Effect of Zinc Oxide Nanoparticles on Capsular Gene Expression in Klebsiella pneumoniae Isolated from Clinical Samples
Abstract
1. Introduction
2. Materials and Methods
2.1. ZnO NP Synthesis
2.2. Bacteria Isolation
2.3. The Coculture of ZnO NPs and Klebsiella Pneumoniae
2.4. Gene Expression
2.4.1. Total RNA Purification
- A—Preparation of the sample
- B—Purification protocol, MagPurix® series
2.4.2. Synthesis of cDNA from RNA Template Protocol
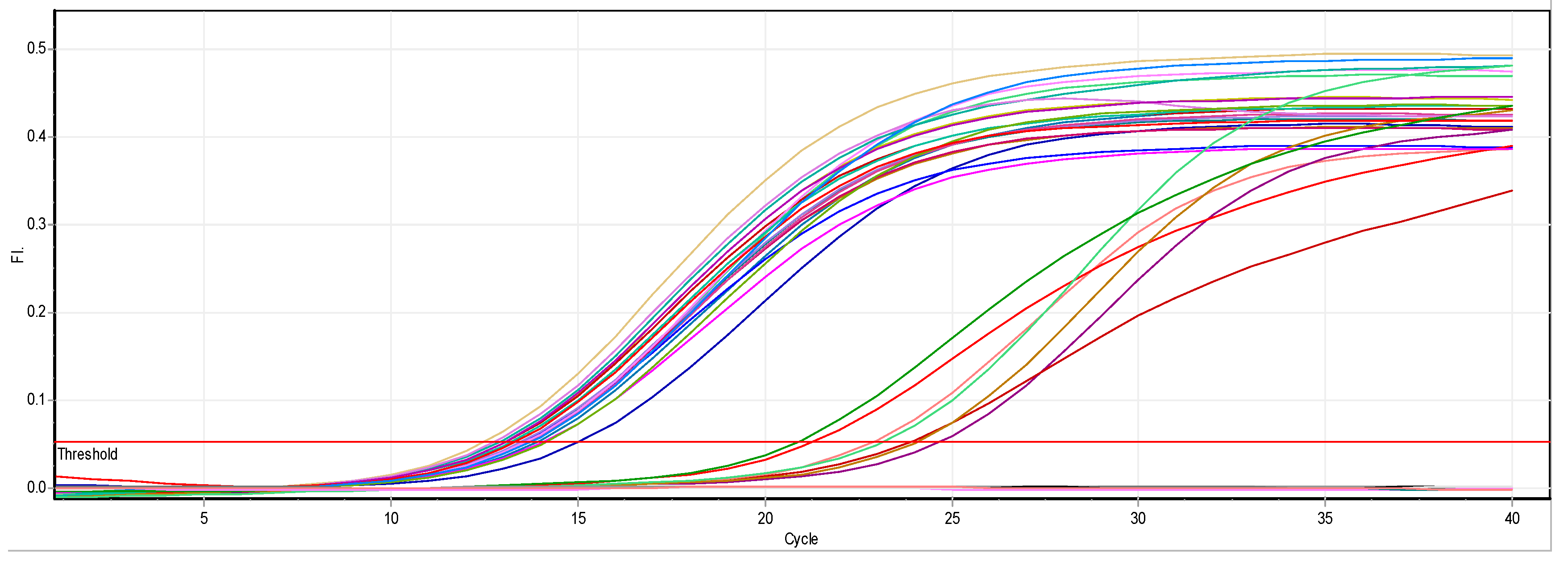
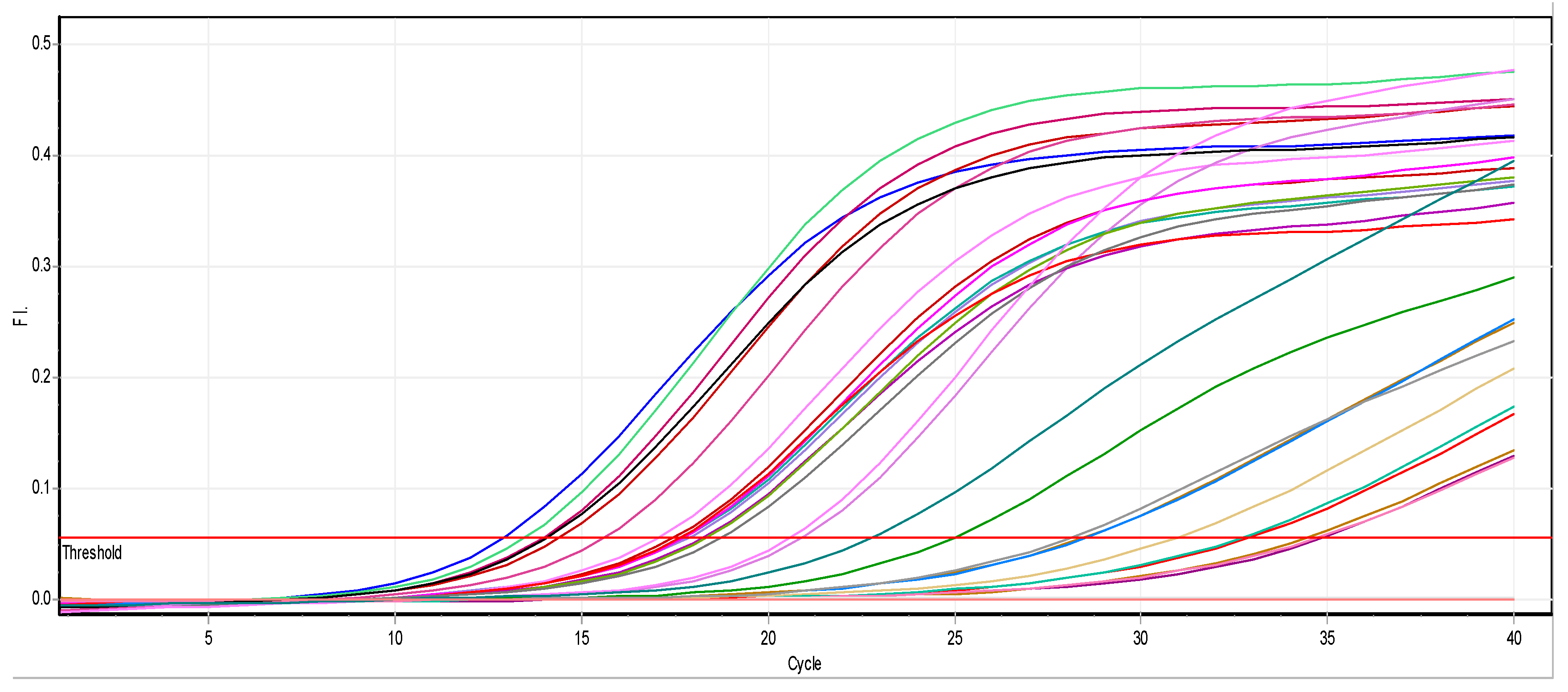
2.4.3. Real-Time Quantitative PCR Assays
- Reaction conditions
| Component | 20 μL Reaction | Final Conc. |
| qPCR Master (SYBR) | 10.0 μL | 1X |
| ROX Dye (50X) * (optional) | 0.4 μL | 1X |
| 10 μM Forward Primer | 0.2~2.0 μL | 0.1~1.0 μM |
| 10 μM Reverse Primer | 0.2~2.0 μL | 0.1~1.0 μM |
| Template DNA | Variable | ≤500 ng/reaction |
| Water, RNase-Free | up to 20 μL | NA |
- 2.
- PCR conditions
| Step | Temp (°C) | Time | Cycle |
| Initial Denaturation | 95 | 5 min | 1 |
| Denature | 95 | 10~30 s | 30~40 |
| Anneal | 55~68 | 10~60 s | 1 |
| Melting Curve Analysis | 65~95 | 2~5 s/step | Cycle |
- 3.
- In this study, we used four primers to detect the effect of ZnO NPs on gene expression. Primers were purchased from Bioneer/South Korea and applied in this work; the table below illustrates the primers and the detected target genes in Klebsiella spp. isolates:
| No. | Target Gene | Primer | Oligo Sequence (5′-3′) | Product Size (bp) | Ref. |
| 1 | magA | magA-F | GGT GCT CTT TAC ATC ATT GC | 1283 | (Turton et al., 2010) |
| magA-R | GCA ATG GCC ATT TGC GTT TGC GTT AG | ||||
| 2 | k2A | k2A-F | CAACCATGGTGGTCGATTAG | 543 | (Rivero et al., 2010) (Doud et al., 2009) |
| k2A-R | TGGTAGCCATATCCCTTTGG | ||||
| 3 | rmpA | rmpA-F | ACT GGG CTA CCT CTG CTT CA | 536 | (Nadasy et al., 2007) (Turton et al., 2010) |
| rmpA-R | CTT GCA TGA GCC ATC TTT CA | ||||
| 4 | kfu | kfu-F | GAAGTGACGCTGTTTCTGGC | 797 | (Yu et al., 2008) |
| kfu-R | TTTCGTGTGGCCAGTGACTC |
3. Results and Discussion
3.1. Antibiotic Resistance of K. pneumoniae Isolates
3.2. Impact of ZnO NPs on Capsular Gene Expression
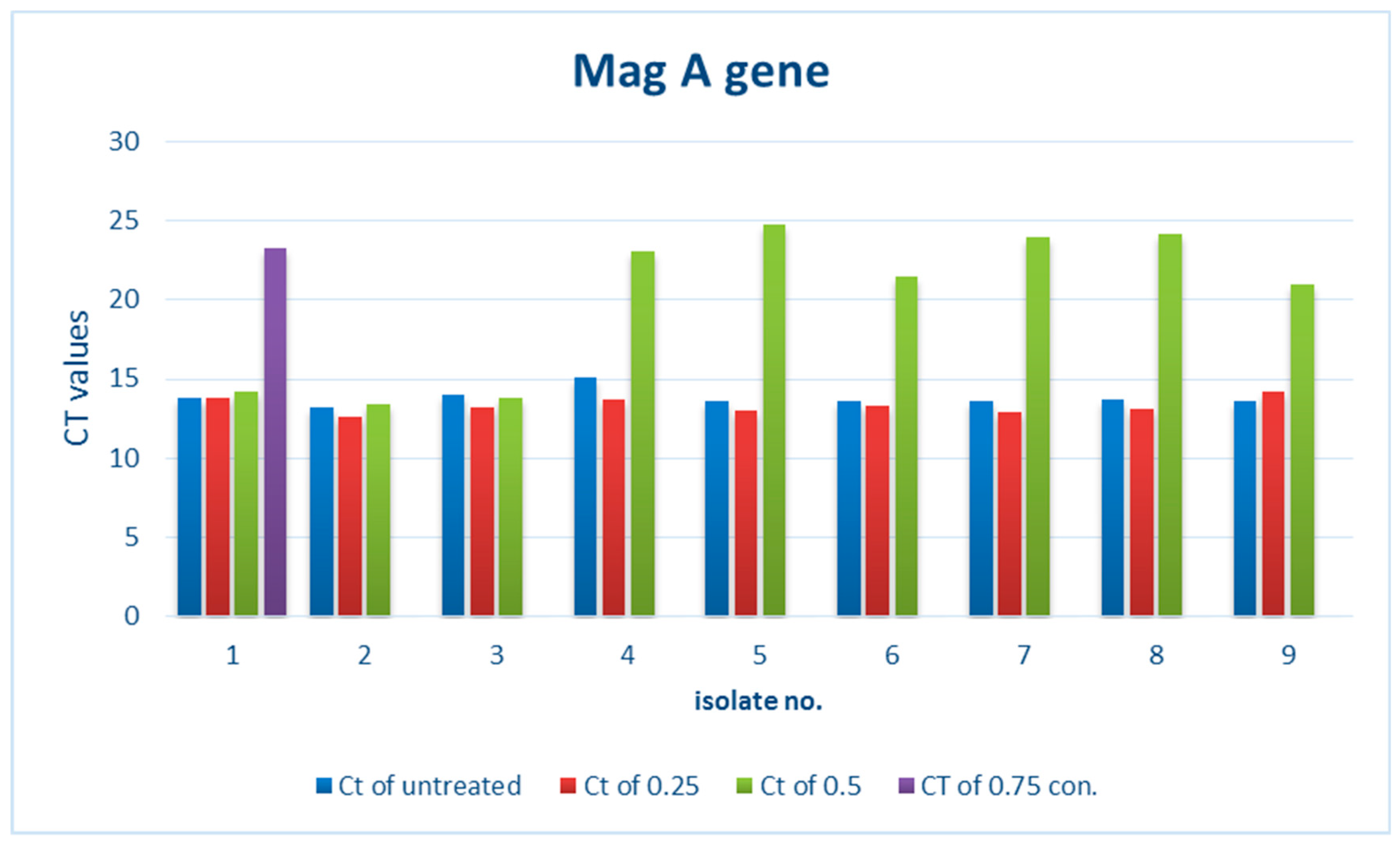
3.2.1. Effect of ZnO NPs on the Rate of magA Gene Expression
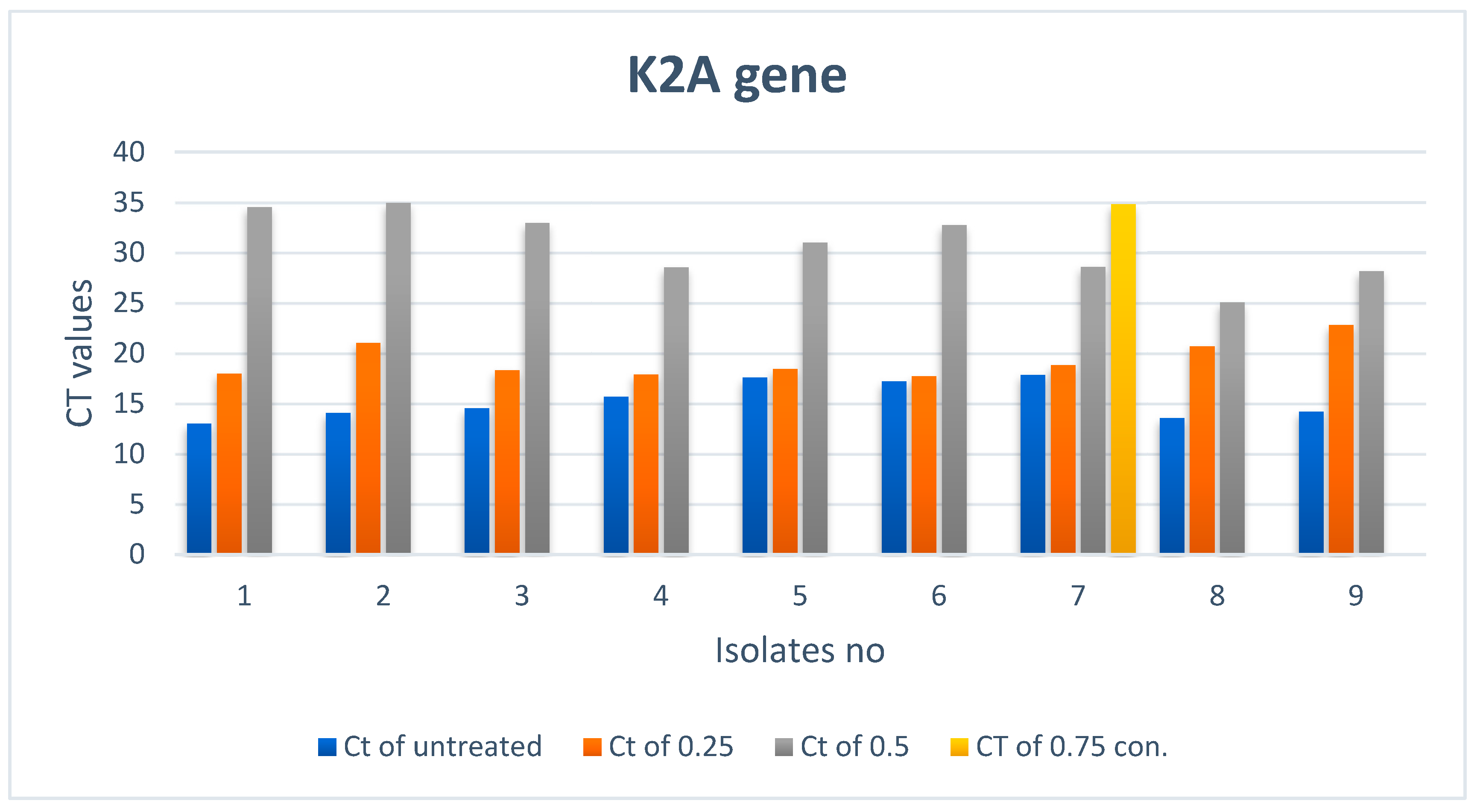
3.2.2. Effect of ZnO NPs on the Rate of k2A Gene Expression
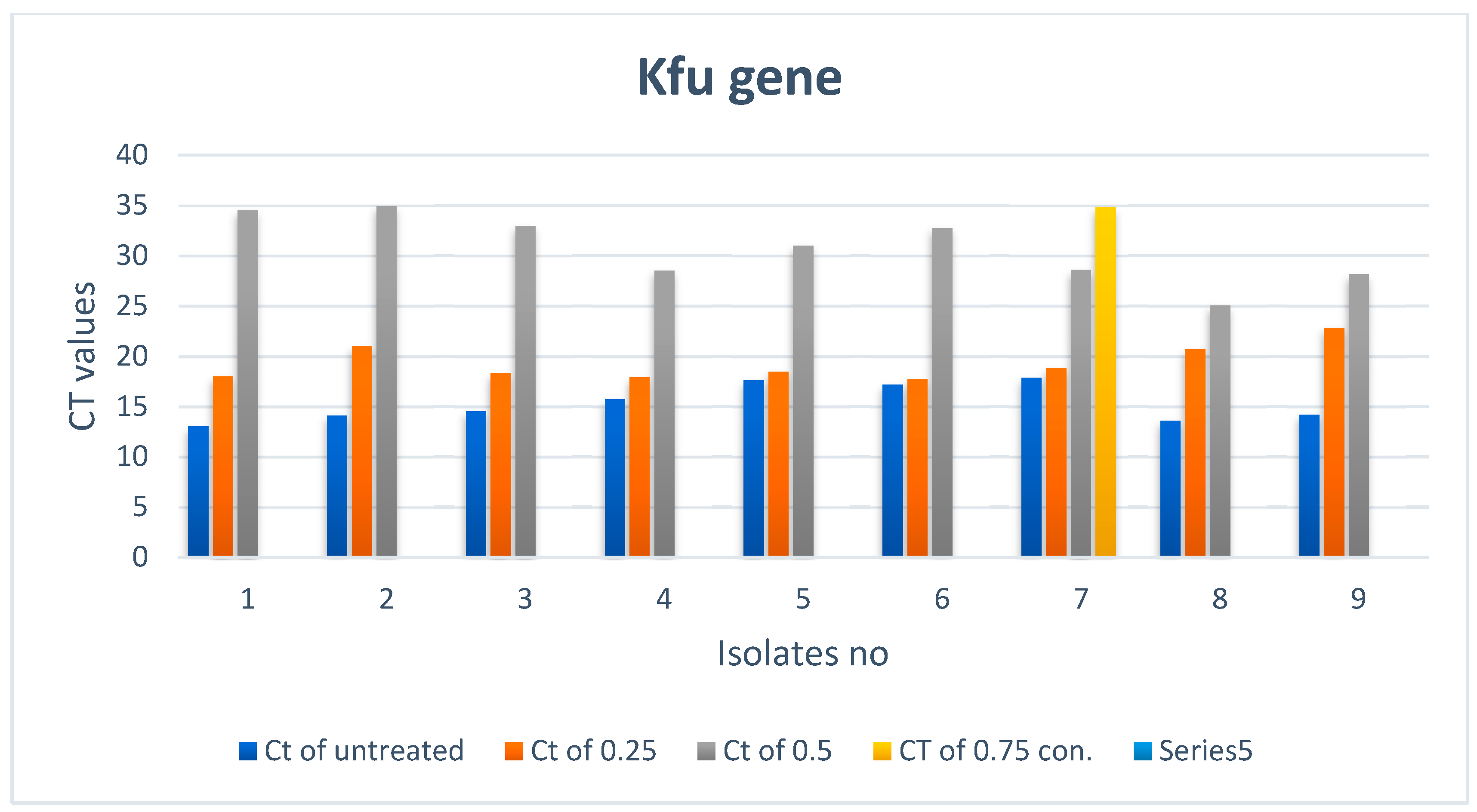
3.2.3. Effect of ZnO NPs on the Rate of kfu Gene Expression
3.2.4. Effect of Zinc Oxide on the Rate of the Gene Expression of rmpA
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chiu, S.; Wu, T.; Chuang, Y.; Lin, J.; Fung, C.; Lu, P.; Wang, J.; Wang, L.; Siu, K.; Yeh, K. National surveillance study on carbapenem Non-susceptible Klebsiella pneumoniae in taiwan: The emergence and rapid dissemination of kpc-2 carbapenemase. PLoS ONE 2013, 8, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Aher, T.; Roy, A.; Kumar, P. Molecular detection of virulence genes associated with pathogenicity of Klebsiella spp. isolated from the respiratory tract of apparently healthy as well as sick goats. J. Vet. Med. 2012, 67, 249–252. [Google Scholar]
- Dubey, D.; Raza, F.; Sawhney, A.; Pandey, A. Klebsiella pneumoniae renal abscess syndrome: A rare case with metastatic involvement of lungs, eye, and brain. Case Rep. Infect. Dis. 2013, 2013, 1–3. [Google Scholar]
- Fang, C.; Lin, Y.; Lin, J.; Chen, T.; Yeh, K.; Chang, F.; Chuang, H.; Wu, H.; Tseng, C.; Siu, L. Klebsiella pneumoniae in Gastrointestinal Tract and Pyogenic Liver Abscess. Emerg. Infect. Dis. 2012, 18, 1322–1325. [Google Scholar]
- Doud, M.; Zeppegno, R.; Molina, E.; Miller, N.; Balachandar, D.; Schneper, L.; Poppiti, R.; Mathee, K. A k2A-positive Klebsiella pneumoniae causes liver and brain abscess in a Saint Kitt’s man. Int. J. Med. Sci. 2009, 6, 301–304. [Google Scholar] [CrossRef]
- Turton, J.; Perry, C.; Elgohari, S.; Hampton, C. PCR characterization and typing of Klebsiella pneumoniae using capsular type-specific, variable number tandem repeat and virulence gene targets. J. Med. Microbiol. 2010, 59, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Azam, A.; Ahmed, A.; Oves, M.; Khan, M.; Memic, A. Size-dependent antimicrobial properties of CuO nanoparticles against Gram-positive and-negative bacterial strains. Int. J. Nanomed. 2012, 7, 3527–3535. [Google Scholar] [CrossRef]
- Padmavathy, N.; Vijayaraghavan, R. Enhanced Bioactivity of ZnO Nanoparticles an Antimicrobial Study. Sci. Technol. Adv. Mater. 2008, 9, 035004. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Pi, J.; Cai, J. The Advancing of Zinc Oxide Nanoparticles for Biomedical Application. Bioinorg. Chem. Appl. 2018, 2018, 1062562. [Google Scholar] [CrossRef]
- Ravishankar Rai, V.; Jamuna Bai, A. Nanoparticles and Their Potential Application as Antimicrobials. In Science against Microbial Pathogens: Communicating Current Research and Technological Advances; Méndez-Vilas, A., Ed.; Microbiology Series Formatex Research Center: Madrid, Spain, 2011; Volume 3, pp. 197–209. [Google Scholar]
- Brayner, R.; Ferrari-Iliou, R.; Brivois, N.; Djediat, S.; Benedetti, M.F.; Fievet, F. Toxicological impact studies based on Escherichia coli bacteria in ultrafine ZnO nanoparticles colloidal medium. Nano Lett. 2006, 6, 866–870. [Google Scholar] [CrossRef]
- Huang, Y.; Liao, H.; Wu, C.; Peng, H. MrkF is a component of type 3 fimbriae in Klebsiella pneumoniae. J. Res. Microbiol. 2009, 160, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Kudaer, N.B.; Yousif, E.; Risan, M.H.; Kadhom, M. Preparation of ZnO Nanoparticles by the Chemical Precipitation Method. J. Serambi Eng. 2022, 7, 3129–3134. [Google Scholar]
- Holt, J.G.; Krieg, N.R. Bergey’s Manual of Determinative Bacteriology, 9th ed.; Williams & Wilkins: Baltimore, MD, USA, 1994. [Google Scholar]
- Hickling, T.P.; Clark, H.; Malhotra, R.; Sim, R.B. Collectins and theirrole in lung immunity. J. Leukoc. Biol. 2004, 75, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Tullus, K. Fecal colonization with P-fimbriated Escherichia coli in newborn children and relation to development of extraintestinal E. coli infections. Acta Paediatr. Scand. 1987, 76, 1–35. [Google Scholar] [CrossRef]
- Stock, I.; Wiedemann, B. Natural antibiotic susceptibility of Klebsiella pneumoniae, K. oxytoca, K. planticola, K. ornithinolytica and K. terrigena strains. J. Med. Microbiol. 2001, 50, 396–406. [Google Scholar] [CrossRef]
- Schurr, J.R.; Young, E.; Byrne, P. Central role of toll-like receptor 4 signaling and host defense in experimental pneumonia caused by Gram-negative bacteria. Infect. Immun. 2005, 73, 532–545. [Google Scholar] [CrossRef]
- Huh, A.J.; Kwon, Y.J. Nanoantibiotics: A new paradigm for treating infectious diseases using nanomaterials in the antibiotics resistant era. J. Control Release 2011, 156, 128–145. [Google Scholar] [CrossRef]
- Mohammed, S.I.; Chandrasekaran, N.; Mukherjee, A. Studies on effect of TiO2 nanoparticles on growth and membrane permeability of Escherichia coli, Pseudomonas aeruginosa, and Bacillus subtilis. Curr. Nanosci. 2010, 6, 381–387. [Google Scholar] [CrossRef]
- Lingling, Z.; Yunhong, J.; Yulon, D. Investigation into the antibacterial behaviour of suspensions of ZnO nanoparticles (ZnO nanofluids). J. Nanopart. Res. 2007, 9, 479–489. [Google Scholar]
- Chung, Y.C.; Su, Y.P.; Chen, C.C. Relationship between antibacterial activity of chitosan and surface characteristics of cell wall. Acta Pharmacol. Sin. 2004, 25, 932–936. [Google Scholar]
- Berry, V.; Gole, A.; Kundu, S. Deposition of CTABterminated nanorods on bacteria to form highly conducting hybrid systems. J. Am. Chem. Soc. 2005, 125, 17600–17601. [Google Scholar] [CrossRef] [PubMed]
- Marsalek, R. Particle size and zeta potential of ZnO. APCBEE Proc. 2014, 9, 13–17. [Google Scholar] [CrossRef]
- Klotz, S. Role of hydrophobic interactions in microbial adhesion to plastics used in medical devices. In Microbial Cell Surface Hydrophobicity; Doyle, R., Rosenberg, M., Eds.; American Society for Microbiology: Washington, DC, USA, 1990; p. 107. [Google Scholar]
- Keisari, Y.; Wang, H.; Mesika, A. Surfactant protein D-coated Klebsiella pneumoniae stimulates cytokine production in mononuclear phagocytes. J. Leukoc. Biol. 2001, 70, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Jalal, R.; Goharshadi, E.; Abareshi, M.; Moosavi, M.; Yousefi, A.; Nancarrow, P. ZnO nanofluids: Green synthesis, characterization, and antibacterial activity. Mater. Chem. Phys. 2010, 121, 198–201. [Google Scholar] [CrossRef]
- Raghupathi, K.; Koodali, R.; Manna, A. Size-Dependent Bacterial Growth Inhibition and Mechanism of Antibacterial Activity of Zinc Oxide Nanoparticles. Langmuir 2011, 27, 4020–4028. [Google Scholar] [CrossRef]

| Antibiotic | Resistant% | Intermediate% | Sensitive% |
|---|---|---|---|
| Piperacillin | 74% | 4% | 22% |
| Cefazolin | 84% | 0% | 16% |
| Cefoxitin | 55% | 0% | 45% |
| Ceftazidime | 63% | 10% | 27% |
| Ceftriaxone | 100% | 0% | 0% |
| Cefepime | 40% | 0% | 60% |
| Imipenem | 21% | 6% | 73% |
| Amikacin | 10% | 10% | 80% |
| Gentamicin | 35% | 5% | 60% |
| Ciprofloxacine | 35% | 0% | 65% |
| Levofloxacin | 34% | 0% | 66% |
| Tigecyclin | 0% | 0% | 100% |
| Nitrofurantoin | 16% | 42% | 42% |
| Trimethoprim | 75% | 0% | 25% |
| Ampicillin | 100% | 0% | 0% |
| Amoxicillin | 97% | 0% | 3% |
| Cephalothin | 100% | 0% | 0% |
| Cephradine | 100% | 0% | 0% |
| Treated Bacteria Isolates with Different con. | Untreated | |||||||
|---|---|---|---|---|---|---|---|---|
| No | Ct of con3 (0.75) | Color | Ct of con2 (0.5) | Color | Ct of con1 (0.25) | Color | Ct | Color |
| 1 | 23.27 |  | 14.24 |  | 13.78 |  | 13.79 |  |
| 2 | Not expressed |  | 13.43 |  | 12.63 |  | 13.2 |  |
| 3 | Not expressed |  | 13.84 |  | 13.2 |  | 13.99 |  |
| 4 | Not expressed |  | 23.01 |  | 13.75 |  | 15.13 |  |
| 5 | Not expressed |  | 24.75 |  | 13.04 |  | 13.65 |  |
| 6 | Not expressed |  | 21.42 |  | 13.32 |  | 13.61 |  |
| 7 | Not expressed |  | 23.96 |  | 12.9 |  | 13.64 |  |
| 8 | Not expressed |  | 24.15 |  | 13.14 |  | 13.69 |  |
| 9 | Not expressed |  | 20.99 |  | 14.18 |  | 13.6 |  |
| Treated Bacteria Isolates with Different con. | Untreated | |||||||
|---|---|---|---|---|---|---|---|---|
| Ct of con3 (0.75) | Color | Ct of con2 (0.5) | Color | Ct of con1 (0.25) | Color | Ct | Color | |
| 1 | Not expressed |  | 34.51 |  | 17.98 |  | 13 |  |
| 2 | Not expressed |  | 34.95 |  | 21.04 |  | 14.09 |  |
| 3 | Not expressed |  | 32.97 |  | 18.33 |  | 14.53 |  |
| 4 | Not expressed |  | 28.53 |  | 17.88 |  | 15.7 |  |
| 5 | Not expressed |  | 31.01 |  | 18.46 |  | 17.58 |  |
| 6 | Not expressed |  | 32.76 |  | 17.72 |  | 17.19 |  |
| 7 | 34.8 |  | 28.59 |  | 18.82 |  | 17.86 |  |
| 8 | Not expressed |  | 25.04 |  | 20.7 |  | 13.58 |  |
| 9 | Not expressed |  | 28.15 |  | 22.83 |  | 14.19 |  |
| Treated Bacteria Isolates with Different con. | Untreated | |||||||
|---|---|---|---|---|---|---|---|---|
| No | Ct of con3 (0.75) | Color | Ct of con2 (0.5) | Color | Ct of con1 (0.25) | Color | Ct | Color |
| 1 | Not expressed |  | 34.51 |  | 17.98 |  | 13 |  |
| 2 | Not expressed |  | 34.95 |  | 21.04 |  | 14.09 |  |
| 3 | Not expressed |  | 32.97 |  | 18.33 |  | 14.53 |  |
| 4 | Not expressed |  | 28.53 |  | 17.88 |  | 15.7 |  |
| 5 | Not expressed |  | 31.01 |  | 18.46 |  | 17.58 |  |
| 6 | Not expressed |  | 32.76 |  | 17.72 |  | 17.19 |  |
| 7 | 34.8 |  | 28.59 |  | 18.82 |  | 17.86 |  |
| 8 | Not expressed |  | 25.04 |  | 20.7 |  | 13.58 |  |
| 9 | Not expressed |  | 28.15 |  | 22.83 |  | 14.19 |  |
| Treated Bacteria Isolates with Different con. | Untreated | |||||||
|---|---|---|---|---|---|---|---|---|
| No | Ct of con3 (0.75) | Color | Ct of con2 (0.5) | Color | Ct of con1 (0.25) | Color | Ct | Color |
| 1 | 22.19 |  | 22.18 |  | 20.73 |  | 14.83 |  |
| 2 | Not expressed |  | 22.63 |  | 18.9 |  | 14.93 |  |
| 3 | Not expressed |  | 23.22 |  | 19.35 |  | 13.24 |  |
| 4 | Not expressed |  | 23.24 |  | 20.4 |  | 13.95 |  |
| 5 | Not expressed |  | 19.36 |  | 19.51 |  | 14.28 |  |
| 6 | Not expressed |  | 23.39 |  | 19.65 |  | 13.89 |  |
| 7 | Not expressed |  | 24.29 |  | 19.59 |  | 18.88 |  |
| 8 | Not expressed |  | 22.48 |  | 19.68 |  | 14.06 |  |
| 9 | Not expressed |  | 22.82 |  | 20.25 |  | 14.44 |  |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kudaer, N.B.; Risan, M.H.; Yousif, E.; Kadhom, M.; Raheem, R.; Salman, I. Effect of Zinc Oxide Nanoparticles on Capsular Gene Expression in Klebsiella pneumoniae Isolated from Clinical Samples. Biomimetics 2022, 7, 180. https://doi.org/10.3390/biomimetics7040180
Kudaer NB, Risan MH, Yousif E, Kadhom M, Raheem R, Salman I. Effect of Zinc Oxide Nanoparticles on Capsular Gene Expression in Klebsiella pneumoniae Isolated from Clinical Samples. Biomimetics. 2022; 7(4):180. https://doi.org/10.3390/biomimetics7040180
Chicago/Turabian StyleKudaer, Nuha B., Mohseen H. Risan, Emad Yousif, Mohammed Kadhom, Rasha Raheem, and Israa Salman. 2022. "Effect of Zinc Oxide Nanoparticles on Capsular Gene Expression in Klebsiella pneumoniae Isolated from Clinical Samples" Biomimetics 7, no. 4: 180. https://doi.org/10.3390/biomimetics7040180
APA StyleKudaer, N. B., Risan, M. H., Yousif, E., Kadhom, M., Raheem, R., & Salman, I. (2022). Effect of Zinc Oxide Nanoparticles on Capsular Gene Expression in Klebsiella pneumoniae Isolated from Clinical Samples. Biomimetics, 7(4), 180. https://doi.org/10.3390/biomimetics7040180








