Characterization of Gene Families Encoding Beta-Lactamases of Gram-Negative Rods Isolated from Ready-to-Eat Vegetables in Mexico City
Abstract
:1. Introduction
2. Material and Methods
2.1. Bioinformatics Analysis and Primer Design
2.2. Sampling
2.3. Bacterial Isolation
2.4. Bacterial Identification
2.5. Antimicrobial Susceptibility Testing
2.6. DNA Extraction
2.6.1. Crude Extract
2.6.2. Genomic DNA Extraction
2.7. Plasmid DNA Extraction
2.8. Endpoint PCR
2.9. Whole Genome Sequencing (WGS) and Genotype Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nguyen-the, C.; Carlin, F. The microbiology of minimally processed fresh fruits and vegetables. Crit. Rev. Food Sci. Nutr. 1994, 34, 371–401. [Google Scholar] [CrossRef] [PubMed]
- Sammarco, M.L.; Ripabelli, G.; Grasso, G.M. Consumer attitude and awareness towards food-related hygienic hazards. J. Food Saf. 1997, 17, 215–221. [Google Scholar] [CrossRef]
- Heit, J. Food Safety. University of Maryland Medical Center, 2015. Available online: http://umm.edu/health/medical/ency/articles/food-safety (accessed on 10 January 2018).
- Harris, L.J.; Farber, J.N.; Beuchat, L.R.; Parish, M.E.; Suslow, T.V.; Garrett, E.H.; Busta, F.F. Outbreaks associated with fresh produce: Incidence, growth, and survival of pathogens in fresh and fresh-cut produce. Compr. Rev. Food Sci. 2003, 2, 78–141. [Google Scholar] [CrossRef]
- Liu, L.; Johnson, H.L.; Cousens, S.; Perin, J.; Scott, S.; Lawn, J.E.; Rudan, I.; Campbell, H.; Cibulskis, R.; Li, M.; et al. Child Health Epidemiology Reference Group of WHO and UNICEF. Global, regional, and national causes of child mortality: An updated systematic analysis for 2010 with time trends since 2000. Lancet 2012, 379, 2151–2156. [Google Scholar] [CrossRef]
- Buys, H.; Muloiwa, R.; Bamford, C.; Eley, B. Klebsiella pneumoniae bloodstream infections at a South African children’s hospital 2006–2011, a cross-sectional study. BMC Infect. Dis. 2016, 16, 570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caltagirone, M.; Nucleo, E.; Spalla, M.; Zara, F.; Novazzi, F.; Marchetti, V.M.; Piazza, A.; Bitar, I.; De Cicco, M.; Paolucci, S.; et al. Occurrence of Extended Spectrum β-Lactamases, KPC-Type, and MCR-1.2-Producing Enterobacteriaceae from Wells, River Water, and Wastewater Treatment Plants in Oltrepò Pavese Area, Northern Italy. Front. Microbiol. 2017, 8, 2232. [Google Scholar] [CrossRef] [PubMed]
- Kpoda, D.S.; Ajayi, A.; Somda, M.; Traore, O.; Guessennd, N.; Ouattara, A.S.; Sangare, L.; Traore, A.S.; Dosso, M. Distribution of resistance genes encoding ESBLs in Enterobacteriaceae isolated from biological samples in health centers in Ouagadougou, Burkina Faso. BMC Res. Notes 2018, 11, 471. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.M.; Ke, S.C.; Li, C.R.; Wu, Y.C.; Chen, T.H.; Lai, C.H.; Wu, X.X.; Wu, L.T. High Diversity of Antimicrobial Resistance Genes, Class 1 Integrons, and Genotypes of Multidrug-Resistant Escherichia coli in Beef Carcasses. Microb. Drug Resist. 2017, 23, 915–924. [Google Scholar] [CrossRef] [PubMed]
- Bush, K.; Jacoby, G.A. Updated functional classification of beta-lactamases. Antimicrob. Agents Chemother. 2010, 54, 969–976. [Google Scholar] [CrossRef] [PubMed]
- Bush, K. The ABCD’s of β-lactamase nomenclature. J. Infect. Chemother. 2013, 19, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Salahuddin, P.; Kumar, A.; Khan, A.U. Structure, Function of Serine and Metallo-β-lactamases and their Inhibitors. Curr. Protein Pept. Sci. 2018, 19, 130–144. [Google Scholar] [CrossRef] [PubMed]
- Bush, K. Past and Present Perspectives on β-Lactamases. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef] [PubMed]
- Jovetic, S.; Zhu, Y.; Marcone, G.L.; Marinelli, F.; Tramper, J. β-Lactam and glycopeptide antibiotics: First and last line of defence? Trends Biotechnol. 2010, 28, 596–604. [Google Scholar] [CrossRef] [PubMed]
- Wilke, M.S.; Lovering, A.L.; Strynadka, N.C.J. b-Lactam antibiotic resistance: A current structural perspective. Curr. Opin. Microbiol. 2005, 8, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Barnaud, G.; Arlet, G.; Danglot, C.; Philippon, A. Cloning and sequencing of the gene encoding the AmpC beta-lactamase of Morganella morganii. FEMS Microbiol. Lett. 1997, 148, 15–20. [Google Scholar] [CrossRef]
- Bush, K.; Jacoby, G.A.; Medeiros, A.A. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 1995, 39, 1211–1233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bebrone, C. Metallo-beta-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochem. Pharmacol. 2007, 74, 1686–1701. [Google Scholar] [CrossRef] [PubMed]
- Parker, A.C.; Smith, C.J. Genetic and biochemical analysis of a novel Ambler class A beta-lactamase responsible for cefoxitin resistance in Bacteroides species. Antimicrob. Agents Chemother. 1993, 37, 1028–1036. [Google Scholar] [CrossRef] [PubMed]
- Queenan, A.M.; Bush, K. Carbapenemases: The versatile beta-lactamases. Clin. Microbiol. Rev. 2007, 20, 440–458. [Google Scholar] [CrossRef] [PubMed]
- Walsh, T.R.; Toleman, M.A.; Poirel, L.; Nordmann, P. Metallo-beta-lactamases: The quiet before the storm? Clin. Microbiol. Rev. 2005, 18, 306–325. [Google Scholar] [CrossRef] [PubMed]
- Jacoby, G.A. AmpC β-Lactamases. Clin. Microbiol. Rev. 2009, 22, 161–182. [Google Scholar] [CrossRef] [PubMed]
- DiBonaventura, M.D.; Meincke, H.; Le Lay, A.; Fournier, J.; Bakker, E.; Ehrenreich, A. Obesity in Mexico: Prevalence, comorbidities, associations with patient outcomes, and treatment experiences. Diabetes Metab. Syndr. Obes. 2017, 11, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Dhroove, G.; Saps, M.; Garcia-Bueno, C.; Leyva Jiménez, A.; Rodriguez-Reynosa, L.; Velasco-Benítez, C.A. Prevalence of functional gastrointestinal disorders in Mexican schoolchildren. Rev. Gastroenterol. Mex. 2017, 82, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence weighting, position specific gap penalties and weighting matrix chose. Nucl. Acids Res. 2014, 22, 4673–4680. [Google Scholar] [CrossRef]
- Martínez-Pérez, F.; Becerra, A.; Valdés, J.; Zinker, S.; Aréchiga, H. A possible molecular ancesor for mollussk APGWamide, insect adipokinetic hormone, and crustacean red pigment concentrating hormone. J. Mol. Evol. 2002, 54, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Pérez, F.; Bendena, W.G.; Chang, B.S.; Tobe, S.S. FGLamide Allatostatin gene in Arthropoda: Introns early or late? Peptides 2009, 30, 1241–1248. [Google Scholar] [CrossRef] [PubMed]
- Marshall, O.J. PerlPrimer: Cross-platform, graphical primer design for standard, bisulphite and real-time PCR. Bioinformatics 2004, 20, 2471–2472. [Google Scholar] [CrossRef] [PubMed]
- FastQC. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 10 September 2018).
- BWA-MEM. Available online: http://sourceforge.net/p/bio-bwa/mailman/message/30894287/ (accessed on 10 September 2018).
- SAMtools. Available online: http://www.codoncode.com/aligner/ (accessed on 10 September 2018).
- Kraken Taxonomic Sequence Classification System. Available online: https://ccb.jhu.edu/software/kraken/ (accessed on 10 September 2018).
- Basic Local Alignment Search Tool. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi/ (accessed on 10 September 2018).
- Hutton, G.; Haller, L.; Bartram, J. Global cost-benefit analysis of water supply and sanitation interventions. J. Water Health 2007, 5, 481–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boehme, S.; Werner, G.; Klare, I.; Reissbrodt, R.; Witte, W. Occurrence of antibiotic resistant enterobacteria in agricultural foodstuffs. Mol. Nutr. Food Res. 2004, 48, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Hamilton-Miller, J.M.; Shah, S. Identity and antibiotic susceptibility of enterobacterial flora of salad vegetables. Int. J. Antimicrob. Agents 2001, 18, 81–83. [Google Scholar] [CrossRef]
- Schmiedel, J.; Falgenhauer, L.; Domann, E.; Bauerfeind, R.; Prenger-Berninghoff, E.; Imirzalioglu, C.; Chakraborty, T. Multiresistant extended-spectrum beta-lactamase-producing Enterobacteriaceae from humans, companion animals and horses in central Hesse, Germany. BMC Microbiol. 2014, 14, 187. [Google Scholar] [CrossRef] [PubMed]
- Philippon, A.; Arlet, G.; Jacoby, G.A. Plasmid-determined AmpC-type beta-lactamases. Antimicrob. Agents Chemother. 2002, 46, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Gilman, R.H.; Koster, F.; Islam, S.; McLaughlin, J.; Rahaman, M.M. Randomized trial of high- and low-dose ampicillin therapy for treatment of severe dysentery due to Shigella dysenteriae type 1. Antimicrob. Agents Chemother. 1980, 17, 402–405. [Google Scholar] [CrossRef] [PubMed]
- Rezaei, F.; Saderi, H.; Boroumandi, S.; Faghihzadeh, S. Relation between Resistance to Antipseudomonal β-Lactams and ampC and mexC Genes of Pseudomonas aeruginosa. Iran. J. Pathol. 2016, 11, 47–53. [Google Scholar] [PubMed]
- Lauretti, L.; Riccio, M.L.; Mazzariol, A.; Cornaglia, G.; Amicosante, G.; Fontana, R.; Rossolini, G.M. Cloning and characterization of blaVIM, a new integron-borne metallo-beta-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 1999, 43, 1584–1590. [Google Scholar] [CrossRef] [PubMed]
- Rangel, W.M.; de Oliveira Longatti, S.M.; Ferreira, P.A.A.; Bonaldi, D.S.; Guimarães, A.A.; Thijs, S.; Weyens, N.; Vangronsveld, J.; Moreira, F.M.S. Leguminosae native nodulating bacteria from a gold mine As-contaminated soil: Multi-resistance to trace elements, and possible role in plant growth and mineral nutrition. Int. J. Phytoremediat. 2017, 19, 925–936. [Google Scholar] [CrossRef] [PubMed]
- Sievert, D.M.; Ricks, P.; Edwards, J.R.; Schneider, A.; Patel, J.; Srinivasan, A.; Kallen, A.; Limbago, B.; Fridkin, S. National Healthcare Safety Network (NHSN) Team and Participating NHSN Facilities. Antimicrobial-resistant pathogens associated with healthcare-associated infections: Summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infect. Control Hosp. Epidemiol. 2013, 34, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention (CDC). Vital Signs: Carbapenem-resistant Enterobacteriaceae. MMWR Morb. Mortal. Wkly. Rep. 2013, 62, 165–170. Available online: https://www.cdc.gov/mmwr/preview/mmwrhtml/mm6209a3.htm (accessed on 10 September 2018).
- Ye, Q.; Wu, Q.; Zhang, S.; Zhang, J.; Yang, G.; Wang, J.; Xue, L.; Chen, M. Characterization of Extended-Spectrum β-Lactamase-Producing Enterobacteriaceae from Retail Food in China. Front. Microbiol. 2018, 9, 1709. [Google Scholar] [CrossRef] [PubMed]
- Laolerd, W.; Akeda, Y.; Preeyanon, L.; Ratthawongjirakul, P.; Santanirand, P. Carbapenemase-Producing Carbapenem-Resistant Enterobacteriaceae from Bangkok, Thailand, and Their Detection by the Carba NP and Modified Carbapenem Inactivation Method Tests. Microb. Drug Resist. 2018, 24, 1006–1011. [Google Scholar] [CrossRef] [PubMed]
- Walther-Rasmussen, J.; Høiby, N. OXA-type carbapenemases. J. Antimicrob. Chemother. 2006, 57, 373–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
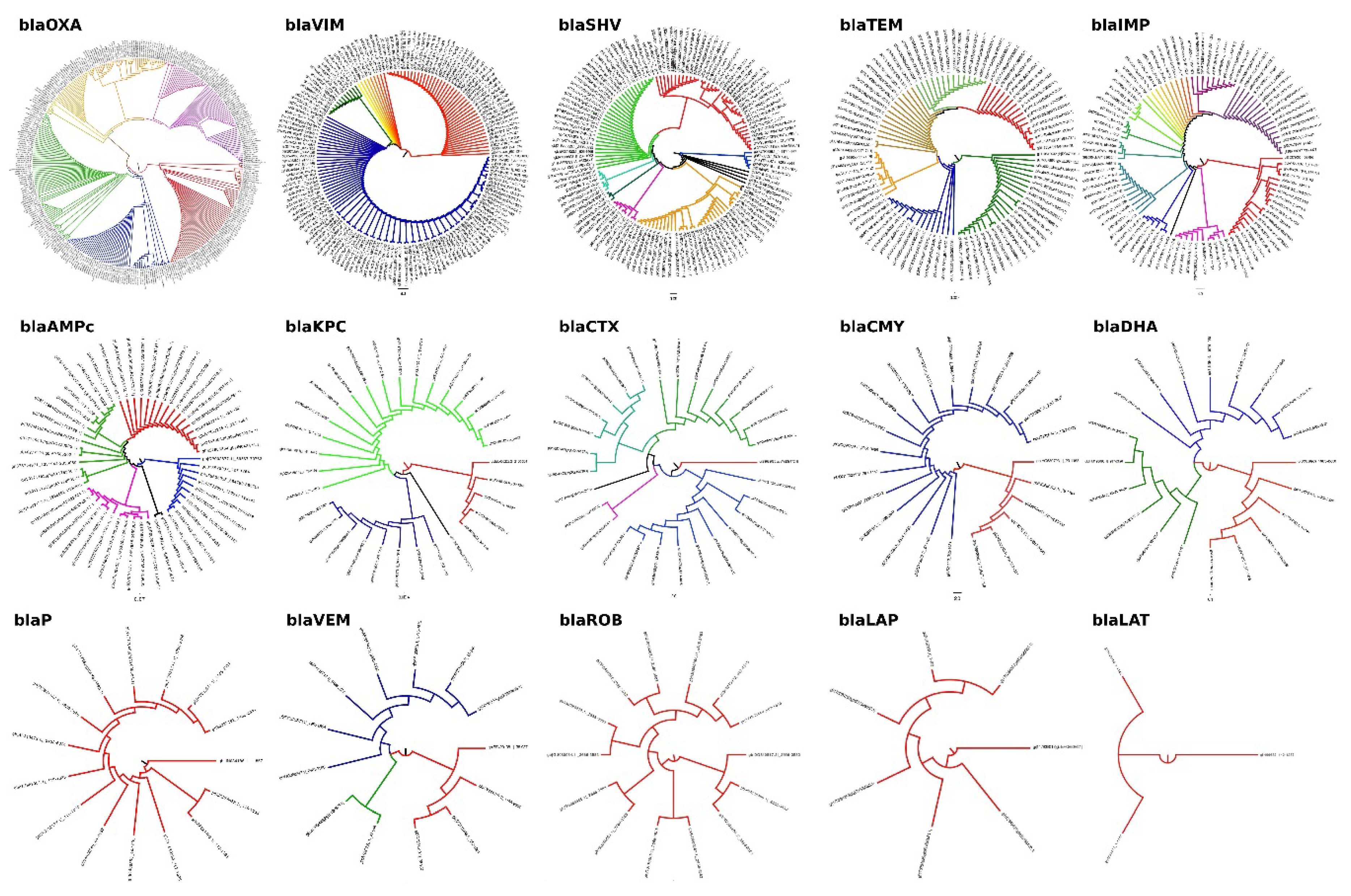
| Gen Family | Sequences | Discarded | Analyzed | Discarded (%) | Analyzed (%) |
|---|---|---|---|---|---|
| blaOXA | 322 | 9 | 313 | 2.80 | 97.20 |
| blaVIM | 186 | 29 | 157 | 13.24 | 71.68 |
| blaSHV | 129 | 3 | 126 | 2.32 | 97.67 |
| blaTEM | 102 | 9 | 93 | 8.82 | 91.18 |
| blaIMP | 100 | 5 | 95 | 5.00 | 95.00 |
| blaROB | 16 | 1 | 15 | 6.25 | 93.75 |
| blaKPC | 33 | 2 | 31 | 6.06 | 93.94 |
| blaCTX | 31 | 2 | 29 | 6.45 | 93.55 |
| blaCMY | 28 | 5 | 23 | 2.87 | 97.13 |
| blaDHA | 21 | 1 | 20 | 4.76 | 95.24 |
| blaP | 16 | 1 | 15 | 6.25 | 93.75 |
| blaCFX | 9 | 1 | 8 | 11.11 | 88.89 |
| blaLAP | 10 | 3 | 7 | 30.00 | 70.00 |
| blaLAT | 3 | 0 | 3 | 0.00 | 100.00 |
| blaBIL | 1 | 0 | 1 | 0.00 | 100.00 |
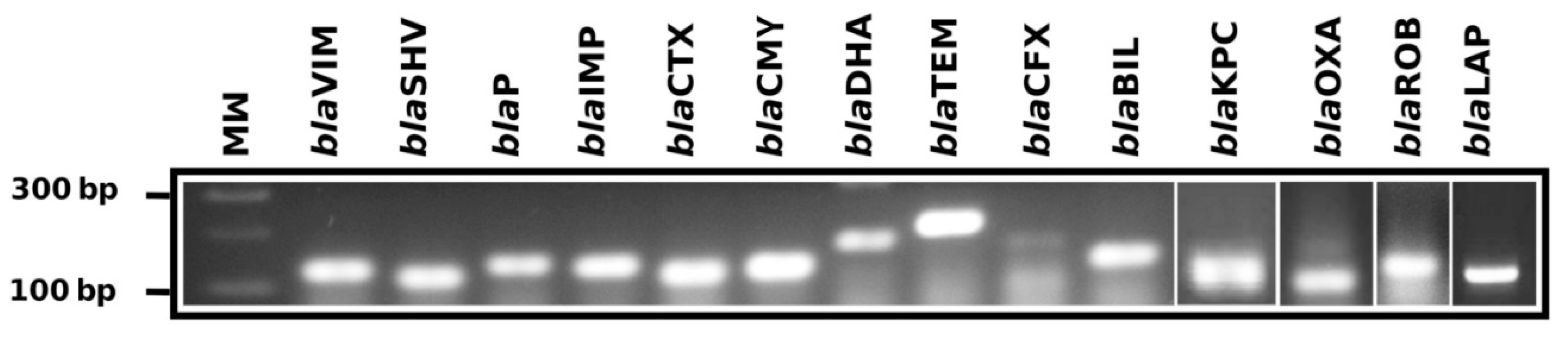
| Gene Family | Primer Name | Primer Sequence (5′ to 3′) | Tm (°C) | Position | Amplicon (bp) |
|---|---|---|---|---|---|
| blaOXA | BlaOXA-FW | GGTTTCGGTAATGCTGAAATTGG | 61.18 | 214–236 | 114 |
| BlaOXA-RW | GCTGTGTATGTGCTAATTGGGA | 61.19 | 327–306 | ||
| blaVIM | BlaVIM-FW | CGACAGTCARCGAAATTCC | 61.39 | 105–123 | 133 |
| BlaVIM-RW | CAATGGTCTSATTGTCCGTG | 61.34 | 238–219 | ||
| blaSHV | BlaSHV-FW1 | CGTAGGCATGATAGAAATGGATC | 61.04 | 133–155 | 106 |
| BlaSHV-RW1 | CGCAGAGCACTACTTTAAAGG | 61.33 | 239–218 | ||
| BlaSHV-FW2 | GCCTCATTCAGTTCCGTTTC | 61.62 | 399–418 | 141 | |
| BlaSHV-RW2 | CCATTACCATGAGCGATAACAG | 61.22 | 540–518 | ||
| blaTEM | BlaTEM-FW | GCCAACTTACTTCTGACAACG | 61.80 | 1699–1719 | 213 |
| BlaTEM-RW | CGTTTGGAATGGCTTCATTC | 60.13 | 1912–1892 | ||
| blaIMP | BlaIMP-FW1 | GGAATAGARTGGCTTAAYTCTCG | 60.92 | 319–332 | 183 |
| BlaIMP-RW1 | CYASTASGTTATCTKGAGTGTG | 62.45 | 502–480 | ||
| BlaIMP-FW2 | GGTGGAATAGARTGGCTTAAYTC | 61.11 | 316–339 | 192 | |
| BlaIMP-RW2 | CCAAACCACTACGTTATCTKGAG | 61.29 | 508–485 | ||
| blaROB | BlaROB-FW | CCAACATCGTGGAAAGTGTAG | 61.27 | 718–739 | 126 |
| BlaROB-RW | GTAAATTGCGTACTCATGATTGC | 60.90 | 844–821 | ||
| blaKPC | BlaKPC-FW | GCTAAACTCGAACAGGACTTTG | 61.79 | 100–121 | 117 |
| BlaKPC-RW | CTTGAATGAGCTGCACAGTG | 61.90 | 216–197 | ||
| blaCTX | BlaCTX-FW1 | GATACCGCAGATAATACGCAG | 60.79 | 161–181 | 116 |
| BlaCTX-RW1 | CGTTTTGCGTTTCACTCTG | 60.28 | 276–258 | ||
| BlaCTX-FW2 | GCTGATTCTGGTCACTTACTTC | 61.02 | 789–810 | 83 | |
| BlaCTX-RW2 | CGCCGACGCTAATACATC | 60.69 | 855–872 | ||
| BlaCTX-FW3 | CTGCTTAACTACAATCCSATTGC | 62.17 | 314–336 | 226 | |
| BlaCTX-RW3 | GGAATGGCGGTATTKAGC | 60.86 | 539–522 | ||
| blaCMY | BlaCMY-FW1 | GTTTGAGCTAGGATCGGTTAG | 60.25 | 337–357 | 123 |
| BlaCMY-RW1 | CTGTTTGCCTGTCAGTTCTG | 61.48 | 460–441 | ||
| BlaCMY-FW2 | GAACGAAGGCTACGTAGCT | 61.71 | 213–231 | 160 | |
| BlaCMY-RW2 | CTGAAACGTGATTCGATCATCA | 61.08 | 372–351 | ||
| blaDHA | BlaDHA-FW1 | GCATATTGATCTGCATATCTCCAC | 61.60 | 399–422 | 200 |
| BlaDHA-RW1 | GCTGCTGTAACTGTTCTGC | 61.62 | 598–580 | ||
| BlaDHA-FW2 | GCGGATCTGCTGAATTTCTATC | 61.54 | 464–485 | 147 | |
| BlaDHA-RW2 | GCAGTCAGCAACTGCTCATAC | 61.05 | 610–591 | ||
| BlaDHA-FW3 | GTAAGATTCCGCATCAAGCTG | 61.74 | 430–450 | 117 | |
| BlaDHA-RW3 | GGGTTATCTCACACCTTTATTACTG | 61.08 | 546–522 | ||
| blaP | BlaP-FW | GGAGAATATTGGGATTACAATGGC | 61.74 | 271–294 | 204 |
| BlaP-RW | CGCATCATCGAGTGTGATTG | 61.80 | 474–455 | ||
| blaCFX | BlaCFX-FW | CCAGTCATATCATTGACAGTGAG | 60.86 | 437–459 | 177 |
| BlaCFX-RW | GACATTTCCTCTTCCGTATAAGC | 61.16 | 613–591 | ||
| blaLAP | BlaLAP-FW | AGGGCTTGAACAACTTGAAC | 61.07 | 249–268 | 126 |
| BlaLAP-RW | GTAATGGCAGCATTGCATAAC | 60.59 | 374–354 | ||
| blaBIL | BlaBIL-FW | GCCGATATCGTTAATCGCAC | 61.65 | 100–119 | 128 |
| BlaBIL-RW | GTTATTGGCGATATCGGCTTTA | 60.98 | 227–206 |
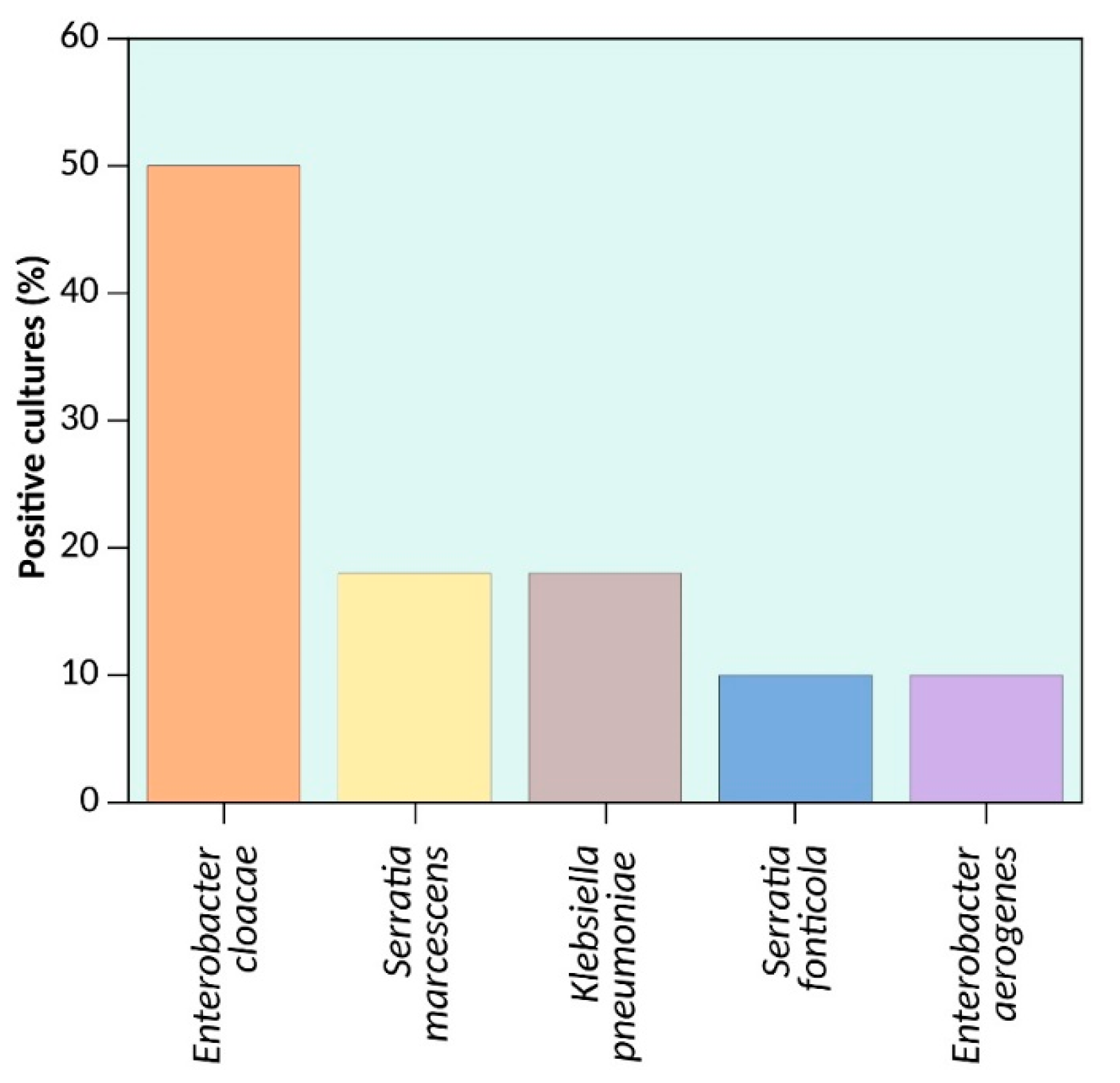
| Bacteria | CDS | Number of Sequence Contigs | Assembled Genome Size (bp) | Reported Genomic Size (bp) | Genomic Size Difference |
|---|---|---|---|---|---|
| Enterobacter cloacae | 4545 | 1484 | 4,982,176 | 4,772,910 | 209,266 |
| Serratia marcescens | 6596 | 8889 | 5,681,210 | 5,241,455 | 439,755 |
| Klebsiella pneumoniae | 5071 | 583 | 5,479,173 | 5,315,120 | 164,053 |
| Serratia fonticola | 5945 | 916 | 6,483,043 | 6,000,511 | 482,532 |
| Enterobacter aerogenes | 4545 | 1484 | 5,578,724 | 5,280,350 | 298,374 |
| Gen Family | n | Frequency |
|---|---|---|
| blaOXA | 2 | 16.7 |
| blaVIM | 4 | 33.3 |
| blaSHV | 11 | 91.6 |
| blaTEM | 1 | 8.3 |
| blaIMP | 2 | 16.6 |
| blaKPC | 1 | 8.3 |
| blaCTX | 10 | 83.3 |
| blaDHA | 9 | 75.0 |
| blaBIL | 11 | 91.6 |
| blaROB | 0 | 0.0 |
| blaCMY | 0 | 0.0 |
| blaP | 0 | 0.0 |
| blaCFX | 0 | 0.0 |
| blaLAP | 0 | 0.0 |
| Bacteria | Carried Beta-Lactamases Gene Families | N | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| blaSHV | blaCTX | blaDHA | blaBIL | blaKPC | blaVIM | blaIMP | blaOXA | blaTEM | ||
| Klebsiella pneumoniae | + | + | + | 1 | ||||||
| Klebsiella pneumoniae | + | + | + | + | + | 1 | ||||
| Enterobacter cloacae | + | + | + | 1 | ||||||
| Enterobacter cloacae | + | + | + | + | 1 | |||||
| Enterobacter cloacae | + | + | + | + | 2 | |||||
| Enterobacter cloacae | + | + | + | + | + | 1 | ||||
| Enterobacter cloacae | + | + | + | + | + | 1 | ||||
| Enterobacter aerogene | + | + | + | + | + | + | 1 | |||
| Serratia fonticola | + | + | + | + | 1 | |||||
| Serratia marcescens | + | + | + | 1 | ||||||
| Serratia marcescens | + | + | + | + | + | 1 | ||||
| NGS Characteristics | Enterobacter Cloacae | Serratia Marcescens | Klebsiella Pneumoniae | Serratia [1 Fonticola] | Enterobacter Aerogenes |
|---|---|---|---|---|---|
| Total Reads | 4,714,939 | 241,072 | 1,523,821 | 3,848,555 | 1,331,424 |
| Classified Reads | 4,541,943 (96.33%) | 183,689 (76.20%) | 1,434,672 (94.15%) | 3,680,364 (95.63%) | 1,200,101 (90.14%) |
| Domain | 4,438,740 (94.14%) | 182,241 (75.60%) | 1,434,058 (94.11%) | 3,679,637 (95.61%) | 1,198,484 (90.02%) |
| Phylum | 4,401,268 (93.35%) | 179,018 (74.26%) | 1,427,130 (93.65%) | 3,678,453 (95.58%) | 1,193,110 (89.61%) |
| Class | 4,397,652 (93.27%) | 177,491 (73.63%) | 1,425,587 (93.55%) | 3,619,438 (94.05%) | 1,190,902 (89.45%) |
| Order | 4,394,905 (93.21%) | 178,775 (74.16%) | 1,419,060 (93.13%) | 3,609,215 (93.78%) | 1,188,531 (89.27%) |
| Family | 4,395,029 (93.21) | 178,735 (74.14%) | 1,419,175 (93.13%) | 3,603,190 (93.62%) | 1,188,519 (89.27%) |
| Gender | 1,730,480 (36.70%) | 176,087 (73.04%) | 1,348,898 (88.52%) | 3,600,628 (93.56%) | 1,141,985 (85.77%) |
| Species | 1,685,267 (35.74%) | 170,381 (70.68%) | 1,219,856 (80.05%) | 3,572,513 (92.83%) | 1,123,080 (84.35%) |
| Bacteria | Beta-Lactamases Families | |||||
|---|---|---|---|---|---|---|
| SHV | CTX | DHA | BIL | KPC/VIM/IMP | OXA | |
| Enterobacter cloacae | blaSHV-12 | blaCTX-M-15 | blaDHA-1 | blaBIL *** | blaKPC-2 | |
| Serratia marcescens | blaSHV-1 | blaCTX-M-3 | blaDHA-1 | blaBIL *** | blaVIM-2 | |
| Klebsiella pneumoniae | blaSHV-12 | blaCTX-M-15 | blaDHA-1 | blaBIL *** | blaIMP *** | |
| Serratia fonticola | blaSHV-12 | blaVIM-1 | blaDHA-1 | blaBIL *** | ||
| Enterobacter aerogene | blaSHV-1 | blaCTX-M-1 | blaDHA-1 | blaBIL *** | blaVIM-2 | blaOXA-9 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vázquez-López, R.; Solano-Gálvez, S.; León-Chávez, B.A.; Thompson-Bonilla, M.R.; Guerrero-González, T.; Gómez-Conde, E.; Martínez-Fong, D.; González-Barrios, J.A. Characterization of Gene Families Encoding Beta-Lactamases of Gram-Negative Rods Isolated from Ready-to-Eat Vegetables in Mexico City. High-Throughput 2018, 7, 36. https://doi.org/10.3390/ht7040036
Vázquez-López R, Solano-Gálvez S, León-Chávez BA, Thompson-Bonilla MR, Guerrero-González T, Gómez-Conde E, Martínez-Fong D, González-Barrios JA. Characterization of Gene Families Encoding Beta-Lactamases of Gram-Negative Rods Isolated from Ready-to-Eat Vegetables in Mexico City. High-Throughput. 2018; 7(4):36. https://doi.org/10.3390/ht7040036
Chicago/Turabian StyleVázquez-López, Rosalino, Sandra Solano-Gálvez, Bertha A. León-Chávez, María R. Thompson-Bonilla, Tayde Guerrero-González, Eduardo Gómez-Conde, Daniel Martínez-Fong, and Juan A. González-Barrios. 2018. "Characterization of Gene Families Encoding Beta-Lactamases of Gram-Negative Rods Isolated from Ready-to-Eat Vegetables in Mexico City" High-Throughput 7, no. 4: 36. https://doi.org/10.3390/ht7040036
APA StyleVázquez-López, R., Solano-Gálvez, S., León-Chávez, B. A., Thompson-Bonilla, M. R., Guerrero-González, T., Gómez-Conde, E., Martínez-Fong, D., & González-Barrios, J. A. (2018). Characterization of Gene Families Encoding Beta-Lactamases of Gram-Negative Rods Isolated from Ready-to-Eat Vegetables in Mexico City. High-Throughput, 7(4), 36. https://doi.org/10.3390/ht7040036





