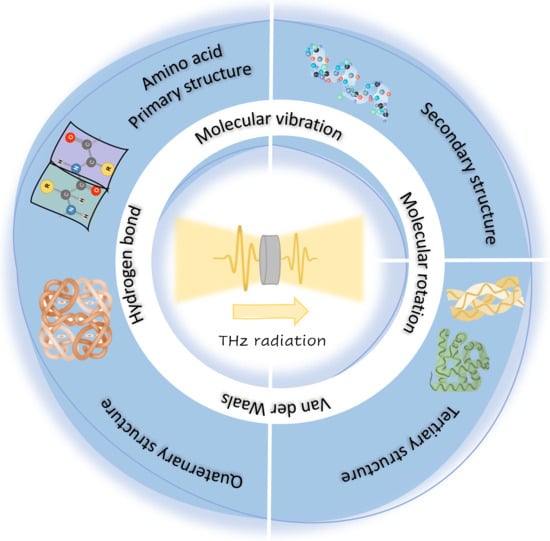
Terahertz Spectroscopic Analysis in Protein Dynamics: Current Status
Abstract
:Simple Summary
Abstract
1. Introduction
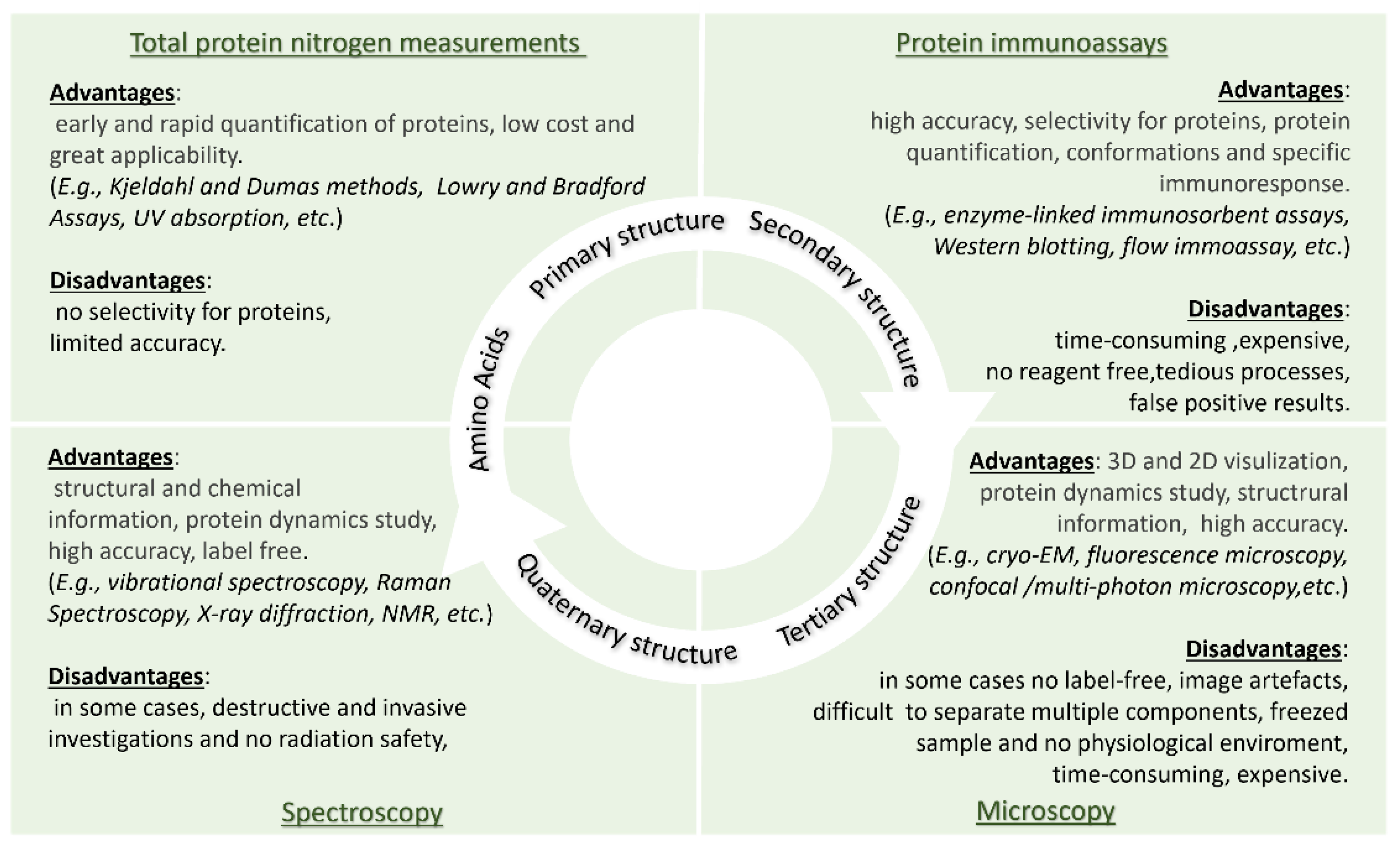
2. Consolidate Technologies for Amino Acid/Protein Detection
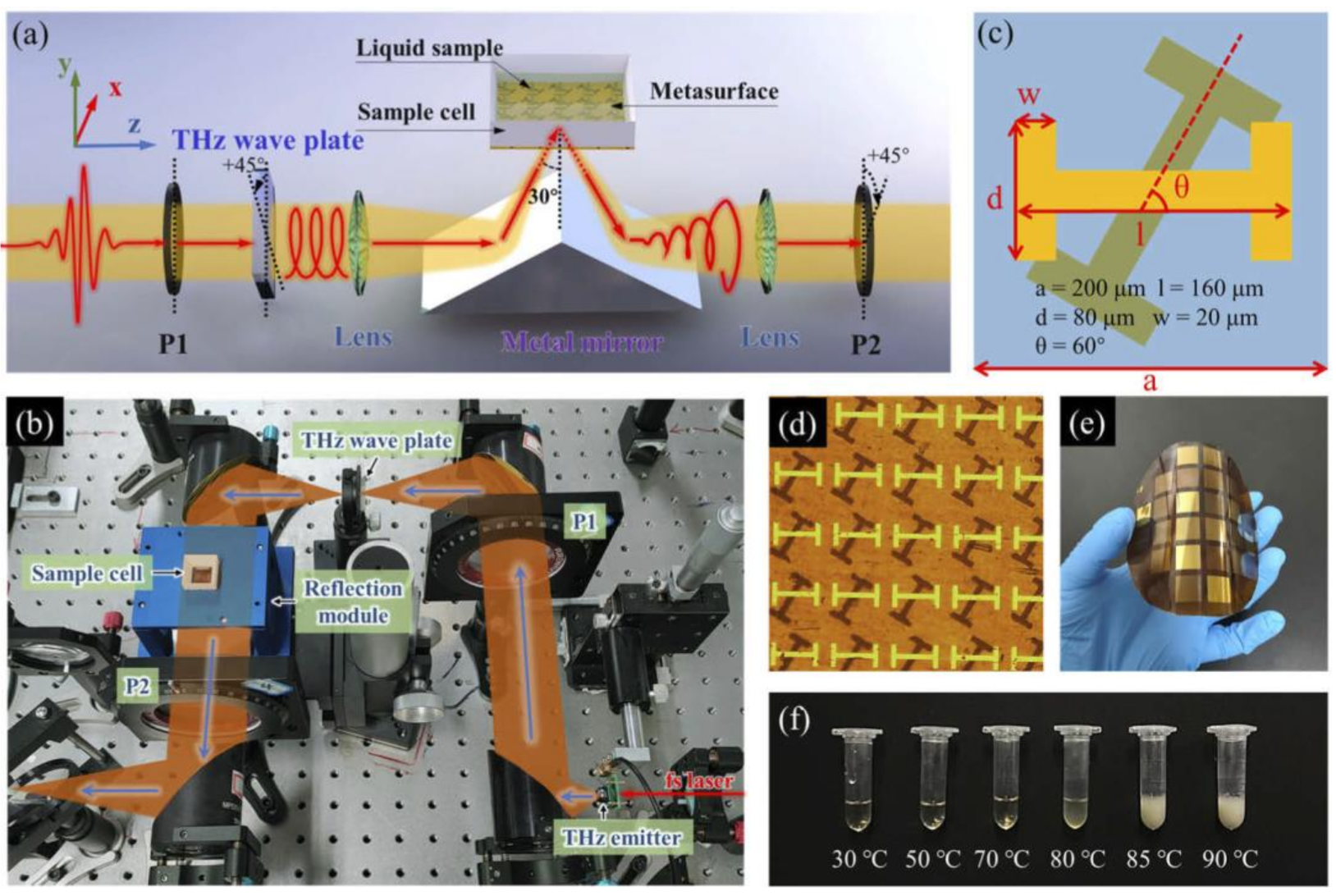
3. THz Technology for Protein Spectroscopy
4. Current Status of THz Spectroscopy for Research on Amino Acids and Short-Chain Peptides
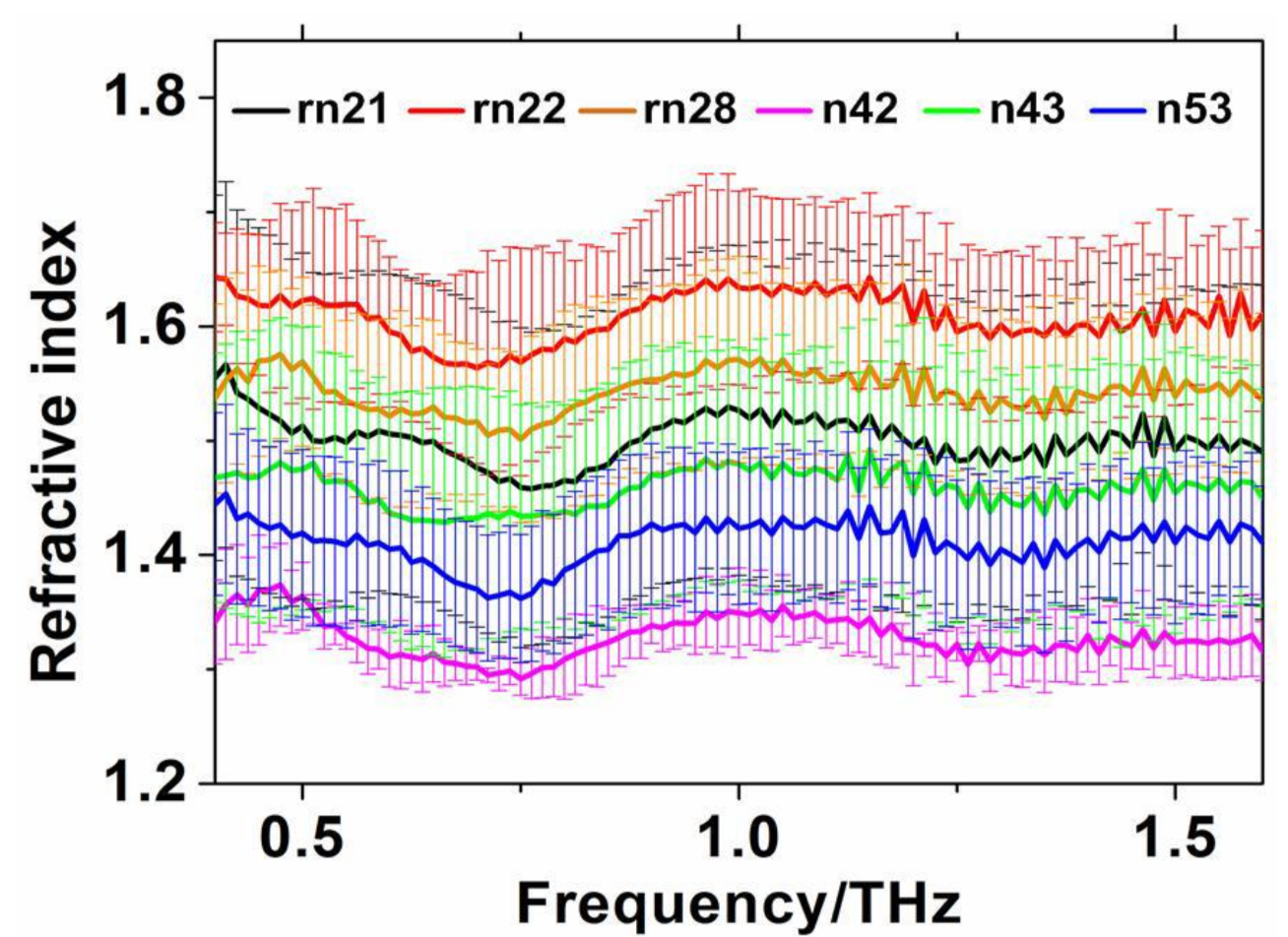
5. Current Status of THz Spectroscopy for Protein Research
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Moore, M.C. Introduction to Western Blotting; Morphosys Ltd.: Oxfordshire, UK, 2009. [Google Scholar]
- Gilda, J.E.; Gomes, A.V. Stain-Free total protein staining is a superior loading control to β-actin for Western blots. Anal. Biochem. 2013, 440, 186–188. [Google Scholar] [CrossRef] [Green Version]
- Moritz, C.P.; Marz, S.X.; Reiss, R.; Schulenborg, T.; Friauf, E. Epicocconone staining: A powerful loading control for Western blots. Proteomics 2014, 14, 162–168. [Google Scholar] [CrossRef]
- Baughman, W.E.; Yokus, H.; Balci, S.; Wilbert, D.S.; Kung, P.; Kim, S.M. Observation of Hydrofluoric Acid Burns on Osseous Tissues by Means of Terahertz Spectroscopic Imaging. IEEE J. Biomed. Health Inform. 2013, 17, 798–805. [Google Scholar] [CrossRef]
- Belouzard, S.; Millet, J.K.; Licitra, B.N.; Whittaker, G.R. Mechanism of Coronavirus Cell Entry Mediated by the Viral Spike Protein. Viruses 2012, 4, 1011–1033. [Google Scholar] [CrossRef] [Green Version]
- Verma, J.; Subbarao, N. A comparative study of human betacoronavirus spike proteins: Structure, function and therapeutics. Arch. Virol. 2021, 166, 697–714. [Google Scholar] [CrossRef]
- Whitford, D. Proteins: Structure and Function, 1st ed.; Wiley: Chichesetr, UK, 2005. [Google Scholar]
- Lesk, A.M. Introduction to Protein Science: Architecture, Function, and Genomics, 2nd ed.; Oxford University Press: Oxford, UK, 2010. [Google Scholar]
- Fields, P.A. Review: Protein function at thermal extremes: Balancing stability and flexibility. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2001, 129, 417–431. [Google Scholar] [CrossRef]
- Johan, K. New method for the determination of nitrogen in organic substances. Z. Anal. Chem. 1883, 22, 366–383. [Google Scholar]
- George, W. New decomposition product of urea. J. Prakt. Chem. 1847, 42, 255–256. [Google Scholar]
- Zor, T.; Selinger, Z. Linearization of the Bradford protein assay increases its sensitivity: Theoretical and experimental studies. Anal. Biochem. 1996, 236, 302–308. [Google Scholar] [CrossRef] [Green Version]
- Everette, J.D.; Bryant, Q.M.; Green, A.M.; Abbey, Y.A.; Wangila, G.W.; Walker, R.B. Thorough study of reactivity of various compound classes toward the Folin-Ciocalteu reagent. J. Agric. Food Chem. 2010, 58, 8139–8144. [Google Scholar] [CrossRef] [Green Version]
- Kurien, B.T.; Scofield, R.H. (Eds.) Western Blotting: An Introduction. Methods Mol. Biol. 2015, 1312, 17–30. [Google Scholar]
- Chang, S.K.C. Food analysis. In Protein Analysis, 3rd ed.; Nielse, S.S., Ed.; Kluwer Academic: New York, NY, USA, 2003; pp. 131–142. [Google Scholar]
- Wilson, P.R. A new instrumentation concept for nitrogen/protein analysis. A challenge to the Kjeldahl method. Asp. Appl. Biol. 1990, 25, 443–446. [Google Scholar]
- Hall, N.G.; Schonfeldt, H.C. Total nitrogen vs. amino-acid profile as indicator of protein content of beef. Food Chem. 2013, 140, 608–612. [Google Scholar] [CrossRef] [Green Version]
- Lee, P.Y.; Costumbrado, J.; Hsu, C.Y.; Kim, Y.H. Agarose gel electrophoresis for the separation of DNA fragments. J. Vis. Exp. 2012, 20, e3923. [Google Scholar] [CrossRef]
- Stellwagen, N.C. Electrophoresis of DNA in agarose gels, polyacrylamide gels and in free solution. Electrophoresis 2009, 30, S188–S195. [Google Scholar] [CrossRef] [Green Version]
- Chattopadhyay, P.K.; Gierahm, T.M.; Roederer, M.; Love, J.C. Single-cell technologies for monitoring immune systems. Nat. Immunol. 2014, 15, 128–135. [Google Scholar] [CrossRef]
- Geering, B.; Fussenegger, M. Synthetic immunology: Modulating the human immune system. Trends Biotechnol. 2015, 33, 65–79. [Google Scholar] [CrossRef]
- Shukla, A.A.; Thommes, J. Recent advances in large-scale production of monoclonal antibodies and related proteins. Trends Biotechnol. 2010, 28, 253–261. [Google Scholar] [CrossRef]
- Tomar, N.; De, R.K. Immunoinformatics: A brief review. Methods Mol. Biol. 2014, 1184, 23–55. [Google Scholar]
- Virgo, P.F.; Gibbs, G.J. Flow cytometry in clinical pathology. Ann. Clin. Biochem. 2012, 49, 17–28. [Google Scholar] [CrossRef] [Green Version]
- Deakin, T. Radisotopic characterization as an analytical tool: Current status, limitations and future challenges. Bioanalysis 2015, 7, 541–555. [Google Scholar] [CrossRef]
- Diao, B.; Wen, K.; Zhang, J.; Chen, J.; Han, C.; Chen, Y.; Wang, S.; Deng, G.; Zhou, H.; Wu, Y. Accuracy of a nucleocapsid protein antigen rapid test in the diagnosis of SARS-CoV-2 infection. Clin. Microbiol. Infect. 2021, 27, 289.e1–289.e4. [Google Scholar] [CrossRef]
- Chen, L.; Zhao, J.; Peng, J.; Li, X.; Deng, X.; Geng, Z.; Shen, Z.; Guo, F.; Zhang, Q.; Jin, Y.; et al. Detection of SARS-CoV-2 in saliva and characterization of oral symptoms in COVID-19 patients. Cell Prolif. 2020, 53, e12923. [Google Scholar] [CrossRef]
- Webb, R.H. Confocal optical microscopy. Rep. Prog. Phys. 1996, 59, 427–471. [Google Scholar] [CrossRef]
- Hoover, E.E.; Squier, J.A. Advances in multiphoton microscopy technology. Nat. Photonics 2013, 7, 93–101. [Google Scholar] [CrossRef] [Green Version]
- Zumbusch, A.; Langbein, W.; Borri, P. Nonlinear vibrational microscopy applied to lipid biology. Prog. Lipid Res. 2013, 52, 615–632. [Google Scholar] [CrossRef] [Green Version]
- D’Arco, A.; Brancati, N.; Ferrara, M.A.; Indolfi, M.; Frucci, M.; Sirleto, L. Subcellular chemical and morphological analysis by stimulated Raman scattering microscopy and image analysis techniques. Biomed. Opt. Express 2016, 7, 1853–1864. [Google Scholar] [CrossRef] [Green Version]
- Cheng, J.X.; Xie, X.S. Vibrational spectroscopic imaging of living systems: An emerging platform for biology and medicine. Science 2015, 50, 62–64. [Google Scholar] [CrossRef]
- D’Arco, A.; Ferrara, M.A.; Indolfi, M.; Tufano, V.; Sirleto, L. Label-free imaging of small lipid droplets by femtosecond-stimulated Raman scattering microscopy. J. Nonlinear Opt. Phys. Mater. 2017, 26, 1750052. [Google Scholar] [CrossRef]
- Min, W.; Freudiger, C.W.; Lu, S.; Xie, X.S. Coherent nonlinear optical imaging: Beyond fluorescence microscopy. Annu. Rev. Phys. Chem. 2011, 62, 507–530. [Google Scholar] [CrossRef] [Green Version]
- Streets, A.M.; Li, A.; Chen, T.; Huang, Y. Imaging without fluorescence: Nonlinear optical microscopy for quantitative cellular imaging. Anal. Chem. 2014, 86, 8506–8513. [Google Scholar] [CrossRef]
- Jepsen, P.U.; Cooke, D.G.; Koch, M. Terahertz spectroscopy and imaging—Modern techniques and applications. Laser Photonics Rev. 2011, 5, 124–166. [Google Scholar] [CrossRef]
- Xie, L.; Gao, W.; Shu, J.; Ying, Y.; Kono, J. Extraordinary sensitivity enhancement by metasurfaces in terahertz detection of antibiotics. Sci. Rep. 2015, 5, 8671. [Google Scholar] [CrossRef]
- Wang, Q.; Ma, Y.H. Qualitative and quantitative identification of nitrogen in terahertz region. Chemom. Intell. Lab. 2013, 127, 43–48. [Google Scholar] [CrossRef]
- Hickman, A.B.; Davies, D.R. Principle of Macromolecular X-ray Crystallography. In Current Protocols in Protein; Chapter 17; Science Wiley Interscience: New York, NY, USA, 2001. [Google Scholar]
- Lipfert, J.; Doniach, S. Small-angle X-ray scattering from RNA, proteins, and protein complexes. Annu. Rev. Biophys. Biomol. Struct. 2007, 36, 307–327. [Google Scholar] [CrossRef] [Green Version]
- Miao, J.; Ishikawa, T.; Shen, Q.; Earnest, T. Extending X-ray crystallography to allow the imaging of noncrystalline materials, cells, and single protein complexes. Annu. Rev. Phys. Chem. 2008, 59, 387–410. [Google Scholar] [CrossRef]
- Neylon, C. Small angle neutron and X-ray scattering in structural biology: Recent examples from the literature. Eur. Biophys. J. 2008, 37, 531–541. [Google Scholar] [CrossRef]
- Blamire, A.M. The technology of MRI: The next 10 years? Br. J. Radiol. Suppl. 2008, 81, 601–617. [Google Scholar] [CrossRef]
- Ishima, R.; Torchia, D.A. Protein dynamics from NMR. Nat. Struct. Biol. 2000, 7, 740–743. [Google Scholar] [CrossRef]
- McDermott, A.; Polenova, T. Solid state NMR: New tools for insight into enzyme function. Curr. Opin. Struct. Biol. 2007, 17, 617–622. [Google Scholar] [CrossRef]
- Bajar, B.T.; Wang, E.S.; Zhang, S.; Lin, M.Z.; Chu, J. A Guide to Fluorescent Protein FRET Pairs. Sensors 2016, 16, 1488. [Google Scholar] [CrossRef]
- Miura, K. An Overview of Current Methods to Confirm Protein-Protein Interactions. Protein Pept. Lett. 2018, 25, 72–733. [Google Scholar] [CrossRef]
- Dale, N.C.; Johnstone, E.K.M.; White, C.W.; Pfleger, K.D.G. NanoBRET: The Bright Future of Proximity-Based Assays. Front. Bioeng. Biotechnol. 2019, 26, 56. [Google Scholar] [CrossRef]
- Lippincott-Schwartz, J.; Snapp, E.L.; Phair, R.D. The Development and Enhancement of FRAP as a Key Tool for Investigating Protein Dynamics. Biophys. J. 2018, 115, 1146–1155. [Google Scholar] [CrossRef] [Green Version]
- Fasman, G.D. Circular Dichroism and the Conformational Analysis of Biomolecules; Plenum Press: New York, NY, USA, 1996. [Google Scholar]
- Hofmann, A.; Simon, A.; Grkovic, T.; Jones, M. Methods of Molecular Analysis in the Life Sciences; Cambridge University Press: Cambridge, UK, 2014. [Google Scholar]
- Pescitelli, G.; Di Bari, L.; Berova, N. Application of electronic circular dichroism in the study of supramolecular systems. Chem. Soc. Rev. 2014, 43, 5211–5233. [Google Scholar] [CrossRef]
- Barth, A. Infrared spectroscopy of proteins. Biochim. Biophys. Acta Bioenerg. 2007, 1767, 1073–1101. [Google Scholar] [CrossRef] [Green Version]
- Marcelli, A.; Cricenti, A.; Kwiatek, W.M.; Petibois, C. Biological applications of synchrotron radiation infrared spectromicroscopy. Biotech. Adv. 2012, 30, 1390–1404. [Google Scholar] [CrossRef]
- Piccirilli, F.; Tardani, F.; D’Arco, A.; Birarda, G.; Vaccari, L.; Sennato, S.; Casciardi, S.; Lupi, S. Infrared Nanospectroscopy Reveals DNA Structural Modifications upon Immobilization onto Clay Nanotubes. Nanomaterials 2021, 11, 1103. [Google Scholar] [CrossRef]
- Auston, D.H.; Nuss, M.C. Electrooptical generation and detection of femtosecond electrical transients. IEEE J. Quantum Electron. 1988, 24, 184–197. [Google Scholar] [CrossRef]
- Mittleman, D.; Gupta, M.; Neelamani, R.; Baraniuk, R.G.; Rudd, J.V.; Koc, M. Recent advances in terahertz imaging. Appl. Phys. B 1999, 68, 1085–1094. [Google Scholar] [CrossRef]
- Siegel, P.H. Terahertz technology in biology and medicine. IEEE Trans. Microw. Theory 2004, 52, 2438–2447. [Google Scholar] [CrossRef]
- Zhang, X.C. Terahertz wave imaging: Horizons and hurdles. Phys. Med. Biol. 2002, 47, 3667–3677. [Google Scholar] [CrossRef]
- Wallace, V.P.; Taday, P.F.; Fitzgerald, A.J.; Woodward, R.M.; Cluff, J.; Pye, R.J.; Arnone, D.D. Terahertz pulsed imaging and spectroscopy for biomedical and pharmaceutical applications. Faraday Discuss. 2004, 126, 255–263. [Google Scholar] [CrossRef]
- Withayachumnankul, W.; Png, G.M.; Yin, X.; Atakaramians, S.; Jones, I.; Lin, H.; Ung, B.S.Y.; Balakrishnan, J.; Ng, B.W.-H.; Ferguson, B.; et al. T-ray sensing and imaging. Proc. IEEE 2007, 95, 1528–1558. [Google Scholar] [CrossRef]
- Cheon, H.; Yang, H.J.; Son, J.-H. Toward Clinical Cancer Imaging Using Terahertz Spectroscopy. IEEE J. Sel. Top. Quantum Electron. 2017, 23, 8600109. [Google Scholar] [CrossRef]
- Mickan, S.; Abbott, D.; Munchb, J.; Zhang, X.C.; van Doornd, T. Analysis of system trade-offs for terahertz imaging. Microelectron. J. 2000, 31, 503–514. [Google Scholar] [CrossRef] [Green Version]
- Mou, S.; D’Arco, A.; Tomarchio, L.; Di Fabrizio, M.; Curcio, A.; Lupi, S.; Petrarca, M. Simultaneous elliptically and radially polarized THz from one-color laser-induced plasma filament. New J. Phys. 2021, 23, 063048. [Google Scholar] [CrossRef]
- Curcio, A.; Petrarca, M. Diagnosing plasmas with wideband terahertz pulses. Opt. Lett. 2019, 44, 1011–1014. [Google Scholar] [CrossRef]
- Curcio, A.; Dolci, V.; Lupi, S.; Petrarca, M. Terahertz-based retrieval of the spectral phase and amplitude of ultrashort laser pulses. Opt. Lett. 2018, 43, 783–786. [Google Scholar] [CrossRef] [Green Version]
- Curcio, A.; Marocchino, A.; Dolci, V.; Lupi, S.; Petrarca, M. Resonant plasma excitation by single-cycle THz pulses. Sci. Rep. 2018, 8, 1052. [Google Scholar] [CrossRef] [Green Version]
- Curcio, A.; Mou, S.; Palumbo, L.; Lupi, S.; Petrarca, M. Selection rules for the orbital angular momentum of optically produced THz radiation. Opt. Lett. 2021, 46, 1514–1517. [Google Scholar] [CrossRef]
- D’Arco, A.; Tomarchio, L.; Dolci, V.; Di Pietro, P.; Perucchi, A.; Mou, S.; Petrarca, M.; Lupi, S. Broadband Anisotropic Optical Properties of the Terahertz Generator HMQ-TMS Organic Crystal. Condens. Matter 2020, 5, 47. [Google Scholar] [CrossRef]
- Lupi, S.; Molle, A. Emerging Dirac materials for THz plasmonics. Appl. Mater. Today 2020, 20, 100732. [Google Scholar] [CrossRef]
- Di Pietro, P.; Ortolani, M.; Limaj, O.; Di Gaspare, A.; Giliberti, V.; Giorgianni, F.; Brahlek, M.; Bansal, N.; Koirala, N.; Oh, S.; et al. Observation of Dirac plasmons in a topological insulator. Nat. Nanotechnol. 2013, 8, 556–560. [Google Scholar] [CrossRef] [Green Version]
- D’Apuzzo, F.; Piacenti, A.R.; Giorgianni, F.; Autore, M.; Cestelli Guidi, M.; Marcelli, A.; Schade, U.; Ito, Y.; Chen, M.; Lupi, S. Terahertz and mid-infrared plasmons in three-dimensional nanoporous graphene. Nat. Commun. 2017, 8, 14885. [Google Scholar] [CrossRef]
- Giorgianni, F.; Chiadroni, E.; Rovere, A.; Cestelli-Guidi, M.; Perucchi, A.; Bellaveglia, M.; Castellano, M.; Di Giovenale, D.; Di Pirro, G.; Ferrario, M.; et al. Strong nonlinear terahertz response induced by Dirac surface states in Bi2Se3 topological insulator. Nat. Commun. 2016, 7, 11421. [Google Scholar] [CrossRef]
- Marcelli, A.; Irizawa, A.; Lupi, S. THz: Research frontiers for new sources, imaging and other advanced technologies. Condens. Matter 2019, 6, 23. [Google Scholar]
- Galstyan, V.; D’Arco, A.; Di Fabrizio, M.; Poli, N.; Lupi, S.; Comini, E. Detection of volatile organic compounds: From chemical gas sensors to terahertz spectroscopy. Rev. Anal. Chem. 2021, 40, 33–57. [Google Scholar] [CrossRef]
- Naftaly, M.; Vieweg, N.; Deninger, A. Industrial Applications of Terahertz Sensing: State of Play. Sensors 2019, 19, 4203. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.; Ryu, S.Y.; Kwon, W.S.; Kim, K.S.; Kim, S. Compound Explosives Detection and Component Analysis via Terahertz Time-Domain Spectroscopy. Korean J. Opt. Photonics 2013, 17, 454–460. [Google Scholar] [CrossRef] [Green Version]
- Smith, R.M.; Arnold, M.A. Selectivity of Terahertz Gas-Phase Spectroscopy. Anal. Chem. 2015, 87, 10679–10683. [Google Scholar] [CrossRef]
- Rothbart, N.; Holz, O.; Koczulla, R.; Schmalz, K.; Hübers, H.W. Analysis of Human Breath by millimeter-Wave/Terahertz Spectroscopy. Sensors 2019, 19, 2719. [Google Scholar] [CrossRef] [Green Version]
- Giuliano, B.M.; Gavdush, A.A.; Müller, B.; Zaytsev, K.I.; Grassi, T.; Ivlev, A.V.; Palumbo, M.E.; Baratta, G.A.; Scirè, C.; Komandin, G.A.; et al. Broadband spectroscopy of astrophysical ice analogues I. Direct measurement of the complex refractive index of CO ice using terahertz time-domain spectroscopy. Astron. Astrophys. 2019, 629, A112. [Google Scholar] [CrossRef] [Green Version]
- D’Arco, A.; Di Fabrizio, M.; Dolci, V.; Marcelli, A.; Petrarca, M.; Della Ventura, G.; Lupi, S. Characterization of volatile organic compounds (VOCs) in their liquid-phase by terahertz time-domain spectroscopy. Biomed. Opt. Express 2020, 11, 1–7. [Google Scholar] [CrossRef]
- Fischer, B.; Hoffmann, M.; Helm, H.; Modjesch, G.; Uhd Jepsen, P. Chemical recognition in terahertz time-domain spectroscopy and imaging. Semicond. Sci. Technol. 2005, 20, S246. [Google Scholar] [CrossRef] [Green Version]
- Stoik, C.D.; Bohn, M.J.; Blackshire, J.L. Nondestructive evaluation of aircraft composites using transmissive terahertz time domain spectroscopy. Opt. Express 2008, 16, 17039–17051. [Google Scholar] [CrossRef]
- Heimbeck, M.S.; Ng, W.R.; Golish, D.R.; Gehm, M.E.; Everitt, H.O. Terahertz digital holographic imaging of voids within visibly opaque dielectrics. IEEE Trans. Terahertz Sci. Technol. 2015, 5, 110–116. [Google Scholar] [CrossRef]
- Tomarchio, L.; Macis, S.; D’Arco, A.; Mou, S.; Grilli, A.; Romani, M.; Cestelli Guidi, M.; Hu, K.; Kukunuri, S.; Jeong, S.; et al. Disordered photonics behavior from terahertz to ultraviolet of a three-dimensional graphene network. NPG Asia Mater. 2021, 13, 73. [Google Scholar] [CrossRef]
- Federici, J.F.; Schulkin, B.; Huang, F.; Gary, D.; Barat, R.; Oliveira, F.; Zimdars, D. THz imaging and sensing for security applications—Explosives, weapons and drugs. Semicond. Sci. Technol. 2005, 20, S266. [Google Scholar] [CrossRef]
- Ergün, S.; Sönmez, S. Terahertz Technology for Military Applications. J. Assoc. Inf. Sci. Technol. 2015, 3, 13–16. [Google Scholar] [CrossRef]
- Liu, H.B.; Zhong, H.; Karpowicz, N.; Chen, Y.; Zhang, X.C. Terahertz spectroscopy and imaging for defense and security applications. Proc. IEEE 2007, 95, 1514–1527. [Google Scholar] [CrossRef]
- D’Arco, A.; Mussi, V.; Petrov, S.; Tofani, S.; Petrarca, M.; Beccherelli, R.; Dimitrov, D.; Marinova, V.; Lupi, S.L.; Zografopoulos, D.C. Fabrication and spectroscopic characterization of graphene transparent electrodes on flexible cyclo-olefin substrates for terahertz electro-optic applications. Nanotechnology 2020, 31, 364006. [Google Scholar] [CrossRef]
- Wang, K.; Sun, D.W.; Pu, H. Emerging non-destructive terahertz spectroscopic imaging technique: Principle and applications in the agri-food industry. Trend Food Sci. Technol. 2017, 67, 93–105. [Google Scholar] [CrossRef]
- Cosentino, A. Terahertz and cultural heritage science: Examination of art and archeology. Technologies 2016, 4, 6. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Yamaguchi, S.; Fukushi, Y.; Kubota, O.; Itsuji, T.; Ouchi, T.; Yamamoto, S. Brain tumor imaging of rat fresh tissue using terahertz spectroscopy. Sci. Rep. 2016, 6, 30124. [Google Scholar] [CrossRef] [Green Version]
- Bajwa, N.; Au, J.; Jarrahy, R.; Sung, S.; Fishbein, M.C.; Riopelle, D.; Ennis, D.B.; Aghaloo, T.; John, M.A.; Grundfest, W.S.; et al. Non-invasive terahertz imaging of tissue water content for flap viability assessment. Biomed. Opt. Express 2017, 8, 460–474. [Google Scholar] [CrossRef] [Green Version]
- Zaytsev, I.; Dolganova, I.N.; Chernomyrdin, N.V.; Katyba, G.M.; Gavdush, A.A.; Cherkasova, O.P.; Komandin, G.A.; Shchedrina, M.A.; Khodan, A.N.; Ponomarev, D.S.; et al. The progress and perspectives of terahertz technology for diagnosis of neoplasms: A review. J. Opt. 2020, 22, 013001. [Google Scholar] [CrossRef]
- Yang, X.; Zhao, X.; Yang, K.; Liu, Y.; Liu, Y.; Fu, W.; Luo, Y. Biomedical Applications of Terahertz Spectroscopy and Imaging. Trends Biotechnol. 2016, 34, 810–824. [Google Scholar] [CrossRef]
- Son, J.H.; Oh, S.J.; Cheon, H. Potential clinical applications of terahertz radiation. J. Appl. Phys. 2019, 125, 190901. [Google Scholar] [CrossRef]
- D’Arco, A.; Di Fabrizio, M.; Dolci, V.; Petrarca, M.; Lupi, S. THz Pulsed Imaging in Biomedical Applications. Condens. Matter 2020, 5, 25. [Google Scholar] [CrossRef] [Green Version]
- Di Fabrizio, M.; D’Arco, A.; Mou, S.; Palumbo, L.; Petrarca, M.; Lupi, S. Performance Evaluation of a THz pulsed Imaging System: Point Spread Function, Broadband THz Beam Visualization and Image Reconstruction. Appl. Sci. 2021, 11, 562. [Google Scholar] [CrossRef]
- Di Fabrizio, M.; Lupi, S.; D’Arco, A. Virus recognition with terahertz radiation: Drawbacks and potentialities. J. Phys. Photonics 2021, 3, 032001. [Google Scholar] [CrossRef]
- Lee, Y.S. Principles of Terahertz Science and Technology; Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Zhang, X.C.; Xu, J. Introduction to THz Wave Photonics; Springer: Berlin/Heidelberg, Germany, 2010. [Google Scholar]
- Walther, M.; Plochocka, P.; Fischer, B.; Helm, H.; Jepsen, P.U. Collective Vibrational Modes in Biological Molecules Investigated by Terahertz Time-domain Spectroscopy. Biopolymers 2002, 67, 310–313. [Google Scholar] [CrossRef]
- Tanabe, T.; Watanabe, K.; Oyama, Y.; Seo, K. Polarization Sensitive THz Absorption Spectroscopy for the Evaluation of Uniaxially Deformed Ultra-high Molecular Weight Polyethylene. NDT & E Int. 2010, 43, 329–333. [Google Scholar]
- Parrott, E.P.J.; Zeitler, J.A. Terahertz Time-Domain and Low-Frequency Raman Spectroscopy of Organic Materials. Appl. Spectrosc. 2015, 69, 1–25. [Google Scholar] [CrossRef]
- Cooksey, C.C.; Greer, B.J.; Heilweil, E.J. Terahertz Spectroscopy of L-Proline in Reverse Aqueous Micelles. Chem. Phys. Lett. 2009, 467, 424–429. [Google Scholar] [CrossRef]
- Yue, W.; Wang, W.; Zhao, G.; Zhang, C.; Yan, H. THz Spectrum of Aromatic Amino Acid. Chin. Phys. Soc. 2005, 54, 3094–3099. [Google Scholar]
- Kutteruf, M.R.; Brown, C.M.; Iwaki, L.K.; Campbell, M.B.; Korter, T.M.; Heilweil, E.J. Terahertz spectroscopy of short-chain polypeptides. Chem. Phys. Lett. 2003, 375, 337–343. [Google Scholar] [CrossRef]
- Manti, L.; D’Arco, A. Cooperative biological effects between ionizing radiation and other physical and chemical agents. Mutat. Res. 2010, 704, 115–122. [Google Scholar] [CrossRef]
- Lee, D.-K.; Kang, J.-H.; Know, J.; Lee, J.-S.; Lee, S.; Woo, D.H.; Kim, J.H.; Song, C.-S.; Park, Q.H.; Seo, M. Nano metamaterials for ultrasensitive terahertz biosensing. Sci. Rep. 2017, 7, 8146. [Google Scholar] [CrossRef]
- Lin, H.; Withayachumnankul, W.; Fischer, B.; Mickan, S.; Abbott, D. Gas recognition with terahertz time-domain spectroscopy and spectral catalog: A preliminary study. Terahertz Photonics 2008, 6840, 68400X. [Google Scholar]
- Kindt, J.T.; Schmuttenmaer, C.A. Far-Infrared Dielectric Properties of Polar Liquids Probed by Femtosecond Terahertz Pulse Spectroscopy. J. Phys. Chem. 1996, 100, 10373–10379. [Google Scholar] [CrossRef]
- Fedulova, E.; Nazarov, M.; Angeluts, A.; Kitai, M.; Sokolov, V.; Shkurinov, A. Studying of dielectric properties of polymers in the terahertz frequency range. In Optical technologies in biophysics and medicine XIII, Proceedings of the Saratov Fall Meeting 2011, Saratov, Russia, 27–30 September 2011; SPIE: Bellingham, WA, USA, 2011. [Google Scholar]
- Rezvani, S.J.; Di Gioacchino, D.; Tofani, S.; D’Arco, A.; Ligi, C.; Lupi, S.; Gatti, C.; Cestelli Guidi, M.; Marcelli, A. A cryogenic magneto-optical device for long wavelength radiation. Rev. Sci. Instrum. 2020, 91, 075103. [Google Scholar] [CrossRef]
- Burford, N.; El-Shenawee, M.O. Review of terahertz photoconductive antenna technology. Opt. Eng. 2017, 56, 010901. [Google Scholar] [CrossRef]
- Dexheimer, S.L. Terahertz Spectroscopy: Principles and Applications; CRC Press: Boca Raton, FL, USA, 2017. [Google Scholar]
- Naftaly, M. Terahertz Metrology; Artech House: London, UK, 2015. [Google Scholar]
- Seo, M.; Park, H.-R. Terahertz Biochemical Molecules-specific sensors. Adv. Opt. Mater. 2019, 8, 1900662. [Google Scholar] [CrossRef]
- Korter, T.M.; Balu, R.; Campbell, M.B.; Beard, M.C.; Gregurick, S.K.; Heilweil, E.J. Terahertz Spectroscopy of Solid Serine and Cysteine. Chem. Phys. Lett. 2006, 418, 65–70. [Google Scholar] [CrossRef]
- Wang, X.; Wang, W. Terahertz Time-domain Spectroscopy of Sulfur-containing Amino Acids. Acta Chim. Sin. 2008, 66, 2248–2252. [Google Scholar]
- Yamamoto, K.; Kabir, M.H.; Tominaga, K. Terahertz Time-domain Spectroscopy of Sulfur-containing Biomolecules. J. Opt. Soc. Am. B 2005, 22, 2417–2426. [Google Scholar] [CrossRef]
- Matei, A.; Drichko, N.; Gompf, B.; Dressel, M. Far-infrared Spectra of Amino Acids. Chem. Phys. 2005, 316, 61–71. [Google Scholar] [CrossRef]
- Wang, W.; Li, H.; Zhang, Y.; Zhang, C. Correlations Between Terahertz Spectra and Molecular Structures of 20 Standard α-Amino Acids. Acta Phys. Chim. Sin. 2009, 25, 2074–2079. [Google Scholar]
- Yan, Z.; Hou, D.; Huang, P.; Cao, B.; Zhang, G.; Zhou, Z. Terahertz Spectroscopic Investigation of L-Glutamic Acid and L-Tyrosine. Meas. Sci. Technol. 2008, 19, 15602–15605. [Google Scholar] [CrossRef]
- Wang, G.; Wang, W. Experimental and Theoretical Investigations on the Terahertz Vibrational Spectroscopy of Alanine Crystal. Acta Phys. Chim. Sin. 2012, 28, 1579–1585. [Google Scholar]
- Zheng, Z.; Fan, W. First Principles Investigation of L-Alanine in Terahertz Region. Biol. Phys. 2012, 38, 405–413. [Google Scholar] [CrossRef] [Green Version]
- Taulbee, A.R.; Heuser, J.A.; Spendel, W.U.; Pacey, G.E. Qualitative Analysis of Collective Mode Frequency Shifts in L-Alanine Using Terahertz Spectroscopy. Anal. Chem. 2009, 81, 2664–2667. [Google Scholar] [CrossRef] [Green Version]
- Taday, P.F.; Bradley, I.V.; Arnone, D.D. Terahertz Pulse Spectroscopy of Biological Materials L-Glutamic Acid. J. Biol. Phys. 2003, 29, 109–115. [Google Scholar] [CrossRef]
- Darkwah, J.; Smith, G.; Ermolina, I.; Mueller-Holtz, M. A THz Spectroscopy Method for Quantifying the Degree of Crystallinity in Freeze-dried Gelatin/Amino Acid Mixtures: An Application for the Development of Rapidly Disintegrating Tablets. Int. J. Pharm. 2013, 455, 357–364. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, Z.; Zhao, X.; Su, H.; Yan, F. Extracting THz Absorption Coefficient Spectrum Based on Accurate Determination of Sample Thickness. Spectrosc. Spect. Anal. 2012, 32, 1043–1046. [Google Scholar]
- Yang, J.; Li, S.; Zhao, H.; Zhang, J.; Yang, N.; Jing, D.; Wang, C.; Han, J. Terahertz Study of L-Asparagine and Its Monohydrate. Acta Phys. Sin. 2014, 63, 133203. [Google Scholar] [CrossRef]
- Nishizawa, J.; Tanno, T.; Yoshida, T.; Suto, K. Consequence of a Defect on the Terahertz Spectra of L-Asparagine Monohydrate. Chem. Lett. 2007, 36, 134–135. [Google Scholar] [CrossRef]
- Chiba, M.; Derreumaux, P.; Vergoten, G. The Use of the Spasiba Spectroscopic Potential for Reproducing the Structures and Vibrational Frequencies of a Series of Acids—Acetic-Acid, Pivalic Acid, Succinic Acid, Adipic Acid And L-Glutamic Acid. J. Mol. Struct. 1994, 317, 171–184. [Google Scholar] [CrossRef]
- Yi, W.; Yu, J.; Xu, Y.; Wang, F.; Yu, Q.; Sun, H.; Xu, L.; Liu, Y.; Jiang, L. Broadband terahertz spectroscopy of amino acids. Instrum. Sci. Technol. 2017, 45, 423–439. [Google Scholar]
- Hufnagle, D.C.; Taulbee-Combs, A.R.; Spendel, W.U.; Pacey, G.E. Collective mode frequency shifts in L-serine and a series of isotopologues in the terahertz regime. Talanta 2012, 88, 61–65. [Google Scholar] [CrossRef]
- Patil, M.R.; Ganorkar, S.B.; Patil, A.S.; Shirkhedkar, A.A. Terahertz Spectroscopy: Encoding the Discovery, Instrumentation, and Applications toward Pharmaceutical Prospectives. Crit. Rev. Anal. Chem. 2020, 1–13. [Google Scholar] [CrossRef]
- Williams, M.R.C.; Aschaffenburg, D.J.; Ofori-Okai, B.K.; Schmuttenmaer, C.A. Intermolecular vibrations in hydrophobic amino acid crystals: Experiments and calculations. J. Phys. Chem. B 2013, 117, 10444–10461. [Google Scholar] [CrossRef]
- King, M.D.; Hakey, P.M.; Korter, T.M. Discrimination of chiral solids: A terahertz spectroscopic investigation of l- and dl-serine. J. Phys. Chem. A 2010, 114, 2945–2953. [Google Scholar] [CrossRef]
- Frommel, C. The apolar surface area of amino acids and its empirical correlation with hydrophobic free energy. J. Theor. Biol. 1984, 111, 247–260. [Google Scholar] [CrossRef]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef] [Green Version]
- Acharya, H.; Vembanur, S.; Jamadagni, S.N.; Garde, S. Mapping hydrophobicity at the nanoscale: Applications to heterogeneous surfaces and proteins. Faraday Discuss. 2010, 146, 353–365. [Google Scholar] [CrossRef]
- Jensen, J.H.; Gordon, M.S. On the number of water molecules necessary to stabilize the glycine zwitterion. J. Am. Chem. Soc. 1995, 117, 8159–8170. [Google Scholar] [CrossRef] [Green Version]
- Gontrani, L.; Mennucci, B.; Tomasi, J. Glycine and alanine: A theoretical study of solvent effects upon energetics and molecular response properties. J. Mol. Struct. THEOCHEM 2000, 500, 113–127. [Google Scholar] [CrossRef]
- Niehues, G.; Heyden, M.; Schmidt, D.A.; Havenith, M. Exploring hydrophobicity by THz absorption spectroscopy of solvated amino acids. Faraday Discuss. 2011, 150, 193. [Google Scholar] [CrossRef] [PubMed]
- Gordon, M.S.; Jensen, J.H. Understanding the hydrogen bond using quantum chemistry. Acc. Chem. Res. 1996, 29, 536–543. [Google Scholar] [CrossRef] [Green Version]
- Hecht, D.; Tadesse, L.; Walters, L. Correlating hydration shell structure with amino acid hydrophobicity. J. Am. Chem. Soc. 1993, 115, 3336–3337. [Google Scholar] [CrossRef]
- Ide, M.; Maeda, Y.; Kitano, H. Effect of hydrophobicity of amino acids on the structure of water. J. Phys. Chem. B 1997, 101, 7022–7026. [Google Scholar] [CrossRef]
- Qvist, J.; Halle, B. Thermal signature of hydrophobic hydration dynamics. J. Am. Chem. Soc. 2008, 130, 10345–10353. [Google Scholar] [CrossRef]
- Balabin, R.M. The First Step in Glycine Solvation: The Glycine−Water Complex. J. Phys. Chem. B 2010, 114, 15075–15078. [Google Scholar] [CrossRef] [PubMed]
- McLain, S.E.; Soper, A.K.; Terry, A.E.; Watts, A. Structure and Hydration of L-Proline in Aqueous Solutions. J. Phys. Chem. B 2007, 111, 4568–4580. [Google Scholar] [CrossRef] [PubMed]
- Pacios, L.F. Distinct molecular surfaces and hydrophobicity of amino acid residues in proteins. J. Chem. Inf. Comput. Sci. 2001, 41, 1427–1435. [Google Scholar] [CrossRef]
- Sato, T.; Buchner, R.; Fernandez, S.; Chiba, A.; Kunz, W. Dielectric relaxation spectroscopy of aqueous amino acid solutions: Dynamics and interactions in aqueous glycine. J. Mol. Liq. 2005, 117, 93–98. [Google Scholar] [CrossRef]
- Rodriguez-Arteche, I.; Cerveny, S.; Alegrıa, A.; Colmenero, J. Dielectric spectroscopy in the GHz region on fully hydrated zwitterionic amino acids. Phys. Chem. Chem. Phys. 2012, 14, 11352–11362. [Google Scholar] [CrossRef]
- Suzuki, M.; Shigematsu, J.; Fukunishi, Y.; Kodama, T. Hydrophobic hydration analysis on amino acid solutions by the microwave dielectric method. J. Phys. Chem. B 1997, 101, 3839–3845. [Google Scholar] [CrossRef]
- Bian, Y.; Zhang, X.; Zhu, Z.; Yang, B. Vibrational modes optimization and terahertz time-domain spectroscopy of L-Lysine and L-Lysine hydrate. J. Mol. Struct. 2021, 1232, 129952. [Google Scholar] [CrossRef]
- Morgante, P.; Peverati, R. The devil in the details: Atutorial review on some undervalued aspects of density functional theory calculations. Int. J. Quant. Chem. 2020, 120, e26332. [Google Scholar] [CrossRef]
- Fox, S.J.; Dziedzic, J.; Fox, T.; Tautermann, C.S.; Skylaris, C.K. Density functional theory calculations on entire proteins for free energies of binging: Application to a model polar binding site. Proteins 2014, 82, 3335–3346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, T.; Li, S.; Zou, T.; Yu, Z.; Zhang, B.; Wang, C.; Zhang, J.; He, M.; Zhao, H. Terahertz spectra of L-phenylalanine and its monohydrate. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2017, 178, 19–23. [Google Scholar] [CrossRef] [Green Version]
- Samanta, N.; Mahanta, D.S.; Choudhury, S.; Barman, A.; Mitra, R.K. Collective hydration dynamics in some amino acid solutions: A combined GHz-THz spectroscopic study. J. Chem. Phys. 2017, 146, 125101. [Google Scholar] [CrossRef]
- Itoh, K.; Shimanouchi, T. Far-infrared spectra of N-methylacetamide and related compounds and hydrogen-bond force constraints. Biopolymers 1965, 5, 921–930. [Google Scholar] [CrossRef]
- Yamamoto, K.; Tominaga, K.; Sasakawa, H.; Tamura, A.; Murakami, H. Terahertz time-domain spectroscopy of amino acids and polypeptides. Biophys. J. 2005, 89, L22–L24. [Google Scholar] [CrossRef] [Green Version]
- Neu, J.; Stone, E.A.; Spies, J.A.; Storch, G.; Hatano, A.S.; Mercado, B.Q.; Miller, S.J.; Schmutternmaer, C.A. Terahertz Spectroscopy of Tetrameric Peptides. CA J. Phys. Chem. Lett. 2019, 10, 2624–2628. [Google Scholar] [CrossRef]
- Yoneyama, H.; Yamashita, M.; Kasai, S.; Kawase, K.; Ueno, R.; Ito, H.; Ouchi, T. Terahertz spectroscopy of native-conformation and thermally denatured bovine serum albumin (BSA). Phys. Med. Biol. 2008, 53, 3543–3549. [Google Scholar] [CrossRef]
- Xie, L.; Yao, Y.; Ying, Y. The application of terahertz spectroscopy to protein detection: A review. Appl. Spectrosc. Rev. 2014, 49, 448–461. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, Y.; Pickwell-Macpherson, E. Investigating antibody interactions with a polar liquid using terahertz pulsed spectroscopy. Biophys. J. 2011, 100, 225–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falconer, R.J.; Markelz, A.G. Terahertz spectroscopic analysis of peptides and proteins. J. Infrared Millim. Terahertz Waves 2012, 33, 973–988. [Google Scholar] [CrossRef]
- Cao, C.; Zhang, Z.; Zhao, X.; Zhang, T. Terahertz spectroscopy and machine learning algorithm for non-destructive evaluation of protein conformation. Opt. Quantum Electron. 2020, 52, 225. [Google Scholar] [CrossRef]
- Markelz, A.G.; Roitberg, A.; Heilweil, E.J. Pulsed terahertz spectroscopy of DNA, bovine serum albumin and collagen between 0.1 and 2.0 THz. Chem. Phys. Lett. 2000, 320, 42–48. [Google Scholar] [CrossRef]
- Markelz, A.; Whitmire, S.; Hillebrecht, J.; Birge, R. THz time domain spectroscopy of biomolecular conformational modes. Phys. Med. Biol. 2002, 47, 3797–3805. [Google Scholar] [CrossRef] [PubMed]
- Paciaroni, A.; Orecchini, A.; Haertlein, M.; Moulin, M.; Nibali, V.C.; Francesco, A.D.; Petrillo, C.; Sacchetti, F. Vibrational collective dynamics of dry proteins in the terahertz region. J. Phys. Chem. B 2012, 116, 3861–3865. [Google Scholar] [CrossRef]
- Sun, Y.; Zhong, J.; Zhang, C.; Zuo, J.; Pickwell-MacPherson, E. Label-free detection and characterization of the binding of hemagglutinin protein and broadly neutralizing monoclonal antibodies using terahertz spectroscopy. J. Biomed. Opt. 2015, 20, 037006. [Google Scholar] [CrossRef]
- Castro-Camus, E.; Johnston, M.B. Conformational changes of photoactive yellow protein monitored by terahertz spectroscopy. Chem. Phys. Lett. 2008, 455, 289–292. [Google Scholar] [CrossRef]
- Markelz, A.G. Terahertz dielectric sensitivity to biomolecular structure and function. IEEE J. Sel. Top. Quantum Electron. 2008, 14, 180–190. [Google Scholar] [CrossRef]
- George, D.K.; Knab, J.R.; He, Y.; Kumauchi, M.; Birge, R.R.; Hoff, W.D.; Markelz, A.G. Photoactive yellow protein terahertz response: Hydration, heating and intermediate states. IEEE Trans. Terahertz Sci. Technol. 2013, 3, 288–294. [Google Scholar] [CrossRef]
- Han, X.; Yan, S.; Zang, Z.; Wie, D.; Cui, H.-L.; Du, C. Label-free protein detection using terahertz time-domain spectroscopy. Biomed. Opt. Express 2018, 9, 994–1005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.Y.; Knab, J.R.; Cerne, J.; Markelz, A.G. Large oxidation dependence observed in terahertz dielectric response for cytochrome c. Phys. Rev. E 2005, 72, 04090. [Google Scholar] [CrossRef] [PubMed]
- Brooks, B.; Karplus, M. Process analytical chemistry: Applications of near infrared spectrometry in environmental and food analysis: An overview. Proc. Natl. Acad. Sci. USA 1985, 82, 4995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kauzmannk, W. Some factors in the interpretation of protein denaturation. Adv. Protein Chem. 1959, 14, 1–63. [Google Scholar]
- Murphy, K.P. Stabilization of protein structure. Methods Mol. Biol. 2001, 168, 1–16. [Google Scholar]
- Freier, E. Thermal denaturation methods in the study of protein folding. Methods Enzymol. 1995, 259, 144–168. [Google Scholar]
- Wijayanti, H.B.; Bansal, N.; Deeth, H.C. Stability of whey proteins during thermal processing: A review. Compr. Rev. Food Sci. F 2014, 13, 1235–1251. [Google Scholar] [CrossRef]
- Wang, B.; Timilsena, Y.P.; Blanch, E.; Adhikari, B. Lactoferrin: Structure, function, denaturation and digestion. Crit. Rev. Food Sci. Nutr. 2019, 59, 580–596. [Google Scholar] [CrossRef]
- Silva, J.L. Pressure stability of proteins. Annu. Rev. Phys. Chem. 1993, 44, 89–113. [Google Scholar] [CrossRef]
- Chirgadze, Y.N.; Ovsepyan, A.M. Observation of small conformational changes in sperm-whale myoglobin by far-infrared spectra. Biopolymers 1973, 12, 637–645. [Google Scholar] [CrossRef]
- Chen, H.; Chen, G.; Li, S.; Wang, L. Reversible conformational change of PsbO Protein detected by terahertz time-domain spectroscopy. Chin. Phys. Lett. 2009, 26, 084204. [Google Scholar]
- Chen, H.; Wang, L.; Qu, Y.G.; Kuang, T.Y.; Li, L.B.; Peng, W.X. Investigation of guanidine hydrochloride induced chlorophyll protein 43 and 47 denaturation in the terahertz frequency range. J. Appl. Phys. 2007, 102, 074701. [Google Scholar] [CrossRef]
- Qu, Y.; Chen, H.; Qin, X.; Wang, L.; Li, L.; Kuang, T. The guanidine hydrochloride-induced denaturation of CP43 and CP47 studied by terahertz time-domain spectroscopy. Sci. China C Life Sci. 2007, 50, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Chen, H.; Qin, X.; Li, L.; Wang, L.; Kuang, T. Thermal denaturation of CP43 studied by Fourier transform-infrared spectroscopy and terahertz time-domain spectroscopy. J. Proteins Proteom. 2007, 1774, 1614–1618. [Google Scholar] [CrossRef]
- He, Y.F.; Pei, P.I.; Knab, J.R.; Chen, J.Y.; Markelz, A.G. Protein dynamical transition does not require protein structure. Phys. Rev. Lett. 2008, 101, 178103. [Google Scholar] [CrossRef] [PubMed]
- Luong, T.Q.; Verma, P.K.; Mitra, R.K.; Havenith, M. Do hydration dynamics follow the structural perturbation during thermal denaturation of a protein: A terahertz absorption study. Biophys. J. 2011, 101, 925–933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- George, D.K.; Chen, J.Y.; He, Y.; Knab, J.R.; Markelz, A.G. Functional-state dependence of picosecond protein dynamics. J. Phys. Chem. B 2021, 125, 11134–11140. [Google Scholar] [CrossRef]
- Zhou, R.; Wang, C.; Wendao, X.; Xie, L. Biological applications of terahertz technology based on nanomaterials and nanostructures. Nanoscale 2019, 11, 3445–3457. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Liang, M.; Yi, L.; Wong, P.K.; Wilmink, G.J.; Zhang, D.D.; Xin, H. Microfluidic devices for terahertz spectroscopy of live cells toward lab-on-a-chip applications. Sensors 2016, 16, 476. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, M.; Wei, D.; Wang, Y.; Yan, S.; Liu, M.; Yang, X.; Yang, K.; Cui, H.; Fu, W. Label-free sensing of the binding state of MUC1 peptide and anti-MUC1 aptamer solution in fluidic chip by terahertz spectroscopy. Biomed. Opt. Express 2017, 8, 4427–4437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, R.; Xu, L.; Yu, X.; Zou, L.; Shen, Y.; Deng, X. High-sensitivity biosensor for identification of protein based on terahertz Fano resonance metasurfaces. Opt. Commun. 2020, 473, 125850. [Google Scholar] [CrossRef]
- Amin, M.; Siddiqui, O.; Abutarboush, H.; Farhat, M.; Ramzan, R. A THz graphene metasurface for polarization selective virus sensing. Carbon 2021, 176, 580–591. [Google Scholar] [CrossRef] [PubMed]
- Ahmadivand, A.; Gerislioglu, B.; Ramezzani, Z.; Kaushik, A.; Manickam, P.; Ghoreishi, A. Functionalized terahertz plasmonic metasensors: Femtomolar-level detection of SARS-CoV-2 spike proteins. Biosens. Bioelectron. 2021, 177, 112971. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Xie, L.; Zhu, J.; Xu, X.; Ye, Z.; Wang, C.; Ma, Y.; Ying, Y. Gold nanoparticle-based terahertz metamaterial sensors: Mechanisms and applications. ACS Photonics 2016, 3, 2308–2314. [Google Scholar] [CrossRef]
- Adak, S.; Tripathi, L.N. Nanoantenna enhanced terahertz interaction of biomolecules. Analyst 2019, 144, 6172–6192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Fan, F.; Shi, W.; Zhang, T.; Chang, S. Terahertz circular polarization sensing for protein denaturation based on a twisted dual-layer metasurface. Biomed. Opt. Express 2022, 13, 209–221. [Google Scholar] [CrossRef]
- Meister, K.; Ebbinghaus, S.; Xu, Y.; Duman, J.G.; DeVries, A.; Gruebele, M.; Leitner, D.M.; Havenith, M. Long-range protein–water dynamics in hyperactive insect antifreeze proteins. Proc. Natl. Acad. Sci. USA 2013, 110, 1617–1622. [Google Scholar] [CrossRef] [Green Version]
- Knab, J.; Chen, J.Y.; Markelz, A. Hydration dependence of conformational dielectric relaxation of lysozyme. Biophys. J. 2006, 90, 2576–2581. [Google Scholar] [CrossRef] [Green Version]
- Heyden, M.; Havenith, M. Combining THz spectroscopy and MD simulations to study protein–hydration coupling. Methods 2010, 52, 74–83. [Google Scholar] [CrossRef]
- Xu, J.; Plaxco, K.W.; Allen, S.J. Probing the collective vibrational dynamics of a protein in liquid water by terahertz absorption spectroscopy. Protein Sci. 2006, 15, 1175–1181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pal, S.; Chattopadhyay, A. Hydration dynamics in biological membranes: Emerging application of terahertz spectroscopy. J. Phys. Chem. Lett. 2021, 12, 9697–9709. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, G.; Xu, D.; Jiang, B.; Ge, M.; Wu, L.; Yang, C.; Mu, N.; Wang, S.; Chang, C.; et al. Terahertz spectroscopic diagnosis of early blast-induced traumatic brain injury in rats. Biomed. Opt. Express 2020, 11, 4085–4098. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Du, P.; Lu, X.; Xie, P.; Qian, Z.; Fan, S.; Zhu, Z. Quantitative characterization of bovine serum albumin thin-films using terahertz spectroscopy and machine learning methods. Biomed. Opt. Express 2018, 9, 2917–2929. [Google Scholar] [CrossRef] [PubMed]


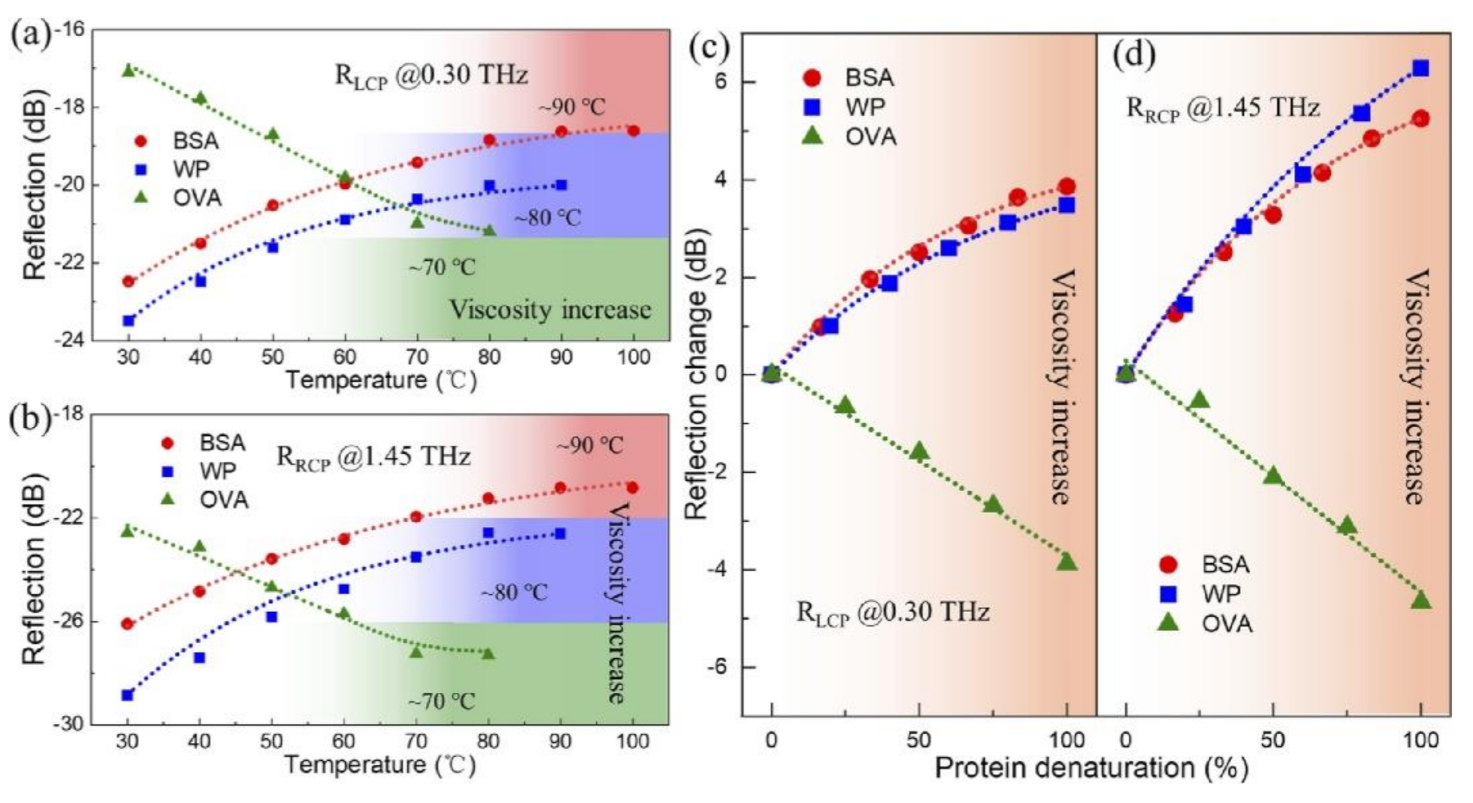
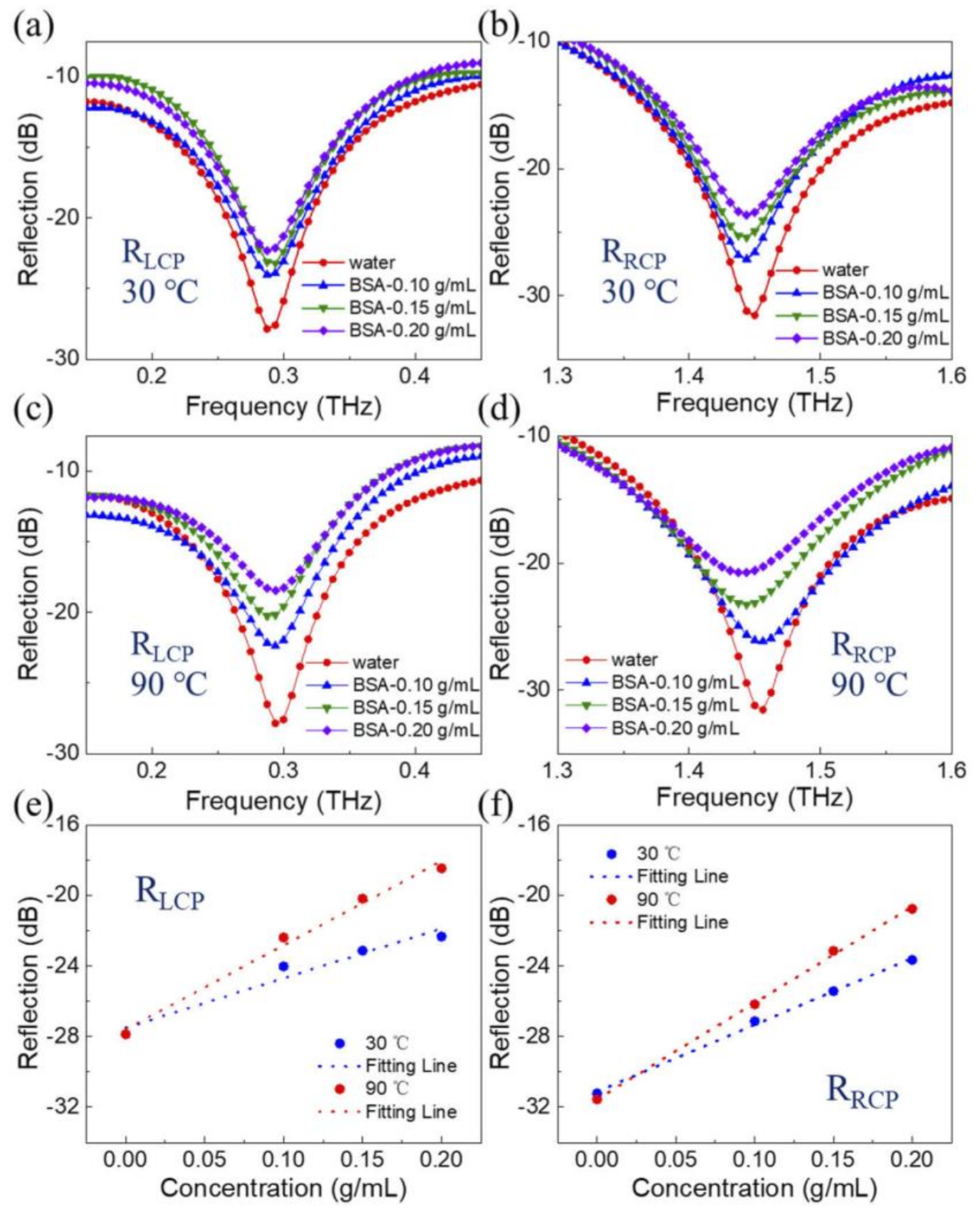
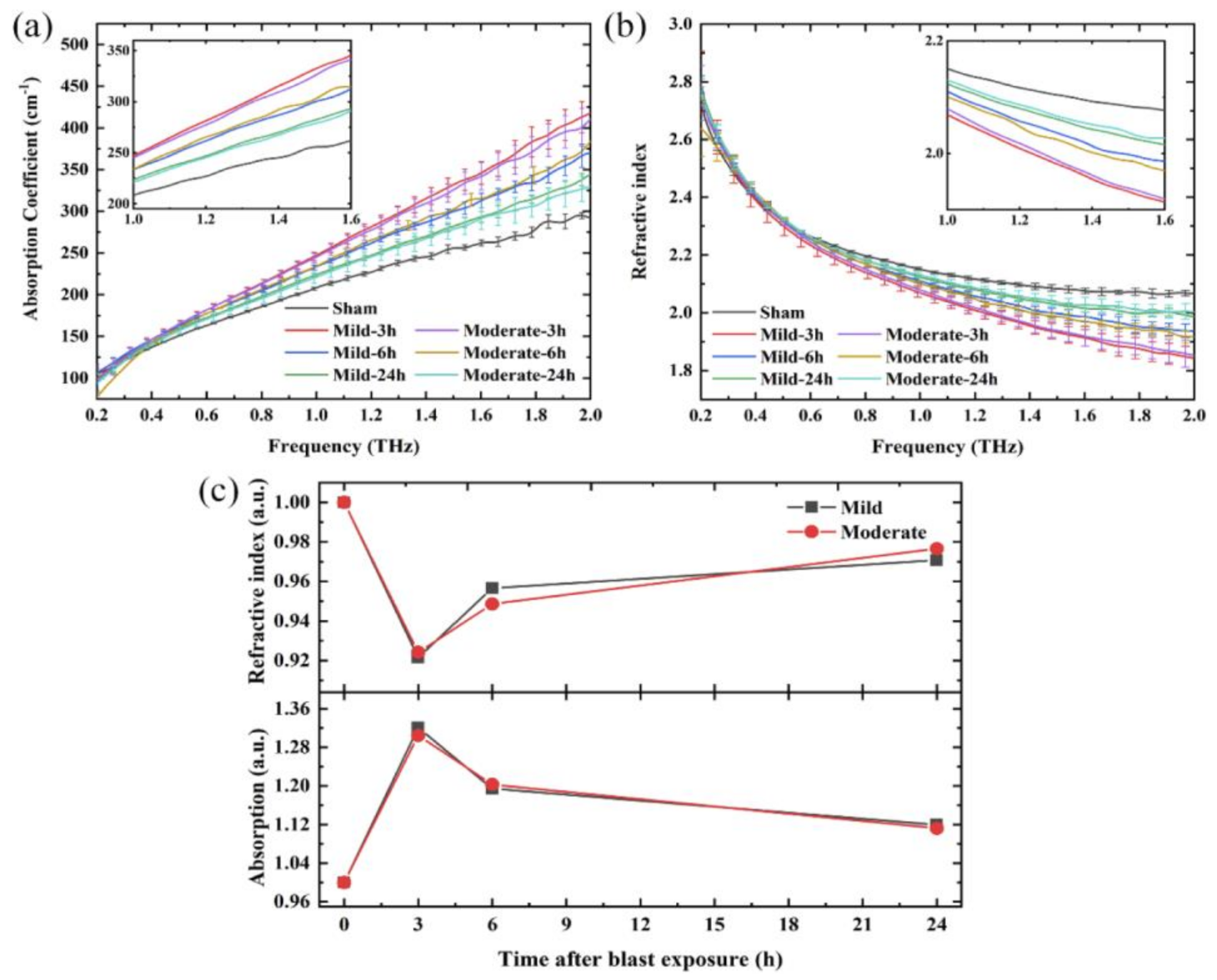
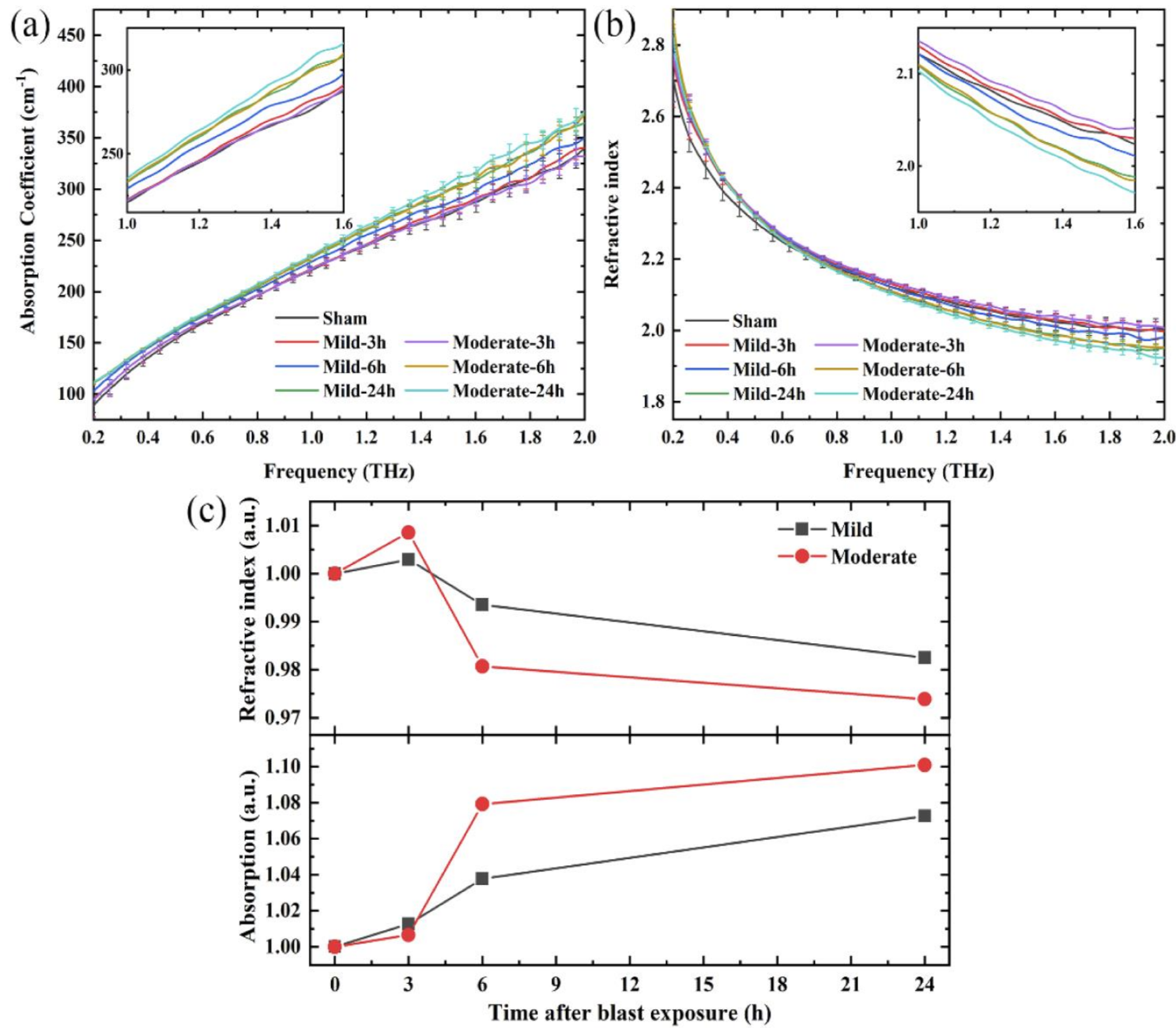
| Amino Acids | THz Absorption Frequencies (THz) |
|---|---|
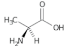 Alanine (Ala) | 2.21, 2.56, 2.72, 2.91, 3.14, 3.37, 4.17 |
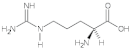 Arginine (Arg) | 0.99, 1.45, 2.02, 2.62, 3.51, 3.77, 4.40 |
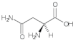 Asparagine (Asn) | 1.64, 1.85, 2.06, 2.26, 2.62, 3.11, 3.57, 3.92, 4.90 |
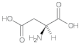 Aspartic acid (Asp) | 1.35, 1.71, 2.58, 3.01, 3.26, 3.98, 4.41, 5.36 |
 Cysteine (Cys) | 1.40, 1.70, 2.33, 2.78, 2.94, 3.23, 3.64, 4.44, 4.78, 5.87 |
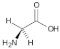 Glycine (Gly) | 1.83, 2.30, 2.51, 2.70, 4.07 |
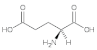 Glutamic acid (Glu) | 1.21, 2.03, 2.23, 2.48, 2.64, 2.80, 3.26, 3.58, 4.0, 4.50 |
 Glutamine (Gln) | 1.70, 2.28, 2.50, 3.37, 4.08, 4.92 |
 Histidine (His) | 0.88, 1.72, 2.08, 2.44, 2.80, 3.00, 3.39, 3.96, 4.33, 5.30 |
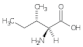 Isoleucine (Iso) | 0.30, 0.85, 1.08, 1.42, 1.72, 2.41, 2.74, 3.54, 4.27, 5.26 |
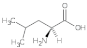 Leucine (Leu) | 0.66, 0.84, 1.46, 1.64, 2.14, 2.56, 2.74, 2.88, 2.96, 3.68, 5.11 |
 Lysine (Lys) | 0.90, 1.26, 1.79, 2.25, 2.64 |
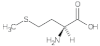 Methionine (Met) | 1.01, 1.79, 2.70, 2.94, 3.77 |
 Phenylalanine (Phe) | 1.25, 2.02, 2.52, 2.76, 4.16 |
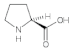 Proline (Pro) | 1.69, 2.00, 2.64, 3.12, 3.62, 4.05, 4.69 |
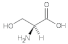 Serine (Ser) | 1.97, 2.41, 2.71, 3.12, 3.98, 4.34 |
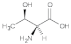 Threonine (Thr) | 1.11, 1.42, 2.12, 2.61, 3.06, 3.33, 3.75, 4.44, 4.98, 5.30 |
 Tryptophan (Trp) | 0.91, 1.19, 1.44, 1.85, 2.26, 2.57, 3.22, 3.69, 4.02, 4.85 |
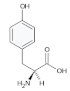 Tyrosine (Tyr) | 0.95, 1.92, 2.06, 2.65, 2.82, 3.31, 3.48, 3.96, 4.32, 4.75, 5.13, 6.22 |
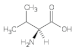 Valine (Val) | 1.11, 1.68, 2.12, 2.22, 2.52, 2.64, 2.84, 3.53, 4.39 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mancini, T.; Mosetti, R.; Marcelli, A.; Petrarca, M.; Lupi, S.; D’Arco, A. Terahertz Spectroscopic Analysis in Protein Dynamics: Current Status. Radiation 2022, 2, 100-123. https://doi.org/10.3390/radiation2010008
Mancini T, Mosetti R, Marcelli A, Petrarca M, Lupi S, D’Arco A. Terahertz Spectroscopic Analysis in Protein Dynamics: Current Status. Radiation. 2022; 2(1):100-123. https://doi.org/10.3390/radiation2010008
Chicago/Turabian StyleMancini, Tiziana, Rosanna Mosetti, Augusto Marcelli, Massimo Petrarca, Stefano Lupi, and Annalisa D’Arco. 2022. "Terahertz Spectroscopic Analysis in Protein Dynamics: Current Status" Radiation 2, no. 1: 100-123. https://doi.org/10.3390/radiation2010008
APA StyleMancini, T., Mosetti, R., Marcelli, A., Petrarca, M., Lupi, S., & D’Arco, A. (2022). Terahertz Spectroscopic Analysis in Protein Dynamics: Current Status. Radiation, 2(1), 100-123. https://doi.org/10.3390/radiation2010008