1. Introduction
Advances in high-throughput molecular analysis have revolutionized our understanding of biological processes. Well-established omics are comprehensive tools for the investigation of different biomolecules including genes, proteins, and metabolites in a given statute and time [
1]. Genomics is the study of the genome and epigenetic modifications. In proteomics, proteins and their post-translational modifications, enzymes, hormones, and other signalling molecules are studied. The intermediary molecules, which enable the information transfer between genes and proteins, for example, RNA, are studied by transcriptomics. Small molecules that are produced as a part of cellular metabolism, including sugars, fatty acids, lipids, and small peptides, are target molecules for metabolomics.
A snapshot of elementary biology in a living system (cells, tissues, organism) at a particular time point can be created with omics, where huge numbers of measurements are procured [
2]. With more such snapshots at different time points, the fate of particular molecules of interest can be followed. This information is then useful in the understanding of cellular responses to external and internal stressors, including ionizing radiation (IR), the development of therapeutic targets, and the discovery of new biomarkers. Integrated omics (a combination of at least two omics approaches) can not only accelerate the search for novel biomarkers for radiation exposure but also cross-validate results obtained from different platforms.
Although more than 3 decades have passed since the publication of the first omics data in the radiation community, the use of omics technology in radiation research is still young. Omics studies address different issues in radiation research including biological effects of radiation exposure on normal and cancer tissues, individual sensitivity, risk assessment and biomarker discovery. A recently conducted systematic review on a broad range of studies in the radiation field discovered few proteins that could serve as biomarkers of IR exposure, but also highlighted a lack of mechanistic knowledge [
3]. The omics offers a comprehensive tool to fulfil these gaps. A good combination of omics and advanced bioinformatics can lead to a correct interpretation of the cellular response to radiation exposure. The generated omics datasets have the potential to be integrated with results from epidemiological and clinical studies to develop a strong strategy for evaluating radiation effects on human health.
In this short commentary, we look back at omics studies performed in radiation research over the last 20 years and highlight the trends in omics research, the most and the least used platforms in the radiation community, and discuss whether omics has met expectations by looking at the knowledge and research gaps in radiation omics.
2. Materials and Methods
All searches were performed on NCBI Pubmed (
https://pubmed.ncbi.nlm.nih.gov/advanced/) [
4] (accessed on 22 October 2021). In this study, genomics and epigenomic studies are included together, which is not always the case with other publications. Epigenomics is a branch of genomics. Just as post-translational modifications (PTMs) were combined with proteomics as PTMs are a part of proteomics, epigenomics and genomics have been combined. It is important to note that PTM or epigenome profiling is not reliable without comparison/normalization to the whole proteome or transcriptome. Similarly, proteomics also includes phosphoproteomics and acetylomics. Metabolomics studies included metabolomics, lipidomics, and hormonal assays. Under transciptomics, miRNA, mRNA, RNA Seq, micronome, and microRNA array were included. Integrated omics was a combination of radiomics, multiomics and interactomics. The search strings for different omics platforms were:
Genomics: ((genom*[Title/Abstract]) OR (epigenom*[Title/Abstract])) AND (ionizing radiation [Title/Abstract]);
Proteomics: ((((((proteom*[Title/Abstract]) OR (phosphoproteom*[Title/Abstract])) OR (secretom*[Title/Abstract])) OR (glycom*[Title/Abstract])) OR (acetylom*[Title/Abstract]))) AND (ionizing radiation [Title/Abstract]);
Metabolomics: (metabolom*[Title/Abstract])) OR lipidom*[Title/Abstract]) OR (hormonal assay [Title/Abstract])) AND (ionizing radiation [Title/Abstract]);
Transcriptomics: (((((((((transcriptom*[Title/Abstract]) OR (miRNA array[Title/Abstract])) OR (mRNA array[Title/Abstract])) OR (DNA array[Title/Abstract])) OR (microarray[Title/Abstract])) OR (RNA Seq[Title/Abstract])) OR (rna seq[Title/Abstract])) OR (microRNA array[Title/Abstract])) OR (micronom*[Title/Abstract])) AND (ionizing radiation[Title/Abstract]);
Integrated omics: (((((radiomic*[Title/Abstract]) OR (integrated omic*[Title/Abstract])) OR (omic*[Title/Abstract])) OR (multiomic*[Title/Abstract])) OR (interactom*[Title/Abstract])) AND (ionizing radiation [Title/Abstract]).
The ‘Results by year’ function was used to filter the papers from 2000 onwards as well as studies published for a particular year. The Pubmed identification (PMID) numbers of retrieved studies were imported into Microsoft Excel for further processing. The Venn diagram was produced via
http://bioinformatics.psb.ugent.be/cgi-bin/liste/Venn/calculate_venn.htpl (accessed on 22 October 2021).
3. Results
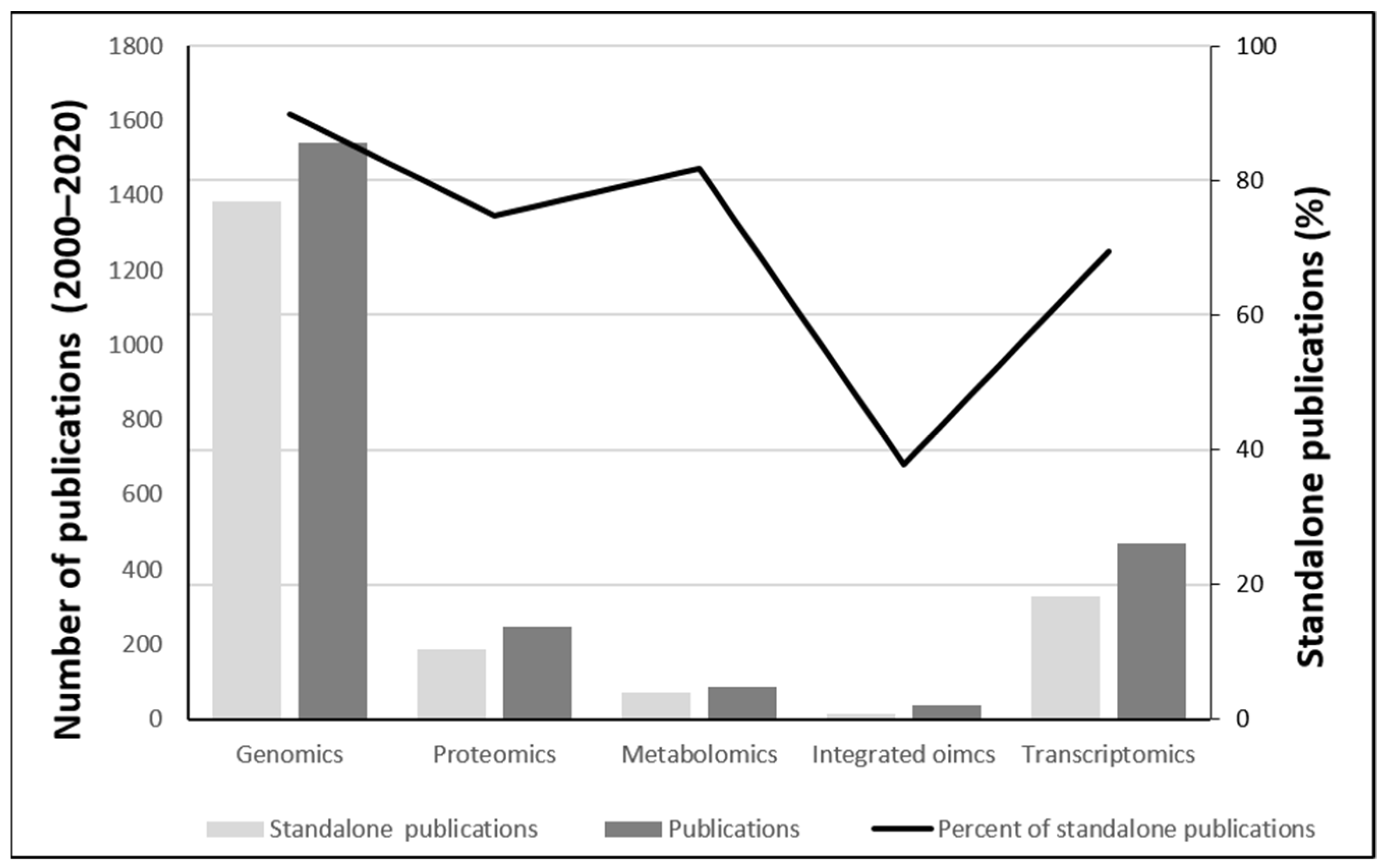
We found 2175 radiobiology studies (1 January 2000–22 October 2021) that met the search criteria. Amongst them, genomics had the highest share of papers, with 1905 papers (1542 without reviews,
Figure 1), followed by transcriptomics (490 papers, including 20 reviews), proteomics (280 papers, including 31 reviews), metabolomics (104 papers, including 14 reviews), and finally integrated omics (49 papers, including 12 reviews). This discrepancy in the popularity of methods is expected as genomics is an established method in general biology for more than half a century, whereas transcriptomics and proteomics were employed around the new millennium. It was also noted that almost 9 out of 10 studies that used genomics did not use any other method, but were published as ‘standalone’ papers. Along the same line, only 7 out of 10 papers employing transcriptomics and 8 out of 10 papers employing metabolomics were standalone papers.
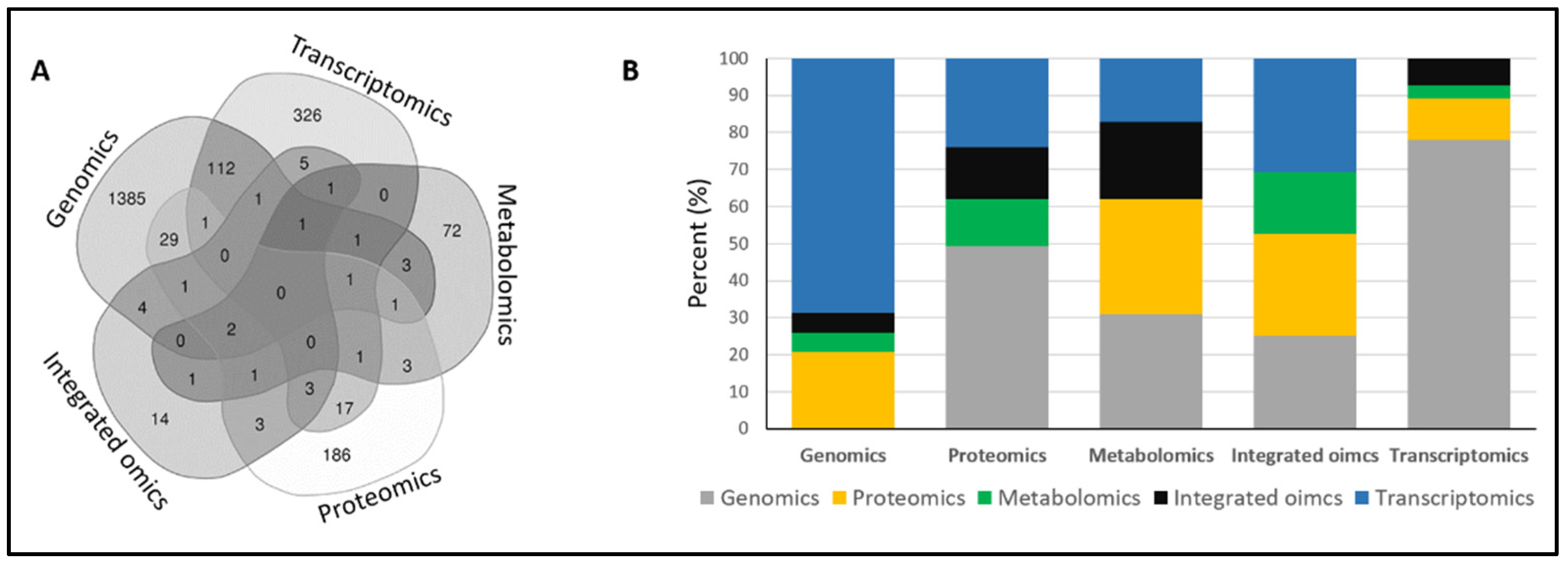
As newer techniques have emerged, radiation biologists have integrated two (117 papers), three (11 papers), or four (four papers) aforementioned techniques together (
Figure 2A). As expected, genomics is the favored additional technique used along with other methods, especially with transcriptomics (
Figure 2B). This could be because transcriptomics, being the intermediary connection between genes and proteins, is vital in connecting the genome to gene function and activity. Only about 5% of genetic code is transcribed into RNA molecules and the information about different mRNA variants produced by the same gene (alternative splicing), or alternative transcription initiation/termination sites are visible only through transcriptomics [
5].
We had assumed that the search results for integrated omics would also be obtained while searching for other omics as it is a combination of the latter. However, there were, surprisingly, 14 standalone papers for the search of integrated omics (
Figure 2A). This is perhaps because the omics terms for primary studies were not included in the title/abstract. Our search also reveals a lack of standardized use of terms for omics in radiation research. For example, the term “protein expression changes” is often used instead of “proteomics”. The authors and scientific journals need to be encouraged to use uniform keywords and standardized terms for omics for easy retrieval of the literature.
The numbers of retrieved studies display the quantity of initially identified studies without further screening for inclusion and therefore amounts may have been overestimated with our search keywords. For example, when we use keywords such as “genom*”, studies that include genomic instability without reporting genomics data may be identified. Similarly, with keyword “proteom*”, studies regarding method development for proteomic purposes might have been retrieved without these studies investigating proteomic changes. However, with this strategy we minimize the risk of losing studies. To overcome such possible sources of errors, we support the development of a unified ontology for the findability of studies. We also encourage authors who wish to write conventional or systematic reviews in omics-biology to formulate inclusion and exclusion criteria for removal of such studies, which would be transparently documented in the writing-process [
6].
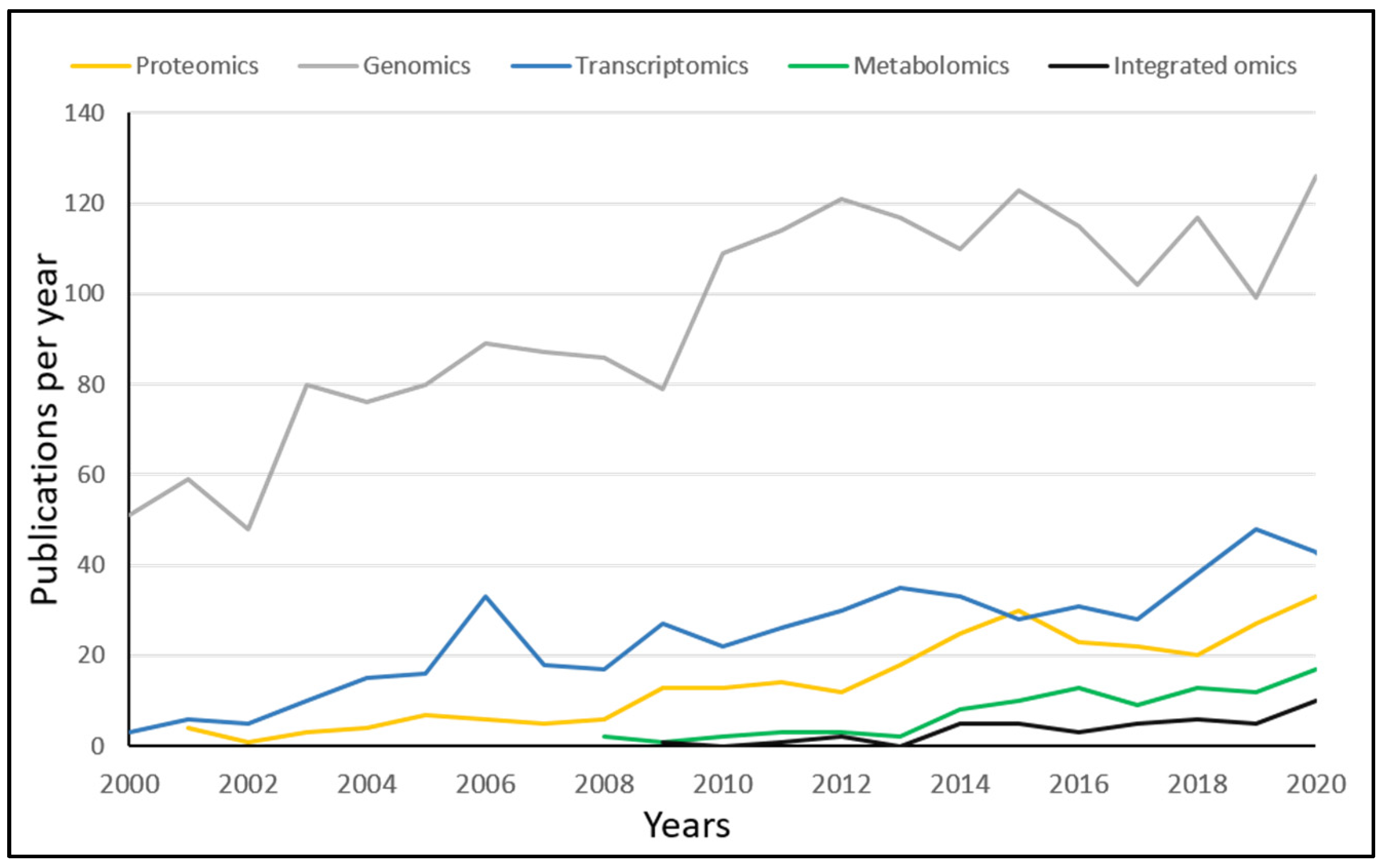
There is a difference in the timeline when the different omics platforms were used to investigate changes in living systems after exposure to IR. To put this into perspective: genomics was already used to study radiobiology (1969) before the British rock band Queen had released their debut album. The first study that employed transcriptomics (1999) happened after the Euro (EUR) was introduced in the financial markets, and it was only after the first iPhone was released (2008) that metabolomics was used to study cellular responses to IR exposure. Despite the gaps that encompass generations, newer methods are, slowly but surely, gaining recognition in the field of radiation biology (
Figure 3). In 2020, there was one transcriptomic paper for every three genomic papers and one proteomic paper for every five genomic papers. Since 2005, omics-studies have a share of 13–14% of total publications in the field of radiation biology.
Omics studies are performed to identify pathways and networks. The main aim of combining omics is the validation of these pathways on different levels. Confirmation of individual molecules is a minor aim for omics. This fact has led to the development of several bioinformatics packages that address this problem. Integrated omics aim to understand the interactions of multiple biological entities rather than validating a single endpoint. In addition, shareable omics data (as is already established and used in cancer research) are aimed to address scientific questions raised by a larger community of researchers.
Combining omics methods, however, remains a challenge in radiation research. For example, our survey showed that, on average, only 1 in 16 papers published during 2000–2020 used two or more techniques. The issue was improved in 2020, with one in nine published studies using two or more omics methods.
The difficulties of combining different omics are well known in the scientific community. The pronounced differences in experimental workflows, analysis criteria, data processing algorithms, and applied statistics in different omics make combining results from these platforms very difficult. An additional problem is the lack of the required optimal funding, which is usually beyond the norm in research institutes and hinders the combined running of multiple omics. An increase in international collaboration could bring research groups with different expertise together, which could help in integrating different omics datasets. Moreover, systems biology, with its paradigms and tools for linking omics data, is still a very young scientific field and needs to be further improved not only for omics analysis but also in the case of integrating omics and nonomics data, including epidemiological and clinical findings.
The application of omics to the large volume of the materials archived in radiation biobanks is also not established as expected. The main reason is that the optimal quality and quantity required for omics analysis is often not foreseen when biomaterials were collected. In addition, protocols for tissue processing and biomolecule extraction are not yet universal and optimized [
7].
Interest in radiobiology had already seeped in after the advent of nuclear warfare in the Second World War but probably had its trigger point after the Chernobyl disaster in 1986, especially in areas of radiological protection. So, we were surprised to have retrieved fewer papers compared to our initial assumption. However, it should be noted that more than 90% of omics studies performed in radiobiology were done between 2000–2020. Only a small group of studies that applied omics (mainly genomics) was reported before 2000.
Moreover, newer omics methods are getting their niche and enabling researchers to integrate omics methods with other analytical platforms to understand the cellular response to IR. In addition to its conventional applications, the recent use of omics platforms in radiation biodosimetry and molecular epidemiology introduces omics into the new research domains of radiation biology. It is envisioned that omics data and its mechanistic link to the adverse outcomes of radiation exposure may attract the attention of decision-makers, stakeholders, and funding agencies in the radiation community to address regulatory issues and identify knowledge and research gaps [
8,
9].
The emerging field of application of omics in radiation research requires better experimental design and optimal sample and data collection following the principles of data findability, accessibility, interoperability, and reusability (FAIR) to facilitate data integrating and sharing across the radiation research society. The available repository databases, including the European Radiobiological Archives (ERA) (
https://era.bfs.de) (accessed on 22 October 2021) [
10,
11] and STOREDB database (
https://www.storedb.org) (accessed on 22 October 2021) hosted by the German Federal Office for Radiation Protection (BfS) (
www.bfs.de) (accessed on 22 October 2021) [
12] provide optimal platforms for managing, sustaining, and sharing data and samples from different disciplines in the research field. A description of various biological and inorganic samples archive is provided by Schofield and co-workers [
12].
An expectation remains that with the inclusion of artificial intelligence and advanced computational datamining in the future to existing omics data sets, the biological effect of radiation exposure can be understood better and biomarkers relevant for clinically as well as for radiation protection purposes will be discovered sooner rather than later.