Metabolomic and Transcriptomic Analysis Reveals the Mechanisms Underlying the Difference in Anthocyanin Accumulation in Apple Fruits at Different Altitudes
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Determination of Total Anthocyanin Content
2.3. RNA Sequencing and Analysis
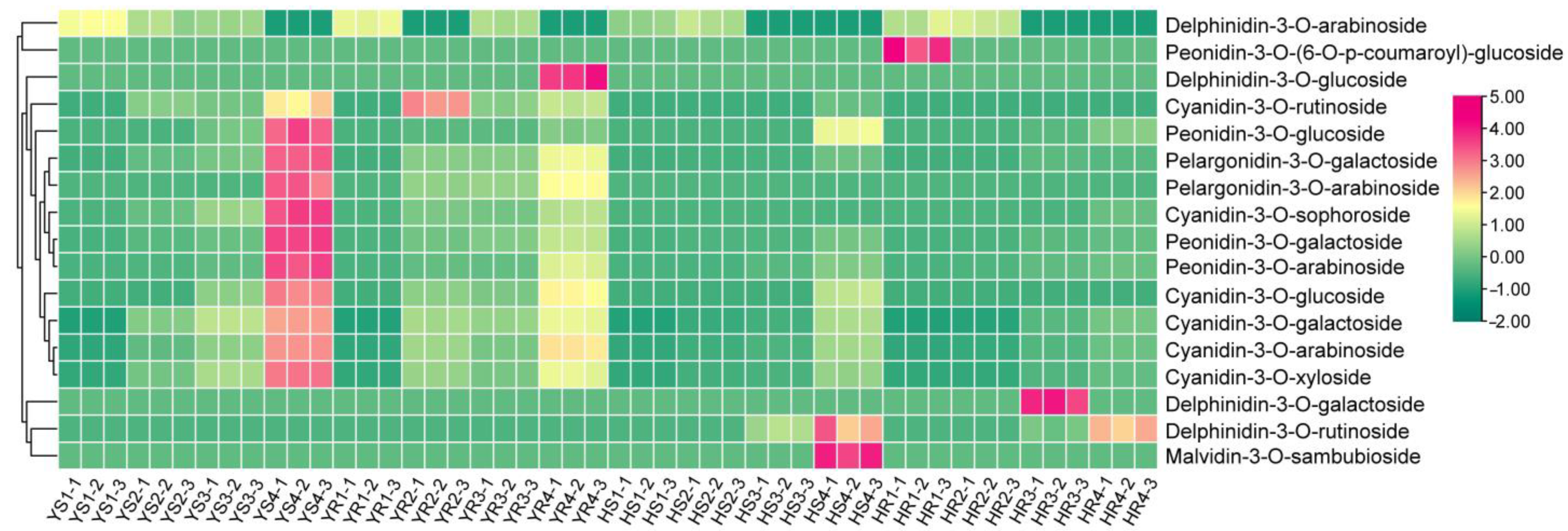
2.4. Measurement of Anthocyanin Metabolites Using HPLC-ESI-MS/MS
2.5. WGCNA and Visualization of Gene Networks
2.6. Quantitative Real-Time PCR (qRT-PCR)
2.7. Function Prediction of Candidate Genes
2.8. Statistical Analyses
3. Results
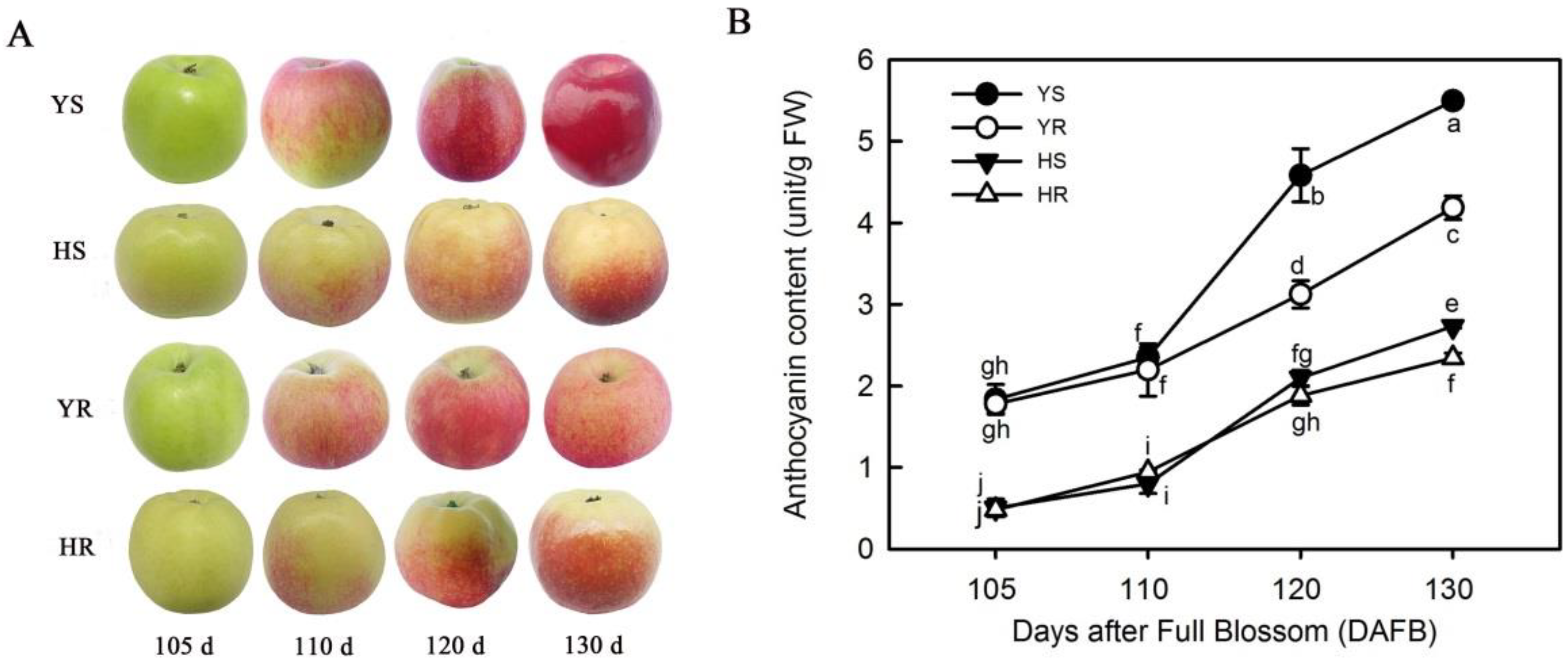
3.1. Relative Content of Anthocyanin in Different Varieties
3.2. Transcriptome Sequencing and Analysis
3.3. WGCNA of Anthocyanin Content and Transcriptome Data
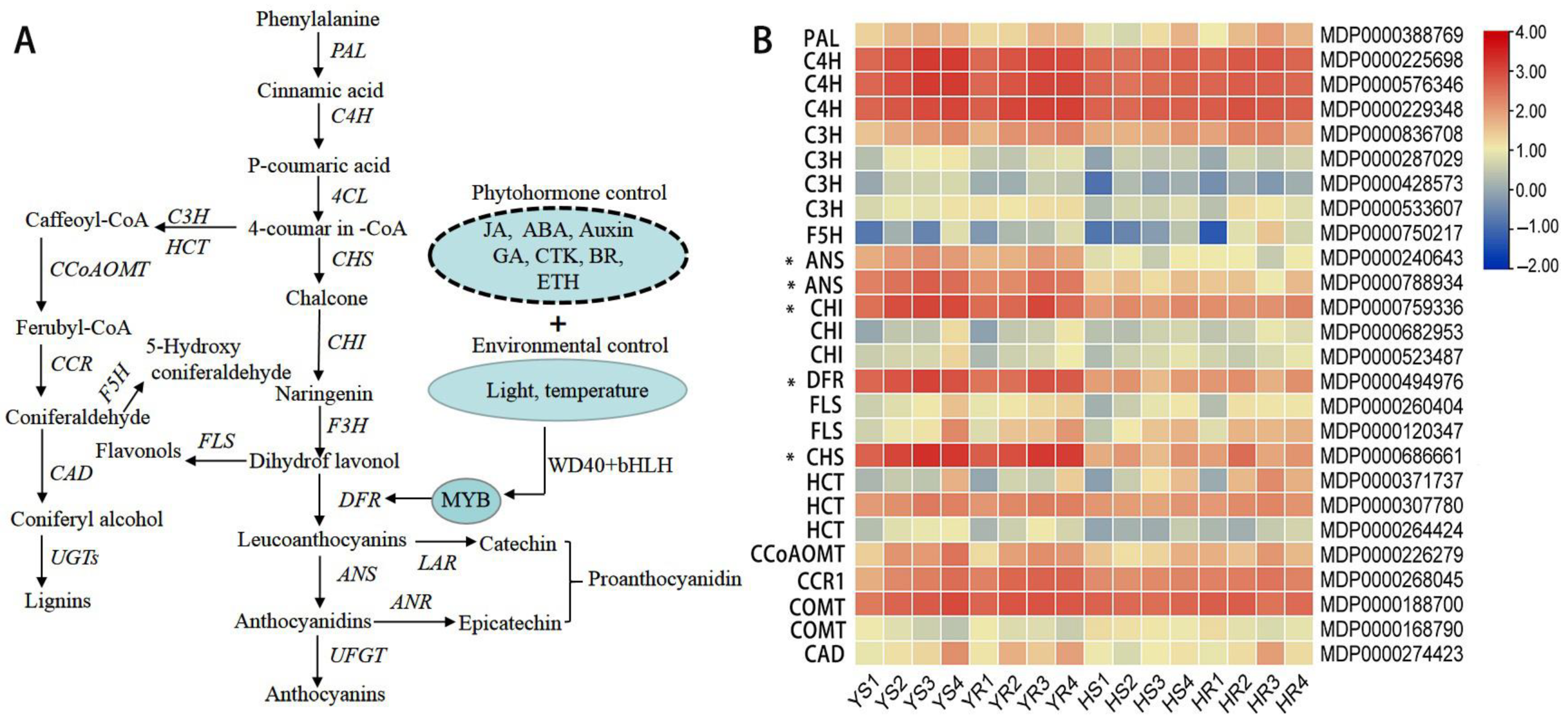
3.4. Expression of Genes Associated with the Regulation of Anthocyanin and Phenylpropanoid Metabolism
3.5. Key Transcription Factors and Structural Genes Associated with Anthocyanin Accumulation and Their Interaction Network
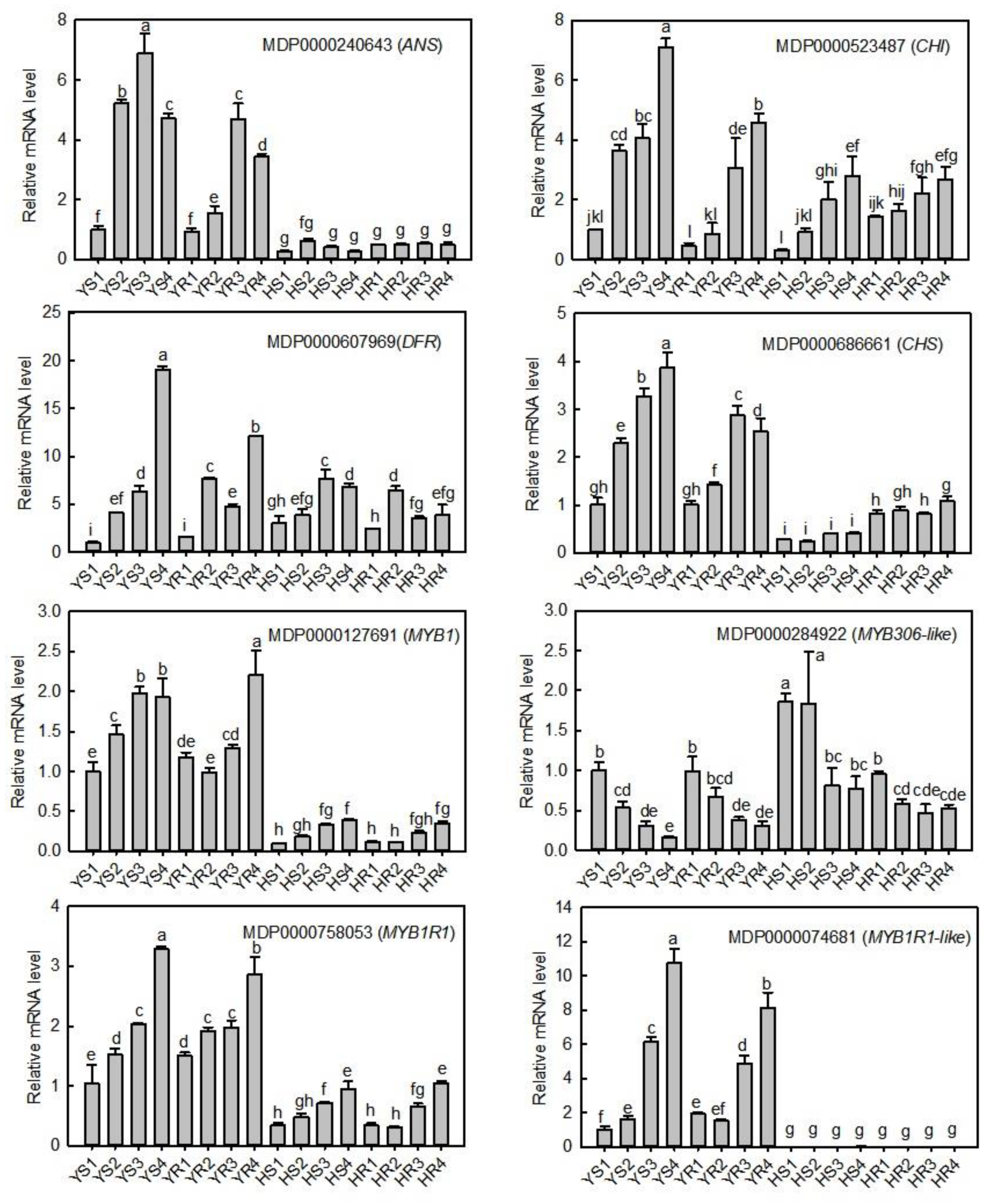
3.6. qRT-PCR Verification of the Expression Levels of Candidate Structural Genes and TFs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tsurunaga, Y.; Takahashi, T.; Katsube, T.; Kudo, A.; Kuramitsu, O.; Ishiwata, M.; Matsumoto, S. Effects of UV-B irradiation on the levels of anthocyanin, rutin and radical scavenging activity of buckwheat sprouts. Food Chem. 2013, 141, 552–556. [Google Scholar] [CrossRef] [PubMed]
- Sivankalyani, V.; Feygenberg, O.; Diskin, S.; Wright, B.; Alkan, N. Increased anthocyanin and flavonoids in mango fruit peel are associated with cold and pathogen resistance. Postharvest Biol. Technol. 2016, 111, 132–139. [Google Scholar] [CrossRef]
- An, J.P.; Zhang, X.W.; Bi, S.Q.; You, C.X.; Wang, X.F.; Hao, Y.J. The ERF transcription factor MdERF38 promotes drought stress-induced anthocyanin biosynthesis in apple. Plant J. 2020, 101, 573–589. [Google Scholar] [CrossRef] [PubMed]
- Lev-Yadun, S.; Gould, K.S. Role of anthocyanins in plant defence. In Anthocyanins; Springer: New York, NY, USA, 2008; pp. 22–28. [Google Scholar]
- Zhang, Y.; Butelli, E.; Martin, C. Engineering anthocyanin biosynthesis in plants. Curr. Opin. Plant Biol. 2014, 19, 81–90. [Google Scholar] [CrossRef]
- Shang, Y.; Wang, W.; Zhu, P.; Ye, Y.; Dai, P.; Zhao, W.; Wang, Y. Anthocyanins: Novel antioxidants in diseases prevention and human health. In Flavonoids—A Coloring Model for Cheering Up Life; IntechOpen: London, UK, 2019; pp. 1–16. [Google Scholar]
- Cakar, U.D.; Petrovic, A.V.; Živković, M.; Vajs, V.; Milovanovic, M.M.; Zeravik, J.; Djordjević, B. Phenolic profile of some fruit wines and their antioxidant properties. Hem. Ind. 2016, 70, 661–672. [Google Scholar] [CrossRef]
- Butelli, E.; Titta, L.; Giorgio, M.; Mock, H.P.; Matros, A.; Peterek, S.; Schijlen, E.G.; Hall, R.D.; Bovy, A.G.; Luo, J. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 2008, 26, 1301–1308. [Google Scholar] [CrossRef]
- Pojer, E.; Mattivi, F.; Johnson, D.; Stockley, C.S. The case for anthocyanin consumption to promote human health: A review. Compr. Rev. Food Sci. Food Saf. 2013, 12, 483–508. [Google Scholar] [CrossRef]
- Fragoso, M.F.; Romualdo, G.R.; Vanderveer, L.A.; Franco-Barraza, J.; Cukierman, E.; Clapper, M.L.; Carvalho, R.F.; Barbisan, L.F. Lyophilized açaí pulp (Euterpe oleracea Mart) attenuates colitis-associated colon carcinogenesis while its main anthocyanin has the potential to affect the motility of colon cancer cells. Food Chem. Toxicol. 2018, 121, 237–245. [Google Scholar] [CrossRef]
- Boyer, J.; Liu, R.H. Apple phytochemicals and their health benefits. Nutr. J. 2004, 3, 5. [Google Scholar] [CrossRef]
- Duan, W.; Sun, P.; Li, J. Expression of genes involved in the anthocyanin biosynthesis pathway in white and red fruits of fragaria pentaphylla and genetic variation in the dihydroflavonol-4-reductase gene. Biochem. Syst. Ecol. 2017, 72, 40–46. [Google Scholar] [CrossRef]
- Gao, H.N.; Jiang, H.; Cui, J.Y.; You, C.X.; Li, Y.Y. Review: The effects of hormones and environmental factors on anthocyanin biosynthesis in apple. Plant Sci. Int. J. Exp. Plant Biol. 2021, 312, 111024. [Google Scholar] [CrossRef] [PubMed]
- Kondo, S.; Hiraoka, K.; Kobayashi, S.; Honda, C.; Terahara, N. Changes in the expression of anthocyanin biosynthetic genes during apple development. J. Am. Soc. Hortic. Sci. Jashs 2002, 127, 971–976. [Google Scholar] [CrossRef]
- Takos, A.M.; Jaffe, F.W.; Jacob, S.R.; Bogs, J.; Robinson, S.P.; Walker, A.R. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 2006, 142, 1216–1232. [Google Scholar] [CrossRef]
- Ma, C.; Liang, B.; Chang, B.; Yan, J.; Liu, L.; Wang, Y.; Yang, Y.; Zhao, Z. Transcriptome profiling of anthocyanin biosynthesis in the peel of ‘granny smith’ apples (Malus domestica) after bag removal. BMC Genom. 2019, 20, 353. [Google Scholar] [CrossRef]
- Lin-Wang, K.; Micheletti, D.; Palmer, J.; Volz, R.; Lozano, L.; Espley, R.; Hellens, R.P.; Chagne, D.; Rowan, D.D.; Troggio, M.; et al. High temperature reduces apple fruit colour via modulation of the anthocyanin regulatory complex. Plant Cell Environ. 2011, 34, 1176–1190. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Fang, H.; Wang, J.; Yue, X.; Su, M.; Mao, Z.; Zou, Q.; Jiang, H.; Guo, Z.; Yu, L.; et al. Ultraviolet b-induced MdWRKY72 expression promotes anthocyanin synthesis in apple. Plant Sci. Int. J. Exp. Plant Biol. 2020, 292, 110377. [Google Scholar] [CrossRef]
- Hu, Y.; Cheng, H.; Zhang, Y.; Zhang, J.; Niu, S.; Wang, X.; Li, W.; Zhang, J.; Yao, Y. The MdMYB16/MdMYB1-miR7125-MdCCR module regulates the homeostasis between anthocyanin and lignin biosynthesis during light induction in apple. New Phytol. 2021, 231, 1105–1122. [Google Scholar] [CrossRef]
- Yu, J.; Qiu, K.; Sun, W.; Yang, T.; Wu, T.; Song, T.; Zhang, J.; Yao, Y.; Tian, J. A long non-coding RNA functions in high-light-induced anthocyanin accumulation in apple by activating ethylene synthesis. Plant Physiol. 2022, 189, 66–83. [Google Scholar] [CrossRef]
- An, J.P.; Qu, F.J.; Yao, J.F.; Wang, X.N.; You, C.X.; Wang, X.F.; Hao, Y.J. The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic. Res. 2017, 4, 17023. [Google Scholar] [CrossRef]
- Bai, S.; Saito, T.; Honda, C.; Hatsuyama, Y.; Ito, A.; Moriguchi, T. An apple B-BOX protein, MdCOL11, is involved in UV-B- and temperature-induced anthocyanin biosynthesis. Planta 2014, 240, 1051–1062. [Google Scholar] [CrossRef]
- Fang, H.; Dong, Y.; Yue, X.; Chen, X.; He, N.; Hu, J.; Jiang, S.; Xu, H.; Wang, Y.; Su, M.; et al. MdCOL4 interaction mediates crosstalk between UV-B and high temperature to control fruit coloration in apple. Plant Cell Physiol. 2019, 60, 1055–1066. [Google Scholar] [CrossRef] [PubMed]
- Ubi, B.E.; Honda, C.; Bessho, H.; Kondo, S.; Wada, M.; Kobayashi, S.; Moriguchi, T. Expression analysis of anthocyanin biosynthetic genes in apple skin: Effect of UV-B and temperature. Plant Sci. 2006, 170, 571–578. [Google Scholar] [CrossRef]
- Wang, N.; Zhang, Z.Y.; Jiang, S.H.; Xu, H.F.; Wang, Y.C.; Feng, S.Q.; Chen, X.S. Synergistic effects of light and temperature on anthocyanin biosynthesis in callus cultures of red-fleshed apple (Malus sieversii f. niedzwetzkyana). Plant Cell Tissue Org. 2016, 127, 217–227. [Google Scholar] [CrossRef]
- Fang, H.; Dong, Y.; Yue, X.; Hu, J.; Jiang, S.; Xu, H.; Wang, Y.; Su, M.; Zhang, J.; Zhang, Z.; et al. The B-box zinc finger protein MdBBX20 integrates anthocyanin accumulation in response to ultraviolet radiation and low temperature. Plant Cell Environ. 2019, 42, 2090–2104. [Google Scholar] [CrossRef]
- Charles, M.; Corollaro, M.L.; Manfrini, L.; Endrizzi, I.; Aprea, E.; Zanella, A.; Corelli Grappadelli, L.; Gasperi, F. Application of a sensory–instrumental tool to study apple texture characteristics shaped by altitude and time of harvest. J. Sci. Food Agric. 2018, 98, 1095–1104. [Google Scholar] [CrossRef]
- Yan, Z.; Zhang, H.; Zhang, R.; Liu, Z.; Guo, G. Performance of new apple variety ‘huashuo’ in 14 producing areas in China. China Fruits 2016, 90–94. [Google Scholar] [CrossRef]
- Shi, C.; Liu, L.; Wei, Z.; Liu, J.; Li, M.; Yan, Z.; Gao, D. Anthocyanin accumulation and molecular analysis of correlated genes by metabolomics and transcriptomics in sister line apple cultivars. Life 2022, 12, 1246. [Google Scholar] [CrossRef]
- Pirie, A.; Mullins, M.G. Changes in anthocyanin and phenolics content of grapevine leaf and fruit tissues treated with sucrose, nitrate, and abscisic acid. Plant Physiol. 1976, 58, 468–472. [Google Scholar] [CrossRef]
- Xu, H.; Zou, Q.; Yang, G.; Jiang, S.; Fang, H.; Wang, Y.; Zhang, J.; Zhang, Z.; Wang, N.; Chen, X. MdMYB6 regulates anthocyanin formation in apple both through direct inhibition of the biosynthesis pathway and through substrate removal. Hortic. Res. 2020, 7, 72. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, J.; Peng, Z.; Shi, M.; Liu, X.; Wen, H.; Jiang, Y.; Cheng, Y.; Xu, J.; Zhang, H. Integrated transcriptomic and metabolomic analysis reveals a transcriptional regulation network for the biosynthesis of carotenoids and flavonoids in ’cara cara’ navel orange. BMC Plant Biol 2021, 21, 29. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. Htseq—A python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Wei, T.; Simko, V. R Package “Corrplot”: Visualization of a Correlation Matrix, Version 0.84. 2017. Available online: https://github.com/taiyun/corrplot (accessed on 15 February 2022).
- Chen, C.; Chen, H.; He, Y.; Xia, R. TBtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. bioRxiv 2018, bioRxiv:289660. [Google Scholar]
- Langfelder, P.; Mehrabian, M.; Schadt, E.E.; Lusis, A.J.; Horvath, S. Weighted gene co-expression network analysis of adipose and liver reveals gene modules related to plasma HDL levels and containing candidate genes at loci identified in genome wide association studies. Am. Heart Assoc. 2008, 118, 327. [Google Scholar]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Franceschini, A.; Szklarczyk, D.; Frankild, S.; Kuhn, M.; Simonovic, M.; Roth, A.; Lin, J.; Minguez, P.; Bork, P.; Von Mering, C. String v9. 1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2012, 41, D808–D815. [Google Scholar] [CrossRef]
- Honda, C.; Moriya, S. Anthocyanin biosynthesis in apple fruit. Hortic. J. 2018, 87, 305–314. [Google Scholar] [CrossRef]
- Wu, B.H.; Cao, Y.G.; Guan, L.; Xin, H.P.; Li, J.H.; Li, S.H. Genome-wide transcriptional profiles of the berry skin of two red grape cultivars (Vitis vinifera) in which anthocyanin synthesis is sunlight-dependent or -independent. PLoS ONE 2014, 9, e105959. [Google Scholar] [CrossRef] [PubMed]
- Honda, C.; Kotoda, N.; Wada, M.; Kondo, S.; Kobayashi, S.; Soejima, J.; Zhang, Z.; Tsuda, T.; Moriguchi, T. Anthocyanin biosynthetic genes are coordinately expressed during red coloration in apple skin. Plant Physiol. Biochem. 2002, 40, 955–962. [Google Scholar] [CrossRef]
- Gleitz, J.; Seitz, H.U. Induction of chalcone synthase in cell suspension cultures of carrot (Daucus carota L. spp. Sativus) by ultraviolet light: Evidence for two different forms of chalcone synthase. Planta 1989, 179, 323–330. [Google Scholar] [PubMed]
- Lancaster, J.; Dougall, D.K. Regulation of skin color in apples. Crit. Rev. Plant Sci. 1992, 10, 487–502. [Google Scholar] [CrossRef]
- Xu, F.; Cao, S.; Shi, L.; Chen, W.; Su, X.; Yang, Z. Blue light irradiation affects anthocyanin content and enzyme activities involved in postharvest strawberry fruit. J. Agric. Food Chem. 2014, 62, 4778–4783. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, X.; Yang, S.; Yuan, Y. Lignin involvement in programmed changes in peach-fruit texture indicated by metabolite and transcriptome analyses. J. Agric. Food Chem. 2018, 66, 12627–12640. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.H.; Cho, J.S.; Bae, E.K.; Choi, Y.I.; Eom, S.H.; Lim, Y.J.; Lee, H.; Park, E.J.; Ko, J.H. PtrMYB120 functions as a positive regulator of both anthocyanin and lignin biosynthetic pathway in a hybrid poplar. Tree Physiol. 2021, 41, 2409–2423. [Google Scholar] [CrossRef]
- Huang, L.N.; Wu, G.B.; Zhang, S.; Kuang, F.Y.; Chen, F.H. The identification and functional verification of the cinnamate 4-hydroxylase gene from wax apple fruit and its role in lignin biosynthesis during nitric oxide-delayed postharvest cottony softening. Postharvest Biol. Technol. 2019, 158, 110964. [Google Scholar] [CrossRef]
- Ring, L.; Yeh, S.Y.; Hucherig, S.; Hoffmann, T.; Blanco-Portales, R.; Fouche, M.; Villatoro, C.; Denoyes, B.; Monfort, A.; Caballero, J.L.; et al. Metabolic interaction between anthocyanin and lignin biosynthesis is associated with peroxidase FaPRX27 in strawberry fruit. Plant Physiol. 2013, 163, 43–60. [Google Scholar] [CrossRef]
- Li, Y.; Nie, P.; Zhang, H.; Wang, L.; Wang, H.; Zhang, L. Dynamic changes of anthocyanin accumulation and endogenous hormone contents in blueberry. J. Beijing For. Univ. 2017, 39, 64–71. [Google Scholar]
- Li, W.F.; Mao, J.; Yang, S.J.; Guo, Z.G.; Ma, Z.H.; Dawuda, M.M.; Zuo, C.W.; Chu, M.Y.; Chen, B.H. Anthocyanin accumulation correlates with hormones in the fruit skin of ‘red delicious’ and its four generation bud sport mutants. BMC Plant Biol. 2018, 18, 363. [Google Scholar] [CrossRef]
- Figueroa, N.E.; Gatica-Meléndez, C.; Figueroa, C.R. Ethylene application at the immature stage of fragaria chiloensis fruit represses the anthocyanin biosynthesis with a concomitant accumulation of lignin. Food Chem. 2021, 358, 129913. [Google Scholar] [CrossRef]
- Shen, X.; Zhao, K.; Liu, L.; Zhang, K.; Yuan, H.; Liao, X.; Wang, Q.; Guo, X.; Li, F.; Li, T. A role for PacMYBA in ABA-regulated anthocyanin biosynthesis in red-colored sweet cherry cv. Hong deng (Prunus avium L.). Plant Cell Physiol. 2014, 55, 862–880. [Google Scholar] [CrossRef]
- Cheng, J.; Yu, K.; Shi, Y.; Wang, J.; Duan, C. Transcription factor VviMYB86 oppositely regulates proanthocyanidin and anthocyanin biosynthesis in grape berries. Front. Plant Sci. 2021, 11, 2263. [Google Scholar] [CrossRef]
- Wei, X.; Ju, Y.; Ma, T.; Zhang, J.; Fang, Y.; Sun, X. New perspectives on the biosynthesis, transportation, astringency perception and detection methods of grape proanthocyanidins. Crit. Rev. Food Sci. Nutr. 2021, 61, 2372–2398. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.; Wang, H. Two homologous ATP-binding cassette transporter proteins, AtMDR1 and AtPGP1, regulate Arabidopsis photomorphogenesis and root development by mediating polar auxin transport. Plant Physiol. 2005, 138, 949–964. [Google Scholar] [CrossRef] [PubMed]
- Ban, Y.; Honda, C.; Hatsuyama, Y.; Igarashi, M.; Bessho, H.; Moriguchi, T. Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol. 2007, 48, 958–970. [Google Scholar] [CrossRef]
- Espley, R.V.; Hellens, R.P.; Putterill, J.; Stevenson, D.E.; Kutty-Amma, S.; Allan, A.C. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2007, 49, 414–427. [Google Scholar] [CrossRef] [PubMed]
- Chagné, D.; Lin-Wang, K.; Espley, R.V.; Volz, R.K.; How, N.M.; Rouse, S.; Brendolise, C.; Carlisle, C.M.; Kumar, S.; De Silva, N. An ancient duplication of apple MYB transcription factors is responsible for novel red fruit-flesh phenotypes. Plant Physiol. 2013, 161, 225–239. [Google Scholar] [CrossRef] [PubMed]
- An, X.H.; Tian, Y.; Chen, K.Q.; Liu, X.J.; Liu, D.D.; Xie, X.B.; Cheng, C.G.; Cong, P.H.; Hao, Y.J. MdMYB9 and MdMYB11 are involved in the regulation of the JA-induced biosynthesis of anthocyanin and proanthocyanidin in apples. Plant Cell Physiol. 2015, 56, 650–662. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Wang, C.; Zhang, W.; Liu, S.; Wang, W.; Yu, X.; Song, T.; Yu, M.; Yu, W.; Qu, S. The R2R3-type MYB transcription factor MdMYB90-like is responsible for the enhanced skin color of an apple bud sport mutant. Hortic. Res. 2021, 8, 156. [Google Scholar] [CrossRef]
- Xu, H.; Wang, N.; Liu, J.; Qu, C.; Wang, Y.; Jiang, S.; Lu, N.; Wang, D.; Zhang, Z.; Chen, X. The molecular mechanism underlying anthocyanin metabolism in apple using the MdMYB16 and MdbHLH33 genes. Plant Mol. Biol. 2017, 94, 149–165. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, Z.; Li, L.X.; Wang, H.B.; Zhou, H.; Chen, X.S.; Feng, S.Q. Apple MdMYB306-like inhibits anthocyanin synthesis by directly interacting with MdMYB17 and MdbHLH33. Plant J. Cell Mol. Biol. 2022, 110, 1021–1034. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, C.; Wei, Z.; Liu, L.; Li, M.; Liu, J.; Gao, D. Metabolomic and Transcriptomic Analysis Reveals the Mechanisms Underlying the Difference in Anthocyanin Accumulation in Apple Fruits at Different Altitudes. Horticulturae 2023, 9, 475. https://doi.org/10.3390/horticulturae9040475
Shi C, Wei Z, Liu L, Li M, Liu J, Gao D. Metabolomic and Transcriptomic Analysis Reveals the Mechanisms Underlying the Difference in Anthocyanin Accumulation in Apple Fruits at Different Altitudes. Horticulturae. 2023; 9(4):475. https://doi.org/10.3390/horticulturae9040475
Chicago/Turabian StyleShi, Caiyun, Zhifeng Wei, Li Liu, Ming Li, Junwei Liu, and Dengtao Gao. 2023. "Metabolomic and Transcriptomic Analysis Reveals the Mechanisms Underlying the Difference in Anthocyanin Accumulation in Apple Fruits at Different Altitudes" Horticulturae 9, no. 4: 475. https://doi.org/10.3390/horticulturae9040475
APA StyleShi, C., Wei, Z., Liu, L., Li, M., Liu, J., & Gao, D. (2023). Metabolomic and Transcriptomic Analysis Reveals the Mechanisms Underlying the Difference in Anthocyanin Accumulation in Apple Fruits at Different Altitudes. Horticulturae, 9(4), 475. https://doi.org/10.3390/horticulturae9040475




