Electrochemical Genosensors as a New Approach to Plant DNA Detection and Quantification for Honey Authentication †
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents and Solutions
2.2. Apparatus and Electrodes
2.3. Oligonucleotides and Real DNA Samples
2.4. Electrochemical Genosensor Design
3. Results and Discussion
3.1. Optimization of the Analytical Parameters
3.2. Analytical Characterization of the Optimized Genosensor
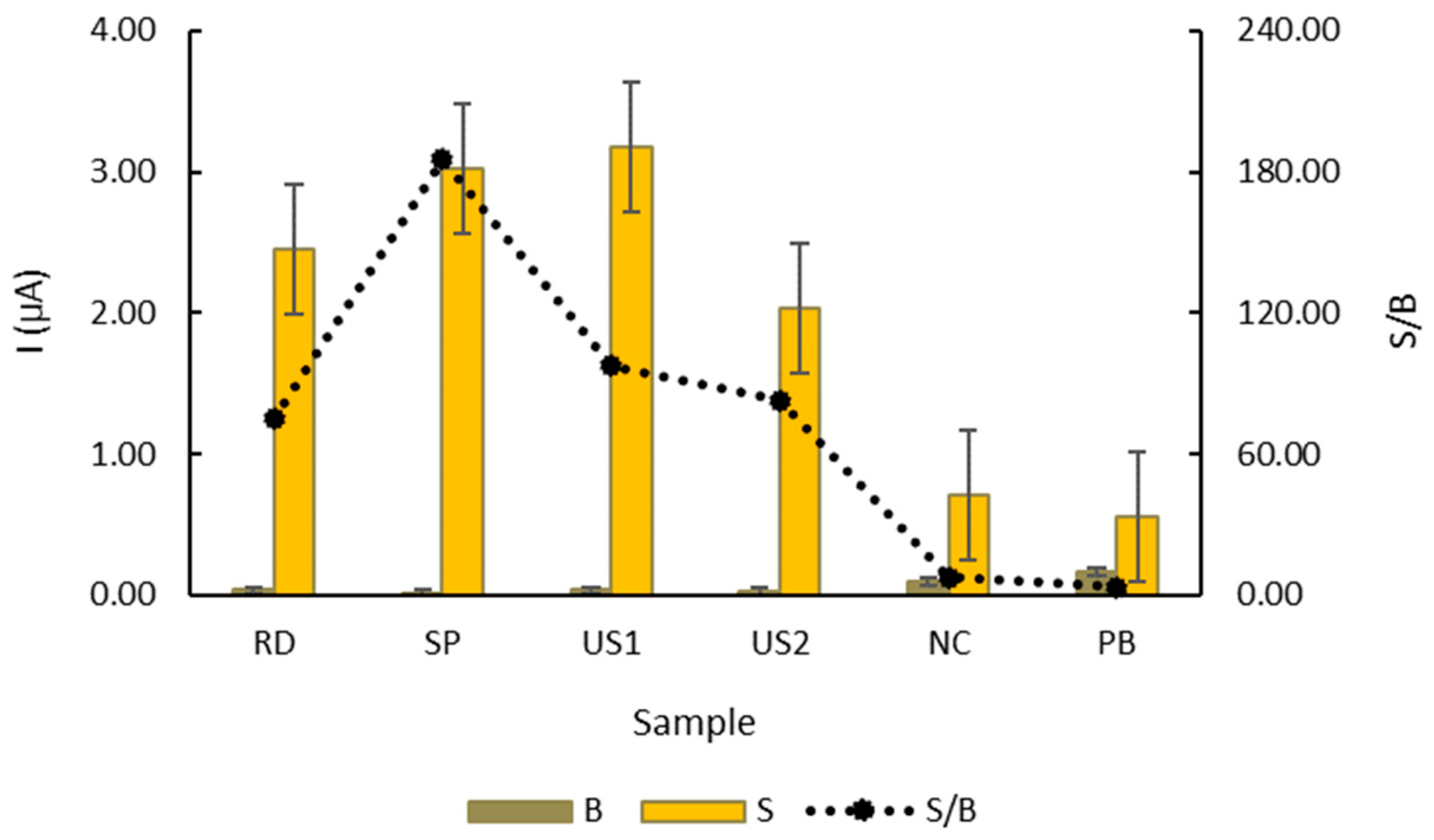
3.3. Evaluation of the Genosensor’s Selectivity
4. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brooks, C.; Parr, L.; Smith, J.M.; Buchanan, D.; Snoich, D.; Hebishy, E. A review of food fraud and food authenticity across the food supply chain, with an examination of the impact of the COVID-19 pandemic and Brexit on food industry. Food Control 2021, 130, 108171. [Google Scholar] [CrossRef]
- Euroactiv. Honeygate: How Europe Is Being Flooded with Fake Honey. 2020. Available online: https://www.euractiv.com/section/agriculture-food/news/honey-gate-how-europe-is-being-flooded-with-fake-honey/ (accessed on 1 September 2024).
- FAO STAT. Food and Agriculture Organization of the United Nations. 2020. Available online: https://www.fao.org/faostat/en/#data (accessed on 1 September 2024).
- Ramanauskiene, K.; Stelmakiene, A.; Briedis, V.; Ivanauskas, L.; Jakštas, V. The quantitative analysis of biologically active compounds in Lithuanian honey. Food Chem. 2012, 132, 1544–1548. [Google Scholar] [CrossRef] [PubMed]
- Tosun, M.; Keles, F. Investigation methods for detecting honey samples adulterated with sucrose syrup. J. Food Comp. Anal. 2021, 101, 103941. [Google Scholar] [CrossRef]
- Zhang, G.; Abdulla, W. On honey authentication and adulterant detection techniques. Food Control 2022, 138, 108992. [Google Scholar] [CrossRef]
- Fakhlaei, R.; Selamat, J.; Khatib, A.; Razis, A.F.A.; Sukor, R.; Ahmad, S.; Babadi, A.A. The Toxic Impact of Honey Adulteration: A Review. Foods 2020, 9, 1538. [Google Scholar] [CrossRef] [PubMed]
- Walker, M.J.; Cowen, S.; Gray, K.; Hancock, P.; Burns, D.T. Honey authenticity: The opacity of analytical reports—Part 1 defining the problem. NPJ Sci. Food 2022, 6, 11. [Google Scholar] [CrossRef] [PubMed]
- Manning, L.; Soon, J.M. Food Safety, Food Fraud, and Food Defense: A Fast-Evolving Literature. J. Food Sci. 2016, 81, 823–834. [Google Scholar] [CrossRef] [PubMed]
- Se, K.W.; Wahab, R.A.; Yaacob, S.N.S.; Ghoshal, S.K. Detection techniques for adulterants in honey: Challenges and recent trends. J. Food Comp. Anal. 2023, 80, 16–32. [Google Scholar] [CrossRef]
- Soares, S.; Rodrigues, F.; Delerue-Matos, C. Towards DNA-Based Methods Analysis for Honey: An Update. Molecules 2023, 28, 2106. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A Tool to Design 463 Target-Specific Primers for Polymerase Chain Reaction. BMC Bioinf. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]

| Probes | 5′→3′ Sequence | Base Pairs |
|---|---|---|
| Capture probe | GAC CTT CTT TTT AGG CCA ACC GAG CAC A | 28 |
| Signaling probe | GAC TGC GTA GCA TGC ACA ACG TGT CGC AGT TTG GCA ACC ACC ACT TGT TGT GAT GTC CGT CAT CAG G | 70 |
| Target probe | TGT GCT CGG TTG GCC TAA AAA GAA GGT CCC TGA TGA CGG ACA TCA CAA CAA GTG GTG GTT GCC AAA CTG TCG CGA CAC GTT GTG CAT GCT ACG CAG TC | 98 |
| Variables | Tested Range | Selected Value |
|---|---|---|
| DNA-capture concentration (μM) | 0.25–10.00 | 1.00 |
| MCH spacer concentration (μM) | 0.00–1.00 | 0.50 |
| MCH spacer incubation time (min) | 5–30 | 5 |
| Homogeneous hybridization incubation time (min) | 15–60 | 30 |
| Temperature (°C) | 25–98 | 25 |
| DNA-signaling concentration probe (μM) | 0.13–0.50 | 0.50 |
| Heterogeneous hybridization incubation time (min) | 30–120 | 60 |
| Antibody concentration (U/mL) | 0.50–3.00 | 2.00 |
| Antibody incubation time (min) | 15–45 | 30 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morais, S.L.; Castanheira, M.; Santos, M.; Domingues, V.F.; Delerue-Matos, C.; Barroso, M.F. Electrochemical Genosensors as a New Approach to Plant DNA Detection and Quantification for Honey Authentication. Eng. Proc. 2024, 82, 79. https://doi.org/10.3390/ecsa-11-20353
Morais SL, Castanheira M, Santos M, Domingues VF, Delerue-Matos C, Barroso MF. Electrochemical Genosensors as a New Approach to Plant DNA Detection and Quantification for Honey Authentication. Engineering Proceedings. 2024; 82(1):79. https://doi.org/10.3390/ecsa-11-20353
Chicago/Turabian StyleMorais, Stephanie L., Michelle Castanheira, Marlene Santos, Valentina F. Domingues, Cristina Delerue-Matos, and M. Fátima Barroso. 2024. "Electrochemical Genosensors as a New Approach to Plant DNA Detection and Quantification for Honey Authentication" Engineering Proceedings 82, no. 1: 79. https://doi.org/10.3390/ecsa-11-20353
APA StyleMorais, S. L., Castanheira, M., Santos, M., Domingues, V. F., Delerue-Matos, C., & Barroso, M. F. (2024). Electrochemical Genosensors as a New Approach to Plant DNA Detection and Quantification for Honey Authentication. Engineering Proceedings, 82(1), 79. https://doi.org/10.3390/ecsa-11-20353








