Bioassessment of Phylogenetic Relatedness and Plant Growth Enhancement of Endophytic Bacterial Isolates from Cowpea (Vigna unguiculata) Plant Tissues
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cowpea (Vigna unguiculata L. Walps) Sampling/Collection
2.1.1. Isolation of Endophytic Microbiota from Leaves, Seeds and Near Root Tissue Zones of Cowpea
2.1.2. Genomic DNA Extraction, PCR of Plant Growth Promoting (PGP) Genes and 16S rRNA Gene Sequencing
2.1.3. Molecular Authentication and Phylogenetic Assessment
2.1.4. An In Vitro Assay of the Endophytic Bacterial Plant Growth-Stimulating Attributes
Potency for Solubilizing Inorganic Phosphate
Potency of Bacterial Isolates to Produce Indole Acetic Acid
Potency of Endophytic Bacterial Isolates to Produce Siderophore
Potency of Endophytic Bacteria Isolates to Promote ACC Deaminase Activity (ACCD)
Potency of Endophytic Bacteria Isolates to Promote Ammonia Production
Potency of Endophytic Bacteria Isolates to Promote Exopolysaccharide Production
Potency of Endophytic Bacteria Isolates to Promote Hydrogen Cyanide Production
2.1.5. Seed Germination and Seedling Growth
2.1.6. Potentials of Endophytic Bacterial Isolates to Inhibit Phytopathogenic Fungal Activity
2.1.7. Potentials of Endophytic Bacterial Isolates to Tolerate Environmental Stress
3. Results
3.1. Morpho-Genotypical Authentication
3.2. Plant Growth-Stimulating Attributes of Endophytic Bacterial Strains
3.3. Tolerance to Environmental Stress
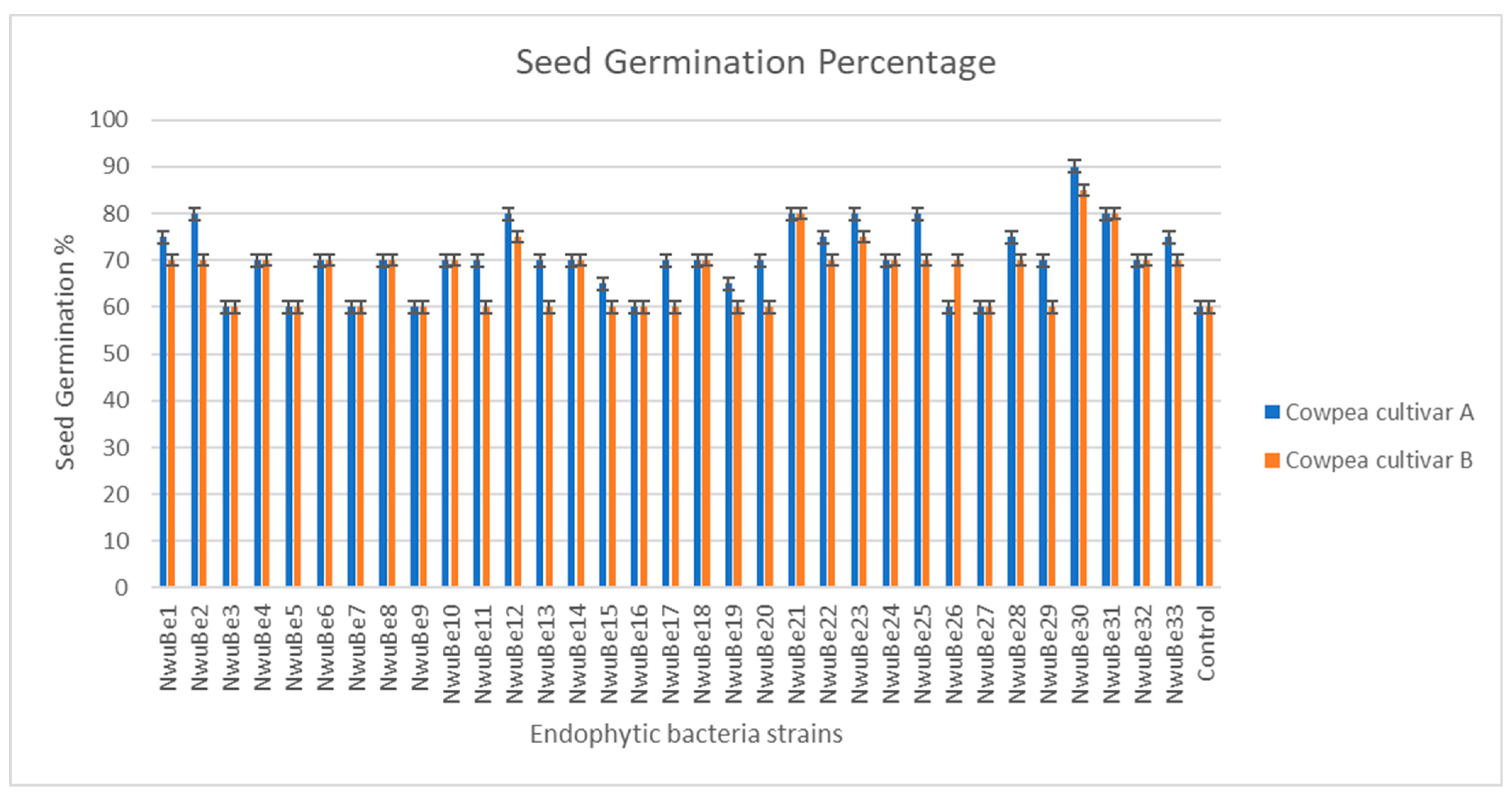
3.4. Influence of Seed Biopriming of Endophytic Bacterial Strains on Cowpea Seed Germination
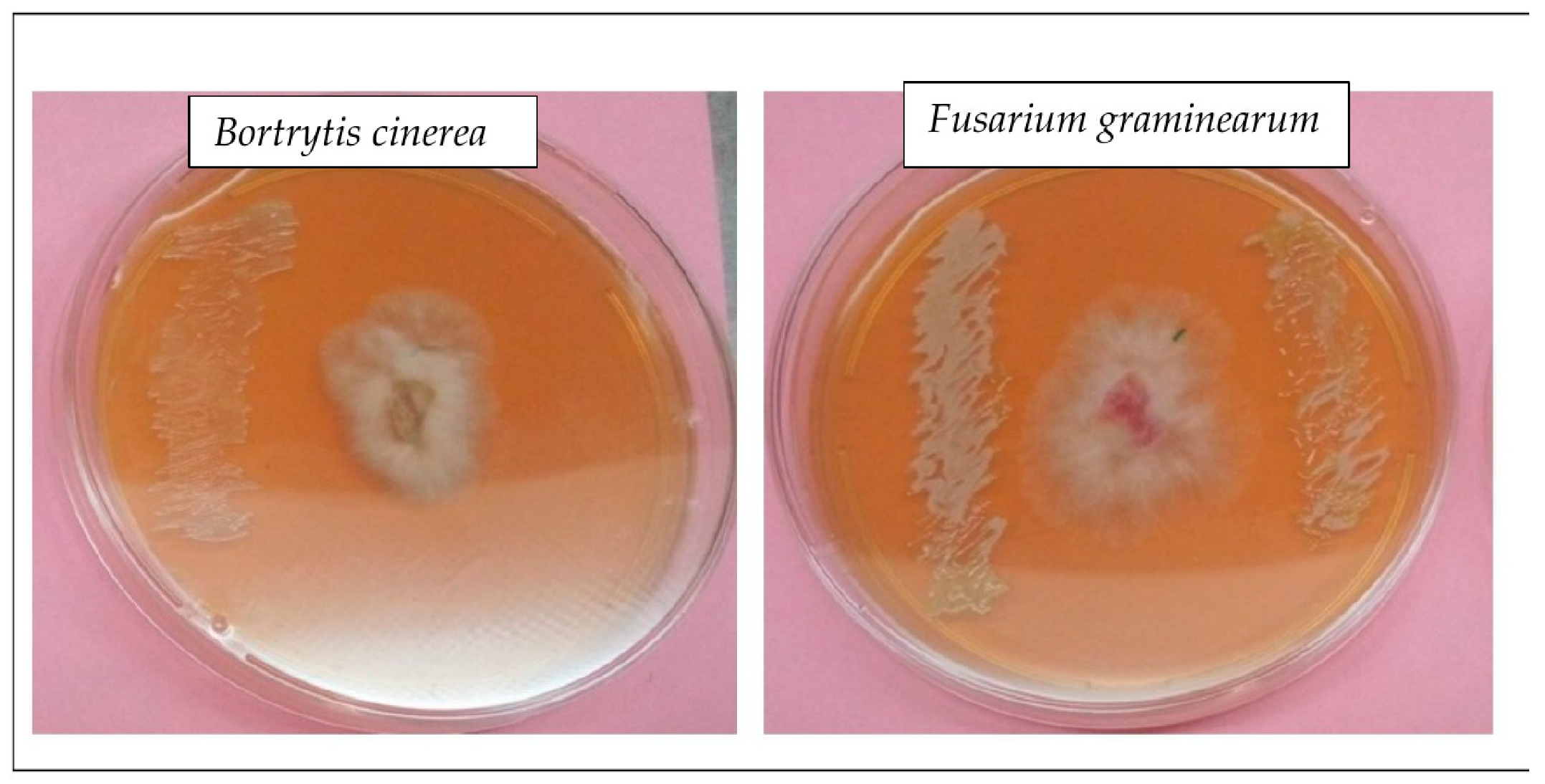
3.5. Endophytic Bacterial Strains Inhibitory Action against Phytopathogenic Fungi
3.6. PCR of the PGP Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gupta, S.; Pandey, S.; Sharma, S. Decoding the Plant Growth Promotion and Antagonistic Potential of Bacterial Endophytes From Ocimum sanctum Linn. Against Root Rot Pathogen Fusarium oxysporum in Pisum sativum. Front. Plant Sci. 2022, 13, 813686. [Google Scholar] [CrossRef] [PubMed]
- Jain, R.; Bhardwaj, P.; Pandey, S.S.; Kumar, S. Arnebia euchroma, a plant species of cold desert in the Himalayas, harbors beneficial cultivable endophytes in roots and leaves. Front. Microbiol. 2021, 12, 696667. [Google Scholar] [CrossRef] [PubMed]
- Omomowo, O.I.; Babalola, O.O. Bacterial and fungal endophytes: Tiny giants with immense beneficial potential for plant growth and sustainable agricultural productivity. Microorganisms 2019, 7, 481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, J.F.; Kingsley, K.L.; Zhang, Q.; Verma, R.; Obi, N.; Dvinskikh, S.; Elmore, M.T.; Verma, S.K.; Gond, S.K.; Kowalski, K.P. Endophytic microbes and their potential applications in crop management. Pest Manage. Sci. 2019, 75, 2558–2565. [Google Scholar] [CrossRef]
- Babalola, O.O.; Sanni, A.I.; Odhiambo, G.D.; Torto, B. Plant growth-promoting rhizobacteria do not pose any deleterious effect on cowpea and detectable amounts of ethylene are produced. World J. Microbiol. Biotechnol. 2007, 23, 747–752. [Google Scholar] [CrossRef]
- Emmanuel, O.C.; Akintola, O.A.; Tetteh, F.M.; Babalola, O.O. Combined Application of Inoculant, Phosphorus and Potassium Enhances Cowpea Yield in Savanna Soils. Agronomy 2021, 11, 15. [Google Scholar] [CrossRef]
- Abd_Allah, E.F.; Alqarawi, A.A.; Hashem, A.; Radhakrishnan, R.; Al-Huqail, A.A.; Al-Otibi, F.O.N.; Malik, J.A.; Alharbi, R.I.; Egamberdieva, D. Endophytic bacterium Bacillus subtilis (BERA 71) improves salt tolerance in chickpea plants by regulating the plant defense mechanisms. J. Plant Interact. 2018, 13, 37–44. [Google Scholar] [CrossRef] [Green Version]
- Bokhari, A.; Essack, M.; Lafi, F.F.; Andres-Barrao, C.; Jalal, R.; Alamoudi, S.; Razali, R.; Alzubaidy, H.; Shah, K.H.; Siddique, S. Bioprospecting desert plant Bacillus endophytic strains for their potential to enhance plant stress tolerance. Sci. Rep. 2019, 9, 18154. [Google Scholar] [CrossRef] [Green Version]
- Shah, S.; Chand, K.; Rekadwad, B.; Shouche, Y.S.; Sharma, J.; Pant, B. A prospectus of plant growth promoting endophytic bacterium from orchid (Vanda cristata). BMC Biotechnol. 2021, 21, 1–9. [Google Scholar] [CrossRef]
- Wang, G.; Meng, J.; Tian, T.; Xiao, X.; Zhang, B.; Xiao, Y. Endophytic Bacillus velezensis strain B-36 is a potential biocontrol agent against lotus rot caused by Fusarium oxysporum. J. Appl. Microbiol. 2020, 128, 1153–1162. [Google Scholar] [CrossRef] [Green Version]
- Potshangbam, M.; Devi, S.I.; Sahoo, D.; Strobel, G.A. Functional characterization of endophytic fungal community associated with Oryza sativa L. and Zea mays L. Front. Microbiol. 2017, 8, 325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahu, P.K.; Singh, S.; Gupta, A.; Singh, U.B.; Brahmaprakash, G.; Saxena, A.K. Antagonistic potential of bacterial endophytes and induction of systemic resistance against collar rot pathogen Sclerotium rolfsii in tomato. Biol. Control 2019, 137, 104014. [Google Scholar] [CrossRef]
- Singh, S.P.; Gaur, R. Endophytic Streptomyces spp. underscore induction of defense regulatory genes and confers resistance against Sclerotium rolfsii in chickpea. Biol. Control 2017, 104, 44–56. [Google Scholar] [CrossRef]
- Ahmad, T.; Bashir, A.; Farooq, S.; Riyaz-Ul-Hassan, S. Burkholderia gladioli E39CS3, an endophyte of Crocus sativus Linn., induces host resistance against corm-rot caused by Fusarium oxysporum. J. Appl. Microbiol. 2022, 132, 495–508. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, H.; Dowarah, B.; Baruah, P.M.; Bordoloi, K.S.; Krishnatreya, D.B.; Agarwala, N. Endophytes from Gnetum gnemon L. can protect seedlings against the infection of phytopathogenic bacterium Ralstonia solanacearum as well as promote plant growth in tomato. Microbiol. Res. 2020, 238, 126503. [Google Scholar] [CrossRef]
- Faria, P.S.A.; de Oliveira Marques, V.; Selari, P.J.R.G.; Martins, P.F.; Silva, F.G.; de Fátima Sales, J. Multifunctional potential of endophytic bacteria from Anacardium othonianum Rizzini in promoting in vitro and ex vitro plant growth. Microbiol. Res. 2021, 242, 126600. [Google Scholar] [CrossRef]
- Compant, S.; Cambon, M.C.; Vacher, C.; Mitter, B.; Samad, A.; Sessitsch, A. The plant endosphere world–bacterial life within plants. Environ. Microbiol. 2021, 23, 1812–1829. [Google Scholar] [CrossRef]
- Hardoim, P.R.; Van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The hidden world within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef] [Green Version]
- Dubey, A.; Saiyam, D.; Kumar, A.; Hashem, A.; Abd_Allah, E.F.; Khan, M.L. Bacterial root endophytes: Characterization of their competence and plant growth promotion in soybean (Glycine max (L.) Merr.) under drought stress. Int. J. Environ. Res. Public Health 2021, 18, 931. [Google Scholar] [CrossRef]
- Morales-Cedeño, L.R.; del Carmen Orozco-Mosqueda, M.; Loeza-Lara, P.D.; Parra-Cota, F.I.; de Los Santos-Villalobos, S.; Santoyo, G. Plant growth-promoting bacterial endophytes as biocontrol agents of pre-and post-harvest diseases: Fundamentals, methods of application and future perspectives. Microbiol. Res. 2021, 242, 126612. [Google Scholar] [CrossRef]
- Xu, W.; Wang, F.; Zhang, M.; Ou, T.; Wang, R.; Strobel, G.; Xiang, Z.; Zhou, Z.; Xie, J. Diversity of cultivable endophytic bacteria in mulberry and their potential for antimicrobial and plant growth-promoting activities. Microbiol. Res. 2019, 229, 126328. [Google Scholar] [CrossRef] [PubMed]
- Santoyo, G.; Moreno-Hagelsieb, G.; del Carmen Orozco-Mosqueda, M.; Glick, B.R. Plant growth-promoting bacterial endophytes. Microbiol. Res. 2016, 183, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Adeleke, B.S.; Babalola, O.O. The endosphere microbial communities, a great promise in agriculture. Int. Microbiol. 2021, 24, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Omomowo, O.I.; Babalola, O.O. Constraints and prospects of improving cowpea productivity to ensure food, nutritional security and environmental sustainability. Front. Plant Sci. 2021, 12, 751731. [Google Scholar] [CrossRef]
- Lonardi, S.; Muñoz-Amatriaín, M.; Liang, Q.; Shu, S.; Wanamaker, S.I.; Lo, S.; Tanskanen, J.; Schulman, A.H.; Zhu, T.; Luo, M.C. The genome of cowpea (Vigna unguiculata [L.] Walp.). Plant J. 2019, 98, 767–782. [Google Scholar] [CrossRef] [Green Version]
- Ravelombola, W.; Qin, J.; Shi, A.; Weng, Y.; Bhattarai, G.; Dong, L.; Morris, J.B. A SNP-based association analysis for plant growth habit in worldwide cowpea (Vigna unguiculata (L.) Walp) Germplasm. Euphytica 2017, 213, 284. [Google Scholar] [CrossRef]
- Muñoz-Amatriaín, M.; Mirebrahim, H.; Xu, P.; Wanamaker, S.I.; Luo, M.; Alhakami, H.; Alpert, M.; Atokple, I.; Batieno, B.J.; Boukar, O. Genome resources for climate-resilient cowpea, an essential crop for food security. Plant J. 2017, 89, 1042–1054. [Google Scholar] [CrossRef] [Green Version]
- Carvalho, M.; Lino-Neto, T.; Rosa, E.; Carnide, V. Cowpea: A legume crop for a challenging environment. J. Sci. Food Agric. 2017, 97, 4273–4284. [Google Scholar] [CrossRef]
- Gonçalves, A.; Goufo, P.; Barros, A.; Domínguez-Perles, R.; Trindade, H.; Rosa, E.A.; Ferreira, L.; Rodrigues, M. Cowpea (Vigna unguiculata L. Walp), a renewed multipurpose crop for a more sustainable agri-food system: Nutritional advantages and constraints. J. Sci. Food Agric. 2016, 96, 2941–2951. [Google Scholar] [CrossRef]
- Kebede, E.; Bekeko, Z. Expounding the production and importance of cowpea (Vigna unguiculata (L.) Walp.) in Ethiopia. Cogent Food Agric. 2020, 6, 1769805. [Google Scholar] [CrossRef]
- Xiong, H.; Shi, A.; Mou, B.; Qin, J.; Motes, D.; Lu, W. Genetic Diversity and Population Structure of Cowpea (Vigna unguiculata L. Walp). PLoS ONE 2016, 11, e0160941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olajide, A.A.; Ilori, C.O. Genetic Variability, Performance and Yield Potentials of Ten Varieties of Cowpea (Vigna unguiculata (L) Walp) under Drought Stress. Legume Genom. Genet. 2017, 8, 17–25. [Google Scholar] [CrossRef]
- Ovalesha, M.; Yadav, B.; Rai, P.K. Effects of polymer seed coating and seed treatment on plant growth, seed yield and quality of Cowpea (Vigna unguiculata). J. Pharmacog. Phytochem. 2017, 6, 106–109. [Google Scholar]
- Fukuda, K.; Ogawa, M.; Taniguchi, H.; Saito, M. Molecular approaches to studying microbial communities: Targeting the 16S ribosomal RNA gene. J. UOEH 2016, 38, 223–232. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Chang, S.; Ye, S.; Chen, M.; Lin, L.; Li, Y.; Li, S.; An, Q. Differentiation of 1-aminocyclopropane-1-carboxylate (ACC) deaminase from its homologs is the key for identifying bacteria containing ACC deaminase. FEMS Microbiol. Ecol. 2015, 91, fiv112. [Google Scholar] [CrossRef] [Green Version]
- Raddadi, N.; Cherif, A.; Boudabous, A.; Daffonchio, D. Screening of plant growth promoting traits of Bacillus thuringiensis. Ann. Microbiol. 2008, 58, 47–52. [Google Scholar] [CrossRef]
- Chen, W.; Yang, F.; Zhang, L.; Wang, J. Organic acid secretion and phosphate solubilizing efficiency of Pseudomonas sp. PSB12: Effects of phosphorus forms and carbon sources. Geomicrobiol. J. 2016, 33, 870–877. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Nautiyal, C.S. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 1999, 170, 265–270. [Google Scholar] [CrossRef]
- Matsuda, R.; Handayani, M.L.; Sasaki, H.; Takechi, K.; Takano, H.; Takio, S. Production of indoleacetic acid by strains of the epiphytic bacteria Neptunomonas spp. isolated from the red alga Pyropia yezoensis and the seagrass Zostera marina. Arch. Microbiol. 2018, 200, 255–265. [Google Scholar] [CrossRef]
- Schwyn, B.; Neilands, J. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Glick, B.R.; Karaturovíc, D.M.; Newell, P.C. A novel procedure for rapid isolation of plant growth promoting pseudomonads. Can. J. Microbiol. 1995, 41, 533–536. [Google Scholar] [CrossRef]
- Cappucino, J.; Sherman, N. Nitrogen cycle. In Microbiology: A Laboratory Manual; Benjamin/Cumming Pub. Co.: New York, NY, USA, 1992. [Google Scholar]
- Khan, N.; Bano, A. Exopolysaccharide producing rhizobacteria and their impact on growth and drought tolerance of wheat grown under rainfed conditions. PLoS ONE 2019, 14, e0222302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dinesh, R.; Anandaraj, M.; Kumar, A.; Bini, Y.K.; Subila, K.P.; Aravind, R. Isolation, characterization, and evaluation of multi-trait plant growth promoting rhizobacteria for their growth promoting and disease suppressing effects on ginger. Microbiol. Res. 2015, 173, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Mohamad, O.A.; Li, L.; Ma, J.-B.; Hatab, S.; Xu, L.; Guo, J.-W.; Rasulov, B.A.; Liu, Y.-H.; Hedlund, B.P.; Li, W.-J. Evaluation of the antimicrobial activity of endophytic bacterial populations from Chinese traditional medicinal plant licorice and characterization of the bioactive secondary metabolites produced by Bacillus atrophaeus against Verticillium dahliae. Front. Microbiol. 2018, 9, 924. [Google Scholar] [CrossRef] [Green Version]
- Vyas, P.; Kaur, R. Culturable stress-tolerant plant growth-promoting bacterial endophytes associated with Adhatoda vasica. J. Soil Sci. Plant Nutr. 2019, 19, 290–298. [Google Scholar] [CrossRef]
- Bhutani, N.; Maheshwari, R.; Sharma, N.; Kumar, P.; Dang, A.S.; Suneja, P. Characterization of halo-tolerant plant growth promoting endophytic Bacillus licheniformis MHN 12. J. Genet. Eng. Biotechnol. 2022, 20, 1–12. [Google Scholar] [CrossRef]
- Phurailatpam, L.; Gupta, A.; Sahu, P.K.; Mishra, S. Insights into the functional potential of bacterial endophytes from the ethnomedicinal plant, Piper longum L. Symbiosis 2022, 87, 165–174. [Google Scholar] [CrossRef]
- Maheshwari, R.; Kumar, P.; Bhutani, N.; Suneja, P. Exploration of plant growth-promoting endophytic bacteria from Pisum sativum and Cicer arietinum from South–West Haryana. J. Basic Microbiol. 2022, 62, 857–874. [Google Scholar] [CrossRef]
- Omomowo, O.I.; Babalola, O.O. Genomic Insights into Two Endophytic Strains: Stenotrophomonas geniculata NWUBe21 and Pseudomonas carnis NWUBe30 from Cowpea with Plant Growth-Stimulating Attributes. Appl. Sci. 2022, 12, 12953. [Google Scholar] [CrossRef]
- ALKahtani, M.D.; Fouda, A.; Attia, K.A.; Al-Otaibi, F.; Eid, A.M.; Ewais, E.E.-D.; Hijri, M.; St-Arnaud, M.; Hassan, S.E.-D.; Khan, N. Isolation and characterization of plant growth promoting endophytic bacteria from desert plants and their application as bioinoculants for sustainable agriculture. Agronomy 2020, 10, 1325. [Google Scholar] [CrossRef]
- Kushwaha, P.; Kashyap, P.L.; Bhardwaj, A.K.; Kuppusamy, P.; Srivastava, A.K.; Tiwari, R.K.; Biotechnology. Bacterial endophyte mediated plant tolerance to salinity: Growth responses and mechanisms of action. World. J. Microbiol. Biotechnol. 2020, 36, 26. [Google Scholar] [CrossRef]
- Vo, Q.; Ballard, R.A.; Barnett, S.J.; Franco, C.M. Isolation and characterisation of endophytic actinobacteria and their effect on the growth and nodulation of chickpea (Cicer arietinum). Plant Soil 2021, 466, 357–371. [Google Scholar] [CrossRef]
- Maheshwari, R.; Bhutani, N.; Kumar, P.; Suneja, P. Plant growth promoting potential of multifarious endophytic Pseudomonas lini strain isolated from Cicer arietinum L. Isr. J. Plant Sci. 2021, 69, 50–60. [Google Scholar] [CrossRef]
- Adeleke, B.S.; Ayangbenro, A.; Babalola, O.O. Effect of endophytic bacterium, Stenotrophomonas maltophilia JVB5 on sunflowers. Plant Prot. Sci. 2022, 58, 185–198. [Google Scholar] [CrossRef]
- Khanna, A.; Raj, K.; Kumar, P.; Wati, L. Antagonistic and growth-promoting potential of multifarious bacterial endophytes against Fusarium wilt of chickpea. Egypt. J. Biol. Pest Control 2022, 32, 17. [Google Scholar] [CrossRef]
- de Almeida Lopes, K.B.; Carpentieri-Pipolo, V.; Fira, D.; Balatti, P.A.; López, S.M.Y.; Oro, T.H.; Stefani Pagliosa, E.; Degrassi, G. Screening of bacterial endophytes as potential biocontrol agents against soybean diseases. J. Appl. Microbiol. 2018, 125, 1466–1481. [Google Scholar] [CrossRef] [Green Version]
- Flores-Duarte, N.J.; Caballero-Delgado, S.; Pajuelo, E.; Mateos-Naranjo, E.; Redondo-Gómez, S.; Navarro-Torre, S.; Rodríguez-Llorente, I.D. Enhanced legume growth and adaptation to degraded estuarine soils using Pseudomonas sp. nodule endophytes. Front. Microbiol. 2022, 13, 1005458. [Google Scholar] [CrossRef]
- Flores-Duarte, N.J.; Pérez-Pérez, J.; Navarro-Torre, S.; Mateos-Naranjo, E.; Redondo-Gómez, S.; Pajuelo, E.; Rodríguez-Llorente, I.D. Improved Medicago sativa Nodulation under Stress Assisted by Variovorax sp. Endophytes. Plants 2022, 11, 1091. [Google Scholar] [CrossRef]
- Jia, H.; Xi, Z.; Ma, J.; Li, Y.; Hao, C.; Lu, M.; Zhang, Z.-Z.; Deng, W.-W. Endophytic bacteria from the leaves of two types of albino tea plants, indicating the plant growth promoting properties. J. P. Growth Regul. 2022, 96, 331–343. [Google Scholar] [CrossRef]
- Bhutani, N.; Maheshwari, R.; Kumar, P.; Suneja, P. Bioprospecting of endophytic bacteria from nodules and roots of Vigna radiata, Vigna unguiculata and Cajanus cajan for their potential use as bioinoculants. Plant Gene 2021, 28, 100326. [Google Scholar] [CrossRef]
- Tran, T.; French, E.; Iyer-Pascuzzi, A.S. In vitro functional characterization predicts the impact of bacterial root endophytes on plant growth. J. Exp. Bot. 2022, 73, 5758–5772. [Google Scholar] [CrossRef]
- Pandey, S.S.; Jain, R.; Bhardwaj, P.; Thakur, A.; Kumari, M.; Bhushan, S.; Kumar, S. Plant Probiotics—Endophytes pivotal to plant health. Microbiol. Res. 2022, 263, 127148. [Google Scholar] [CrossRef] [PubMed]
- Afzal, I.; Shinwari, Z.K.; Sikandar, S.; Shahzad, S. Plant beneficial endophytic bacteria: Mechanisms, diversity, host range and genetic determinants. Microbiol. Res. 2019, 221, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Aeron, A.; Dubey, R.C.; Maheshwari, D.K. Next-Generation biofertilizers and novel biostimulants: Documentation and validation of mechanism of endophytic plant growth-promoting rhizobacteria in tomato. Arch. Microbiol. 2021, 203, 3715–3726. [Google Scholar] [CrossRef]
- Gorai, P.S.; Ghosh, R.; Mandal, S.; Ghosh, S.; Chatterjee, S.; Gond, S.K.; Mandal, N.C. Bacillus siamensis CNE6-a multifaceted plant growth promoting endophyte of Cicer arietinum L. having broad spectrum antifungal activities and host colonizing potential. Microbiol. Res. 2021, 252, 126859. [Google Scholar] [CrossRef] [PubMed]
- Etminani, F.; Harighi, B. Isolation and identification of endophytic bacteria with plant growth promoting activity and biocontrol potential from wild pistachio trees. Plant Pathol. J. 2018, 34, 208. [Google Scholar] [CrossRef] [PubMed]
- Eid, A.M.; Fouda, A.; Abdel-Rahman, M.A.; Salem, S.S.; Elsaied, A.; Oelmüller, R.; Hijri, M.; Bhowmik, A.; Elkelish, A.; Hassan, S.E.-D. Harnessing bacterial endophytes for promotion of plant growth and biotechnological applications: An overview. Plants 2021, 10, 935. [Google Scholar] [CrossRef] [PubMed]
- Brígido, C.; Singh, S.; Menéndez, E.; Tavares, M.J.; Glick, B.R.; Félix, M.d.R.; Oliveira, S.; Carvalho, M. Diversity and functionality of culturable endophytic bacterial communities in chickpea plants. Plants 2019, 8, 42. [Google Scholar] [CrossRef] [Green Version]
- Adhikari Dhungana, S.; Adachi, F.; Hayashi, S.; Raj Puri, R.; Itoh, K. Plant growth promoting effects of Nepalese sweet potato endophytes. Horticulturae 2018, 4, 53. [Google Scholar] [CrossRef] [Green Version]
- Bhutani, N.; Maheshwari, R.; Negi, M.; Suneja, P. Optimization of IAA production by endophytic Bacillus spp. from Vigna radiata for their potential use as plant growth promoters. Isr. J. Plant Sci. 2018, 65, 83–96. [Google Scholar] [CrossRef]
- Lucero, C.T.; Lorda, G.S.; Anzuay, M.S.; Ludueña, L.M.; Taurian, T. Peanut endophytic phosphate solubilizing bacteria increase growth and P content of soybean and maize plants. Curr. Microbiol. 2021, 78, 1961–1972. [Google Scholar] [CrossRef] [PubMed]
- Mei, C.; Chretien, R.L.; Amaradasa, B.S.; He, Y.; Turner, A.; Lowman, S. Characterization of phosphate solubilizing bacterial endophytes and plant growth promotion in vitro and in greenhouse. Microorganisms 2021, 9, 1935. [Google Scholar] [CrossRef]
- Chen, C.; Xin, K.; Liu, H.; Cheng, J.; Shen, X.; Wang, Y.; Zhang, L. Pantoea alhagi, a novel endophytic bacterium with ability to improve growth and drought tolerance in wheat. Sci. Rep. 2017, 7, 41564. [Google Scholar] [CrossRef] [Green Version]
- Adeleke, B.S.; Ayangbenro, A.S.; Babalola, O.O. In vitro Screening of Sunflower Associated Endophytic Bacteria With Plant Growth-Promoting Traits. Front. Sustain. Food Syst. 2022, 6, 903114. [Google Scholar] [CrossRef]
- Kumar, N.; Dubey, R.C. Plant growth-promoting attributes of an endophyte Enterobacter roggenkampii BLS02 isolated from Barleria lupulina Lindl. Org. Agric. 2022, 12, 137–145. [Google Scholar] [CrossRef]

| Target Gene | Primer Name | Primer Sequence (5′→3′) | Product Size (bp) | Reference |
|---|---|---|---|---|
| 16S | 341-F 907-R | AGAGTTTGATCCTGGCTCAG AAGGAGGTGATCCAGCCGCA | 1300–1500 | [34] |
| 1-Aminocyclopropane-1-carboxylic acid deaminase (ACDS) | ACDS-F3 ACDS-R3 | ATCGGCGGCATCCAGWSNAAYCANAC GTGCATCGACTTGCCCTCRTANACNGGRT | 800–950 | [35] |
| Indo-pyruvate decarboxylase (IPDC) | IPDC-F IPDC-R | CAYTTGAAAACKCAMTATACTG AAGAATTTGYWKGCCGAATCT | 1715–1809 | [36] |
| Acid phosphatase (ACPHO) | ACPHO-F ACPHO-R | AAGAGGGGCATTACCACTTTATTA CGCCTTCCCAATCRCCATACAT | 828 | [36] |
| Glucose dehydrogenase (GCD) | GCD-F GCD-R | GACCTGTGGGACATGGACGT GTCCTTGCCGGTGTAGSTCATC | 875 | [37] |
| S/N | Culture Code | Molecularly Identified Strains | Similarity (%) | Bacteria Phyla | GenBank Accession Number |
|---|---|---|---|---|---|
| 1. | Be1 | Stenotrophomonas maltophilia strain NwuBe01 | 100 | Proteobacteria | OK050078 |
| 2. | Be2 | Stenotrophomonas pavanii strain NwuBe02 | 100 | Proteobacteria | OK050079 |
| 3. | Be3 | Brevundimonas bullata strain NwuBe03 | 100 | Proteobacteria | OK050080 |
| 4. | Be4 | Bacillus wiedmannii strain NwuBe04 | 100 | Firmicute | OK050081 |
| 5. | Be5 | Bacillus anthracis strain NwuBe05 | 100 | Firmicute | OK050082 |
| 6. | Be6 | Micrococcus luteus strain NwuBe06 | 100 | High G + C | OK050083 |
| 7. | Be7 | Myroides pelagicus strain NwuBe07 | 100 | CFB | OK050084 |
| 8. | Be8 | Bacillus tropicus strain NwuBe08 | 100 | Firmicute | OK050085 |
| 9. | Be9 | Ignatzschineria indica strain NwuBe09 | 100 | Proteobacteria | OK050086 |
| 10. | Be10 | Bacillus thuringiensis strain NwuBe10 | 100 | Firmicute | OK050087 |
| 11. | Be11 | Staphylococcus saprophyticus strain NwuBe11 | 100 | Firmicute | OK050088 |
| 12. | Be12 | Staphylococcus edaphicus strain NwuBe12 | 100 | Firmicute | OK050089 |
| 13. | Be13 | Staphylococcus nepalensis strain NwuBe13 | 100 | Firmicute | OK050090 |
| 14. | Be14 | Staphylococcus xylosus strain NwuBe14 | 100 | Firmicute | OK050091 |
| 15. | Be15 | Staphylococcus cohnii strain NwuBe15 | 100 | Firmicute | OK050092 |
| 16. | Be16 | Mammaliicoccus stepanovicii strain NwuBe16 | 100 | Firmicute | OK050093 |
| 17. | Be17 | Staphylococcus succinus strain NwuBe17 | 100 | Firmicute | OK050094 |
| 18. | Nwa1 | Staphylococcus casei strain NwuBe18 | 100 | Firmicute | OK050095 |
| 19. | Nwa2 | Staphylococcus arlettae strain NwuBe19 | 100 | Firmicute | OK050096 |
| 20. | Nwa3 | Staphylococcus saprophyticus strain NwuBe20 | 100 | Firmicute | OK050097 |
| 21. | Nwa5 | Lysinibacillus xylanilyticus strain NwuBe21 | 100 | Firmicute | OK050098 |
| 22. | Nwa6 | Bacillus altitudinis strain NwuBe22 | 100 | Firmicute | OK050099 |
| 23. | Nwa7 | Paenibacillus illinoisensis strain NwuBe23 | 100 | Firmicute | OK050100 |
| 24. | Nwa8 | Brevundimonas bullata strain NwuBe24 | 100 | Proteobacteria | OK050101 |
| 25. | Nwa9 | Bacillus tropicus strain NwuBe25 | 100 | Firmicute | OK050102 |
| 26. | Nwa11 | Brevundimonas terrae strain NwuBe26 | 100 | Proteobacteria | OK050103 |
| 27. | Nwa12 | Myroides odoratimimus strain NwuBe27 | 100 | CFB | OK050104 |
| 28. | Nwa13 | Bacillus paramycoides strain NwuBe28 | 100 | Firmicute | OK050105 |
| 29. | Nwa16 | Brevundimonas bullata strain NwuBe29 | 100 | Proteobacteria | OK050106 |
| 30. | Nwa17 | Pseudomonas fluorescens strain NwuBe30 | 100 | Proteobacteria | OK050107 |
| 31. | Nwa18 | Bacillus cereus strain NwuBe31 | 100 | Firmicute | OK050108 |
| 32. | Nwa21 | Bacillus bingmayongensis strain NwuBe32 | 100 | Firmicute | OK050109 |
| 33. | Nwa22 | Staphylococcus kloosii strain NwuBe33 | 100 | Firmicute | OK050110 |
| Bacteria Code | ACDS Production | Ammonia Production | Auxin Production | Exopolysaccharide Production | HCN Production | Phosphate Solubilization | Siderophore Production |
|---|---|---|---|---|---|---|---|
| Be1 | − | ++ | ++ | + | ++ | − | ++ |
| Be2 | + | +++ | +++ | + | ++ | ++++ | ++ |
| Be3 | + | +++ | +++ | + | ++ | ++ | ++ |
| Be4 | + | ++ | ++ | + | ++ | − | ++ |
| Be5 | + | ++ | +++ | + | ++ | ++ | ++ |
| Be6 | − | ++ | ++ | + | − | − | − |
| Be7 | − | ++ | ++ | + | − | − | − |
| Be8 | − | ++ | ++ | + | − | +++ | ++ |
| Be9 | − | ++ | ++ | + | − | − | - |
| Be10 | + | +++ | +++ | + | ++ | ++ | ++ |
| Be11 | − | ++ | ++ | + | ++ | − | ++ |
| Be12 | + | +++ | +++ | + | +++ | +++ | ++ |
| Be13 | − | ++ | ++ | + | ++ | − | − |
| Be14 | + | +++ | +++ | + | +++ | ++ | ++ |
| Be15 | − | ++ | ++ | + | − | − | − |
| Be16 | − | ++ | ++ | + | − | − | ++ |
| Be17 | − | ++ | ++ | + | − | − | − |
| Nwa1 | − | ++ | ++ | + | − | − | ++ |
| Nwa2 | − | ++ | ++ | + | − | − | − |
| Nwa3 | − | ++ | ++ | + | − | − | ++ |
| Nwa5 | + | ++ | ++++ | + | ++ | ++ | ++ |
| Nwa6 | + | +++ | +++ | + | ++ | ++ | − |
| Nwa7 | + | +++ | +++ | + | ++ | ++ | ++ |
| Nwa8 | − | ++ | ++ | + | − | − | ++ |
| Nwa9 | + | +++ | +++ | + | ++ | − | − |
| Nwa11 | − | ++ | ++ | + | − | − | − |
| Nwa12 | − | ++ | ++ | + | − | − | ++ |
| Nwa13 | + | +++ | +++ | + | − | − | − |
| Nwa16 | − | ++ | ++ | + | − | − | − |
| Nwa17 | + | +++ | +++ | + | ++ | ++ | ++ |
| Nwa18 | + | +++ | +++ | + | ++ | ++ | ++ |
| Nwa21 | + | +++ | +++ | + | ++ | ++ | ++ |
| Nwa22 | − | ++ | ++ | + | − | ++ | ++ |
| S/N | Endophytic Bacteria Strains | pH 5 | pH 7 | pH 10 |
|---|---|---|---|---|
| 1 | Stenotrophomonas maltophilia strain NwuBe01 | ++ | +++ | + |
| 2 | Stenotrophomonas pavanii strain NwuBe02 | ++ | ++ | + |
| 3 | Brevundimonas bullata strain NwuBe03 | + | ++ | + |
| 4 | Bacillus wiedmannii strain NwuBe04 | + | ++ | + |
| 5 | Bacillus anthracis strain NwuBe05 | ++ | ++ | + |
| 6 | Micrococcus luteus strain NwuBe06. | ++ | ++ | + |
| 7 | Myroides pelagicus strain NwuBe07 | + | ++ | + |
| 8 | Bacillus tropicus strain NwuBe08 | ++ | +++ | ++ |
| 9 | Ignatzschineria indica strain NwuBe09 | ++ | + | + |
| 10 | Bacillus thuringiensis strain NwuBe10 | ++ | ++ | + |
| 11 | Staphylococcus saprophyticus strain NwuBe11 | + | + | + |
| 12 | Staphylococcus edaphicus strain NwuBe12 | ++ | +++ | ++ |
| 13 | Staphylococcus nepalensis strain NwuBe13 | + | + | + |
| 14 | Staphylococcus xylosus strain NwuBe14 | ++ | + | + |
| 15 | Staphylococcus cohnii strain NwuBe15 | + | ++ | + |
| 16 | Mammaliicoccus stepanovicii strain NwuBe16 | + | + | + |
| 17 | Staphylococcus succinus strain NwuBe17 | + | + | + |
| 18 | Staphylococcus casei strain NwuBe18 | ++ | + | + |
| 19 | Staphylococcus arlettae strain NwuBe19 | + | + | + |
| 20 | Staphylococcus saprophyticus strain NwuBe20 | ++ | ++ | + |
| 21 | Lysinibacillus xylanilyticus strain NwuBe21 | ++ | +++ | ++ |
| 22 | Bacillus altitudinis strain NwuBe22 | ++ | ++ | ++ |
| 23 | Paenibacillus illinoisensis strain NwuBe23 | ++ | +++ | ++ |
| 24 | Brevundimonas bullata strain NwuBe24 | + | + | + |
| 25 | Bacillus tropicus strain NwuBe25 | ++ | ++ | ++ |
| 26 | Brevundimonas terrae strain NwuBe26 | + | + | + |
| 27 | Myroides odoratimimus strain NwuBe27 | + | + | + |
| 28 | Bacillus paramycoides strain NwuBe28 | ++ | ++ | + |
| 29 | Brevundimonas bullata strain NwuBe29 | ++ | + | + |
| 30 | Pseudomonas fluorescens strain NwuBe30 | ++ | +++ | ++ |
| 31 | Bacillus cereus strain NwuBe31 | ++ | ++ | + |
| 32 | Bacillus bingmayongensis strain NwuBe32 | + | ++ | + |
| 33 | Staphylococcus kloosii strain NwuBe33 | ++ | ++ | + |
| S/N | Endophytic Bacteria Strains | NaCl 1% | NaCl 3% | NaCl 5% |
|---|---|---|---|---|
| 1 | Stenotrophomonas maltophilia strain NwuBe01 | + | + | + |
| 2 | Stenotrophomonas pavanii strain NwuBe02 | ++ | + | + |
| 3 | Brevundimonas bullata strain NwuBe03 | + | + | − |
| 4 | Bacillus wiedmannii strain NwuBe04 | ++ | + | + |
| 5 | Bacillus anthracis strain NwuBe05 | ++ | + | + |
| 6 | Micrococcus luteus strain NwuBe06 | ++ | + | + |
| 7 | Myroides pelagicus strain NwuBe07 | + | + | − |
| 8 | Bacillus tropicus strain NwuBe08 | ++ | ++ | + |
| 9 | Ignatzschineria indica strain NwuBe09 | ++ | + | − |
| 10 | Bacillus thuringiensis strain NwuBe10 | ++ | + | + |
| 11 | Staphylococcus saprophyticus strain NwuBe11 | + | + | − |
| 12 | Staphylococcus edaphicus strain NwuBe12 | ++ | + | + |
| 13 | Staphylococcus nepalensis strain NwuBe13 | + | + | − |
| 14 | Staphylococcus xylosus strain NwuBe14 | ++ | + | − |
| 15 | Staphylococcus cohnii strain NwuBe15 | ++ | + | − |
| 16 | Mammaliicoccus stepanovicii strain NwuBe16 | + | + | − |
| 17 | Staphylococcus succinus strain NwuBe17 | + | + | − |
| 18 | Staphylococcus casei strain NwuBe18 | ++ | + | − |
| 19 | Staphylococcus arlettae strain NwuBe19 | + | + | − |
| 20 | Staphylococcus saprophyticus strain NwuBe20 | ++ | + | + |
| 21 | Lysinibacillus xylanilyticus strain NwuBe21 | ++ | ++ | + |
| 22 | Bacillus altitudinis strain NwuBe22 | ++ | + | + |
| 23 | Paenibacillus illinoisensis strain NwuBe23 | ++ | + | + |
| 24 | Brevundimonas bullata strain NwuBe24 | + | + | − |
| 25 | Bacillus tropicus strain NwuBe25 | ++ | + | + |
| 26 | Brevundimonas terrae strain NwuBe26 | + | + | − |
| 27 | Myroides odoratimimus strain NwuBe27 | + | + | − |
| 28 | Bacillus paramycoides strain NwuBe28 | ++ | + | − |
| 29 | Brevundimonas bullata strain NwuBe29 | ++ | + | − |
| 30 | Pseudomonas fluorescens strain NwuBe30 | ++ | ++ | + |
| 31 | Bacillus cereus strain NwuBe31 | ++ | + | + |
| 32 | Bacillus bingmayongensis strain NwuBe32 | ++ | + | + |
| 33 | Staphylococcus kloosii strain NwuBe33 | ++ | + | − |
| S/N | Endophytic Bacteria Strains | 25 °C | 30 °C | 40 °C |
|---|---|---|---|---|
| 1 | Stenotrophomonas maltophilia strain NwuBe01 | + | + | + |
| 2 | Stenotrophomonas pavanii strain NwuBe02 | ++ | + | + |
| 3 | Brevundimonas bullata strain NwuBe03 | + | + | − |
| 4 | Bacillus wiedmannii strain NwuBe04 | ++ | + | + |
| 5 | Bacillus anthracis strain NwuBe05 | ++ | + | + |
| 6 | Micrococcus luteus strain NwuBe06. | ++ | + | + |
| 7 | Myroides pelagicus strain NwuBe07 | + | + | − |
| 8 | Bacillus tropicus strain NwuBe08 | ++ | ++ | + |
| 9 | Ignatzschineria indica strain NwuBe09 | ++ | + | − |
| 10 | Bacillus thuringiensis strain NwuBe10 | ++ | + | + |
| 11 | Staphylococcus saprophyticus strain NwuBe11 | + | + | − |
| 12 | Staphylococcus edaphicus strain NwuBe12 | ++ | + | + |
| 13 | Staphylococcus nepalensis strain NwuBe13 | + | + | − |
| 14 | Staphylococcus xylosus strain NwuBe14 | ++ | + | − |
| 15 | Staphylococcus cohnii strain NwuBe15 | ++ | + | − |
| 16 | Mammaliicoccus stepanovicii strain NwuBe16 | + | + | − |
| 17 | Staphylococcus succinus strain NwuBe17 | + | + | − |
| 18 | Staphylococcus casei strain NwuBe18 | ++ | + | − |
| 19 | Staphylococcus arlettae strain NwuBe19 | + | + | − |
| 20 | Staphylococcus saprophyticus strain NwuBe20 | ++ | + | + |
| 21 | Lysinibacillus xylanilyticus strain NwuBe21 | ++ | ++ | + |
| 22 | Bacillus altitudinis strain NwuBe22 | ++ | + | + |
| 23 | Paenibacillus illinoisensis strain NwuBe23 | ++ | + | + |
| 24 | Brevundimonas bullata strain NwuBe24 | + | + | − |
| 25 | Bacillus tropicus strain NwuBe25 | ++ | + | + |
| 26 | Brevundimonas terrae strain NwuBe26 | + | + | − |
| 27 | Myroides odoratimimus strain NwuBe27 | + | + | − |
| 28 | Bacillus paramycoides strain NwuBe28 | ++ | + | − |
| 29 | Brevundimonas bullata strain NwuBe29 | ++ | + | − |
| 30 | Pseudomonas fluorescens strain NwuBe30 | ++ | ++ | + |
| 31 | Bacillus cereus strain NwuBe31 | ++ | + | + |
| 32 | Bacillus bingmayongensis strain NwuBe32 | ++ | + | + |
| 33 | Staphylococcus kloosii strain NwuBe33 | ++ | + | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Omomowo, O.I.; Babalola, O.O. Bioassessment of Phylogenetic Relatedness and Plant Growth Enhancement of Endophytic Bacterial Isolates from Cowpea (Vigna unguiculata) Plant Tissues. Horticulturae 2023, 9, 332. https://doi.org/10.3390/horticulturae9030332
Omomowo OI, Babalola OO. Bioassessment of Phylogenetic Relatedness and Plant Growth Enhancement of Endophytic Bacterial Isolates from Cowpea (Vigna unguiculata) Plant Tissues. Horticulturae. 2023; 9(3):332. https://doi.org/10.3390/horticulturae9030332
Chicago/Turabian StyleOmomowo, Olawale Israel, and Olubukola Oluranti Babalola. 2023. "Bioassessment of Phylogenetic Relatedness and Plant Growth Enhancement of Endophytic Bacterial Isolates from Cowpea (Vigna unguiculata) Plant Tissues" Horticulturae 9, no. 3: 332. https://doi.org/10.3390/horticulturae9030332
APA StyleOmomowo, O. I., & Babalola, O. O. (2023). Bioassessment of Phylogenetic Relatedness and Plant Growth Enhancement of Endophytic Bacterial Isolates from Cowpea (Vigna unguiculata) Plant Tissues. Horticulturae, 9(3), 332. https://doi.org/10.3390/horticulturae9030332








