Contrast-Enhanced CT Texture Analysis in Colon Cancer: Correlation with Genetic Markers
Abstract
1. Introduction
2. Materials and Methods
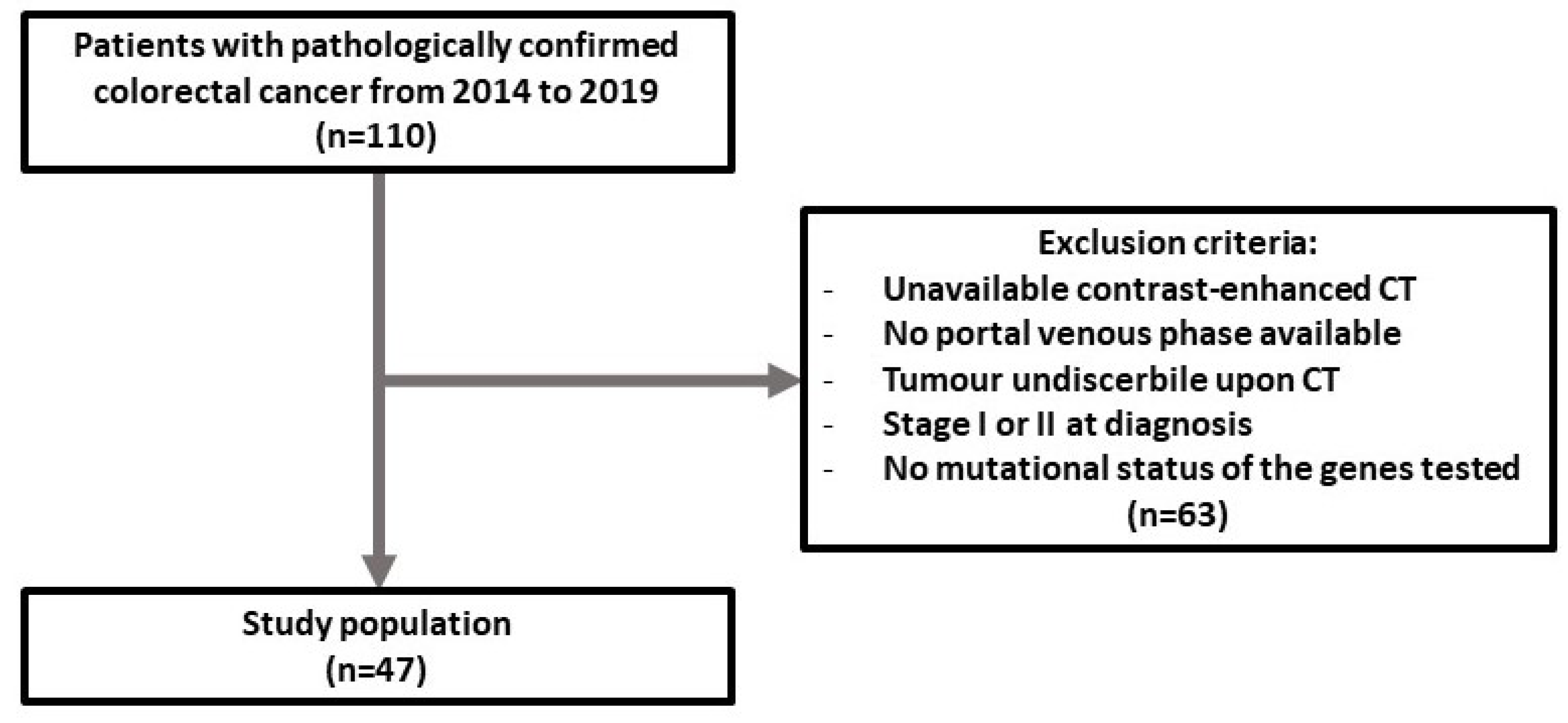
2.1. Patient Selection
2.2. CT Image Acquisition
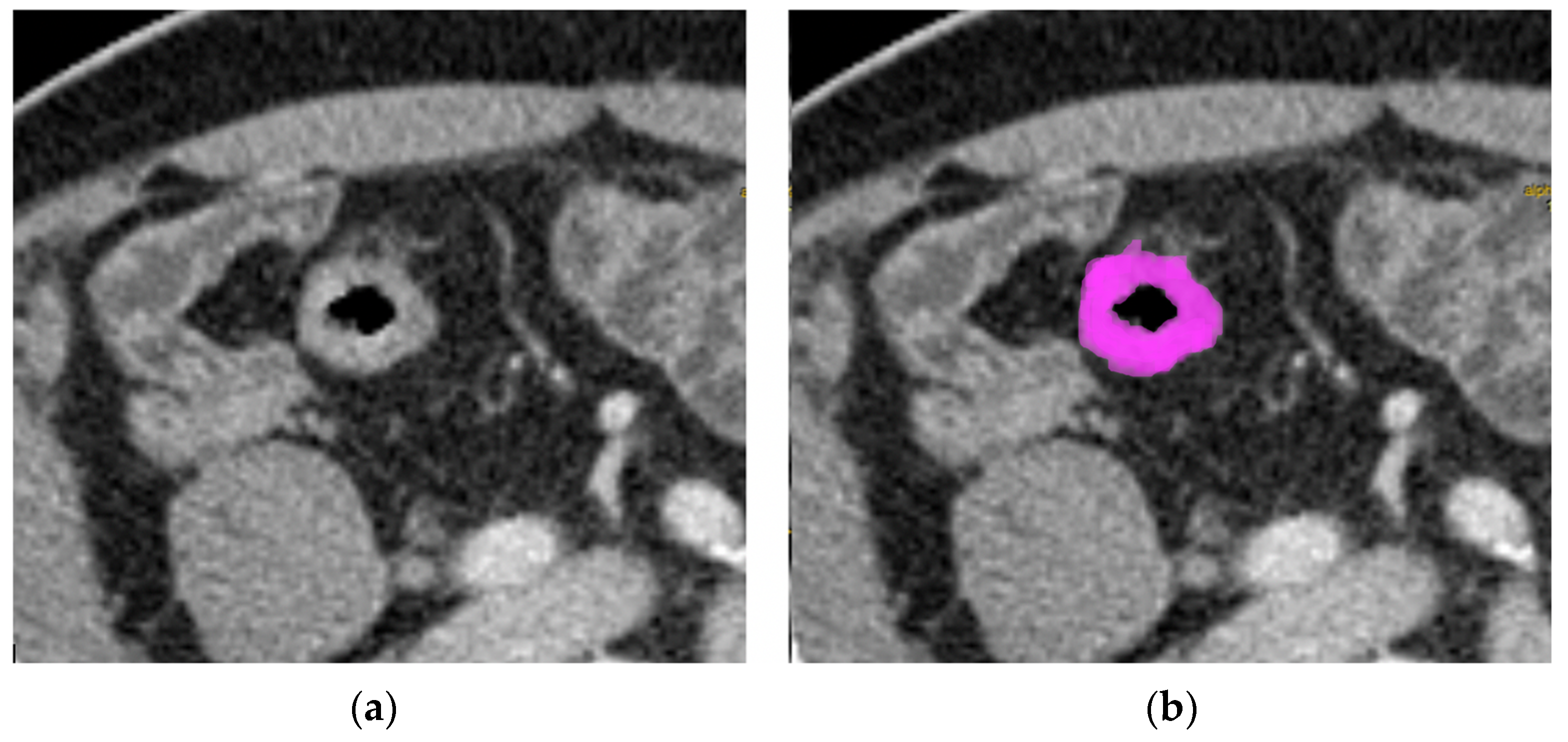
2.3. Texture Analysis
2.4. Statistical Analysis
3. Results
3.1. Patient Characteristics
3.2. Texture Values
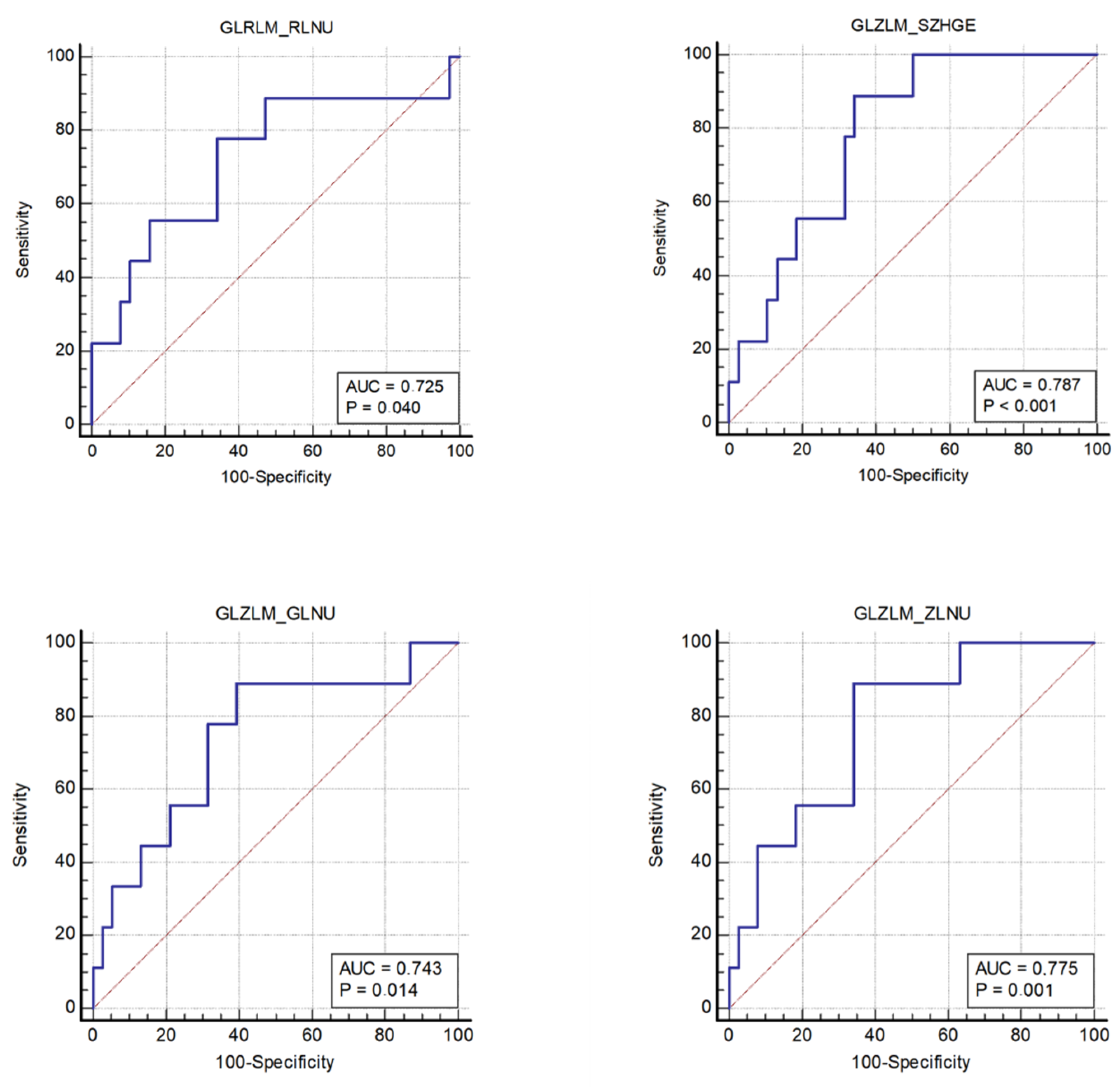
- The nonuniformity of the lengths of the homogeneous runs (GLRLM RLNU), which was significantly higher in patients with MSI (AUC 0.725, sensitivity 77.8%, specificity 65.8%);
- The distribution of the short homogeneous zones with high gray levels (GLZLM SZHGE), which was significantly lower in patients with MSI (AUC 0.787, sensitivity 88.9%, specificity 65.8%);
- The nonuniformity of the gray levels (GLZLM GLNU), which was significantly higher in patients with MSI (AUC 0.743, sensitivity 88.9%, specificity 60.5%);
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Rawla, P.; Sunkara, T.; Barsouk, A. Epidemiology of colorectal cancer: Incidence, mortality, survival, and risk factors. Gastroenterol. Rev. 2019, 14, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Valderrama-Treviño, A.I.; Barrera-Mera, B.; Ceballos-Villalva, J.C.; Montalvo-Javé, E.E. Hepatic metastasis from colorectal cancer. Euroasian J. Hepato-Gastroenterol. 2017, 7, 166–175. [Google Scholar]
- Murphy, P.; Petersen, G.; Thibodeau, S.; Fishel, R. Genetic testing for colon cancer: Joint statement of the American College of Medical Genetics and American Society of Human Genetics. Joint Test and Technology Transfer Committee Working Group. Genet. Med. 2000, 2, 362–366. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Boland, C.R.; Goel, A. Microsatellite instability in colorectal cancer. Gastroenterology 2010, 138, 2073–2087. [Google Scholar] [CrossRef]
- Xie, Y.-H.; Chen, Y.-X.; Fang, J.-Y. Comprehensive review of targeted therapy for colorectal cancer. Signal Transduct. Target. Ther. 2020, 5, 22. [Google Scholar]
- Afrăsânie, V.-A.; Marinca, M.V.; Alexa-Stratulat, T.; Gafton, B.; Păduraru, M.; Adavidoaiei, A.M.; Miron, L.; Rusu, C. KRAS, NRAS, BRAF, HER2 and microsatellite instability in metastatic colorectal cancer—Practical implications for the clinician. Radiol. Oncol. 2019, 53, 265–274. [Google Scholar] [CrossRef]
- Testa, U.; Pelosi, E.; Castelli, G. Colorectal cancer: Genetic abnormalities, tumor progression, tumor heterogeneity, clonal evolution and tumor-initiating cells. Med. Sci. 2018, 6, 31. [Google Scholar] [CrossRef]
- Kocak, B.; Durmaz, E.S.; Ates, E.; Kilickesmez, O. Radiomics with artificial intelligence: A practical guide for beginners. Diagn. Interv. Radiol. 2019, 25, 485–495. [Google Scholar] [CrossRef]
- Ognerubov, N.A.; Shatov, I.A.; Shatov, A.V. Radiogenomics and radiomics in the diagnostics of malignant tumours: A literary review. Tambov Univ. Rep. Ser. Nat. Tech. Sci. 2017, 22, 1453–1460. [Google Scholar] [CrossRef]
- Lubner, M.G.; Smith, A.D.; Sandrasegaran, K.; Sahani, D.V.; Pickhardt, P.J. CT texture analysis: Definitions, applications, biologic correlates, and challenges. RadioGraphics 2017, 37, 1483–1503. [Google Scholar] [CrossRef] [PubMed]
- Weiser, M.R. AJCC 8th edition: Colorectal cancer. Ann. Surg. Oncol. 2018, 25, 1454–1455. [Google Scholar] [CrossRef] [PubMed]
- Nioche, C.; Orlhac, F.; Boughdad, S.; Reuzé, S.; Goya-Outi, J.; Robert, C.; Pellot-Barakat, C.; Soussan, M.; Frouin, F.; Buvat, I. LIFEx: A freeware for radiomic feature calculation in multimodality imaging to accelerate advances in the characterization of tumor heterogeneity. Cancer Res. 2018, 78, 4786–4789. [Google Scholar] [CrossRef] [PubMed]
- Schirripa, M.; Cremolini, C.; Loupakis, F.; Morvillo, M.; Bergamo, F.; Zoratto, F.; Salvatore, F.; Antoniotti, C.; Marmorino, F.; Sensi, E.; et al. Role of NRAS mutations as prognostic and predictive markers in metastatic colorectal cancer. Int. J. Cancer 2015, 136, 83–90. [Google Scholar] [CrossRef] [PubMed]
- de Smedt, L.; Lemahieu, J.; Palmans, S.; Govaere, O.; Tousseyn, T.; Van Cutsem, E.; Prenen, H.; Tejpar, S.; Spaepen, M.; Matthijs, G.; et al. Microsatellite instable vs stable colon carcinomas: Analysis of tumour heterogeneity, inflammation and angiogenesis. Br. J. Cancer 2015, 113, 500–509. [Google Scholar] [CrossRef]
- Remo, A.; Fassan, M.; Vanoli, A.; Bonetti, L.R.; Barresi, V.; Tatangelo, F.; Gafà, R.; Giordano, G.; Pancione, M.; Grillo, F.; et al. Morphology and molecular features of rare colorectal carcinoma histotypes. Cancers 2019, 11, 1036. [Google Scholar] [CrossRef]
- Kawada, K.; Toda, K.; Nakamoto, Y.; Iwamoto, M.; Hatano, E.; Chen, F.; Hasegawa, S.; Togashi, K.; Date, H.; Uemoto, S.; et al. Relationship between 18F-FDG PET/CT scans and KRAS mutations in metastatic colorectal cancer. J. Nucl. Med. 2015, 56, 1322–1327. [Google Scholar] [CrossRef]
- Krikelis, D.; Skoura, E.; Kotoula, V.; Rondogianni, P.; Pianou, N.; Samartzis, A.; Xanthakis, I.; Fountzilas, G.; Datseris, I.E. Lack of association between KRAS mutations and 18F-FDG PET/CT in Caucasian metastatic colorectal cancer patients. Anticancer Res. 2014, 34, 2571–2579. [Google Scholar]
- Lovinfosse, P.; Koopmansch, B.; Lambert, F.; Jodogne, S.; Kustermans, G.; Hatt, M.; Visvikis, D.; Seidel, L.; Polus, M.; Albert, A.; et al. 18F-FDG PET/CT imaging in rectal cancer: Relationship with the RAS mutational status. Br. J. Radiol. 2016, 89, 1063. [Google Scholar] [CrossRef]
- Kim, S.-J.; Pak, K.; Kim, K. Diagnostic performance of F-18 FDG PET/CT for prediction of KRAS mutation in colorectal cancer patients: A systematic review and meta-analysis. Abdom. Radiol. 2019, 44, 1703–1711. [Google Scholar] [CrossRef]
- Taguchi, N.; Oda, S.; Yokota, Y.; Yamamura, S.; Imuta, M.; Tsuchigame, T.; Nagayama, Y.; Kidoh, M.; Nakaura, T.; Shiraishi, S.; et al. CT texture analysis for the prediction of KRAS mutation status in colorectal cancer via a machine learning approach. Eur. J. Radiol. 2019, 118, 38–43. [Google Scholar] [CrossRef] [PubMed]
- González-Castro, V.; Cernadas, E.; Huelga, E.; Fernández-Delgado, M.; Porto, J.; Antunez, J.R.; Souto-Bayarri, M. CT radiomics in colorectal cancer: Detection of KRAS mutation using texture analysis and machine learning. Appl. Sci. 2020, 10, 6214. [Google Scholar] [CrossRef]
- Yang, L.; Dong, D.; Fang, M.; Zhu, Y.; Zang, Y.; Liu, Z.; Zhang, H.; Ying, J.; Zhao, X.; Tian, J. Can CT-based radiomics signature predict KRAS/NRAS/BRAF mutations in colorectal cancer? Eur. Radiol. 2018, 28, 2058–2067. [Google Scholar] [CrossRef] [PubMed]
- Pernicka, J.S.G.; Gagniere, J.; Chakraborty, J.; Yamashita, R.; Nardo, L.; Creasy, J.M.; Petkovska, I.; Do, R.R.K.; Bates, D.D.B.; Paroder, V.; et al. Radiomics-based prediction of microsatellite instability in colorectal cancer at initial computed tomography evaluation. Abdom. Radiol. 2019, 44, 3755–3763. [Google Scholar]
- Fan, S.; Li, X.; Cui, X.; Zheng, L.; Ren, X.; Ma, W.; Ye, Z. Computed tomography-based radiomic features could potentially predict microsatellite instability status in stage II colorectal cancer: A preliminary study. Acad. Radiol. 2019, 26, 1633–1640. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, Q.; Zhao, Y.; Liu, Y.; Chen, A.; Li, X.; Wu, T.; Li, J.; Guo, Y.; Liu, A. Radiomics analysis of iodine-based material decomposition images with dual-energy computed tomography imaging for preoperatively predicting microsatellite instability status in colorectal cancer. Front. Oncol. 2019, 9, 1250. [Google Scholar] [CrossRef]
- Zhang, W.; Huang, Z.; Zhao, J.; He, D.; Li, M.; Yin, H.; Tian, S.; Zhang, H.; Song, B. Development and validation of magnetic resonance imaging-based radiomics models for preoperative prediction of microsatellite instability in rectal cancer. Ann. Transl. Med. 2021, 9, 134. [Google Scholar] [CrossRef]
- Ganeshan, B.; Miles, K.A. Quantifying tumour heterogeneity with CT. Cancer Imaging 2013, 13, 140–149. [Google Scholar] [CrossRef]
- Taieb, J.; Jung, A.; Sartore-Bianchi, A.; Peeters, M.; Seligmann, J.; Zaanan, A.; Burdon, P.; Montagut, C.; Laurent-Puig, P. The evolving biomarker landscape for treatment selection in metastatic colorectal cancer. Drugs 2019, 79, 1375–1394. [Google Scholar] [CrossRef]
- Lakatos, G.; Köhne, C.H.; Bodoky, G. Current therapy of advanced colorectal cancer according to RAS/RAF mutational status. Cancer Metastasis Rev. 2020, 39, 1143–1157. [Google Scholar]
| Characteristics | N (%) |
|---|---|
| Sex | |
| Males | 27 (57%) |
| Females | 20 (43%) |
| Age (years) | |
| Median | 70 |
| IQR | 26–87 |
| Body mass index (kg/m2) | |
| Median | 24.6 |
| IQR | 19.1–31.8 |
| Stages | |
| III | 16 (33%) |
| IV | 31 (66%) |
| Tumor locations | |
| Rectum-sigma | 18 (38%) |
| Ascending colon | 8 (17%) |
| Tranverse | 8 (17%) |
| Descending | 7 (15%) |
| Ciecum | 6 (13%) |
| Genetic mutations | |
| BRAF | 7 (15%) |
| KRAS | 18 (38%) |
| NRAS | 3 (6%) |
| MMR | 9 (19%) |
| Median (IQR) | p-Value | SE | SP | AUC (95% CI) | p | |
|---|---|---|---|---|---|---|
| Group 0 NRAS | Group 1 NRAS | |||||
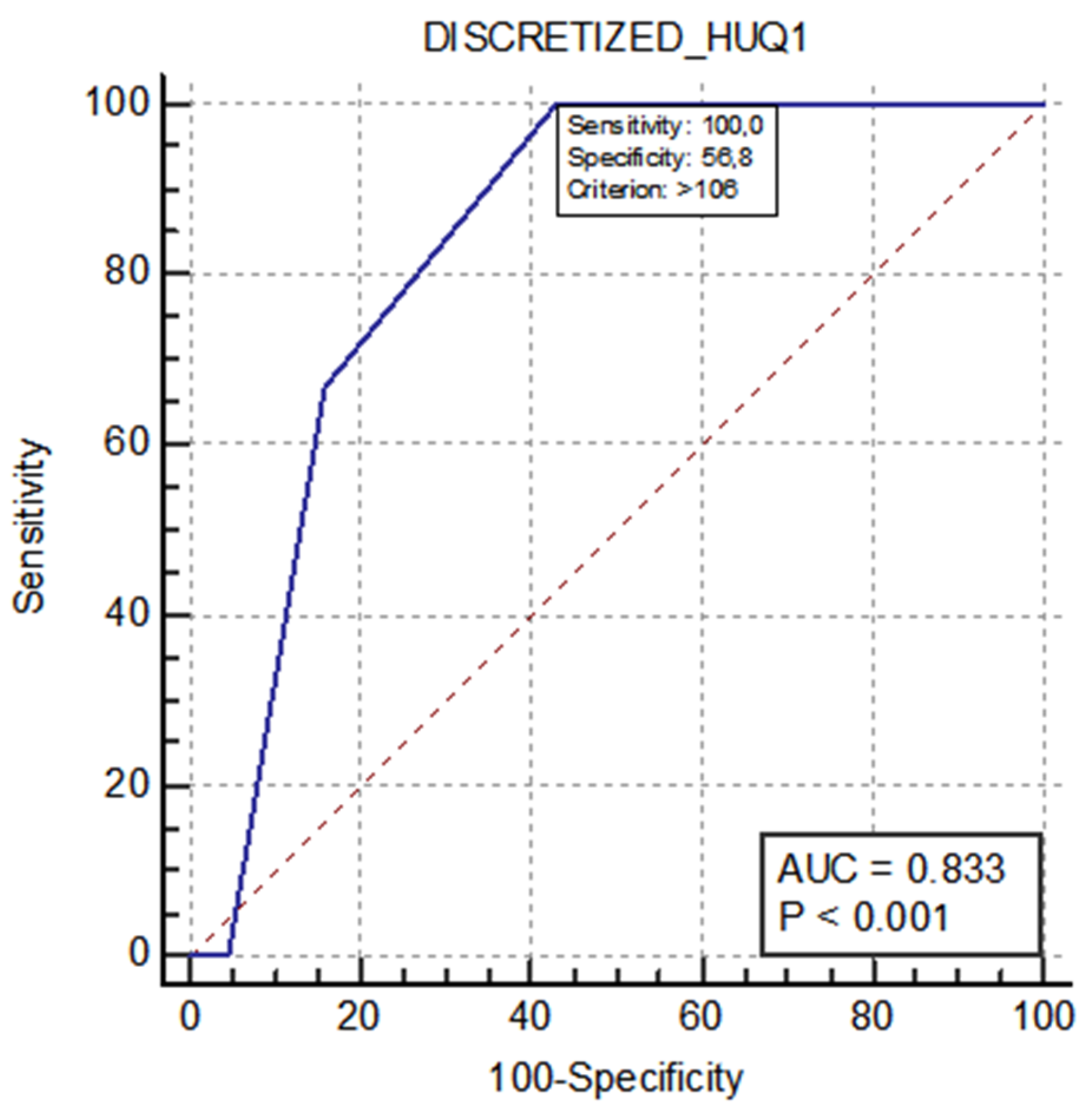
| 106 (105–107) | 108 | 0.049 | 100% | 56.8% | 0.833 (0.696–0.926) | <0.001 |
| CT-TA Parameters | Group 0 (MSS) Median (IQR) | Group 1 (MSI) Median (IQR) | p | YI | SE (%) | SP (%) | AUC (95% CI) | p |
|---|---|---|---|---|---|---|---|---|
| GLRLM RLNU | 4419 (2811–9267) | 11829 (5918–21721) | 0.037 | 0.44 | 77.8 | 65.8 | 0.725 (0.575–0.845) | 0.040 |
| GLZLM SZHGE | 7334 (7114–7457) | 7070 (6937–7192) | 0.0081 | 0.55 | 88.9 | 65.8 | 0.787 (0.643–0.892) | 0.001 |
| GLZLM GLNU | 97.39 (62.29–177.79) | 186.42 (133.31–290.23) | 0.025 | 0.49 | 88. 9 | 60.5 | 0.743 (0.594–0.859) | 0.014 |
| GLZLM ZLNU | 378.96 (304.16–763.17) | 920.71 (546.78–1378.71) | 0.011 | 0.55 | 88.9 | 65.8 | 0.775 (0.630–0.884) | 0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Crimì, F.; Zanon, C.; Cabrelle, G.; Luong, K.D.; Albertoni, L.; Bao, Q.R.; Borsetto, M.; Baratella, E.; Capelli, G.; Spolverato, G.; et al. Contrast-Enhanced CT Texture Analysis in Colon Cancer: Correlation with Genetic Markers. Tomography 2022, 8, 2193-2201. https://doi.org/10.3390/tomography8050184
Crimì F, Zanon C, Cabrelle G, Luong KD, Albertoni L, Bao QR, Borsetto M, Baratella E, Capelli G, Spolverato G, et al. Contrast-Enhanced CT Texture Analysis in Colon Cancer: Correlation with Genetic Markers. Tomography. 2022; 8(5):2193-2201. https://doi.org/10.3390/tomography8050184
Chicago/Turabian StyleCrimì, Filippo, Chiara Zanon, Giulio Cabrelle, Kim Duyen Luong, Laura Albertoni, Quoc Riccardo Bao, Marta Borsetto, Elisa Baratella, Giulia Capelli, Gaya Spolverato, and et al. 2022. "Contrast-Enhanced CT Texture Analysis in Colon Cancer: Correlation with Genetic Markers" Tomography 8, no. 5: 2193-2201. https://doi.org/10.3390/tomography8050184
APA StyleCrimì, F., Zanon, C., Cabrelle, G., Luong, K. D., Albertoni, L., Bao, Q. R., Borsetto, M., Baratella, E., Capelli, G., Spolverato, G., Fassan, M., Pucciarelli, S., & Quaia, E. (2022). Contrast-Enhanced CT Texture Analysis in Colon Cancer: Correlation with Genetic Markers. Tomography, 8(5), 2193-2201. https://doi.org/10.3390/tomography8050184










