Genetic Variation and Association Analysis of Phenolic Compounds in Rapeseed (Brassica napus L.) Mutant Lines Using Genotyping-by-Sequencing (GBS)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Library Construction and Genotyping-by-Sequencing
2.3. Sequence Preprocessing and Alignment to Reference Genome Sequence
2.4. Raw SNP Detection and Consensus Sequence Extraction
2.5. Generate SNP Matrix
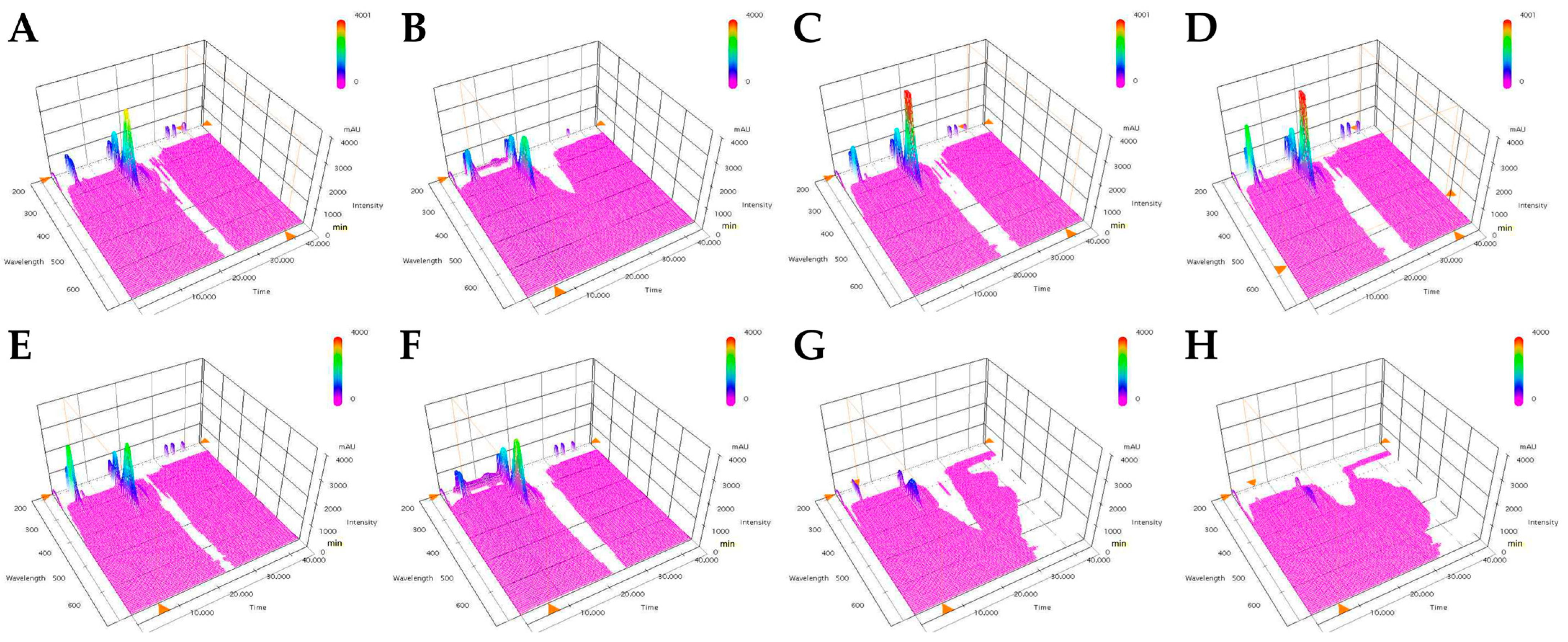
2.6. Ultra-High-Performance Liquid Chromatography (UPLC) and Total Phenolic Content Analysis
2.7. Association and Networking Study
3. Results
3.1. Genotyping-by-Sequencing of Rapeseed Genotypes
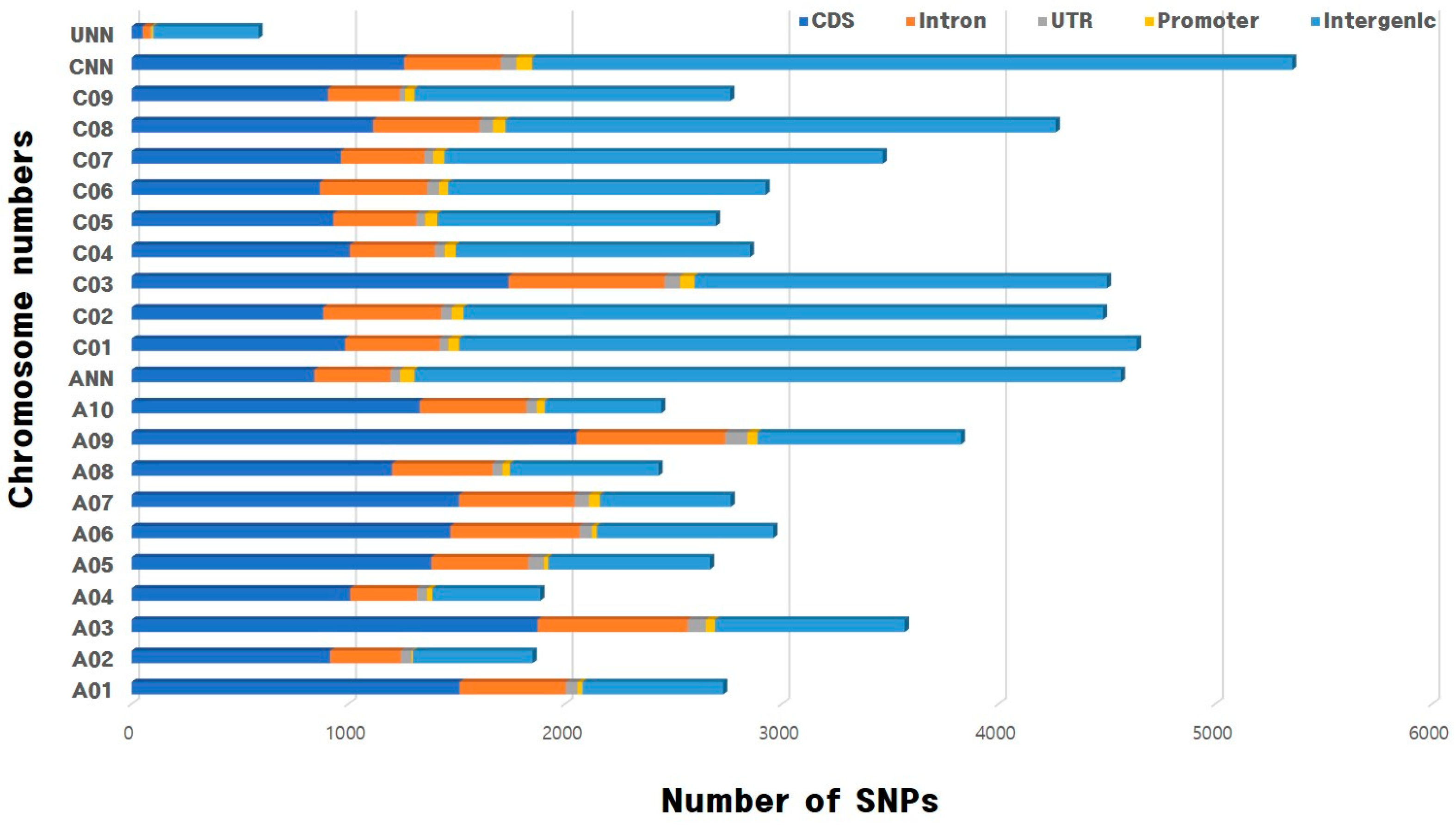
3.2. Identification of SNPs
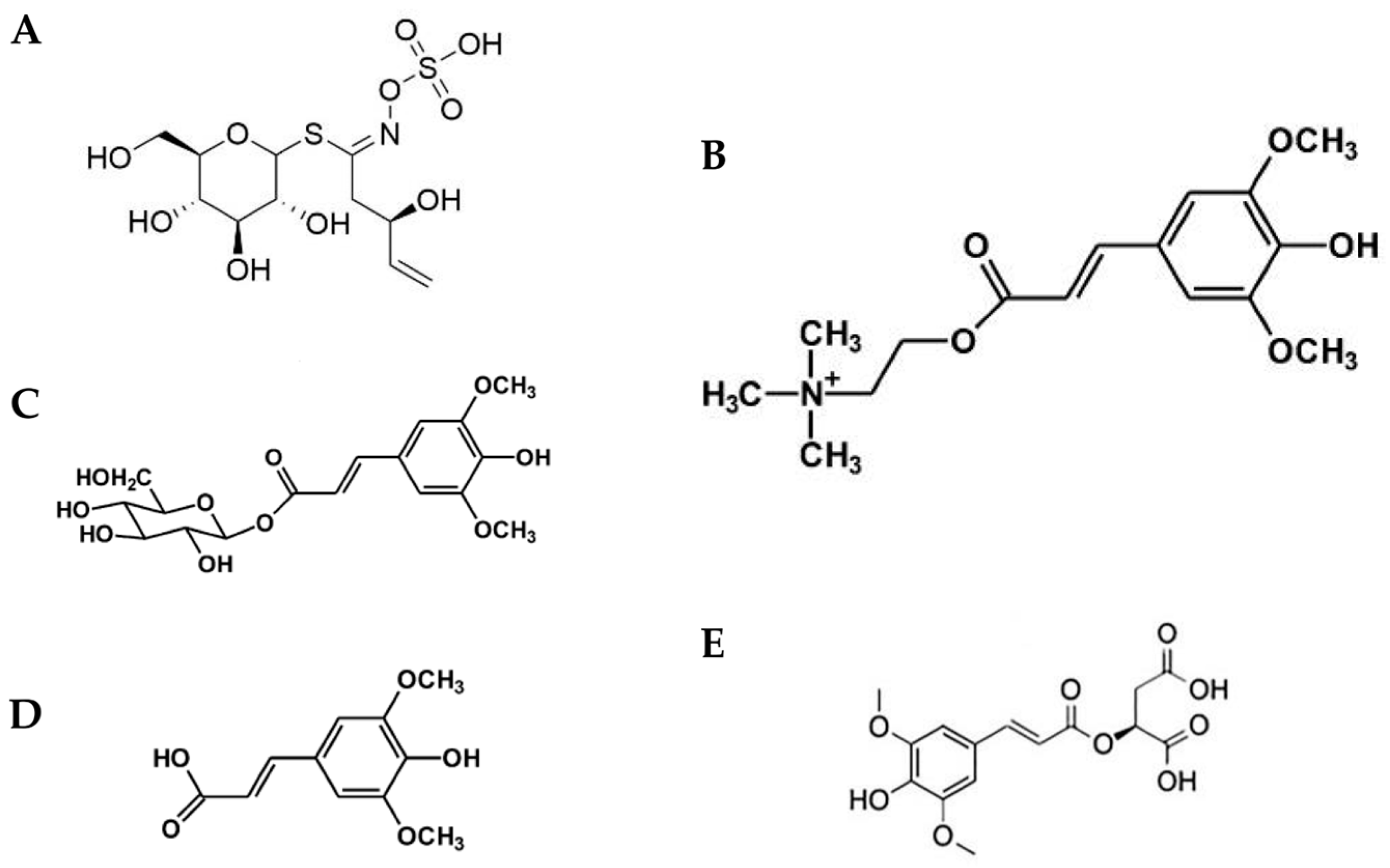
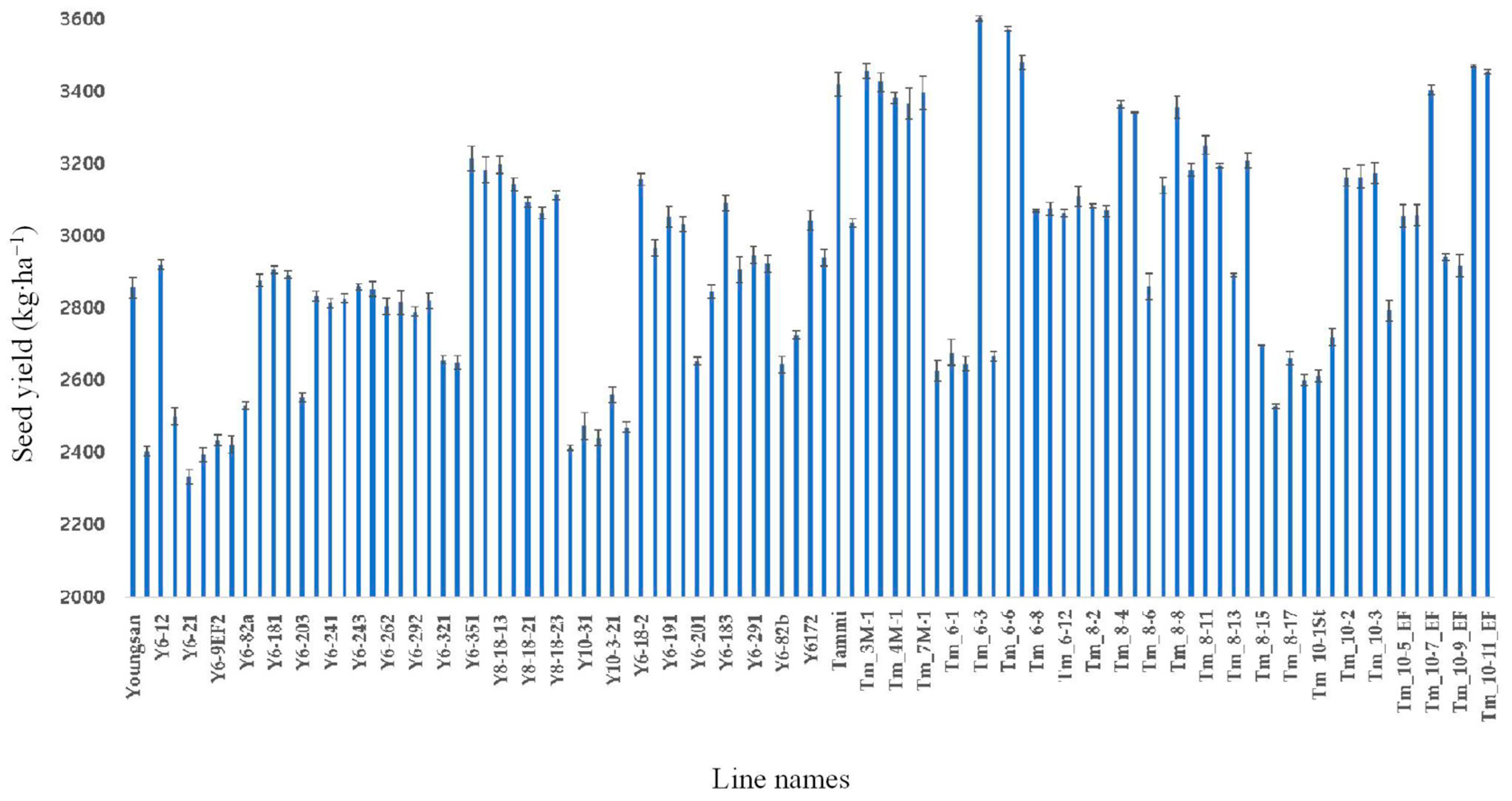
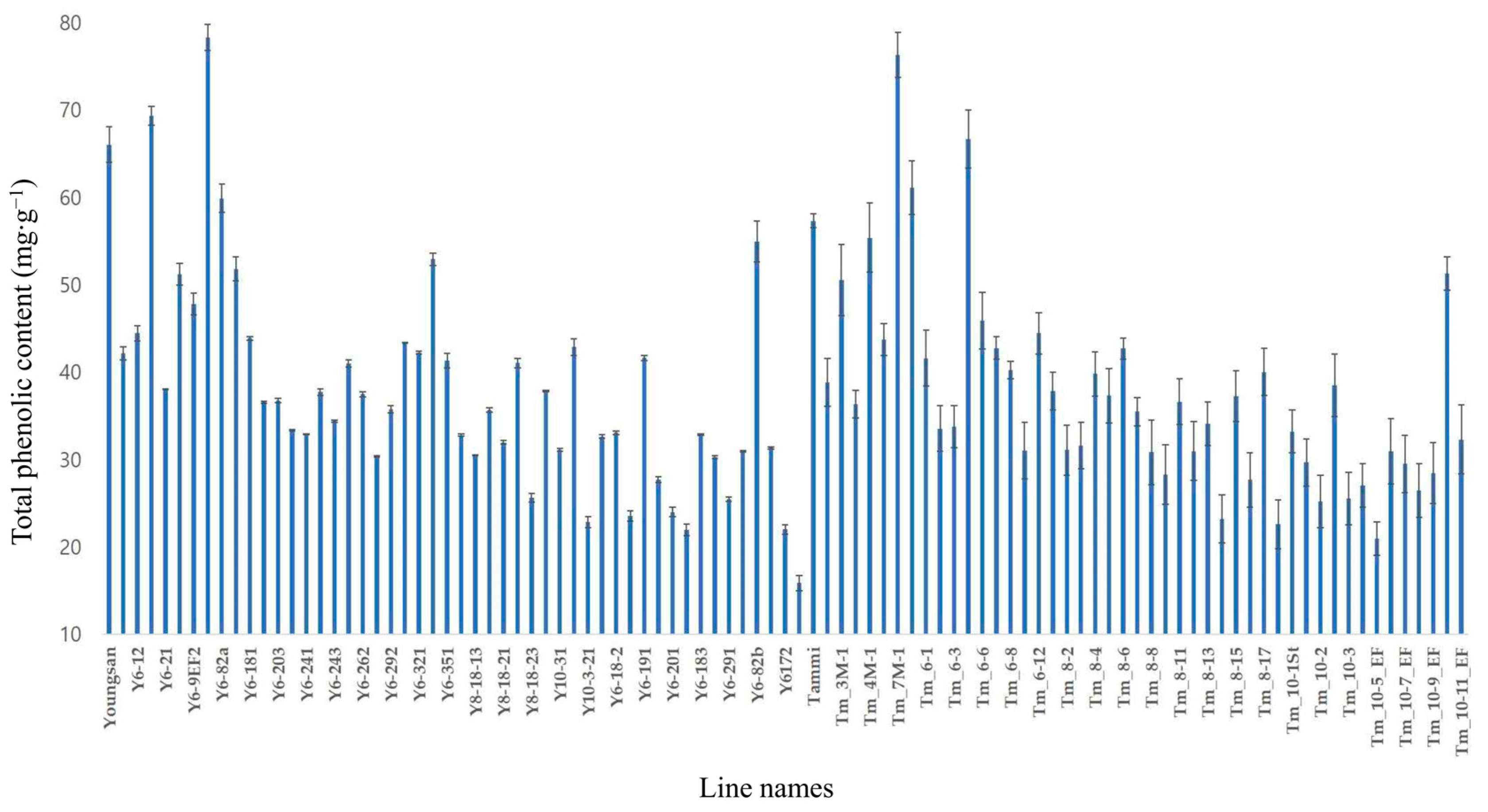
3.3. Phenolic Compound Content and Seed Yield
3.4. Association Analysis
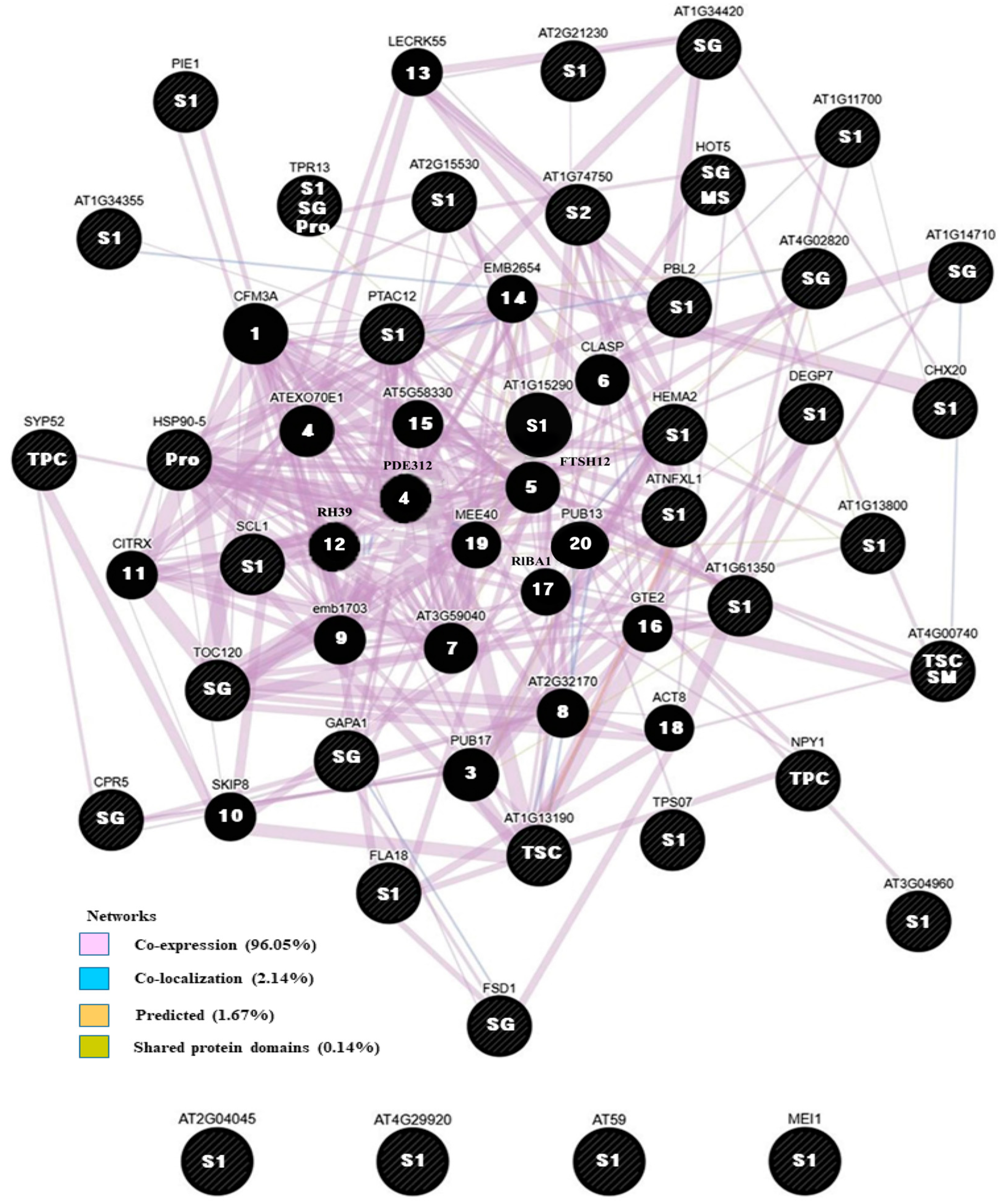
3.5. Networking Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hu, Q.; Hua, W.; Yin, Y.; Zhang, X.; Liu, L.; Shi, J.; Zhao, Y.; Qin, L.; Chen, C.; Wang, H. Rapeseed research and production in Chaina. Crop J. 2017, 5, 127–135. [Google Scholar] [CrossRef]
- Delgado, M.; Felix, M.; Bengoechea, C. Development of bioplastic materials: From rapeseed oil industry by products to added-value biodegradable biocomposite materials. Ind. Crops Prod. 2018, 125, 401–407. [Google Scholar] [CrossRef]
- Nega, T.; Woldes, Y. Review on nutritional limitations and opportunities of using rapeseed meal and other rapeseed by-products in animal feeding. J. Nutr. Health Food Eng. 2018, 8, 43–48. [Google Scholar] [CrossRef]
- Wolfram, K.; Schmidt, J.; Wray, V.; Milkowski, C.; Schliemann, W.; Strack, D. Profiling of phenylpropanoids in transgenic low-sinapine oilseed rapeseed (Brassica napus). Phytochem. 2010, 71, 1076–1084. [Google Scholar] [CrossRef]
- Cartea, M.E.; Francisco, M.; Soengas, P.; Velasco, P. Phenolic compounds in Brassica vegetables. Molecules 2011, 16, 251–280. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Meng, G.; Chen, S.; Chen, Y.; Jiang, J.; Wang, Y.P. Correlation analysis of phenolic contents and antioxidation in Yellow- and black-seeded Brassica napus. Molecules 2018, 23, 1815. [Google Scholar] [CrossRef]
- Obied, H.K.; Song, Y.; Foley, S.; Loughlin, M.; Reman, A.; Mailer, R.; Masud, T.; Agboola, S. Biophenols and antioxidant properties of Australian canola meal. J. Agric. Food Chem. 2013, 61, 9176–9184. [Google Scholar] [CrossRef]
- Tolra, R.P.; Alonso, R.; Poschenrieder, C.; Barcelo, D.; Barcelo, J. Determination of glucosinolates in rapeseed and Thlaspi caerulescens plants by liquid chromatography–atmospheric pressure chemical ionization mass spectrometry. J. Chromatogr. A 2000, 889, 75–81. [Google Scholar] [CrossRef]
- Bischoff, K.L. Glucosinolates. In Nutraceuticals; Academic Press: Cambridge, MA, USA, 2016; pp. 551–554. [Google Scholar]
- Miklavcic Visnjevec, A.; Tamayo Tenorio, A.; Steenkjaer Hastrup, A.C.; Hansen, N.M.L.; Peeters, K.; Schwarzkopf, M. Glucosinolates and isothiocyantes in processed rapeseed determined by HPLC-DAD-qTOF. Plants 2021, 10, 2548. [Google Scholar] [CrossRef]
- Kim, J.I.; Zhang, X.; Pascuzzi, P.E.; Liu, C.J.; Chapple, C. Glucosinolate and phenylpropanoid biosynthesis are linked by proteasome-dependent degradation of PAL. New Phytol. 2020, 225, 154–168. [Google Scholar] [CrossRef]
- Melim, C.; Lauro, M.R.; Pires, I.M.; Oliveira, P.J.; Cabral, C. The role of glucosinolates from cruciferous vegetables (Brassicaceae) in gastrointestinal cancers: From prevention to therapeutics. Pharmaceutics 2022, 14, 190. [Google Scholar] [CrossRef] [PubMed]
- Bhinder, G.; Sharma, S.; Kaur, H.; Akhater, J.; Mittal, M.; Sandhu, S. Genomic regions associated with seed meal quality traits in Brassica napus germplasm. Front. Plant Sci. 2022, 13, 882766. [Google Scholar] [CrossRef] [PubMed]
- Spencer-Lopes, M.M.; Forster, B.P.; Jankuloski, L. Manual on mutation. In Food and Agriculture Organization of the United Nations, 3rd ed.; FAO/IAEA: Rome, Italy, 2018; pp. 187–189. [Google Scholar]
- Singh, S.; Verma, A.K. A review on efforts of induced mutagenesis for qualitative and quantitative improvement of oilseed brassicas. J. Pharm. Phytochem. 2015, 4, 298–302. [Google Scholar]
- Ryu, J.; Nam, B.; Kim, B.R.; Kim, S.H.; Jo, Y.D.; Ahn, J.W.; Kim, J.B.; Jin, C.H.; Han, A.R. Comparative analysis of phytochemical composition of gamma-irradiated mutant cultivars of Chrysanthemum morifolium. Molecules 2019, 24, 3003. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Banks, T.W.; Cloutier, S. SNP discovery through next generation sequencing and its applications. Int. J. Plant Genom. 2012, 2012, 831460. [Google Scholar] [CrossRef]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple genotyping-by- sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef]
- Zhu, M.C.; Ran, H.; Zhao, H.Y.; Tang, Y.S.; Shi, X.T.; Jiang, H.Y.; Zhang, Z.Y.; Fu, F.Y.; Xu, X.F.; Tang, Z.L.; et al. Identification of quantitative trait loci and candidate genes controlling seed pigments of rapeseed. J. Interg. Agric. 2021, 20, 2862–2879. [Google Scholar] [CrossRef]
- Cai, D.; Xiao, Y.; Yang, W.; Ye, W.; Wang, B.; Younas, M.; Wu, J.; Liu, K. Association mapping of six yield-related traits in rapeseed (Brassica napus L.). Theor. Appl. Genet. 2014, 127, 85–96. [Google Scholar] [CrossRef]
- Ryu, J.; Lyu, J.I.; Kim, D.G.; Koo, K.M.; Yang, B.; Jo, Y.D.; Kim, S.H.; Kwon, S.J.; Ha, B.K.; Kang, S.Y.; et al. Single nucleotide polymorphism (SNP) discovery and association study of rapeseed (Brassica napus L.) mutant lines using genotyping-by- sequencing (GBS). Agronomy 2021, 11, 508. [Google Scholar] [CrossRef]
- Khalifa, A.M.; Abd-Elshafy, E.; Abu-Khudir, R.; Gaafar, R.M. Infuence of gamma radiation and phenylalanine on secondary metabolites in callus cultures of milk thistle (Silybum marianum L.). J. Genetic Eng. Biotech. 2022, 20, 166. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–13. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence align- ment/map format and SAM tools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.E.; Kim, J.E.; Kim, J.; Lee, M.H.; Lee, K.; Kim, T.H.; Jo, S.H.; Lee, J.H. Development of an SNP marker set for marker-assisted backcrossing using genotyping-by-sequencing in tetraploid perilla. Mol. Genet. Genom. 2023, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Diosady, L.L. Rapid method for total phenolic acid determination in rapeseed/canola meals. Food Resear. Inter. 1997, 30, 571–574. [Google Scholar] [CrossRef]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet. 2016, 12, e1005767. [Google Scholar] [CrossRef]
- Raboanatahiry, N.; Li, H.; Yu, L.; Li, M. Rapeseed (Brassica napus): Processing, utilization, and genetic improvement. Agronomy 2021, 11, 1776. [Google Scholar] [CrossRef]
- Guirrou, I.; El Harrak, A.; El Antari, A.; Hssaini, L.; Hanine, H.; El Fechtali, M.; Nabloussi, A. Bioactive compounds assessment in six Moroccan rapeseed (Brassica napus L.) varieties grown in two contrasting environments. Agronomy 2023, 13, 460. [Google Scholar] [CrossRef]
- Ryu, J.; Kim, W.J.; Im, J.; Kim, S.H.; Lee, K.S.; Jo, H.J.; Kim, E.Y.; Kang, S.Y.; Lee, J.H.; Ha, H.K. Genotyping-by-sequencing based single nucleotide polymorphisms enabled kompetitive allele specific PCR marker development in mutant Rubus genotypes. Electron. J. Biotechnol. 2018, 35, 57–62. [Google Scholar] [CrossRef]
- Lim, G.H.; Kim, S.W.; Ryu, J.; Kang, S.Y.; Kim, J.B.; Kim, S.H. Upregulation of the MYB2 transcription factor is associated with increased accumulation of anthocyanin in the leaves of Dendrobium bigibbum. Int. J. Mol. Sci. 2020, 21, 5653. [Google Scholar] [CrossRef]
- Lee, T.S.; Lee, Y.H.; Kim, K.S.; Lee, H.K.; Jang, Y.S.; Choi, I.H.; Kim, K.S. Effect of sowing time on oil content and fatty acid composition characteristics in rapeseed cultivars. Korean J. Plant Resour. 2014, 27, 202–208. [Google Scholar] [CrossRef][Green Version]
- Naczk, M.; Amarowicz, R.; Sullivan, A.; Shahidi, F. Current research development on polyphenolics of rapeseed/canola: A review. Food Chem. 1998, 62, 489–502. [Google Scholar] [CrossRef]
- Felde, T.Z.; Baumert, A.; Strack, D.; Becker, H.C.; Mollers, C. Genetic variation for sinapate ester content in winter rapeseed (Brassica napus L.) and development of NIRS calibration equations. Plant Breed. 2007, 126, 291–296. [Google Scholar] [CrossRef]
- Yang, S.C.; Arasu, M.V.; Chun, J.H.; Jang, Y.S.; Lee, Y.H.; Kim, I.H.; Lee, K.T.; Hong, S.T.; Kim, S.J. Identification and determination of phenolic compounds in rapeseed meals (Brassica napus L.). J. Agric. Chem. Environ. 2015, 4, 14–23. [Google Scholar] [CrossRef]
- Chen, G.; Geng, J.F.; Rahman, M.; Liu, X.P.; Tu, J.X.; Fu, T.D.; Li, G.Y.; McVetty, P.B.E.; Tahir, M. Identification of QTL for oil content, seed yield, and flowering time in oilseed rape (Brassica napus). Euphytica 2010, 175, 161–174. [Google Scholar] [CrossRef]
- Lu, K.; Wei, L.; Li, X.; Wang, Y.; Wu, M.; Liu, M.; Zhang, C.; Chen, Z.; Xiao, Z.; Jian, H.; et al. Whole-genome resequencing reveals Brassica napus origin and genetic loci involved in its improvement. Nat. Commun. 2019, 10, 1154. [Google Scholar] [CrossRef]
- Soundararajan, P.; Kim, J.S. Anti-carcinogenic glucosinolates in cruciferous vegetables and their antagonistic effects on prevention of cancers. Molecules 2018, 23, 2983. [Google Scholar] [CrossRef]
- Jeschke, V.; Weber, K.; Moore, S.S.; Burow, M. Coordination of glucosinolate biosynthesis and turnover under different nutrient conditions. Front. Plant Sci. 2019, 10, 1560. [Google Scholar] [CrossRef]
- Gangurde, S.S.; Nayak, S.N.; Joshi, P.; Purohit, S.; Sudini, H.K.; Chitikineni, A.; Hong, Y.; Guo, B.; Chen, X.; Pandey, M.K.; et al. Comparative transcriptome analysis identified candidate genes for late leaf spot resistance and cause of defoliation in groundnut. Int. J. Mol. Sci. 2021, 22, 4491. [Google Scholar] [CrossRef]
- Zhang, X.; Gou, M.; Liu, C.J. Arabidopsis Kelch repeat F-Box proteins regulate phenylpropanoid biosynthesis via controlling the turnover of phenylalanine ammonia-lyase. Plant Cell 2013, 25, 4994–5010. [Google Scholar] [CrossRef]
- Qu, C.; Li, J.; Zhu, K.; Liang, D.; Liu, J.; Hu, Q. Metabolic engineering of fatty acids for breeding of new oilseed crops: Strategies, progress, and perspectives. Engineering 2017, 3, 27–35. [Google Scholar]
- Zhu, Q.; Dugardeyn, J.; Zhang, C.; Takenaka, M.; Kristina Kuhn, K.; Craddock, C.; Smalle, J.; Karampelias, M.; Denecke, J.; Peters, J.; et al. SLO2, a mitochondrial pentatricopeptide repeat protein affecting several RNA editing sites, is required for energy metabolism. Plant J. 2012, 71, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Chateigner-Boutin, A.L.; des Francs-Small, C.C.; Delannoy, E.; Kahlau, S.; Tanz, S.K.; de Longevialle, A.F.; Sota Fujii, S.; Small, L. OTP70 is a pentatricopeptide repeat protein of the E subgroup involved in splicing of the plastid transcript rpoC1. Plant J. 2012, 65, 532–542. [Google Scholar] [CrossRef] [PubMed]
- Eren, E.; Arguello, J.M. Arabidopsis HMA2, a divalent heavy metal-transporting PIB-type ATPase, is involved in cytoplasmic Zn2+ homeostasis. Plant Physiol. 2004, 136, 3712–3723. [Google Scholar] [CrossRef]
- Yuan, Y.; Wu, H.; Wang, N.; Li, J.; Zhao, W.; Du, J.; Wang, D.; Ling, H.Q. FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res. 2008, 18, 385–397. [Google Scholar] [CrossRef]
- Nguyen, V.P.T.; Stewart, J.D.; Ioannou, I.; Allais, F. Sinapic acid and sinapate esters in Brassica: Innate accumulation, biosynthesis, accessibility via chemical synthesis or recovery from biomass, and biological activities. Front. Chem. 2021, 9, 664602. [Google Scholar] [CrossRef]
- Strehmel, N.; Hoehenwarter, W.; Mönchgesang, S.; Majovsky, P.; Krüger, S.; Scheel, D.; Lee, J. Stress-related mitogen-activated protein kinases stimulate the accumulation of small molecules and proteins in Arabidopsis thaliana root exudates. Front. Plant Sci. 2017, 8, 1292. [Google Scholar] [CrossRef]
- Wang, Z.; Bao, L.L.; Zhao, F.Y.; Tang, M.Q.; Chen, T.; Li, Y.; Wang, B.X.; Fu, B.; Fang, H.; Li, G.Y.; et al. BnaMPK3 Is a key regulator of defense responses to the devastating plant pathogen Sclerotinia sclerotiorum in oilseed rape. Front. Plant Sci. 2019, 10, 91. [Google Scholar] [CrossRef]
- Tohge, T.; Watanabe, M.; Hoefgen, R.; Fernie, A.R. The evolution of phenylpropanoid metabolism in the green lineage. CRBMol. Biol. Rev. 2013, 2, 123–152. [Google Scholar] [CrossRef]
- Vogt, T. Phenylpropanoid biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef]
- Kim, D.G.; Lyu, J.I.; Kim, J.M.; Seo, J.S.; Choi, H.I.; Jo, Y.D.; Kim, S.H.; Eom, S.H.; Ahn, J.W.; Bae, C.H.; et al. Identification of loci governing agronomic traits and mutation hotspots via a gbs-based genome-wide association study in a soybean mutant diversity pool. Int. J. Mol. Sci. 2022, 23, 10441. [Google Scholar] [CrossRef] [PubMed]
| Total | Average/Plant | |
|---|---|---|
| Raw data | ||
| Reads | 1,088,590,054 | 11,222,578 |
| Bases (bp) | 164,377,098,154 | 1,694,609,259 |
| After trimming | ||
| Reads | 967,244,884 | 9,971,597 |
| Bases (bp) | 107,921,780,499 | 1,112,595,675 |
| Mapped reads on reference genome 1 | ||
| Mapped reads | 740,184,076 | 7,630,764 |
| Bases (bp) | 1,765,446,134 | 18,200,476 |
| Reference genome coverage (%) | 2.14 | |
| Reference genome depth | 15.82 | |
| No. | Line Names | Dihexose | Progoitrin | trans- Sinapine 1 | trans- Sinapine 2 | Sinapine (4-O-8′) Guaiacyl | Sinapoyl Malate | Methyl Sinapate | Total |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Youngsan | 0.70 ± 0.29 | 0.15 ± 0.06 | 16.03 ± 0.04 | 17.65 ± 2.00 | 0.14 ± 0.08 | 0.94 ± 0.11 | 0.05 ± 0.01 | 35.66 ± 2.69 |
| 2 | Y6-3 | 0.49 ± 0.21 | 0.24 ± 0.07 | 13.25 ± 2.00 | 11.77 ± 1.04 | 0.15 ± 0.02 | 0.22 ± 0.05 | 0.05 ± 0.01 | 26.18 ± 3.50 |
| 3 | Y6-12 | 0.56 ± 0.20 | 0.18 ± 0.02 | 14.00 ± 2.04 | 12.50 ± 1.13 | 0.16 ± 0.01 | 0.27 ± 0.06 | 0.06 ± 0.02 | 27.73 ± 3.58 |
| 4 | Y6-11 | 0.52 ± 0.23 | 0.13 ± 0.07 | 18.46 ± 0.30 | 22.89 ± 0.88 | 0.16 ± 0.06 | 0.30 ± 0.06 | 0.04 ± 0.01 | 42.50 ± 1.71 |
| 5 | Y6-21 | 0.38 ± 0.13 | 0.16 ± 0.06 | 12.41 ± 1.04 | 09.11 ± 0.55 | 0.21 ± 0.04 | 0.04 ± 0.01 | 0.18 ± 0.07 | 22.48 ± 1.99 |
| 6 | Y6-9EF1 | 0.60 ± 0.24 | 0.13 ± 0.07 | 14.74 ± 0.74 | 14.18 ± 1.47 | 0.20 ± 0.06 | 0.68 ± 0.13 | 0.04 ± 0.01 | 30.57 ± 2.82 |
| 7 | Y6-9EF2 | 0.67 ± 0.30 | 0.26 ± 0.05 | 14.87 ± 0.25 | 13.54 ± 1.40 | 0.18 ± 0.03 | 0.32 ± 0.06 | 0.04 ± 0.01 | 29.88 ± 2.21 |
| 8 | Y6-81a | 1.48 ± 0.66 | 0.40 ± 0.00 | 18.07 ± 1.13 | 24.77 ± 1.45 | 0.21 ± 0.03 | 0.81 ± 0.15 | 0.29 ± 0.08 | 46.03 ± 3.71 |
| 9 | Y6-82a | 1.23 ± 0.55 | 0.36 ± 0.01 | 15.70 ± 0.88 | 16.85 ± 1.65 | 0.23 ± 0.04 | 0.69 ± 0.12 | 0.25 ± 0.07 | 35.31 ± 3.43 |
| 10 | Y6-171 | 1.11 ± 0.48 | 0.34 ± 0.02 | 13.82 ± 0.24 | 14.19 ± 1.55 | 0.20 ± 0.02 | 0.63 ± 0.12 | 0.22 ± 0.07 | 30.51 ± 2.60 |
| 11 | Y6-181 | 0.40 ± 0.13 | 0.15 ± 0.05 | 13.72 ± 0.88 | 10.19 ± 0.82 | 0.08 ± 0.00 | 0.34 ± 0.10 | 0.12± 0.04 | 25.01 ± 2.01 |
| 12 | Y6-192 | 0.43 ± 0.16 | 0.15 ± 0.05 | 11.25 ± 1.12 | 09.12 ± 0.57 | 0.07 ± 0.00 | 0.34 ± 0.09 | 0.15 ± 0.05 | 21.51 ± 2.04 |
| 13 | Y6-203 | 0.29 ± 0.10 | 0.14 ± 0.06 | 12.06 ± 0.58 | 09.78 ± 0.72 | 0.17 ± 0.03 | 0.02 ± 0.01 | 0.12 ± 0.04 | 22.59 ± 1.54 |
| 14 | Y6-221 | 0.33 ± 0.12 | 0.15 ± 0.05 | 10.95 ± 0.55 | 08.72 ± 0.50 | 0.18 ± 0.05 | 0.02 ± 0.01 | 0.13 ± 0.04 | 20.49 ± 1.31 |
| 15 | Y6-241 | 0.32 ± 0.12 | 0.15 ± 0.05 | 10.81 ± 0.03 | 08.85 ± 0.53 | 0.19 ± 0.05 | 0.02 ± 0.00 | 0.12 ± 0.04 | 20.47 ± 0.83 |
| 16 | Y6-242 | 0.28 ± 0.10 | 0.14 ± 0.06 | 12.77 ± 0.23 | 10.14 ± 0.82 | 0.16 ± 0.03 | 0.02 ± 0.01 | 0.14 ± 0.04 | 23.64 ± 1.29 |
| 17 | Y6-243 | 0.29 ± 0.10 | 0.14 ± 0.05 | 11.63 ± 1.47 | 09.39 ± 0.64 | 0.14 ± 0.03 | 0.02 ± 0.00 | 0.11 ± 0.03 | 21.72 ± 2.34 |
| 18 | Y6-251 | 0.90 ± 0.40 | 0.27 ± 0.03 | 13.43 ± 1.24 | 10.93 ± 1.05 | 0.28 ± 0.08 | 0.01 ± 0.00 | 0.11 ± 0.03 | 25.93 ± 2.83 |
| 19 | Y6-262 | 0.40 ± 0.12 | 0.16 ± 0.04 | 11.94 ± 0.17 | 10.17 ± 0.86 | 0.19 ± 0.05 | 0.01 ± 0.00 | 0.14 ± 0.04 | 23.01 ± 1.28 |
| 20 | Y6-271 | 0.33 ± 0.11 | 0.10 ± 0.02 | 09.74 ± 1.40 | 08.46 ± 0.48 | 0.13 ± 0.03 | 0.02 ± 0.01 | 0.08 ± 0.03 | 18.87 ± 2.07 |
| 21 | Y6-292 | 0.25 ± 0.06 | 0.13 ± 0.05 | 10.97 ± 1.22 | 09.28 ± 0.77 | 0.07 ± 0.01 | 0.05 ± 0.01 | 0.14 ± 0.06 | 20.89 ± 2.20 |
| 22 | Y6-294 | 0.29 ± 0.09 | 0.13 ± 0.05 | 12.69 ± 0.08 | 10.62 ± 1.02 | 0.11 ± 0.04 | 0.01 ± 0.00 | 0.18 ± 0.07 | 24.03 ± 1.34 |
| 23 | Y6-321 | 0.30 ± 0.11 | 0.09 ± 0.02 | 12.08 ± 1.45 | 09.54 ± 0.88 | 0.13 ± 0.02 | 0.04 ± 0.01 | 0.10 ± 0.04 | 22.27 ± 2.54 |
| 24 | Y6-322 | 0.44 ± 0.17 | 0.10 ± 0.01 | 16.71 ± 1.53 | 13.38 ± 1.61 | 0.15 ± 0.03 | 0.04 ± 0.01 | 0.17 ± 0.06 | 30.99 ± 3.43 |
| 25 | Y6-351 | 0.46 ± 0.22 | 0.09 ± 0.04 | 12.26 ± 0.02 | 12.78 ± 1.35 | 0.11 ± 0.02 | 0.19 ± 0.04 | 0.04 ± 0.01 | 25.94 ± 1.70 |
| 26 | Y8-18-12 | 0.33 ± 0.13 | 0.11 ± 0.01 | 10.64 ± 1.65 | 09.07 ± 0.67 | 0.08 ± 0.02 | 0.03 ± 0.01 | 0.12 ± 0.04 | 20.37 ± 2.53 |
| 27 | Y8-18-13 | 0.31 ± 0.11 | 0.10 ± 0.02 | 09.82 ± 1.63 | 08.41 ± 0.48 | 0.06 ± 0.02 | 0.04 ± 0.01 | 0.08 ± 0.02 | 18.82 ± 2.28 |
| 28 | Y8-18-14 | 0.41 ± 0.17 | 0.13 ± 0.00 | 11.24 ± 0.22 | 09.69 ± 0.77 | 0.07 ± 0.02 | 0.04 ± 0.01 | 0.13 ± 0.06 | 21.72 ± 1.25 |
| 29 | Y8-18-21 | 0.31 ± 0.11 | 0.09 ± 0.02 | 10.01 ± 1.55 | 08.37 ± 0.50 | 0.07 ± 0.02 | 0.02 ± 0.01 | 0.10 ± 0.03 | 18.98 ± 2.24 |
| 30 | Y8-18-22 | 0.35 ± 0.13 | 0.11 ± 0.02 | 13.04 ± 1.33 | 10.94 ± 1.08 | 0.09 ± 0.02 | 0.03 ± 0.01 | 0.16 ± 0.07 | 24.71 ± 2.65 |
| 31 | Y8-18-23 | 0.25 ± 0.08 | 0.09 ± 0.02 | 07.66 ± 0.60 | 06.74 ± 0.18 | 0.05 ± 0.02 | 0.02 ± 0.01 | 0.11 ± 0.04 | 14.91 ± 0.95 |
| 32 | Y10-12 | 0.37 ± 0.13 | 0.16 ± 0.04 | 10.26 ± 0.82 | 08.99 ± 0.77 | 0.09 ± 0.03 | 0.06 ± 0.02 | 0.18 ± 0.07 | 20.11 ± 1.88 |
| 33 | Y10-31 | 0.23 ± 0.08 | 0.09 ± 0.02 | 09.57 ± 0.22 | 08.11 ± 0.51 | 0.07 ± 0.03 | 0.03 ± 0.01 | 0.14 ± 0.05 | 18.24 ± 0.93 |
| 34 | Y10-32 | 0.31 ± 0.11 | 0.09 ± 0.02 | 12.85 ± 0.69 | 10.45 ± 1.31 | 0.14 ± 0.04 | 0.05 ± 0.01 | 0.17 ± 0.08 | 24.06 ± 2.28 |
| 35 | Y10-3-21 | 0.33 ± 0.11 | 0.14 ± 0.04 | 07.20 ± 0.57 | 06.05 ± 0.07 | 0.06 ± 0.01 | 0.04 ± 0.01 | 0.07 ± 0.02 | 13.89 ± 0.84 |
| 36 | Y10-1b2 | 0.43 ± 0.18 | 0.12 ± 0.00 | 10.15 ± 0.12 | 08.20 ± 0.51 | 0.19 ± 0.05 | 0.02 ± 0.00 | 0.11 ± 0.04 | 19.22 ± 0.91 |
| 37 | Y6-18-2 | 0.27 ± 0.09 | 0.09 ± 0.03 | 09.79 ± 0.49 | 07.94 ± 0.49 | 0.04 ± 0.01 | 0.02 ± 0.00 | 0.11 ± 0.03 | 18.25 ± 1.14 |
| 38 | Y6-81b | 0.40 ± 0.14 | 0.15 ± 0.04 | 07.39 ± 0.72 | 06.18 ± 0.09 | 0.07 ± 0.01 | 0.03 ± 0.01 | 0.05 ± 0.01 | 14.27 ± 1.03 |
| 39 | Y6-191 | 0.37 ± 0.12 | 0.10 ± 0.02 | 13.00 ± 0.23 | 09.84 ± 0.95 | 0.07 ± 0.03 | 0.02 ± 0.01 | 0.14 ± 0.05 | 23.54 ± 1.41 |
| 40 | Y8-18-11 | 0.25 ± 0.10 | 0.09 ± 0.02 | 08.53 ± 0.57 | 07.26 ± 0.32 | 0.06 ± 0.01 | 0.02 ± 0.01 | 0.13 ± 0.05 | 16.35 ± 1.08 |
| 41 | Y6-201 | 0.51 ± 0.20 | 0.14 ± 0.01 | 07.18 ± 0.50 | 06.28 ± 0.13 | 0.15 ± 0.04 | 0.02 ± 0.01 | 0.08 ± 0.02 | 14.37 ± 0.91 |
| 42 | Y6-202 | 0.20 ± 0.07 | 0.08 ± 0.02 | 07.05 ± 0.07 | 05.78 ± 0.06 | 0.08 ± 0.02 | 0.02 ± 0.01 | 0.11 ± 0.03 | 13.32 ± 0.27 |
| 43 | Y6-183 | 0.46 ± 0.15 | 0.10 ± 0.02 | 09.16 ± 0.58 | 07.80 ± 0.50 | 0.04 ± 0.02 | 0.02 ± 0.01 | 0.14 ± 0.04 | 17.73 ± 1.32 |
| 44 | Y6-252 | 0.37 ± 0.12 | 0.10 ± 0.01 | 09.76 ± 0.53 | 08.12 ± 0.54 | 0.15 ± 0.04 | 0.01 ± 0.00 | 0.07 ± 0.02 | 18.59 ± 1.26 |
| 45 | Y6-291 | 0.29 ± 0.09 | 0.10 ± 0.02 | 06.85 ± 0.05 | 06.69 ± 0.26 | 0.04 ± 0.01 | 0.10 ± 0.02 | 0.11 ± 0.03 | 14.17 ± 0.49 |
| 46 | Y6-222 | 0.37 ± 0.15 | 0.10 ± 0.01 | 09.73 ± 0.47 | 08.30 ± 0.62 | 0.13 ± 0.03 | 0.02 ± 0.01 | 0.12 ± 0.05 | 18.77 ± 1.34 |
| 47 | Y6-82b | 1.41 ± 0.70 | 0.29 ± 0.08 | 13.32 ± 0.82 | 16.32 ± 2.64 | 0.09 ± 0.03 | 0.71 ± 0.16 | 0.25 ± 0.07 | 32.39 ± 4.50 |
| 48 | Y6-22 | 0.41 ± 0.17 | 0.11 ± 0.01 | 08.91 ± 0.35 | 07.69 ± 0.55 | 0.17 ± 0.03 | 0.02 ± 0.01 | 0.14 ± 0.06 | 17.44 ± 1.18 |
| 49 | Y6172 | 0.32 ± 0.11 | 0.10 ± 0.01 | 06.41 ± 0.54 | 05.50 ± 0.11 | 0.05 ± 0.01 | 0.02 ± 0.00 | 0.06 ± 0.02 | 12.47 ± 0.80 |
| 50 | Y6-29-30 | 0.39 ± 0.13 | 0.15 ± 0.03 | 04.31 ± 0.64 | 04.01 ± 0.16 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.04 ± 0.01 | 08.96 ± 0.99 |
| 51 | Tammi | 0.21 ± 0.04 | 0.08 ± 0.01 | 07.64 ± 1.63 | 22.22 ± 1.48 | 0.12 ± 0.03 | 1.08 ± 0.27 | 0.09 ± 0.03 | 31.45 ± 3.50 |
| 52 | Tm2M-1 | 0.30 ± 0.06 | 0.05 ± 0.01 | 03.67 ± 0.66 | 17.14 ± 1.32 | 0.05 ± 0.02 | 0.29 ± 0.07 | 0.04 ± 0.01 | 21.54 ± 2.15 |
| 53 | Tm3M-1 | 0.69 ± 0.13 | 0.45 ± 0.19 | 03.36 ± 0.55 | 17.96 ± 1.40 | 0.01 ± 0.00 | 0.35 ± 0.09 | 0.07 ± 0.03 | 22.89 ± 2.39 |
| 54 | Tm3M-2 | 0.07 ± 0.01 | 0.08 ± 0.01 | 18.85 ± 1.74 | 12.29 ± 2.72 | 0.05 ± 0.01 | 0.30 ± 0.08 | 0.02 ± 0.01 | 31.65 ± 4.58 |
| 55 | Tm4M-1 | 0.48 ± 0.04 | 0.35 ± 0.13 | 04.50 ± 0.77 | 19.64 ± 1.35 | 0.07 ± 0.03 | 0.86 ± 0.20 | 0.07 ± 0.02 | 25.97 ± 2.54 |
| 56 | Tm4M-2 | 0.34 ± 0.03 | 0.10 ± 0.01 | 04.15 ± 0.67 | 19.71 ± 1.55 | 0.07 ± 0.03 | 0.41 ± 0.10 | 0.06 ± 0.02 | 24.84 ± 2.41 |
| 57 | Tm7M-1 | 0.57 ± 0.12 | 0.41 ± 0.09 | 07.09 ± 0.97 | 25.67 ± 1.22 | 0.12 ± 0.04 | 1.01 ± 0.26 | 0.31 ± 0.09 | 35.18 ± 2.79 |
| 58 | Tm7M-2 | 0.52 ± 0.11 | 0.37 ± 0.07 | 04.39 ± 1.14 | 19.56 ± 1.30 | 0.09 ± 0.03 | 0.78 ± 0.20 | 0.24 ± 0.07 | 25.94 ± 2.91 |
| 59 | Tm6-1 | 0.03 ± 0.01 | 0.08 ± 0.01 | 11.41 ± 1.82 | 12.92 ± 2.77 | 0.07 ± 0.02 | 0.02 ± 0.01 | 0.02 ± 0.01 | 24.55 ± 4.65 |
| 60 | Tm6-2 | 0.06 ± 0.00 | 0.13 ± 0.02 | 06.34 ± 0.99 | 11.16 ± 2.59 | 0.04 ± 0.02 | 0.02 ± 0.01 | 0.02 ± 0.01 | 17.78 ± 3.63 |
| 61 | Tm6-3 | 0.05 ± 0.00 | 0.14 ± 0.01 | 03.80 ± 0.50 | 12.30 ± 2.88 | 0.05 ± 0.02 | 0.03 ± 0.01 | 0.02 ± 0.01 | 16.38 ± 3.43 |
| 62 | Tm6-4 | 0.54 ± 0.10 | 0.48 ± 0.00 | 04.82 ± 1.14 | 21.71 ± 0.90 | 0.10 ± 0.03 | 0.87 ± 0.22 | 0.27 ± 0.08 | 28.79 ± 2.47 |
| 63 | Tm6-6 | 0.21 ± 0.03 | 0.11 ± 0.00 | 04.79 ± 0.94 | 24.41 ± 1.38 | 0.07 ± 0.02 | 0.38 ± 0.09 | 0.01 ± 0.00 | 29.98 ± 2.47 |
| 64 | Tm6-7 | 0.43 ± 0.07 | 0.50 ± 0.05 | 04.54 ± 0.80 | 11.23 ± 2.65 | 0.05 ± 0.02 | 0.03 ± 0.01 | 0.02 ± 0.01 | 16.80 ± 3.61 |
| 65 | Tm6-8 | 0.39 ± 0.06 | 0.35 ± 0.04 | 03.10 ± 0.35 | 11.21 ± 2.65 | 0.04 ± 0.01 | 0.02 ± 0.01 | 0.02 ± 0.01 | 15.13 ± 3.12 |
| 66 | Tm6-10 | 0.03 ± 0.01 | 0.07 ± 0.00 | 02.77 ± 0.43 | 12.22 ± 2.71 | 0.04 ± 0.01 | 0.02 ± 0.01 | 0.02 ± 0.01 | 15.16 ± 3.18 |
| 67 | Tm6-12 | 0.55 ± 0.09 | 0.40 ± 0.03 | 04.86 ± 0.98 | 16.93 ± 1.01 | 0.07 ± 0.02 | 0.02 ± 0.01 | 0.02 ± 0.01 | 22.85 ± 2.15 |
| 68 | Tm6-13 | 0.13 ± 0.01 | 0.08 ± 0.00 | 04.06 ± 0.81 | 13.70 ± 2.66 | 0.06 ± 0.02 | 0.03 ± 0.01 | 0.03 ± 0.01 | 18.09 ± 3.52 |
| 69 | Tm8-2 | 0.10 ± 0.04 | 0.07 ± 0.01 | 02.85 ± 0.54 | 11.25 ± 2.53 | 0.04 ± 0.01 | 0.03 ± 0.01 | 0.02 ± 0.01 | 14.37 ± 3.14 |
| 70 | Tm8-3 | 0.04 ± 0.00 | 0.06 ± 0.00 | 09.94 ± 2.35 | 11.65 ± 2.66 | 0.05 ± 0.01 | 0.07 ± 0.02 | 0.03 ± 0.01 | 21.84 ± 5.06 |
| 71 | Tm8-4 | 0.13 ± 0.02 | 0.06 ± 0.01 | 13.21 ±3.34 | 13.43 ± 2.66 | 0.09 ± 0.02 | 0.05 ± 0.01 | 0.04 ± 0.01 | 27.01 ± 6.07 |
| 72 | Tm8-5 | 0.03 ± 0.01 | 0.06 ± 0.01 | 20.81 ± 1.32 | 11.85 ± 2.86 | 0.07 ± 0.02 | 0.06 ± 0.02 | 0.02 ± 0.01 | 32.91 ± 4.23 |
| 73 | Tm8-6 | 0.09 ± 0.02 | 0.07 ± 0.00 | 22.15 ± 1.57 | 18.04 ± 0.30 | 0.10 ± 0.02 | 0.06 ± 0.02 | 0.04 ± 0.01 | 40.55 ± 1.94 |
| 74 | Tm8-7 | 0.14 ± 0.03 | 0.08 ± 0.01 | 02.74 ± 0.69 | 17.10 ± 1.03 | 0.09 ± 0.02 | 0.25 ± 0.06 | 0.01 ± 0.00 | 20.40 ± 1.84 |
| 75 | Tm8-8 | 0.09 ± 0.00 | 0.07 ± 0.00 | 04.85 ± 1.03 | 11.00 ± 2.70 | 0.07 ± 0.02 | 0.04 ± 0.01 | 0.03 ± 0.01 | 16.17 ± 3.77 |
| 76 | Tm8-10 | 0.07 ± 0.03 | 0.06 ± 0.00 | 04.34 ± 0.86 | 10.71 ± 2.47 | 0.05 ± 0.01 | 0.06 ± 0.02 | 0.02 ± 0.01 | 15.31 ± 3.39 |
| 77 | Tm8-11 | 0.17 ± 0.04 | 0.31 ± 0.04 | 05.54 ± 1.20 | 12.38 ± 2.43 | 0.07 ± 0.02 | 0.05 ± 0.01 | 0.04 ± 0.01 | 18.55 ± 3.75 |
| 78 | Tm8-12 | 0.08 ± 0.02 | 0.06 ± 0.00 | 04.74 ± 1.17 | 10.31 ± 2.38 | 0.05 ± 0.02 | 0.03 ± 0.01 | 0.03 ± 0.01 | 15.30 ± 3.60 |
| 79 | Tm8-13 | 0.18 ± 0.01 | 0.27 ± 0.03 | 07.81 ± 0.38 | 15.66 ± 0.69 | 0.08 ± 0.02 | 0.04 ± 0.01 | 0.05 ± 0.02 | 24.09 ±1.16 |
| 80 | Tm8-14 | 0.08 ± 0.00 | 0.05 ± 0.01 | 04.38 ± 1.09 | 07.92 ± 2.14 | 0.04 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 | 12.53 ± 3.27 |
| 81 | Tm8-15 | 0.17 ± 0.01 | 0.28 ± 0.02 | 08.00 ± 1.97 | 10.37 ± 2.43 | 0.08 ± 0.02 | 0.08 ± 0.02 | 0.05 ± 0.02 | 19.03 ±4.49 |
| 82 | Tm8-16 | 0.09 ± 0.00 | 0.06 ± 0.00 | 03.30 ± 0.79 | 09.37 ± 2.33 | 0.12 ± 0.02 | 0.03 ± 0.01 | 0.04 ± 0.01 | 13.02 ± 3.16 |
| 83 | Tm8-17 | 0.17 ± 0.01 | 0.31 ± 0.00 | 08.42 ± 1.18 | 11.47 ± 2.51 | 0.26 ± 0.04 | 0.07 ± 0.02 | 0.06 ± 0.02 | 20.74 ± 3.78 |
| 84 | Tm10-1 | 0.08 ± 0.01 | 0.06 ± 0.01 | 03.28 ± 0.73 | 07.32 ± 2.03 | 0.10 ± 0.02 | 0.06 ± 0.01 | 0.02 ± 0.01 | 10.92 ± 2.81 |
| 85 | Tm10-1St | 0.16 ± 0.02 | 0.30 ± 0.03 | 05.39 ± 0.42 | 09.93 ± 2.28 | 0.34 ± 0.06 | 0.04 ± 0.00 | 0.03 ± 0.01 | 16.20 ± 2.82 |
| 86 | Tm10-1Lin | 0.13 ± 0.01 | 0.16 ± 0.03 | 07.08 ± 0.83 | 09.06 ± 2.15 | 0.03 ± 0.01 | 0.02 ± 0.01 | 0.03 ± 0.01 | 16.52 ± 3.05 |
| 87 | Tm10-2 | 0.09 ± 0.02 | 0.07 ± 0.00 | 02.12 ± 0.33 | 07.52 ± 2.04 | 0.12 ± 0.02 | 0.04 ± 0.01 | 0.02 ± 0.01 | 09.98 ± 2.43 |
| 88 | Tm10Oel | 0.14 ± 0.01 | 0.22 ± 0.03 | 11.30 ± 1.75 | 12.54 ± 2.19 | 0.13 ± 0.02 | 0.02 ± 0.01 | 0.05 ± 0.02 | 24.40 ± 4.03 |
| 89 | Tm10-3 | 0.08 ± 0.00 | 0.05 ± 0.00 | 25.85 ± 1.32 | 01.11 ± 0.25 | 0.05 ± 0.01 | 0.03 ± 0.01 | 0.04 ± 0.02 | 27.21 ± 1.60 |
| 90 | Tm10-4EF | 0.51 ± 0.08 | 0.20 ± 0.04 | 03.11 ± 0.58 | 07.95 ± 2.03 | 0.30 ± 0.06 | 0.03 ± 0.01 | 0.02 ± 0.01 | 12.13 ± 2.81 |
| 91 | Tm10-5EF | 0.10 ± 0.01 | 0.08 ± 0.03 | 01.86 ± 0.36 | 06.93 ± 1.92 | 0.14 ± 0.02 | 0.03 ± 0.01 | 0.03 ± 0.01 | 09.18 ± 2.35 |
| 92 | Tm10-6EF | 0.08 ± 0.01 | 0.04 ± 0.00 | 08.02 ± 1.80 | 10.28 ± 2.09 | 0.10 ± 0.03 | 0.03 ± 0.01 | 0.05 ± 0.02 | 18.60 ± 3.95 |
| 93 | Tm10-7EF | 0.09 ± 0.01 | 0.06 ± 0.01 | 04.51 ± 0.26 | 10.40 ± 2.13 | 0.31 ± 0.06 | 0.02 ± 0.00 | 0.02 ± 0.01 | 15.39 ± 2.47 |
| 94 | Tm10-8EF | 0.08 ± 0.01 | 0.04 ± 0.00 | 08.61 ± 0.54 | 08.03 ± 2.03 | 0.06 ± 0.01 | 0.13 ± 0.03 | 0.04 ± 0.01 | 16.99 ± 2.64 |
| 95 | Tm10-9EF | 0.28 ± 0.10 | 0.03 ± 0.01 | 04.36 ± 0.30 | 10.81 ± 2.00 | 0.27 ± 0.06 | 0.03 ± 0.01 | 0.04 ± 0.01 | 15.83 ± 2.49 |
| 96 | Tm10-10EF | 0.06 ± 0.01 | 0.22 ± 0.02 | 04.88 ± 0.27 | 20.93 ± 0.87 | 0.53 ± 0.12 | 0.80 ± 0.16 | 0.27 ± 0.07 | 27.68 ± 1.53 |
| 97 | Tm10-11EF | 0.31 ± 0.11 | 0.04 ± 0.01 | 11.82 ± 0.83 | 09.59 ± 2.16 | 0.33 ± 0.08 | 0.05 ± 0.01 | 0.05 ± 0.02 | 22.20 ± 3.21 |
| Compounds | SNP No. | SNP Loci |
|---|---|---|
| Dihexose | 5 | BnaC02g20420D, BnaA03g40510D, BnaA03g18260D, BnaC03g32440D, BnaC03g20990D |
| Progoitrin | 15 | BnaC08g30570D, BnaC06g02290D, BnaC06g02290D, BnaA06g31740D, BnaC01g10570D, BnaA01g13400D, BnaC04g51220D, BnaA08g28090D, BnaC07g03420D, BnaC01g13560D, BnaC01g04810D, BnaC05g24360D, BnaAnng10510D, BnaA08g24650D, BnaC09g27750D |
| trans-sinapine 1 | 27 | BnaCnng39930D, BnaC02g35370D, BnaC06g07520D, BnaA09g13790D, BnaAnng26190D, BnaC05g11590D, BnaC08g35930D, BnaA09g19450D, BnaA06g07300D, BnaA06g08830D, BnaCnng18490D, BnaC06g38030D, BnaA08g09430D, BnaC06g08200D, BnaCnng23620D, BnaA10g22100D, BnaCnng58110D, BnaA01g20360D, BnaA07g37760D, BnaC04g10570D, BnaA09g24030D, BnaA06g09240D, BnaC04g10570D, BnaA08g25950D, BnaC05g46690D, BnaA07g31720D, BnaC03g31950D |
| trans-sinapine 2 | 1 | BnaA07g31720D |
| Total sinapine content | 5 | BnaC06g38030D, BnaC05g08990D, BnaC03g31950D, BnaAnng09880D, BnaA06g09180D |
| Sinapine(4-O-8′)guaiacyl | 13 | BnaA09g16810D, BnaA05g23210D, BnaC07g23750D, BnaCnng31150D, BnaC09g23720D, BnaC09g22600D, BnaC08g16340D, BnaC03g49370D, BnaC01g17020D, BnaA08g06400D, BnaC02g26880D, BnaC09g54330D, BnaC07g23750D |
| Methyl sinapate | 41 | BnaCnng46600D, BnaCnng42240D, BnaCnng31900D, BnaCnng20310D, BnaCnng13750D, BnaC09g41550D, BnaC09g26810D, BnaC09g24670D, BnaC09g17390D, BnaC07g04160D, BnaC06g18890D, BnaC06g18590D, BnaC06g07440D, BnaC06g07070D, BnaC06g06790D, BnaC05g04580D, BnaC04g56040D, BnaC03g70630D, BnaC03g50270D, BnaC03g32630D, BnaC03g09080D, BnaC02g16010D, BnaC01g41960D, BnaC01g21850D, BnaC01g16590D, BnaC01g05320D, BnaAnng07310D, BnaA10g14470D, BnaA10g11420D, BnaA10g00800D, BnaA09g16810D, BnaA08g12900D, BnaA04g14130D, BnaA03g31990D, BnaA01g27460D, BnaA01g25280D, BnaA01g19380D, BnaA01g19380D, BnaA01g19380D, BnaA01g19380D, BnaA01g06890D |
| Sinapoyl malate | 1 | BnaC03g31950D |
| Total phenolic content | 2 | BnaC01g06630D, BnaAnng27700D |
| Triats | Chr_Position | LOG10(P) | Transcript:Feature | Description | TAIR ID | Allele |
|---|---|---|---|---|---|---|
| DI a | C02_16859648 | 4.70 | BnaC02g20420D:CDS | Ulp1 protease family protein | AT5G45570 | T/G |
| MS b | A01_ 19200278 | 6.74 | BnaA01g06890D:CDS | Heavy metal atpase 2 | AT4G30110 | C/T |
| Pro c | C08_30292042 | 5.66 | BnaC08g30570D:CDS | ARABIDILLO-2 | AT3G60350 | A/T |
| Pro | A06_21272025 | 4.68 | BnaA06g31740D:CDS | Tetratricopeptide repeat (TPR)-like superfamily protein | AT3G27960 | C/T |
| SG d | A09_10109576 | 6.52 | BnaA09g16810D:Intron | GroES-like zinc-binding dehydrogenase family protein | AT5G43940 | T/C |
| S1 e | CNN_38500012 | 6.42 | BnaCnng39930D:CDS | RING/U-box superfamily protein | AT2G15530 | G/A |
| S2 f | A07_22086512 | 4.59 | BnaA07g31720D:CDS | Pentatricopeptide repeat (PPR) superfamily protein | AT1G79590 | G/T |
| TSC g | ANN_10591701 | 4.47 | BnaAnng09880D:CDS | No annotated function | - | A/T |
| TSC | SC06_35793928 | 4.33 | BnaC06g38030D:CDS | Transcription coactivators | AT1G77320 | G/T |
| TSC | C03_19651313 | 4.25 | BnaC03g31950D:Intron | S-adenosyl-L-methionine-dependent methyltransferases superfamily protein | AT4G00740 | T/G |
| SM h | 4.59 | |||||
| TSC | C05_4843154 | 4.21 | BnaC05g08990D:promoter | RNA-binding (RRM/RBD/RNP motifs) family protein | G/A | |
| TSC | A06_4969999 | 4.07 | BnaA06g09180D:Intron | Protein kinase 2A | A/T | |
| TPC i | ANN_31705069 | 5.39 | BnaAnng27700D:promoter | Syntaxin of plants 52 | AT1G79590 | G/T |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, D.-G.; Ryu, J.; Yang, B.; Lee, Y.-J.; Kim, J.H.; Kim, J.; Kim, W.J.; Kim, S.H.; Kwon, S.-J.; Kim, J.-B.; et al. Genetic Variation and Association Analysis of Phenolic Compounds in Rapeseed (Brassica napus L.) Mutant Lines Using Genotyping-by-Sequencing (GBS). Horticulturae 2023, 9, 1204. https://doi.org/10.3390/horticulturae9111204
Kim D-G, Ryu J, Yang B, Lee Y-J, Kim JH, Kim J, Kim WJ, Kim SH, Kwon S-J, Kim J-B, et al. Genetic Variation and Association Analysis of Phenolic Compounds in Rapeseed (Brassica napus L.) Mutant Lines Using Genotyping-by-Sequencing (GBS). Horticulturae. 2023; 9(11):1204. https://doi.org/10.3390/horticulturae9111204
Chicago/Turabian StyleKim, Dong-Gun, Jaihyunk Ryu, Baul Yang, Ye-Jin Lee, Jae Hoon Kim, Juyoung Kim, Woon Ji Kim, Sang Hoon Kim, Soon-Jae Kwon, Jin-Baek Kim, and et al. 2023. "Genetic Variation and Association Analysis of Phenolic Compounds in Rapeseed (Brassica napus L.) Mutant Lines Using Genotyping-by-Sequencing (GBS)" Horticulturae 9, no. 11: 1204. https://doi.org/10.3390/horticulturae9111204
APA StyleKim, D.-G., Ryu, J., Yang, B., Lee, Y.-J., Kim, J. H., Kim, J., Kim, W. J., Kim, S. H., Kwon, S.-J., Kim, J.-B., Kang, S.-Y., Lyu, J. I., Bae, C.-H., & Ahn, J.-W. (2023). Genetic Variation and Association Analysis of Phenolic Compounds in Rapeseed (Brassica napus L.) Mutant Lines Using Genotyping-by-Sequencing (GBS). Horticulturae, 9(11), 1204. https://doi.org/10.3390/horticulturae9111204











