar] [CrossRef]
- Valera, A.; Lopez-Guillermo, A.; Cardesa-Salzmann, T.; Climent, F.; Gonzalez-Barca, E.; Mercadal, S.; Espinosa, I.; Novelli, S.; Briones, J.; Mate, J.L.; et al. MYC protein expression and genetic alterations have prognostic impact in patients with diffuse large B-cell lymphoma treated with immunochemotherapy. Haematologica 2013, 98, 1554–1562. [Google Scholar] [CrossRef]
- Horlad, H.; Ma, C.; Yano, H.; Pan, C.; Ohnishi, K.; Fujiwara, Y.; Endo, S.; Kikukawa, Y.; Okuno, Y.; Matsuoka, M.; et al. An IL-27/Stat3 axis induces expression of programmed cell death 1 ligands (PD-L1/2) on infiltrating macrophages in lymphoma. Cancer Sci. 2016, 107, 1696–1704. [Google Scholar] [CrossRef]
- Wada, N.; Zaki, M.A.; Hori, Y.; Hashimoto, K.; Tsukaguchi, M.; Tatsumi, Y.; Ishikawa, J.; Tominaga, N.; Sakoda, H.; Take, H.; et al. Tumour-associated macrophages in diffuse large B-cell lymphoma: A study of the Osaka Lymphoma Study Group. Histopathology 2012, 60, 313–319. [Google Scholar] [CrossRef]
- Carreras, J.; Kikuti, Y.Y.; Bea, S.; Miyaoka, M.; Hiraiwa, S.; Ikoma, H.; Nagao, R.; Tomita, S.; Martin-Garcia, D.; Salaverria, I.; et al. Clinicopathological characteristics and genomic profile of primary sinonasal tract diffuse large B cell lymphoma (DLBCL) reveals gain at 1q31 and RGS1 encoding protein; high RGS1 immunohistochemical expression associates with poor overall survival in DLBCL not otherwise specified (NOS). Histopathology 2017, 70, 595–621. [Google Scholar] [CrossRef]
- Alizadeh, A.A.; Eisen, M.B.; Davis, R.E.; Ma, C.; Lossos, I.S.; Rosenwald, A.; Boldrick, J.C.; Sabet, H.; Tran, T.; Yu, X.; et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000, 403, 503–511. [Google Scholar] [CrossRef]
- Gutierrez-Garcia, G.; Cardesa-Salzmann, T.; Climent, F.; Gonzalez-Barca, E.; Mercadal, S.; Mate, J.L.; Sancho, J.M.; Arenillas, L.; Serrano, S.; Escoda, L.; et al. Gene-expression profiling and not immunophenotypic algorithms predicts prognosis in patients with diffuse large B-cell lymphoma treated with immunochemotherapy. Blood 2011, 117, 4836–4843. [Google Scholar] [CrossRef]
- Rosenwald, A.; Wright, G.; Chan, W.C.; Connors, J.M.; Campo, E.; Fisher, R.I.; Gascoyne, R.D.; Muller-Hermelink, H.K.; Smeland, E.B.; Giltnane, J.M.; et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N. Engl. J. Med. 2002, 346, 1937–1947. [Google Scholar] [CrossRef]
- Shipp, M.A.; Ross, K.N.; Tamayo, P.; Weng, A.P.; Kutok, J.L.; Aguiar, R.C.; Gaasenbeek, M.; Angelo, M.; Reich, M.; Pinkus, G.S.; et al. Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat. Med. 2002, 8, 68–74. [Google Scholar] [CrossRef]
- Carreras, J.; Hamoudi, R.; Nakamura, N. Artificial intelligence analysis of gene expression data predicted the prognosis of patients with diffuse large B-cell Lymphoma. Tokai J. Exp. Clin. Med. 2020, 45, 37–48. [Google Scholar]
- Carreras, J.; Kikuti, Y.Y.; Miyaoka, M.; Hiraiwa, S.; Tomita, S.; Ikoma, H.; Kondo, Y.; Ito, A.; Shiraiwa, S.; Hamoudi, R.; et al. A single gene expression set derived from artificial intelligence predicted the prognosis of several lymphoma subtypes; and high immunohistochemical expression of TNFAIP8 associated with poor prognosis in diffuse large B-cell lymphoma. AI 2020, 1, 342–360. [Google Scholar] [CrossRef]
- Kumar, D.; Gokhale, P.; Broustas, C.; Chakravarty, D.; Ahmad, I.; Kasid, U. Expression of SCC-S2, an antiapoptotic molecule, correlates with enhanced proliferation and tumorigenicity of MDA-MB 435 cells. Oncogene 2004, 23, 612–616. [Google Scholar] [CrossRef]
- Kumar, D.; Whiteside, T.L.; Kasid, U. Identification of a novel tumor necrosis factor-alpha-inducible gene, SCC-S2, containing the consensus sequence of a death effector domain of fas-associated death domain-like interleukin- 1beta-converting enzyme-inhibitory protein. J. Biol. Chem. 2000, 275, 2973–2978. [Google Scholar] [CrossRef] [PubMed]
- UniProt, C. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Moratz, C.; Kang, V.H.; Druey, K.M.; Shi, C.S.; Scheschonka, A.; Murphy, P.M.; Kozasa, T.; Kehrl, J.H. Regulator of G protein signaling 1 (RGS1) markedly impairs Gi alpha signaling responses of B lymphocytes. J. Immunol. 2000, 164, 1829–1838. [Google Scholar] [CrossRef] [PubMed]
- Cardesa-Salzmann, T.M.; Colomo, L.; Gutierrez, G.; Chan, W.C.; Weisenburger, D.; Climent, F.; Gonzalez-Barca, E.; Mercadal, S.; Arenillas, L.; Serrano, S.; et al. High microvessel density determines a poor outcome in patients with diffuse large B-cell lymphoma treated with rituximab plus chemotherapy. Haematologica 2011, 96, 996–1001. [Google Scholar] [CrossRef]
- Lenz, G.; Wright, G.; Dave, S.S.; Xiao, W.; Powell, J.; Zhao, H.; Xu, W.; Tan, B.; Goldschmidt, N.; Iqbal, J.; et al. Stromal gene signatures in large-B-cell lymphomas. N. Engl. J. Med. 2008, 359, 2313–2323. [Google Scholar] [CrossRef]
- Carreras, J.; Yukie Kikuti, Y.; Miyaoka, M.; Hiraiwa, S.; Tomita, S.; Ikoma, H.; Kondo, Y.; Shiraiwa, S.; Ando, K.; Sato, S.; et al. Genomic profile and pathologic features of diffuse large B-cell lymphoma subtype of methotrexate-associated lymphoproliferative disorder in rheumatoid arthritis patients. Am. J. Surg. Pathol. 2018, 42, 936–950. [Google Scholar] [CrossRef]
- Ogura, G.; Kikuti, Y.Y.; Kikuchi, T.; Carreras, J.; Sato, T.; Nakamura, N. MYD88 (L265P) Mutation in malignant lymphoma using formalin-fixed paraffin-embedded section. J. Clin. Exp. Hematop. 2013, 53, 175–177. [Google Scholar] [CrossRef][Green Version]
- Cheson, B.D.; Pfistner, B.; Juweid, M.E.; Gascoyne, R.D.; Specht, L.; Horning, S.J.; Coiffier, B.; Fisher, R.I.; Hagenbeek, A.; Zucca, E.; et al. Revised response criteria for malignant lymphoma. J. Clin. Oncol. 2007, 25, 579–586. [Google Scholar] [CrossRef]
- Driscoll, J.J.; Rixe, O. Overall survival: Still the gold standard: Why overall survival remains the definitive end point in cancer clinical trials. Cancer J. 2009, 15, 401–405. [Google Scholar] [CrossRef]
- Carreras, J.; Kikuti, Y.Y.; Miyaoka, M.; Hiraiwa, S.; Tomita, S.; Ikoma, H.; Kondo, Y.; Ito, A.; Nakamura, N.; Hamoudi, R. Artificial intelligence analysis of the gene expression of follicular lymphoma predicted the overall survival and correlated with the immune microenvironment response signatures. Mach. Learn. Knowl. Extract. 2020, 2, 647–671. [Google Scholar] [CrossRef]
- Carreras, J.Y.K.Y.; Miyaoka, M.; Hiraiwa, S.; Tomita, S.; Ikoma, H.; Kondo, Y.; Ito, A.; Nakamura, N.; Hamoudi, R. A Combination of multilayer perceptron, radial basis function artificial neural networks, and machine learning image segmentation for the dimension reduction and the prognosis assessment of diffuse large B-cell lymphoma. AI 2021, 2, 106–134. [Google Scholar] [CrossRef]
- Tummers, B.; Green, D.R. Caspase-8: Regulating life and death. Immunol. Rev. 2017, 277, 76–89. [Google Scholar] [CrossRef]
- Chaitanya, G.V.; Steven, A.J.; Babu, P.P. PARP-1 cleavage fragments: Signatures of cell-death proteases in neurodegeneration. Cell Commun. Signal 2010, 8, 31. [Google Scholar] [CrossRef]
- Cryns, V.; Yuan, J. Proteases to die for. Genes Dev. 1998, 12, 1551–1570. [Google Scholar] [CrossRef] [PubMed]
- Thornberry, N.A.; Lazebnik, Y. Caspases: Enemies within. Science 1998, 281, 1312–1316. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Medeiros, L.J.; Xu-Monette, Z.Y.; Li, J.; Young, K.H. Dysregulation of cell survival in diffuse large B cell lymphoma: Mechanisms and therapeutic targets. Front. Oncol. 2019, 9, 107. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.R.; Uddin, S.; Ahmed, M.; Bu, R.; Ahmed, S.O.; Abubaker, J.; Sultana, M.; Ajarim, D.; Al-Dayel, F.; Bavi, P.P.; et al. Prognostic significance of XIAP expression in DLBCL and effect of its inhibition on AKT signalling. J. Pathol. 2010, 222, 180–190. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Xu-Monette, Z.Y.; Cao, X.; Manyam, G.C.; Wang, X.; Tzankov, A.; Xia, Y.; Li, X.; Visco, C.; Sun, R.; et al. Prognostic and biological significance of survivin expression in patients with diffuse large B-cell lymphoma treated with rituximab-CHOP therapy. Mod. Pathol. 2015, 28, 1297–1314. [Google Scholar] [CrossRef] [PubMed]
- Nikas, J.B.; Mitanis, N.T.; Nikas, E.G. Whole exome and transcriptome RNA-Sequencing model for the diagnosis of prostate cancer. ACS Omega 2019, 5, 481–486. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).