Abstract
Grapevine (Vitis vinifera subsp. vinifera) is one of the most widespread and economically important perennial fruit crops in the world. Viticulture has changed over the years in response to changing environmental conditions and market demands, triggering the development of new and improved varieties to ensure the crop’s sustainability. The aim of this review is to provide a perspective on the recent developments in biotechnology and molecular biology and to establish the potential of these technologies for the genetic improvement of grapevine. The following aspects are discussed: (i) the importance of molecular marker-based methods for proper cultivar identification and how NGS-based high-throughput technologies have greatly benefited the development of genotyping techniques, trait mapping, and genomic selection; (ii) the recent advances in grapevine regeneration, genetic transformation, and genome editing, such as new breeding technology approaches for enhanced grapevine yield, quality improvement, and the selection of valuable varieties and cultivars. The specific problems and challenges linked to grapevine biotechnology, along with the importance of integrating classical and new technologies, are highlighted.
1. Introduction
Domesticated grapevine (Vitis vinifera subsp. vinifera), one of the most economically important perennial fruit crops in the world [,], had a worldwide production of approximately 78 million tons in 2020 []. In 2020, the International Organization of Vine and Wine (OIV) estimated the world area under vines at 7.3 million ha []. The European Union (EU) had 3.2 million hectares of vines in 2020, equivalent to approximately 44% of the world’s total wine-growing areas []. Over half of these areas were cultivated with red wine varieties. Most of the harvested grapes are used for wine production but also for consumption as fresh fruit, raisins, juices, vinegar, seed oils, and spirit drinks [,]. Grape extracts are also used as food additives, in cosmetics, and in the pharmaceutical industry, while some species are grown for ornamental purposes [,].
Grapevine cultivars are susceptible to biotic (bacteria, fungi, viruses, and insects) and abiotic stressors (drought, extreme temperatures, and salinity) that reduce both the yield and lifespan of vineyards, causing substantial economic losses to grape production and the wine industry []. Vegetative propagation and growth on rootstocks ensure the grapevine tolerance to phylloxera, an insect pest that almost destroyed European viticulture in the late nineteenth century []. Other pathogens, such as gray mold, downy, and powdery mildew, as well as bacterial diseases (e.g., black rot and Pierce’s disease), cause serious yield losses [,]. Due to continuous climate change, viticulture faces outbreaks of these diseases, while thermal stress and severe dryness registered during the last decade harmfully impacted the whole winemaking sector, especially in southern European winemaking regions [,,]. Thus, a reduction in table quality vines and wine grape production was anticipated []. All these threats triggered the need to improve viticulture’s sustainability by imposing new varieties versus traditional varieties [,,].
Conventional breeding, based on the hybridization of valuable grapevine genotypes, is difficult due to the long life cycle and heterozygosity []. Moreover, numerous diseases require specific solutions found in the natural variations of the Vitis genus [] but also in genetic improvements via molecular manipulation. Thus, in recent years, biotechnology research proved to be a powerful complement to conventional breeding methods playing an increasing role in the improvement of the existing grapevine varieties and rootstocks. The novel approaches include the development of molecular markers for fingerprinting, genetic mapping, genetic diversity assessments in populations, gene tagging for breeding purposes (marker-assisted selection), and gene cloning; these technologiesaim to improve on the current plant transformation strategies and genetic editing to enhance disease tolerance and improve berry quality [,].
In the precision breeding of grapevine, only genetic elements encoding desirable traits are used; thus, the results are more predictable than conventional breeding. In the last decades, whole genome sequencing by next-generation sequencing (NGS) platforms and bioinformatics have allowed the rapid selection of plants for propagation and manipulation for different purposes.
Although modern techniques of precision breeding have taken a large scale and numerous valuable varieties have been obtained, genetically modified plants, however, are not easily accepted on the market. Edited plants were considered a solution to genetically transformed plants, mostly because the edited plants did not contain a transgene; thus, these plants were not subject to legal restrictions. Depending on the specific legislations in different countries, such as the USA, Argentina, Australia, and Brazil, even genetically modified organisms (GMOs) are accepted on the market because it is considered that edited plants do not contain foreign genes; thus, risk assessments are unnecessary. On the other hand, the European Union also considers that organisms obtained via mutagenesis, as well as those obtained via genome editing, are GMOs, as stated in the GMO European Directive 2001/18/EC [] and should pass through the regulatory process of classical GMOs [,]. It is important to mention that point mutations induced by gene editing are practically impossible to distinguish from the natural ones or those induced by mutagenesis. This uncertainty could have negative consequences not only for agriculture but also for the economy; most researchers claim that organisms with only small genetic modifications without any foreign genetic material should not be considered GMOs [,]. Nevertheless, the latest technologies, i.e., genetic transformation and genome editing, are currently developing, the main limitation in grapevine being related to the complexity of some important agronomic traits [].
In this review, we discuss the most recent achievements, specific problems, and challenges linked to grapevine biotechnology, along with the importance of integrating classical and new technologies for the genetic improvement of grapevine.
2. Grapevine Genetic Diversity and Molecular Markers Used in the Identification of Cultivars
Archaeological and archaeobotanical data showed that grapevine domestication began 6000–8000 years ago in the Transcaucasian region []. The dispersal of cultivated varieties from this primary center throughout the Near East and Europe relied upon (i) cultivars, the later clonal selection, and (ii) vegetative propagation []. In addition, secondary domestication events in other areas were also reported [,,,].
The genus Vitis [2n = 38] exhibits significant genetic diversity among cultivars, wild subspecies, and hybrids [,]. This is mainly due to their asexual reproduction, wide range of suitable planting, and frequent communication among grape accessions []. There are approximately 60–70 inter-fertile wild Vitis species widespread throughout Eurasia and Northern America []. Vitis vinifera subsp. sylvestris, the only wild Vitis taxon native to Europe and the Near East, is considered the ancestor of almost 10,000 domesticated grapevine cultivars []. Moreover, approximately 1200 commercial grapevine cultivars are interspecific hybrids of the domesticated grapevines and other wild Vitis species []. Consequently, there is a huge number of named cultivars (15,000), including several synonyms (identical genotypes but different names) and homonyms (same names but different genotypes) []. The working group on Vitis, referring to the European cooperative programme for plant genetic resources (ECPGR), reports 27,000 accessions of grapes held only in European collections []. In spite of the passport data available for approximately 35,000 accessions from European countries, the problem of cultivar synonyms and the presence of duplications need to be solved []. Therefore, a proper identification system, cultivar registration and protection, seed certification, and plant variety rights are essential in grapevine germplasm management for breeding programs but also for economic interests, trade, and scientific knowledge [,].
2.1. Morphological Markers
Grapevine cultivars have traditionally been identified based on their morphological characteristics (ampelography) jointly provided by the International Organization of Vine and Wine (OIV), the International Union for the Protection of New Varieties of Plants (UPOV), and the International Plant Genetic Resources Institute (IPGRI) []. Although efficient for the assessment of qualitative traits, the application of morphological markers is limited in the evaluation of quantitative traits. Moreover, this method is time-consuming, requiring extensive field trials and the identification of closely related cultivars is difficult [].
To complement the morphological identification of grapevine varieties and overcome classification errors and double designations, cytogenetic, biochemical, as well as DNA and RNA-based technologies were developed for the analysis of the existing grape germplasm diversity [].
2.2. Cytological Markers
In earlier studies, following morphological markers, cytological markers (karyotypes, banding patterns, deletions, repeats, translocations, and inversions) were developed. The chromosome number and morphology, and the DNA amount and composition, are characterized with Giemsa staining, fluorochrome banding, silver staining, and fluorescence in situ hybridisation (FISH) []. However, only a few cytogenetic studies are available in grapevine, mainly due to the large number of small chromosomes and the difficulty of obtaining good chromosome preparations from the roots or anthers []. Sequential silver nitrate staining and FISH were used to study ribosomal DNA (rDNA) loci [,,] to localize the retrotransposon Gret1 [], BAC clones [], and telomeric sequences []. However, FISH is considered a niche technique because it allows the analysis of only a few samples at a time, and its accuracy is highly dependent on excellent quality confocal microscopy and image analysis procedures []. However, with the advent of sequencing technologies (next-generation sequencing (NGS)) and the availability of high-quality de novo reference genomes for grapes, new horizons were opened for modern cytogenomics []. Thus, the physical mapping of DNA sequences on chromosomes facilitated comparative plant genomics, improving the genome and chromosome assemblies.
2.3. Molecular Markers
Molecular markers include protein-based markers (products of gene expression) and DNA-based markers derived from the direct analysis of polymorphisms in DNA sequences []. Depending on the detection method used, DNA markers are categorized as hybridization-based markers and polymerase chain reaction (PCR) and DNA sequence-dependent molecular markers.
Since their discovery, different molecular markers, such as (RFLPs) [], random amplified polymorphic DNA (RAPD) [], sequence characterized amplified regions (SCARs) [,], simple sequence repeats (SSRs) [,], inter-simple sequence repeats (ISSRs) [,], amplified fragment length polymorphisms (AFLPs) [], single nucleotide polymorphisms (SNPs) [], expressed sequence tags (ESTs) [,], and random amplified microsatellite polymorphisms (RAMPs) [] have been widely used for the genetic diversity characterization of grapevine cultivars, molecular mapping, parentage analysis, clone’s identification, and the detection of synonymies.
Among these, SSRs and SNPs have become the preferred markers for the characterization of grapevine genetic resources and varietal identification in germplasm collections []. Several studies reported nuclear and chloroplastidial SSR loci (nSSR and cpSSR) as useful to demonstrate the multiple origins of V. vinifera spp. sativa (cultivated grapevine), to reveal synonymies, homonymies, as well as inter and intra-specific genetic variations and phylogenetic relationships among wild and cultivated grapevines [,,,,,,]. In recent years, SNP markers have also gained high popularity for evaluating allelic variations throughout grapevine genomes and dissecting complex traits via QTL (quantitative trait loci) for breeding programs [,]. SNPs are highly abundant across plant genomes and offer higher reproducibility than microsatellite data, facilitating the integration and interpretation of genotyping data throughout grape gene banks and databases [,]. The rise of NGS and resequencing techniques have facilitated the release of an extensive number of SNPs [,] and the development of reliable platforms, such as VitisGDB (the multifunctional database for grapevine breeding and genetics), for comparing and mining Vitis genomic information []. There are currently many reference Vitis databases, including simple genetic information or only descriptive information (i. e., the species name, country of origin, cultivar names, usage, etc.) Among these, we mention the International Variety Catalogue (VIVC) [], Instituto de Ciencias de la Vid y del Vino [], the European Vitis database [], the Greek Vitis database [], the Italian Vitis database [], the molecular pathway database, the transcriptome database, grape sRNA atlas [], etc.
In the last years, DNA marker systems used in grapevine characterization have evolved from interrogating small numbers of loci and individuals to tens of thousands of loci in studies of large populations []. Moreover, Vitis spp. genome sequencing has led to significant progress in the development of large-scale high-throughput DNA markers and the identification of QTL, allowing the confirmation of candidate genes and the development of breeding programs based on marker-assisted selection (MAS). The earliest method, restriction site-associated DNA [RAD] sequencing, was successfully applied to identify significant traits in elite grape cultivars [,]. Other sequencing-based methods, such as whole genome resequencing approaches, have been applied for the characterization of somaclonal variations within cultivars [,]. Moreover, RNA-seq has been widely applied in Vitis vinifera to study different aspects such as bud development [], berry development and ripening [], and the response to disease or pathogens []. Recently, a set of 2000 DNA low-cost marker panels transferrable across the entire Vitis genus was designed and implemented using rhAmpSeq (RNase H2 enzyme-dependent amplicon sequencing), which could be easily adapted for other taxa for ecological and evolutionary studies, QTL mapping, a genome-wide association study (GWAS), and molecular breeding [].
In general, each technology provides results with a different resolution and accuracy, and the degree of detected genetic divergence depends on the marker system applied and the scope and type of plant samples used. The different applications of molecular markers in grapevine are presented in Table 1.

Table 1.
Application of the most widely used molecular markers in grapevine.
Although high-throughput sequencing technologies provide enormous potential to improve our way of understanding and accessing grapevine biodiversity, downstream bioinformatic analysis requires reliability to be ensured. There is not just one perfect array for all different research questions; therefore, the choice of the genotyping tool should be based on the purpose, sample size, resolution, accuracy, and budget available.
3. Grapevine Plant Regeneration Methods
Biotechnology research offers the potential to improve the yield and quality of grapes. The development of in vitro plant regeneration methods is essential to overcome the difficulties in conventional breeding studies, preserve and propagate valuable genotypes, as well as to increase genetic variability through genetic engineering (transgenic, cisgenic, and gene-edited plants) [,]. In addition, in vitro mass multiplication represents an alternative to the current greenhouses or outdoor repositories and allows the exposure of genotypes to in vitro-induced stresses (i.e., biotic and abiotic risks) [].
To date, grapevine regeneration has been obtained with two fundamental pathways of propagation and regeneration through organogenesis and somatic embryogenesis [,]. Organogenesis is based on the ability of competent tissues to form whole plants directly from the meristematic regions of explants (direct organogenesis) or intervened with callus formation (indirect organogenesis). Indirect organogenesis induces somaclonal variation involving both genetic and epigenetic changes in in vitro regenerated plants. In applied studies involving commercial scale multiplication or transgenic plants, the plants must be “true to type”, meaning high genetic uniformity of the regenerated plants [,]. Therefore, to demonstrate the genetic fidelity as well as the somaclonal variation of the in vitro plants, molecular marker approaches were applied. Somaclonal variation is an alternative source of genetic variability in horticultural crops with a narrow genetic base or difficult breeding [].
Grapevine, similar to other woody species, has revealed a genotype and explant type dependent recalcitrance to in vitro regeneration techniques [,]. Many studies have reported that rootstock varieties reveal higher organogenesis and somatic embryogenesis potential than hybrids and varieties belonging to the V. vinifera species []. However, several other factors were reported to influence the efficiency of grapevine in vitro plant regeneration, such as culture medium composition, especially the type and concentrations of plant growth regulators (PGRs) [,]; explants’ developmental stage [] and phyllotactic position []; light regime []; pH value [], etc. Therefore, over the years, numerous studies were focused mainly on optimizing protocols for efficient regeneration across different grapevine genotypes. A thorough review of successful reports via organogenesis and somatic embryogenesis in several grapevine cultivars and rootstock species using different explants was published by Zhang et al. []. Further on, we briefly discuss some aspects related to the applications of in vitro regeneration systems in grapevine improvements.
The first attempts in grapevine biotechnologies started in the 1960s with in vitro propagation for mass production and healthy plant regeneration []. Over the years, different studies have focused on inflorescence culture [,] to study the mechanisms of floral induction [], hairy root cultures [,,] to study plant-pathogen interactions, the efficiency against nematodes [,], or phylloxera [,], and shoot tip culture combined with thermotherapy, chemotherapy, or cryotherapy to eliminate viruses [,,].
Nowadays, the rapid technological advancements in molecular and cell biology include a wide range of new plant breeding technologies (NPBTs), which in association with the new genomic data available, offer the opportunity: (i) to develop new grapevine varieties with enhanced yields, quality, stress tolerance, and disease resistance through genetic manipulation [] and (ii) to have a clearer picture of the molecular regulation of plant cells, tissue culture, and regeneration processes [].
Due to their morphogenetic competence, embryogenic tissues are mostly preferred in genetic transformation studies for the application of new genomic techniques, such as cisgenesis and intragenesis, genome editing, and RNAi [], and these could be a possible tool for virus and viroid elimination []. Moreover, somatic embryogenesis could prevent the development of chimerism, allowing the regeneration of genetically transformed embryos under selective culture conditions []. However, some genotypes have proven to be very recalcitrant to somatic embryogenesis; thus, genetic engineering techniques are difficult to apply [].
The acquisition of embryogenic competence is related to different patterns of gene expression involving internal cell reprogramming leading to a reversion of the differentiation state [,,]. Somatic embryos may be obtained through the development of an embryogenic callus (indirect embryogenesis) followed by the emergence of pro-embryogenic masses (PEMs), from which new somatic embryos are formed [], or may occur directly from the explants without the callus developmental phase. In most cases, direct somatic embryogenesis is used for clonal propagation rather than indirect somatic embryogenesis, which is characterized by a high incidence of somaclonal variation []. The explants mostly employed for somatic embryogenesis induction are anthers, ovaries, leaves, petioles, tendrils, and nodal sections []. It was demonstrated that, amongst other factors, the type and concentrations of plant growth regulators (PGRs) play a crucial role in the induction of an embryogenic callus []. In particular, 2,4-dichlorophenoxyacetic acid (2,4-D) was reported as the most effective compound for the induction of somatic embryos [,].
Considering the importance of somatic embryogenesis in the genetic improvement of grapevine and the critical factors affecting its success, the development of efficient and reproducible genotype-specific protocols for all major grapevine, table grape cultivars, and rootstocks is required.
4. Somaclonal Variation
Plants cultivated in vitro could develop different modified characteristics and genetic variability due to somaclonal variation []. The genetic bases of somaclonal variation are gene mutations and also the rearrangements of chromosomes, karyotype changes [,,], or epigenetic modifications driven by hyper or hypo-methylation []. Most of these are induced by oxidative stress [], as shown in Figure 1.
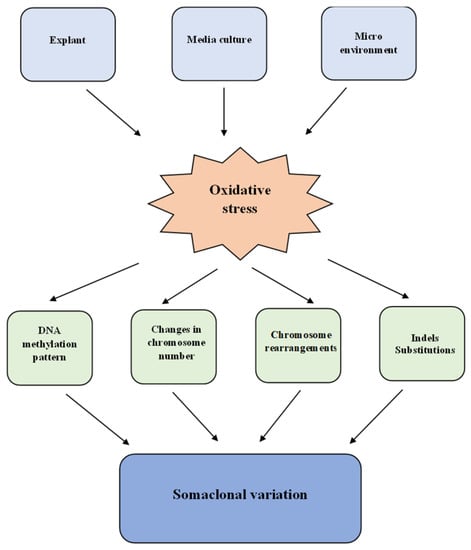
Figure 1.
Mechanism of somaclonal variation induced by oxidative stress in plants cultivated in vitro.
The genetic variation of micro-propagated plants is generally considered obstructive, and the loss of genetic fidelity was often observed. However, increased genetic variability has applications in the improvement of horticultural crops. The most important advantage of somaclonal variations is the reduced time and space for the screening of valuable genitors and traits than the crossing of perennial crops. Moreover, different somaclones could be used in breeding applications and genetic improvements with in vitro selection [].
Several in vitro procedures imposing oxidative stress, such as protoplast culture, callus induction, or somatic embryogenesis, are more frequently followed by somaclonal variation due to the epigenetic changes in plant tissues []. Thus, polyploids were obtained with somatic embryogenesis in six Spanish V. vinifera cultivars []. High methylation was detected in two cultivars during somatic embryogenesis, and the AFLP markers showed higher variability in these plants, but the SSR patterns were similar in plants derived from somatic embryos and control plants []. Somaclonal variation in grapevine could also appear by several major genetic changes by the spatial arrangement of periclinal, mericlinal, and sectorial cell layers that are genetically different []. Periclinal chimeras originated from mutations in one of the cell layers, are stable plants producing axillary buds and have the same apical organization as the terminal meristem from which they were generated []. Somatic embryos in grapevine derive from a single cell; thus, the clonal characteristics are not transferred to the progeny, as shown with microsatellite markers in Pinot Meunier. Nevertheless, different accessions of Pinot Meunier have three alleles in the VVS2 locus and the accessions of Pinot Noir, Pinot Gris, and Pinot Blanc have two alleles in this locus, explained by a mutation in one of the two alleles in one of the cell layers in Pinot Meunier and its maintenance through the vegetative propagation of a periclinal chimera []. A similar variation was also observed in the VVS19 locus in the embryogenic callus induced from the cell layers of anther filaments in Primmitivo ist [,].
Other studies showed that the intracultivar variability within Pinot Noir and chardonnay are also due to mutations in the by a mutation in one of the two alleles in one of the cell layers in Pinot Meunier and its maintenance through vegetative propagation of a periclinal chimera [,]. Thus, somatic embryogenesis is a valuable tool to understand the origins, genetic structures, and relationships between ancient cultivars and should be considered before using them for Micropropagation, genetic conservation, or transformation [].
5. Genome Sequencing and Applications
Grapevine is not only an important fruit crop but also a plant model for genetic studies due to its small genome size of 475–500 Mb and 38 chromosomes (n = 19). Most of the Vitis species are diploids, but there are also fertile interspecies hybrids []. The international grape genome program (IGGP) generated the first genome sequence for the Pinot Noir clone ENTAV 115 with Sanger and shotgun sequencing, which was important to understand the genome organization and the 19 linkage groups of Vitis vinifera. The genomic sequences of Pinot Noir clone ENTAV 115 (477.1 Mb) were assembled in 2093 metacontigs (approximately 28,352 genes and pseudogenes), of which 96.1% were assigned to linkage groups and candidate genes encoding relevant traits were predicted. In the NCBI taxonomy web portal for V. vinifera, there are 29,971 listed unique coding genes, and the information about these genes and the metabolic pathways in which they are involved are available at the TIGR site []. A consensus map was developed based on the genetic maps [,,,,] and physical maps [] previously developed. Other facilities are also available for different genomic and transcriptomic analyses, such as the grape BAC library from the French national resources center for plant genomics []; 14,000 transcripts from V. vinifera and 1700 transcripts from other Vitis species were released by The GeneChip® Vitis vinifera genome array (Affymetrix). An array-ready oligo set contains 14,562 probes of 70-mers representing grape gene transcripts released by Qiagen (http://www1.qiagen.com, accessed on 12 September 2022). The design of the grape oligo set was based on the sequence information from TIGR’s grape gene index (http://www.tigr.org/tdb/tgi, accessed on 12 September 2022) [].
Advancements in NGS technology allowed the development of genome-wide approaches for the genetic characterization of complex traits or for marker-assisted selection, such as genome-wide association studies (GWAS) or genomic selection (GS).
GWAS was used to understand the genetic bases of the important traits and to identify the polymorphic molecular markers associated with these traits [] that could be further used in marker-assisted selection programs.
In contrast to the GWAS method, which identifies polymorphisms linked to the variations for selected traits, GS allows the prediction of a breeding value for the genotypes tested [] based on large sets of markers. Thus, GS could significantly reduce costs for the marker-assisted selection of valuable variants by limiting the size and number of field experiments. Genotype-based prediction also allows selection in breeding schemes when the phenotyping of breeding candidates is impossible or difficult [,].
Unfortunately, GWAS and GS methods, which use genome-wide marker data for phenotype prediction, are difficult to use in highly heterozygous species such as grapevine []. Moreover, the efficiency of GWAS also depends on the genetic architecture of the trait; thus, the detection of molecular markers associated with polygenic traits depends on the size of the sample and the density of the molecular marker used []. In grapevine, there are no available valuable lines from complex breeding schemes; the breeders use highly diverse and heterozygous parental genotypes (H = 0.76) []. This is the result of strong inbreeding depression and vegetative propagation [,]. In addition, this panel of parental genotypes is also characterized by a low level of linkage disequilibrium (LD) between marker loci (r2~0.2 at 5–10 Kb) [,]; most cultivars are interconnected by a series of first-degree relationships (i.e., Pinot Noir—Chardonnay—Gouais Blanc, Cabernet Franc—Merlot [,]), but the number of connected generations is relatively low [,]. However, GWAS and GS have become more relevant in grapevine since the number of molecular tools is constantly increasing due to the high demand for new grapevine cultivars adapted to climate change [,].
6. Genetic Transformation
As conventional culture and the selection of new valuable varieties are time and resource-consuming, genetic transformation provided an alternative for developing new varieties with increased productivity, higher quality, and tolerance to different stress factors. Conventional breeding cannot provide resistance to diseases or pests to elite cultivars of Vitis []; thus, these cultivars are currently maintained through vegetative propagation [] and require the frequent use of pesticides to control diseases []. To overcome these concerns, modern biotechnology proposed so-called precision breeding [] and the genetic improvement of elite cultivars, which was previously known as cisgenic or intragenic improvements [].
Unfortunately, the grapevine is considered a recalcitrant species in terms of genetic transformation due to several aspects, including (i) genes involved in grapevine transformation, (ii) vectors used for gene delivery and protocols for grapevine transformation, and (iii) protocols for transgenic plant regeneration [].
The insertion of specific genes into plants with different methods and vectors was developed for over thirty years in perennial crops. In grapevine, physical and chemical delivery methods were tested over the years, and transgene delivery was mediated by Agrobacterium and viruses [,,,,] Several grapevine varieties were transformed with biolistic bombardment [] and Agrobacterium-mediated transformation []. There are also several other methods of transformation, such as electroporation or protoplast transfection [,]. Viral vectors were used for the heterologous gene expression or the silencing of host genes (i.e., virus-induced gene silencing -VIGS) []. Cloning strategies and tools for the genetic engineering of grapevine were detailed and reviewed by [,].
Several grapevine infectious viruses, such as Vitivirus Grapevine Virus A (GVA) [], the Closterovirus Grapevine leafroll-associated virus-2 (GLRaV-2) [], and the Foveavirus Grapevine rupestris stem pitting-associated virus (GRSPaV) [] were used for the silencing of PDS (phytoene desaturase) or ChlI (subunit I of magnesium protoporphyrin IX chelatase), which was observed with the development of the albino phenotype [,]. A diagram showing the most important steps in genetic manipulation with transformation or editing of grapevine is shown in Figure 2.
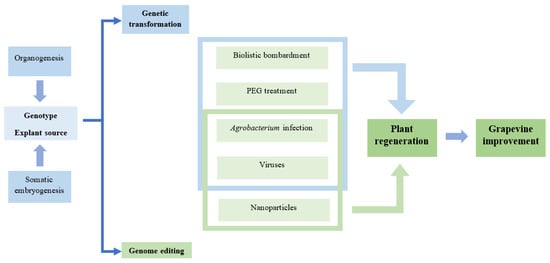
Figure 2.
Diagram of grapevine genetic manipulation with transformation or editing.
The success of a transformation is facilitated by the marker genes as selectable marker genes and reporter genes that ensure the rapid and accurate selection of modified cells from non-modified cells []. Some of the marker genes encode resistance to antibiotics or herbicides, and the reporter genes encode proteins such as green fluorescence protein (GFP) or glucuronidase (GUS) []. Previously, most transgenic plants were obtained with vectors containing the NPTII gene as a selectable marker gene and GFP as a reporter gene, ensuring the visual selection of transformed plants []. For the elimination of the marker gene from transgenic plants, a complex system is used, which consists of two strains of Agrobacterium, one of them carrying a binary vector which contains the target gene, and another containing a binary vector with the selection marker genes as codA/nptII. The cells containing the target gene and the codA/nptII genes grow on the media with kanamycin, and then the cells containing the marker gene are eliminated with negative selection based on the codA function [].
The analysis of the gene expression was performed using the VvMybA1 gene involved in the anthocyanin biosynthesis pathway []. This gene allows the visual identification of transformed cells without kanamycin selection []. Unfortunately, the transformed plants of grapevines (Thompson Seedless) containing this visual reporter gene were not vigorous due to the intense pigmentation and curly and highly brittle leaves [], and the viability was reduced. These inconveniences were mitigated by placing the VvMybA1 gene under the control of tissue-specific and developmentally regulated promoters, for example, the promoter of the Dc3 gene, expressed in late embryogenesis in carrots (Daucus carota), followed by the production of anthocyanin exclusively in embryos []. Moreover, being a visible marker, VvMybA1 could be used for monitoring transgene expression in the whole plant. Transgene expression could also be monitored by reporter genes such as GUS and GFP []. Anthocyanin has a pink-to-red color which is easily discerned, and it is suitable for the analysis of gene expression in experiments involving the selection of hundreds of transgenic plants. []. The marker gene VvMybA1, placed under the control of the ubiquitin gene promoter, was used for the high-throughput analysis of genes and promoters [].
The employment of appropriate promoters highly influences the development of valuable traits. Unfortunately, the progress of functional analyses in grapevine is reduced in comparison with Arabidopsis and rice, mainly due to the limited ability to obtain enough explants and difficulties in the analysis of the gene expression. The functional annotation of at least 18,725 genes in the grapevine allowed the evaluation of their promoters, as well as the gene expression under developmental and environmental factors. For proper uses, a detailed characterization of these promoters is required regarding their sequences and activation. First, constitutive viral promoters were used for most of the transgenes in grapevines [] and then after, these promoters were replaced by ubiquitin gene promoters. The gene expression was monitored with an anthocyanin-based assay mentioned previously [].
Precision breeding requires appropriate promoters for gene expression in particular tissues or certain stages of plant development. Thus, the discovery and characterization of potential promoters are extremely important for precision breeding in grapevine [].
Successful genetic transformation also depends on the efficient regeneration of transgenic plants. Factors such as genotype, explant source, acceptor material, culture medium, bacterial strains, selectable markers, and selection methods affect the efficiency of plant transformation and regeneration. The genetic transformation of several grapevine cultivars, such as Thompson Seedless, Silcora, and Chardonnay, was obtained with shoot organogenesis from meristematic tissue [,,]. The in vitro organogenesis of some grapevine cultivars and rootstocks was obtained from different types of explants, such as petioles, leaf internodes, and shoot apices [,,,,]. One limitation of direct organogenesis is the regeneration of chimeras explained by the induction of adventitious buds from multiple cells []. Plant regeneration from somatic embryos induced from a single cell could be used to avoid such chimeras. Somatic embryogenesis was also used for grapevine micropropagationand genetic transformation. Unfortunately, the induction of somatic embryogenesis is generally low and dependent on the type of explants []. Moreover, the maintenance of embryogenic masses on calluses and somatic embryos is also very important []. Therefore, transgenesis in grapevine is mostly based on the Agrobacterium system, and the regeneration of transformants is generally achieved with somatic embryogenesis. Improved Agrobacterium-mediated transformation protocols were published to enhance the fruit quality and tolerance to different stress factors [,].
Several factors influence the regeneration of transformants via somatic embryogenesis, such as the grapevine genotype [], explant source [,,], and culture medium [,,]. All of these factors were reviewed by Zhang et al. [].
Important achievements in grapevine using direct transformation methods and Agrobacterium-mediated transformations are shown in Table 2. Several experiments were also carried out in order to develop an efficient transformation method; these were reviewed by Zhang et al. [].

Table 2.
Grapevine improvements with genetic transformations.
7. Genome Editing
Genome editing consists of precise genetic modifications with different purposes, such as gene inactivation, that allow the possibility to explore the function of a particular gene and the insertion or replacement of genes at specific sites for genetic improvements. Strategies involving genome editing are known as new breeding technologies []. Artificially engineered nucleases, such as zinc-finger nucleases (ZFNs) [], transcription activator-like effector nucleases (TALENs) [,] and clustered regularly interspersed short palindromic repeats (CRISPR) in association with the Cas9 nuclease [] are capable of inducing specific double-stranded breaks (DSBs) of DNA molecules. The DBS are repaired with natural mechanisms present in all cells, such as non-homologous end joining (NHEJ), which is followed by point mutations due to the insertion or deletion of some nucleotides (INDELS) in the target gene, or homologous recombination (HDR) if a DNA sequence is available for recombination. CRISPR-Cas9 technology is considered the most efficient, among the genome editing tools, due to the high specificity and minimal nontarget effects []. Gene editing with the CRISPR-Cas9 system requires a guide RNA (gRNA) containing a spacer sequence complementary with the desired DNA sequence. The complex formed by guide RNA and Cas9 scans the genome, searching for complementary double-stranded DNA []. The nuclease recognizes the protospacer-adjacent motif (PAM) and generates a DSB in the specific gene sequence. Thus, genome editing through CRISPR-Cas9 technology requires the PAM sequence downstream of the target gene and proper guide RNAs, designed based on the gene sequences encoding important traits []. The description of the genome editing technologies based on ZFNs, TALENs and the CRISPR-Cas9 system was provided by Bortesi and Fischer [] and Butiuc-Keul et al. []. Despite the many advantages of CRISPR-Cas9 technology over ZFNs and TALENs, the occurrence of off-target mutations is one of the shortcomings [,], being influenced by different parameters, such as the recognition of the target, the design of guide RNAs, the frequency of repair events with homologous recombination, and anti-CRISPR proteins that inactivate Cas9 [].
Another limitation of using CRISPR-Cas9 technology is related to the delivery of the system in plant tissues. Generally, gRNAs and Cas9 are delivered into plant cells by Agrobacterium, viral vectors, PEG-mediated transformation, biolistic methods, and nanoparticles []. The CRISPR-Cas9 system could be released in tissues in its DNA, mRNA, or ribonucleoprotein forms and incorporated in different biomaterials for proper delivery [], but these studies were carried out mostly in animal and human cells for cancer therapies. The structure of the plant cell wall limits the delivery of the system; thus, the most used system for delivery into plant tissues is Agrobacterium []. Nevertheless, the direct delivery of the purified Cas9 protein and gRNAs was also applied for the editing of the plant genome [,].
Most of the studies regarding the genome editing of grapevine were conducted in order to increase the resistance to powdery mildew [,,,] and Botrytis cinerea [], the production of tartaric acid [], the manipulation of the carotenoid biosynthesis pathway, and the induction of the albino phenotype [,] [Table 3].

Table 3.
Grapevine improvements with CRIPSR-Cas9 technology (modified from Zhang et al. []; Butiuc-Keul et al. []).
8. Conclusions and Perspectives
The selection of grapevine rootstocks and scion varieties with improved fruit qualities, resistance to herbicides, and tolerance to biotic and abiotic stressors requires different biotechnologies, such as in vitro plant regeneration and multiplication, mutagenesis, the induction of somaclonal variability and selection of new valuable genotypes, and genetic transformation and genome editing. The strategies based on genome editing have the advantage of speeding up crop improvement and reducing the cost of the process, but the implementation of these technologies needs government support. Usually, the edited plants are not considered transgenic even though the delivery of the CRISPR-Cas9 system is mediated by Agrobacterium. Thus, edited plants could be easily accepted on the market. Improving grapevine tolerance to diseases and pests is the most promising contribution of new breeding technologies because little is known about genes encoding disease resistance and their functions, and the QTLs are not identified. The development of a multi-resistant genotype is complicated, and V. vinifera is susceptible to different fungi. Thus, new breeding technology could be considered a significant alternative to the classical selection and breeding of grapevine varieties resistant to biotic and abiotic stress.
Author Contributions
Conceptualization, A.B.-K. and A.C.; writing—original draft preparation, A.B.-K. and A.C.; writing—review and editing, A.B.-K. and A.C.; supervision, A.B.-K.; funding acquisition, A.B.-K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the grant AGC nr.332066/23.06.2022 and Ministry of Research, Innovation and Digitization through the Core Project BioClimpact nr.7/30.12.2022, code 23020401.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We would like to thank Matei Dobrescu, for his support in formatting the figures.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Basso, M.F.; Fajardo, T.V.; Saldarelli, P. Grapevine virus diseases: Economic impact and current advances in viral prospection and management. Rev. Bras. Frutic. 2017, 39, e-411. [Google Scholar] [CrossRef]
- Bi, W.L.; Pan, C.; Hao, X.Y.; Cui, Z.H.; Kher, M.M.; Marković, Z.; Wang, Q.C.; Teixeira da Silva, J.A. Cryopreservation of grapevine [Vitis spp.]–A review. Vitr. Cell. Dev. Biol. Plant. 2017, 53, 449–460. [Google Scholar] [CrossRef]
- FAOSTAT. 2020. Available online: https://www.fao.org/faostat/en/#data (accessed on 5 January 2021).
- OIV 2020. State of the World Vitivinicultural Sector in 2020. Available online: https://www.oiv.int/ (accessed on 13 April 2021).
- EUROSTAT 2020. Available online: https://ec.europa.eu/eurostat/statistics-explained/index.php?title=Vineyards_in_the_EU_-_statistics&oldid=566726. (accessed on 5 March 2021).
- Reynolds, A.G. The Grapevine, Viticulture, and Winemaking: A Brief Introduction. In Grapevine Viruses: Molecular Biology, Diagnostics and Management; Meng, B., Martelli, G., Golino, D., Fuchs, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 3–29. [Google Scholar] [CrossRef]
- Campos, G.; Chialva, C.; Miras, S.; Lijavetzky, D. New technologies and strategies for grapevine breeding through genetic transformation. Front. Plant. Sci. 2021, 25, 767522. [Google Scholar] [CrossRef] [PubMed]
- Abiri, K.; Rezaei, M.; Tahanian, H.; Heidari, P.; Khadivi, A. Morphological and pomological variability of a grape (Vitis vinifera L.) germplasm collection. Sci. Hortic. 2020, 266, 1–12. [Google Scholar] [CrossRef]
- Kambiranda, D.; Obuya, J.; Snowden, J. Grapevine Improvement through Biotechnology. In Genetic Transformation in Crops; To, K.-Y., Ed.; IntechOpen: London, UK, 2020. [Google Scholar] [CrossRef]
- Chacón-Vozmediano, J.L.; Gramaje, D.; León, M.; Armengol, J.; Moral, J.; Izquierdo-Cañas, P.M.; Martínez-Gascueña, J. Cultivar Susceptibility to Natural Infections Caused by Fungal Grapevine Trunk Pathogens in La Mancha Designation of Origin (Spain). Plants 2021, 10, 1171. [Google Scholar] [CrossRef]
- Galet, P. Apoplexie. In: Les maladies et les parasites de la vigne. Imp. Paysan du Midi Montp. 1977, 1, 409–430. [Google Scholar]
- Santos, R.B.; Figueiredo, A. Two sides of the same story in grapevine-pathogen interactions. J. Exp. Bot. 2021, 72, 3367–3380. [Google Scholar] [CrossRef]
- Cardell, M.; Amengual, A.; Romero, R. Future effects of climate change on the suitability of wine grape production across Europe. Reg. Environ. Change 2019, 19, 1–12. [Google Scholar] [CrossRef]
- Santos, J.A.; Fraga, H.; Malheiro, A.C.; Moutinho-Pereira, J.; Dinis, L.-T.; Correia, C.; Moriondo, M.; Leolini, L.; Dibari, C.; Costafreda-Aumedes, S.; et al. A Review of the Potential Climate Change Impacts and Adaptation Options for European Viticulture. Appl. Sci. 2020, 10, 3092. [Google Scholar] [CrossRef]
- Töpfer, R.; Trapp, O. A cool climate perspective on grapevine breeding: Climate change and sustainability are driving forces for changing varieties in a traditional market. Appl. Genet. 2022, 7, 1–4. [Google Scholar] [CrossRef]
- Fraga, H. Climate Change: A New Challenge for the Winemaking Sector. Agronomy 2020, 10, 1465. [Google Scholar] [CrossRef]
- Zhang, X.-M.; Wu, Y.-F.; Li, Z.; Song, C.-B.; Wang, X.-P. Advancements in plant regeneration and genetic transformation of grapevine (Vitis spp.). J. Integrat. Agric. 2021, 20, 1407–1434. [Google Scholar] [CrossRef]
- Gray, D.J.; Meredith, C.P. Biotechnology of Perennial Fruit Crops; Hammerschlag, F.A., Litz, R.E., Eds.; CAB International: Wallingford, UK, 1992; pp. 229–262. [Google Scholar]
- Vezzulli, S.; Doligez, A.; Bellin, D. Molecular Mapping of Grapevine Genes. In The Grape Genome. Compendium of Plant Genomes; Cantu, D., Walker, M., Eds.; Springer: Cham, Switzerland, 2019. [Google Scholar] [CrossRef]
- Directive 2001/18/ec of the European Parliament and of the Council of 12 March 2001 on the Deliberate Release into the Environment of Genetically Modified Organisms and Repealing Council Directive 90/220/EEC (OJ L 106, 17.4.2001, pp. 1–60). Available online: https://www.legislation.gov.uk/eudr/2001/18/contents (accessed on 21 September 2022).
- Regulation (EC) No 1829/2003 of the European Parliament and of the Council of 22 September 2003 on Genetically Modified Food and Feed (Text with EEA Relevance). Official Journal L 268, 18/10/2003 P. 0001–0023. Available online: https://eur-lex.europa.eu/legal-content/EN/ALL/?uri=CELEX%3A32003R1829 (accessed on 21 September 2022).
- Regulation (EC) No 1830/2003 of the European Parliament and of the Council of 22 September 2003 Concerning the Traceability and Labelling of Genetically Modified Organisms and the Traceability of Food and Feed Products Produced from Genetically Modified Organisms and Amending Directive 2001/18/EC. Official Journal L 268, 18/10/2003 P. 0024–0028. Available online: https://eur-lex.europa.eu/legal-content/EN/ALL/?uri=CELEX%3A32003R1829 (accessed on 21 September 2022).
- Dalla Costa, L.; Malnoy, M.; Lecourieux, D.; Deluc, L.; Ouaked-Lecourieux, F.; Deluc, L.; Ouaked-Lecourieux, F.; Thomas, M.; Torregrosa, L.J.-M. The state-of-the-art of grapevine biotechnology and new breeding technologies (NBTS). OENO One 2019, 53, 189–212. [Google Scholar] [CrossRef]
- Grassi, F.; De Lorenzis, G. Back to the origins: Background and perspectives of grapevine domestication. Int. J. Mol. Sci. 2021, 22, 4518. [Google Scholar] [CrossRef] [PubMed]
- Rivera Nuñez, D.; Walker, M.J. A review of palaeobotanical findings of early Vitis in the Mediterranean and the origins of cultivated grape-vines, with special reference to new pointers to prehistoric exploitation in the Western Mediterranean. Rev. Palaeobot. Palynol. 1989, 61, 205–237. [Google Scholar] [CrossRef]
- Grassi, F.; Labra, M.; Imazio, S.; Spada, A.; Sgorbati, S.; Scienza, A.; Sala, F. Evidence of secondary grapevine domestication centre detected by SSR analysis. Theor. App. Genet. 2003, 107, 1315–1320. [Google Scholar] [CrossRef]
- Arroyo-García, R.; Ruiz-García, L.; Bolling, L.; Ocete, R.; López, M.A.; Arnold, C.A.; Ergul, A.; Söylemezoglu, G.; Uzun, H.I.; Cabello, F.; et al. Multiple origins of cultivated grapevine (Vitis vinifera L. ssp. sativa) based on chloroplast DNA polymorphisms. Mol. Ecol. 2006, 15, 3707–3714. [Google Scholar] [CrossRef]
- Terral, J.F.; Tabard, E.; Bouby, L.; Ivorra, S.; Pastor, T.; Figueiral, I.; Picq, S.; Chevance, J.B.; Jung, C.; Fabre, L.; et al. Evolution and history of grapevine (Vitis vinifera) under domestication: New morphometric perspectives to understand seed domestication syndrome and reveal origins of ancient European cultivars. Ann. Bot. 2010, 105, 443–455. [Google Scholar] [CrossRef]
- Rusjan, D. Genetic and phenotypic diversity and relations between grapevine varieties: Slovenian germplasm. In The Mediterranean Genetic Code-Grapevine and Olive; Poljuha, D., Sladonja, B., Eds.; IntechOpen: London, UK, 2013. [Google Scholar] [CrossRef][Green Version]
- Villano, C.; Aiese Cigliano, R.; Esposito, S.; D’Amelia, V.; Iovene, M.; Carputo, D.; Aversano, R. DNA-Based Technologies for grapevine biodiversity exploitation: State of the art and future perspectives. Agronomy 2022, 12, 491. [Google Scholar] [CrossRef]
- Emanuelli, F.; Lorenzi, S.; Grzeskowiak, L.; Catalano, V.; Stefanini, M.; Troggio, M.; Myles, S.; Martinez-Zapater, J.M.; Zyprian, E.; Moreira, F.M.; et al. Genetic diversity and population structure assessed by SSR and SNP markers in a large germplasm collection of grape. BMC Plant. Biol. 2013, 13, 39. [Google Scholar] [CrossRef]
- Zecca, G.; Grassi, F.; Tabidze, V.; Pipia, I.; Kotorashvili, A.; Kotaria, N.; Beridze, T. Dates and rates in grape’s plastomes: Evolution in slow motion. Curr. Genet. 2020, 66, 123–140. [Google Scholar] [CrossRef]
- Lacombe, T.; Audeguin, L.; Boselli, M.; Bucchetti, B.; Cabello, F.; Chatelet, P.; Crespan, M.; D’Onofrio, C.; Eiras Dias, J.; Ercisli, S.; et al. Grapevine European catalogue: Towards a comprehensive list. Vitis 2011, 50, 65–68. [Google Scholar] [CrossRef]
- Jackson, R.S. Grape Species and Varieties. In Food Science and Technology, Wine Science, 4th ed.; Ronald, S.J., Ed.; Academic Press: Cambridge, MA, USA, 2014; pp. 21–67. ISBN 9780123814685. [Google Scholar] [CrossRef]
- Panara, F.; Bergamini, C.; Palliotti, A.; Calderini, O. Use of Molecular Markers (Ssrs) and Public Databases in Vitis vinifera L. as the Main Case of Efficient Crop Cultivar Identification. JOJ Hortic. Arboric. 2018, 2, 555576. [Google Scholar] [CrossRef]
- Sefc, K.; Pejić, I.; Maletić, E.; Thomas, M.; Lefort, F. Microsatellite Markers for Grapevine: Tools for Cultivar Identification & Pedigree Reconstruction. In Grapevine Molecular Physiology & Biotechnology; Roubelakis-Angelakis, K.A., Ed.; Springer: Dordrecht, The Netherland, 2009. [Google Scholar] [CrossRef]
- Nasiri, A.; Taheri-Garavand, A.; Fanourakis, D.; Zhang, Y.-D.; Nikoloudakis, N. Automated Grapevine Cultivar Identification via Leaf Imaging and Deep Convolutional Neural Networks: A Proof-of-Concept Study Employing Primary Iranian Varieties. Plants 2021, 10, 1628. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.H.; Noh, J.H.; Park, S.J.; Kim, S.H.; Kim, D.; Chun, J.A. Development of sequence characterized amplified region markers for the identification of grapevine cultivars. Hortic. Sci. 2015, 50, 1744–1750. [Google Scholar] [CrossRef]
- Castro, C.; Carvalho, A.; Pavia, I.; Leal, F.; Moutinho-Pereira, J.; Lima-Brito, J. Nucleolar Activity and Physical Location of Ribosomal DNA Loci in Vitis vinifera L. by Silver Staining and Sequential FISH. Sci. Hortic. 2018, 232, 57–62. [Google Scholar] [CrossRef]
- Pereira, H.S.; Barão, A.; Delgado, M.; Morais-Cecílio, L.; Viegas, W. Genomic Analysis of Grapevine Retrotransposon 1 (Gret1) in Vitis vinifera. Theor. Appl. Genet. 2005, 111, 871–878. [Google Scholar] [CrossRef]
- Falistocco, E.; Passeri, V.; Marconi, G. Investigations of 5S RDNA of Vitis vinifera L.: Sequence Analysis and physical mapping. Genome 2007, 50, 927–938. [Google Scholar] [CrossRef]
- Giannuzzi, G.; D’Addabbo, P.; Gasparro, M.; Martinelli, M.; Carelli, F.N.; Antonacci, D.; Ventura, M. Analysis of High-Identity Segmental Duplications in the Grapevine Genome. BMC Genom. 2011, 12, 436. [Google Scholar] [CrossRef]
- Pereira, H.S.; Delgado, M.; Avó, A.P.; Barão, A.; Serrano, I.; Viegas, W. Pollen grain development is highly sensitive to temperature stress in Vitis vinifera. Aust. J. Grape Wine Res. 2014, 20, 474–484. [Google Scholar] [CrossRef]
- Goswami, M.; Attri, K.; Goswami, I. Applications of Molecular Markers in Fruit Crops: A Review. IJEP 2022, 9, 121–126. Available online: http://www.pphouse.org/ijep-article-details.php?art=323 (accessed on 12 September 2022). [CrossRef]
- Bourquin, J.; Otten, L.; Walter, B. PCR-RFLP analysis of Vitis, Ampelopsis and Parthenocissus and its application to the identification of rootstocks. Vitis 1995, 34, 103–108. [Google Scholar]
- Karataş, H.; Ağaoğlu, Y.S. RAPD analysis of selected local Turkish grape cultivars (Vitis vinifera). GMR 2010, 9, 1980–1986. [Google Scholar] [CrossRef]
- Vidal, J.; Delavault, P.; Coarer, M.; Defontaine, A. Design of grapevine (Vitis vinifera L.) cultivar-specific SCAR primers for PCR fingerprinting. Appl Genet. 2000, 101, 1194–1201. [Google Scholar] [CrossRef]
- Pollastro, S.; Dongiovanni, C.; Abbatecola, A.; de Guido, M.A.; de Miccolis Angelini, R.M.; Faretra, F. Specific SCAR Primers for Fungi Associated with Wood Decay of Grapevine. Phytopathol. Mediterr. 2001, 40, S362–S368. [Google Scholar]
- Villano, C.; Carputo, D.; Frusciante, L.; Santoro, X.; Aversano, R. Use of SSR and Retrotransposon-Based Markers to Interpret the Population Structure of Native Grapevines from Southern Italy. Mol. Biotechnol. 2014, 56, 1011–1020. [Google Scholar] [CrossRef] [PubMed]
- Karimi, M.R.; Dehvari, V.; Hajiyan, M. Genetics Diversity of Some Grape Genotypes by ISSR and RAPD Markers. Eur. J. Hortic.Sci. 2011, 76, 201–207. [Google Scholar]
- Nookaraju, A.; Agrawal, D.C. Genetic homogeneity of in vitro raised plants of grapevine cv. Crimson Seedless revealed by ISSR and microsatellite markers. S. Afr. J. Bot. 2012, 78, 302–306. [Google Scholar] [CrossRef][Green Version]
- Stajner, N.; Jakse, J.; Javornik, B.; Masuelli, R.W.; Martínez, L.E. Highly variable AFLP and S-SAP markers for the identification of ‘Malbec’ and ‘Syrah’ clones. Vitis 2009, 48, 145–150. [Google Scholar]
- Péros, J.P.; Cousins, P.; Launay, A.; Cubry, P.; Walker, A.; Prado, E.; Peressotti, E.; Wiedemann-Merdinoglu, S.; Laucou, V.; Merdinoglu, D.; et al. Genetic diversity and population structure in Vitis species illustrate phylogeographic patterns in eastern North America. Mol. Ecol. 2021, 30, 2333–2348. [Google Scholar] [CrossRef]
- Peng, F.Y.; Reid, K.E.; Liao, N.; Schlosser, J.; Lijavetzky, D.; Holt, R.; Martínez Zapater, J.M.; Jones, S.; Marra, M.; Bohlmann, J.; et al. Generation of ESTs in Vitis vinifera wine grape (Cabernet Sauvignon) and table grape (Muscat Hamburg) and discovery of new candidate genes with potential roles in berry development. Gene 2007, 402, 40–50. [Google Scholar] [CrossRef]
- Ji, X.N.; Li, F.; Yang, C.J.; Li, B.; Wang, J.; Zhang, W. Expressed sequence tags (ESTs) analysis of the ripening Vitis amurensis cv. Shuang Hong berry skins. J. For. Res. 2013, 24, 495–502. [Google Scholar] [CrossRef]
- Lü, X.-L.; Zhang, G.-L.; Liao, M.-A.; Gong, R.-G.; Zeng, X.-L. Random Amplified Microsatellite Polymorphism (RAMP) Analysis of Grape Breeds Genetic Relationships. JSAU 2004, 22, 133–137. [Google Scholar] [CrossRef]
- Arroyo-García, R.; Lefort, F.; de Andrés, M.T.; Ibáñaez, J.; Borrego, J.; Jouve, N.; Cabello, F.; Martínez-Zapater, J.M. Chloroplast microsatellite polymorphisms in Vitis species. Genome 2002, 45, 1142–1149. [Google Scholar] [CrossRef]
- Veloso, M.M.; Almadanim, M.C.; Baleiras-Couto, M.; Pereira, H.S.; Carneiro, L.C.; Fevereiro, P.; Eiras-Dias, J.E. Microsatellite database of grapevine (Vitis vinifera L.) cultivars used for wine production in Portugal. Cien. Tecn. Vitivinic. 2010, 25, 53–61. [Google Scholar]
- Castro, I.; Martín, J.P.; Ortiz, J.M.; Pinto-Carnide, O. Varietal discrimination and genetic relationships of Vitis vinifera L. cultivars from two major controlled appellation (DOC) regions in Portugal. Sci. Hortic. 2011, 127, 507–514. [Google Scholar] [CrossRef]
- Castro, I.; Pinto-Carnide, O.; Ortiz, J.M.; Martin, J.P. Chloroplast genome diversity in Portuguese grapevine (Vitis vinifera L.) cultivars. Mol. Biotechnol. 2013, 54, 528–540. [Google Scholar] [CrossRef]
- Ferreira, V.; Pinto-Carnide, O.; Mota, T.; Martín, J.P.; Ortiz, J.M.; Castro, I. Identification of minority grapevine cultivars from Vinhos Verdes Portuguese DOC Region. Vitis 2015, 54, 53–58. [Google Scholar] [CrossRef]
- De Michele, R.; La Bella, F.; Gristina, A.S.; Fontana, I.; Pacifico, D.; Garfi, G.; Motisi, A.; Crucitti, D.; Abbate, L.; Carimi, F. Phylogenetic Relationship Among Wild and Cultivated Grapevine in Sicily: A Hotspot in the Middle of the Mediterranean Basin. Front. Plant. Sci. 2019, 10, 1506. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, D.; Brancadoro, L.; De Lorenzis, G. Genetic Diversity and Population Structure in a Vitis spp. Core Collection Investigated by SNP Markers. Diversity 2020, 12, 103. [Google Scholar] [CrossRef]
- Flutre, T.; Le Cunff, L.; Fodor, A.; Launay, A.; Romieu, C.; Berger, G.; Bertrand, Y.; Terrier, N.; Beccavin, I.; Bouckenooghe, V.; et al. A genome-wide association and prediction study in grapevine deciphers the genetic architecture of multiple traits and identifies genes under many new QTLs. G3 Genes|Genomes|Genet. 2022, 12, jkac103. [Google Scholar] [CrossRef] [PubMed]
- Ghaffari, S.; Hasnaoui, N.; Zinelabidine, L.H.; Ferchichi, A.; Martínez-Zapater, J.M.; Ibáñez, J. Genetic diversity and parentage of Tunisian wild and cultivated grapevines (Vitis vinifera L.) as revealed by single nucleotide polymorphism (SNP) markers. Tree Genet. Genomes 2014, 10, 1103–1112. [Google Scholar] [CrossRef]
- Augusto, D.; Ibáñez, J.; Pinto-Sintra, A.L.; Falco, V.; Leal, F.; Martínez-Zapater, J.M.; Oliveira, A.A.; Castro, I. Grapevine Diversity and Genetic Relationships in Northeast Portugal Old Vineyards. Plants 2021, 10, 2755. [Google Scholar] [CrossRef] [PubMed]
- Lijavetzky, D.; Cabezas, J.A.; Ibáñez, A.; Rodríguez, V.; Martínez-Zapater, J.M. High throughput SNP discovery and genotyping in grapevine (Vitis vinifera L.) by combining a re-sequencing approach and SNPlex technology. BMC Genom. 2007, 8, 424. [Google Scholar] [CrossRef]
- Dong, X.; Chen, W.; Liang, Z.; Li, X.; Nick, P.; Chen, S.; Dong, Y.; Li, S.; Sheng, J. VitisGDB: The Multifunctional Database for Grapevine Breeding and Genetics. Mol. Plant. 2020, 13, 1098–1100. [Google Scholar] [CrossRef]
- Maul, E.; Töpfer, R. Vitis International Variety Catalogue (V IVC): A cultivar database referenced by genetic profiles and morphology. BIO Web Conf. 2015, 5, 01009. [Google Scholar] [CrossRef]
- Cabezas, J.A.; Cabezas, M.T.; Cervera, M.T.; Cervera, L.; Ruiz-García, L.; Ruiz-García, J.; Carreño, J.; Martínez-Zapater, C.J.M. A genetic analysis of seed and berry weight in grapevine. Genome 2006, 49, 1572–1585. [Google Scholar] [CrossRef]
- Maul, E.; Sudharma, K.N.; Kecke, S.; Marx, G.; Müller, C.; Audeguin, L.; Boselli, M.; Boursiquot, J.M.; Bucchetti, B.; Cabello, F.; et al. The European Vitis Database (www.Eu-Vitis.De): A Technical Innovation through An Online Uploading and Interactive Modification System. Vitis 2012, 51, 79–85. [Google Scholar]
- Lefort, F.; Roubelakis-Angelakis, K.A. The Greek Vitis Database: A MultimediaWeb-Backed Genetic Database for Germplasm Management of Vitis Resources in Greece. J. Wine Res. 2000, 11, 233–242. [Google Scholar] [CrossRef]
- Davey, J.W.; Hohenlohe, P.A.; Etter, P.D.; Boone, J.Q.; Catchen, J.M.; Blaxter, M.L. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat. Rev. Genet. 2011, 12, 499–510. [Google Scholar] [CrossRef]
- Marrano, A.; Birolo, G.; Prazzoli, M.L.; Lorenzi, S.; Valle, G.; Grando, M.S. SNP-Discovery by RAD-Sequencing in a Germplasm Collection of Wild and Cultivated Grapevines (V. vinifera L.). PLoS ONE 2017, 12, e0170655. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Guo, Y.; Su, K.; Liu, Z.; Ren, Z.; Li, K.; Guo, X. Construction of a highly saturated genetic map for Vitis by Next-generation restriction site-associated DNA sequencing. BMC Plant. Biol. 2018, 18, 347. [Google Scholar] [CrossRef] [PubMed]
- Gambino, G.; Dal Molin, A.; Boccacci, P.; Minio, A.; Chitarra, W.; Avanzato, C.G.; Tononi, P.; Perrone, I.; Raimondi, S.; Schneider, A. Whole-Genome Sequencing and SNV Genotyping of ‘Nebbiolo’ (Vitis vinifera L.) Clones. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Carbonell-Bejerano, P.; Royo, C.; Torres-Pérez, R.; Grimplet, J.; Fernandez, L.; Franco-Zorrilla, J.M.; Lijavetzky, D.; Baroja, E.; Martínez, J.; García-Escudero, E. Catastrophic unbalanced genome rearrangements cause somatic loss of berry color in grapevine. Plant. Physiol. 2017, 175, 786–801. [Google Scholar] [CrossRef]
- Pucker, B.; Schwandner, A.; Becker, S.; Hausmann, L.; Viehöver, P.; Töpfer, R.; Weisshaar, B.; Holtgräwe, D. RNA-Seq time series of Vitis vinifera bud development reveals correlation of expression patterns with the local temperature profile. Plants 2020, 9, 1548. [Google Scholar] [CrossRef]
- Ma, Q.; Yang, J. Transcriptome profiling and identification of the functional genes involved in berry development and ripening in Vitis vinifera. Gene 2019, 680, 84–96. [Google Scholar] [CrossRef] [PubMed]
- Toffolatti, S.L.; De Lorenzis, G.; Brilli, M.; Moser, M.; Shariati, V.; Tavakol, E.; Maddalena, G.; Passera, A.; Casati, P.; Pindo, M. Novel aspects on the interaction between grapevine and Plasmopara Viticola: Dual-RNA-Seq analysis highlights gene expression dynamics in the pathogen and the plant during the battle for infection. Genes 2020, 11, 261. [Google Scholar] [CrossRef] [PubMed]
- Zou, C.; Karn, A.; Reisch, B.; Nguyen, A.; Sun, Y.; Bao, Y.; Campbell, M.S.; Church, D.; Williams, S.; Xu, X. Haplotyping the Vitis collinear core genome with rhAmpSeq improves marker transferability in a diverse genus. Nat. Commun. 2020, 11, 413. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh, M.; Singh, S.K.; Jhang, T.; Sharma, T.R. Inter Simple Sequence Repeat Analysis to Confirm Genetic Stability of Micropropagated Plantlets in Three Grape (Vitis spp.) Rootstock Genotypes. J. Plant. Biochem. Biotechnol. 2008, 17, 77–80. [Google Scholar] [CrossRef]
- Hameed, U.K.A.; Abdelaziz, K.; El Sherif, N. Genetic diversity of grapevine (Vitis vinifera L.) cultivars in Al-Madinah Al-Munawara based on molecular markers and morphological traits. Bangladesh J. Plant. Taxon. 2020, 27, 113–127. [Google Scholar] [CrossRef]
- Guo, Y.; Lin, H.; Liu, Z.; Zhao, Y.; Guo, X.; Li, K. SSR and SRAP marker-based linkage map of Vitis vinifera L. Biotechnol. Biotechnol. Equip. 2014, 28, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Gong, P.; Shi, Y.; Wang, Y.; Zhang, C. Genetic inter-relationships among Chinese wild grapes based on SRAP marker analyses. Vitis 2018, 57, 151–157. [Google Scholar] [CrossRef]
- Pelsy, F. Untranslated leader region polymorphism of Tvv1, a retrotransposon family, is a novel marker useful for analyzing genetic diversity and relatedness in the genus Vitis. Appl. Genet. 2007, 116, 15–27. [Google Scholar] [CrossRef]
- Pelsy, F.; Bevilacqua, L.; Blanc, S.; Merdinoglu, D.A. Molecular marker set combining a retrotransposon insertion and SSR polymorphisms is useful for assessing diversity in Vitis. OENO One 2021, 55, 403–414. [Google Scholar] [CrossRef]
- Roach, M.J.; Johnson, D.L.; Bohlmann, J.; van Vuuren, H.J.J.; Jones, S.J.M.; Pretorius, I.S.; Schmidt, S.A.; Borneman, A.R. Population sequencing reveals clonal diversity and ancestral inbreeding in the grapevine cultivar Chardonnay. PLoS Genet. 2018, 14, e1007807. [Google Scholar] [CrossRef] [PubMed]
- Butiuc-Keul, A.; Crăciunaş, C.; Coste, A.; Farago, M. Discrimination and genetic polymorphism in several cultivar of grapevine by RAPD markers. Rom. Biotechnol. Lett. 2010, 15, 110–115. [Google Scholar]
- Cretazzo, E.; Meneghetti, S.; De Andrés, M.T.; Gaforio, L.; Frare, E.; Cifre, J. Clone Differentiation and Varietal Identification by means of SSR, AFLP, SAMPL and M-AFLP in order to assess the clonal selection of grapevine: The case study of Manto Negro, Callet and Moll, autochthonous cultivars of Majorca. Ann. Appl. Biol. 2010, 157, 213–227. [Google Scholar] [CrossRef]
- Coste, A.; Postolache, D.; Popescu, F.; Butiuc-Keul, A. Authentication of valuable grapevine varieties from Romania, through molecular markers. Rom. Biotechnol. Lett. 2010, 15, 3–10. [Google Scholar]
- Baránková, K.; Sotolář, R.; Baránek, M. Identification of Rare Traditional Grapevine Cultivars Using SSR Markers and Their Geographical Location within the Czech Republic. Czech. J. Genet. Plant. Breed. 2020, 56, 71–78. [Google Scholar] [CrossRef]
- Jiménez-Cantizano, A.; Muñoz-Martín, A.; Amores-Arrocha, A.; Sancho-Galán, P.; Palacios, V. Identification of red grapevine cultivars (Vitis vinifera L.) preserved in ancient vineyards in Axarquia (Andalusia, Spain). Plants 2020, 9, 1572. [Google Scholar] [CrossRef]
- Ocaña, J.; Walter, B.; Schellenbaum, P. Stable MSAP markers for the distinction of Vitis vinifera cv Pinot Noir clones. Mol. Biotechnol. 2013, 55, 236–248. [Google Scholar] [CrossRef]
- Emanuelli, F.; Sordo, M.; Lorenzi, S.; Battilana, J.U.R.I.; Grando, M. Development of user-friendly functional molecular markers for VvDXS gene conferring muscat flavor in grapevine. Mol. Breed. 2014, 33, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Castro, I.; D’Onofrio, C.; Martín, J.P.; Ortiz, J.M.; De Lorenzis, G.; Ferreira, V.; Pinto-Carnide, O. Effectiveness of AFLPs and retrotransposon-based markers for the identification of Portuguese grapevine cultivars and clones. Mol. Biotechnol. 2012, 52, 26–39. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fan, X.C.; Chu, J.Q.; Liu, C.H.; Sun, X.; Fang, J.G. Identification of grapevine rootstock cultivars using expressed sequence tag-simple sequence repeats. GMR 2014, 13, 7649–7657. [Google Scholar] [CrossRef] [PubMed]
- D’Onofrio, C.; De Lorenzis, G.; Giordani, T.; Natali, L.; Cavallini, A.; Scalabrelli, G. Retrotransposon-based molecular markers for grapevine species and cultivars identification. Tree Genet. Genomes 2010, 6, 451–466. [Google Scholar] [CrossRef]
- Ulanovsky, S.; Gogorcena, Y.; Martinez de Toda, F.; Ortiz, J. Use of molecular markers in detection of synonymies and homonymies in grapevines (Vitis vinifera L.). Sci. Hortic. 2002, 92, 241–254. [Google Scholar] [CrossRef]
- Moreno-Sanz, P.; Suárez, B.; Loureiro, M.D. Identification of synonyms and homonyms in grapevine cultivars (Vitis vinifera L.) from Asturias (Spain). J. Hortic. Sci. Biotechnol. 2008, 83, 683–688. [Google Scholar] [CrossRef][Green Version]
- Castro, I.; Martín, J.P.; Ortiz, J.M.; Mota, M.T.; Pinto-Carnide, O.; Martin, J.; Ortiz, J.M.; Mota, M.T.; Pinto-Carnide, O. The Portuguese grapevine cultivar ‘Amaral’: Synonymies, homonymies and misnames. Vitis 2012, 51, 61–63. [Google Scholar] [CrossRef]
- Crespan, M.; Migliaro, D.; Larger, S.; Pindo, M.; Palmisano, M.; Manni, A.; Manni, E.; Polidori, E.; Sbaffi, F.; Silvestri, Q.; et al. Grapevine (Vitis vinifera L.) varietal assortment and evolution in the Marche region (central Italy). OENO One 2021, 55, 4628. [Google Scholar] [CrossRef]
- D’Onofrio, C.; Tumino, G.; Gardiman, M.; Crespan, M.; Bignami, C.; de Palma, L.; Barbagallo, M.G.; Muganu, M.; Morcia, C.; Novello, V.; et al. Parentage atlas of Italian grapevine varieties as inferred from SNP genotyping. Front. Plant. Sci. 2021, 11, 2265. [Google Scholar] [CrossRef]
- Nebish, A.; Tello, J.; Ferradás, Y.; Aroutiounian, R.; Martínez-Zapater, J.M.; Ibáñez, J. SSR and SNP Genetic Profiling of Armenian Grape Cultivars Gives Insights into Their Identity and Pedigree Relationships. OENO One 2021, 55, 101–114. [Google Scholar] [CrossRef]
- Botstein, D.; White, R.; Skolnick, M.; Davis, R. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar] [PubMed]
- Vervalle, J.A.; Costantini, L.; Lorenzi, S.; Pindo, M.; Mora, R.; Bolognesi, G.; Marini, M.; Lashbrooke, J.G.; Tobutt, K.R.; Vivier, M.A.; et al. A high-density integrated map for grapevine based on three mapping populations genotyped by the Vitis18K SNP chip. TAG. Theor. Appl. Genet. 2022, 21, 1–20. [Google Scholar] [CrossRef]
- Duduk, B.; Botti, S.; Ivanović, M.; Krstić, B.; Dukić, N.; Bertaccini, A. Identification of Phytoplasmas associated with grapevine yellows in Serbia. J. Phytopathol. 2004, 152, 575–579. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Y.; Cao, X.; Wu, D.; Hui, M.; Han, X.; Yao, F.; Li, Y.; Li, H.; Wang, H. Screening and validation of ssr molecular markers for identification of downy mildew resistance in intraspecific hybrid F1 progeny (V. vinifera). Horticulturae 2022, 8, 706. [Google Scholar] [CrossRef]
- Goszczynski, D.E.; Jooste, A.E.C. The application of single-strand conformation polymorphism (SSCP) technique for the analysis of molecular heterogeneity of grapevine virus A. Vitis 2002, 41, 77–82. [Google Scholar]
- Aroca, A.; Raposo, R. PCR-based strategy to detect and identify species of Phaeoacremonium causing grapevine diseases. Appl. Environ. Microbiol. 2007, 73, 2911–2918. [Google Scholar] [CrossRef]
- Wang, X.C.; Guo, L.; Shangguan, L.F.; Wang, C.; Yang, G.; Qu, S.C.; Fang, J.G. Analysis of expressed sequence tags from grapevine flower and fruit and development of simple sequence repeat markers. Mol. Biol. Rep. 2012, 39, 6825–6834. [Google Scholar] [CrossRef]
- Crăciunaş, C.; Coste, A.; Farago, M.; Iliescu, M.; Iuoraş, R.; Butiuc-Keul, A. Genetic stability of several cultivars of grapevine cultivated in vitro. Acta Hortic. 2009, 812, 515–520. [Google Scholar] [CrossRef]
- Schneider, S.; Reustle, G.; Zyprian, E. Detection of somaclonal variation in grapevine regenerants from protoplasts by RAPD-PCR. Vitis 1996, 35, 99–100. [Google Scholar]
- Baránek, M.; Raddová, J.; Krizan, B.; Pidra, M. Genetic changes in grapevine genomes after stress induced by in vitro cultivation, thermotherapy and virus infection, as revealed by AFLP. Genet. Mol. Biol. 2009, 32, 834–839. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gribaudo, I.; Torello Marinoni, D.; Gambino, G.; Mannini, F.; Akkak, A.; Botta, R. Assessment of genetic fidelity in regenerants from two Vitis vinifera cultivars. Acta Hortic. 2009, 827, 131–136. [Google Scholar] [CrossRef]
- Aljuaid, B.S.; Ismail, I.A.; Attia, A.O.; El Dessoky, S. Genetic Stability of in vitro Propagated Grapevine (Vitis vinifera L.) cv. Al-Bayadi. J. Agric. Crops 2022, 8, 12–19. [Google Scholar] [CrossRef]
- Schellenbaum, P.; Mohler, V.; Wenzel, G.; Walter, B. Variation in DNA methylation patterns of grapevine somaclones (Vitis vinifera L.). BMC Plant. Biol. 2008, 8, 78. [Google Scholar] [CrossRef] [PubMed]
- Dalbò, M.A. Genetic mapping, Qtl Analysis, and Marker-Assisted Selection for Disease Resistance Loci in Grapes. Ph.D. Thesis, Cornell University, Ithaca, NY, USA, August 1998. [Google Scholar]
- Doligez, A.; Bouquet, A.; Danglot, Y.; Lahogue, F.; Riaz, S.; Meredith, P.; Edwards, J.; This, P. Genetic mapping of grapevine (Vitis vinifera L.) applied to the detection of QTLs for seedlessness and berry weight. TAG. Theoretical and applied genetics. Theor. Und Angew. Genet. 2002, 105, 780–795. [Google Scholar] [CrossRef]
- Fanizza, G.; Lamaj, F.; Costantini, L.; Chaabane, R.; Grando, M.S. QTL analysis for fruit yield components in table grapes (Vitis vinifera). Appl Genet. 2005, 111, 658–664. [Google Scholar] [CrossRef] [PubMed]
- Costantini, L.; Moreira, F.; Zyprians, E.; Martínez-Zapater, J.; Grando, M. Molecular Maps, Qtl Mapping & Association Mapping In Grapevine Molecular Physiology & Biotechnology; Roubelakis-Angelakis, K.A., Ed.; Springer: Dordrecht, The Netherland, 2009. [Google Scholar] [CrossRef]
- Demmings, E.M.; Williams, B.R.; Lee, C.-R.; Barba, P.; Yang, S.; Hwang, C.-F.; Reisch, B.I.; Chitwood, D.H.; Londo, J.P. Quantitative trait locus analysis of leaf morphology indicates conserved shape loci in grapevine. Front. Plant. Sci. 2019, 10, 1373. [Google Scholar] [CrossRef]
- Su, K.; Guo, Y.; Zhong, W.; Lin, H.; Liu, Z.; Li, K.; Li, Y.; Guo, X. High-Density Genetic Linkage Map Construction and White Rot Resistance Quantitative Trait Loci Mapping for Genus Vitis Based on Restriction Site-Associated DNA Sequencing. Phytopathology 2021, 111, 659–670. [Google Scholar] [CrossRef]
- Su, K.; Xing, H.; Guo, Y.; Zhao, F.; Liu, Z.; Li, K.; Li, Y.; Guo, X. High-density genetic linkage map construction and cane cold hardiness QTL mapping for Vitis based on restriction site-associated DNA sequencing. BMC Genom. 2020, 21, 419. [Google Scholar] [CrossRef]
- Reshef, N.; Karn, A.; Manns, D.C.; Mansfield, A.K.; Cadle-Davidson, L.; Reisch, B.; Sacks, G.L. Stable QTL for malate levels in ripe fruit and their transferability across Vitis species. Hortic. Res. 2022, 9, uhac009. [Google Scholar] [CrossRef]
- Wang, H.; Yan, A.; Sun, L.; Zhang, G.; Wang, X.; Ren, J.; Xu, H. Novel stable QTLs identification for berry quality traits based on high-density genetic linkage map construction in table grape. BMC Plant. Biol. 2020, 20, 411. [Google Scholar] [CrossRef] [PubMed]
- Fu, P.; Wu, W.; Lai, G.; Li, R.; Peng, Y.; Yang, B.; Wang, B.; Yin, L.; Qu, J.; Song, S.; et al. Identifying Plasmopara viticola resistance loci in grapevine (Vitis amurensis) via genotyping-by-sequencing-based QTL mapping. PPB 2020, 154, 75–84. [Google Scholar] [CrossRef]
- Fehér, A. Callus, dedifferentiation, totipotency, somatic embryogenesis: What these terms mean in the era of molecular plant biology? Front. Plant. Sci. 2019, 10, 536. [Google Scholar] [CrossRef] [PubMed]
- Nuzzo, F.; Gambino, G.; Perrone, I. Unlocking grapevine in vitro regeneration: Issues and perspectives for genetic improvement and functional genomic studies. Plant. Physiol. Biochem. 2022, 193, 99–109. [Google Scholar] [CrossRef]
- Butiuc-Keul, A.; Coste, A.; Oltean, B.; Crăciunaş, C.; Halmagyi, A.; Deliu, C.; Farago, M.; Iliescu, M.; Iuoraş, R. In vitro clonal propagation of several grapevine cultivars. Acta Hortic. 2009, 843, 151–156. [Google Scholar] [CrossRef]
- Butiuc-Keul, A.; Coste, A.; Crăciunaş, C. Molecular characterization and in vitro preservation of some grapevine cultivars. Rom. Biotechnol. Lett. 2011, 16, 6226–6233. [Google Scholar]
- Krishna, H.; Alizadeh, M.; Singh, D.; Singh, U.; Chauhan, N.; Eftekhari, M.; Sadh, R.K. Somaclonal variations and their applications in horticultural crops improvement. 3 Biotech 2016, 6, 54. [Google Scholar] [CrossRef] [PubMed]
- Atak, A. New Perspectives in Grapevine (Vitis spp.) Breeding. In Plant Breeding-New Perspectives; Wang, H., Ed.; IntechOpen: London, UK, 2022; Available online: https://www.intechopen.com/online-first/82151 (accessed on 15 November 2022). [CrossRef]
- Mukherjee, P.; Husain, N.; Misra, S.C.; Rao, V.S. In vitro propagation of a grape rootstock, deGrasset Vitis champinii Planch. Effects of medium compositions and plant growth regulators. Sci. Hortic. 2010, 126, 13–19. [Google Scholar] [CrossRef]
- Yerbolova, L.S.; Ryabushkina, N.A.; Oleichenko, S.N.; Kampitova, G.A.; Galiakparov, N. The effect of growth regulators on in vitro culture of some Vitis vinifera L. cultivars. World Appl. Sci. J. 2013, 23, 76–80. [Google Scholar] [CrossRef]
- Capriotti, L.; Baraldi, E.; Mezzetti, B.; Limera, C.; Sabbadini, S. Biotechnological approaches: Gene overexpression, gene silencing, and genome editing to control fungal and oomycete diseases in grapevine. Int. J. Molec. Sci. 2020, 21, 5701. [Google Scholar] [CrossRef]
- Nicholson, K.L.; Tarlyn, N.; Armour, T.; Swanson, M.E.; Dhingra, A. Effect of phyllotactic position and cultural treatments toward successful direct shoot organogenesis in dwarf ‘Pixie’ grapevine (Vitis vinifera L.). Plant. Cell Tiss. Organ. Cult. 2012, 111, 123–129. [Google Scholar] [CrossRef]
- Zhang, P.; Yu, Z.Y.; Cheng, Z.M.; Zhang, Z.; Tao, J.M. In vitro explants regeneration of the grape ‘Wink’ (Vitis vinifera L. ‘Wink’). J. Plant Breed. Crop Sci. 2011, 3, 276–282. [Google Scholar]
- Park, H.J.; Lee, H.R.; Pyee, J.; Cha, H.C. Regeneration of grape (Vitis labruscana cv. Kyoho) by shoot-tip culture. J. Plant. Biol. 2001, 44, 185–192. [Google Scholar] [CrossRef]
- Pool, R.M. Effect of cytokinin on in vitro development of ‘concord’ flowers. Am. J. Enol. Vitic. 1975, 26, 43–46. [Google Scholar]
- Lilov, D.; Isvorska, N. Flower bud initiation from isolated meristem tissues of grapevine tendrils. Fisiol. Rast. 1978, 4, 73–78. [Google Scholar]
- Barreto, M.S. In Vitro Plant Regeneration and Transformation Studies in Grape (Vitis vinifera L.). Ph.D. Thesis, University of Pune, Pune, India.
- Torregrosa, L. Culture in vitro et Transformation Genetique de la Vigne. These, Ecole Nationale Superieure Agronomique de Montpellier, France, 1994. Available online: https://www.theses.fr/1994ENSA0024 (accessed on 12 September 2022).
- Torregrosa, L.; Bouquet, A. Agrobacterium tumefaciens and A. rhizogenes-rhizogenes cotransformation to obtain grapevine hair roots producing coat protin of grapevine chrome mosaic nepo virus. Plant. Cell Tiss. Org. Cult. 1997, 49, 59–63. [Google Scholar] [CrossRef]
- Loubser, J.T.; Meyer, A.J. Dual cultures of Meloidogyne javanica and grapevine rootstocks on artificial media. S. Afr. J. Enol. Vitic. 1990, 11, 42–45. [Google Scholar] [CrossRef][Green Version]
- Bavaresco, L.; Walker, M.A. Techniques for successfully establishing Xiphinema index in dual culture with grapes. Am. J. Enol. Vitic. 1994, 45, 273–277. [Google Scholar]
- Forneck, A.; Merkt, N.; Blaich, R. A tripartite asceptic culture system for grapes (Vitis spp.), phylloxera (Daktulosphaera vitifoliae) and mites (Tarsanemus sp.). Vitis 1998, 37, 95–96. [Google Scholar]
- Kellow, A.V. A Study of The Interaction between Susceptible and Resistant Grapevine and Phylloxera. Ph.D. Thesis, Natural and Agricultural Resource Sciences, Department of Horitculture, Vitculture and Oenology, The University of Adelaide, Adelaide, Australia, June 2000. [Google Scholar]
- Saygaç, S.; Önder, S. Effects of shoot tip size on in vitro regeneration and virus elimination of grapevine cv. Superior Seedless. Plant. Prot. Bull. 2021, 61, 5–9. [Google Scholar] [CrossRef]
- Mourad, A. Elimination of grapevine bois noir phytoplasma by tissue culture coupled or not with heat therapy or hot water treatment. Adv. Crop. Sci. Technol. 2013, 1, 1–4. [Google Scholar] [CrossRef]
- Miljanić, V.; Rusjan, D.; Škvarč, A.; Chatelet, P.; Štajner, N. Elimination of eight viruses and two viroids from preclonal candidates of six grapevine varieties (Vitis vinifera L.) through in vivo thermotherapy and in vitro meristem tip micrografting. Plants 2022, 11, 1064. [Google Scholar] [CrossRef] [PubMed]
- Malenica, N.; Jagić, M.; Pavletić, B.; Bauer, N.; Vončina, D.; Zdunić, G.; Levanić, D.L. Somatic embryogenesis as a tool for virus elimination in Croatian indigenous grapevine cultivars. Acta Bot. Croat. 2020, 79, 26–34. [Google Scholar] [CrossRef]
- Pérez-Núñez, M.T.; Chan, J.L.; Sáenz, L.; González, T.; Verdeil, J.L.; Oropeza, C. Improved somatic embryogenesis from Cocos nucifera (L.) plumule explants. Vitr. Cell. Dev. Biol. Plant. 2006, 42, 37–43. [Google Scholar] [CrossRef]
- Forleo, L.R.; D’Amico, M.; Basile, T.; Marsico, A.D.; Cardone, M.F.; Maggiolini, F.A.M.; Velasco, R.; Bergamini, C. Somatic embryogenesis in Vitis for genome editing: Optimization of protocols for recalcitrant genotypes. Horticulturae 2021, 7, 511. [Google Scholar] [CrossRef]
- Dal Santo, S.; De Paoli, E.; Pagliarani, C.; Amato, A.; Celii, M.; Boccacci, P.; Zenoni, S.; Gambino, G.; Perrone, I. Stress responses and epigenomic instability mark the loss of somatic embryogenesis competence in grapevine. Plant. Physiol. 2022, 188, 490–508. [Google Scholar] [CrossRef]
- Capriotti, L.; Limera, C.; Mezzetti, B.; Ricci, A.; Sabbadini, S. From induction to embryo proliferation: Improved somatic embryogenesis protocol in grapevine for Italian cultivars and hybrid Vitis rootstocks. Plant. Cell Tiss Organ. Cult. 2022, 151, 221–233. [Google Scholar] [CrossRef]
- Horstman, A.; Bemer, M.; Boutilier, K. A transcriptional view on somatic embryogenesis. Regeneration 2017, 4, 201–216. [Google Scholar] [CrossRef]
- Ghadirzadeh-Khorzoghi, E.; Jahanbakhshian-Davaran, Z.; Seyedi, S.M. Direct somatic embryogenesis of drought resistance pistachio (Pistacia vera L.) and expression analysis of somatic embryogenesis-related genes. S. Afr. J. Bot. 2019, 121, 558–567. [Google Scholar] [CrossRef]
- Catalano, C.; Abbate, L.; Motisi, A.; Crucitti, D.; Cangelosi, V.; Pisciotta, A.; Di Lorenzo, R.; Carimi, F.; Carra, A. Autotetraploid emergence via somatic embryogenesis in Vitis vinifera induces marked morphological changes in shoots, mature leaves, and stomata. Cells 2021, 10, 1336. [Google Scholar] [CrossRef] [PubMed]
- Corredoira, E.; Merkle, S.A.; Martínez, M.T.; Toribio, M.; Canhoto, J.M.; Correia, S.I.; Ballester, A.; Vieitez, A. Non-zygotic embryogenesis in hardwood species. Crit Rev. Plant. Sci. 2019, 38, 29–97. [Google Scholar] [CrossRef]
- Maillot, P.; Lebel, S.; Schellenbaum, P.; Jacques, A.; Walter, B. Differential regulation of SERK, LEC1-like and pathogenesis-related genes during indirect secondary somatic embryogenesis in grapevine. Plant. Physiol. Biochem. 2009, 47, 743–752. [Google Scholar] [CrossRef]
- Ricci, A.; Sabbadini, S.; Prieto, H.; Padilla, I.M.; Dardick, C.; Li, Z.; Scorza, R.; Limera, C.; Mezzetti, B.; Perez-Jimenez, M.; et al. Genetic transformation in peach (Prunus persica L.): Challenges and ways forward. Plants 2020, 9, 971. [Google Scholar] [CrossRef]
- Phillips, R.L.; Kaeppler, S.M.; Olhoft, P. Genetic instability of plant tissue cultures: Breakdown of normal controls. Proc. Natl. Acad. Sci. USA 1994, 91, 5222–5226. [Google Scholar] [CrossRef] [PubMed]
- Muler, E.; Brown, P.T.H.; Hartke, S.; Lorz, H. DNA variation in tissue-culture-derived rice plants. Theor. Appl. Genet. 1990, 80, 673–679. [Google Scholar] [CrossRef]
- Kuksova, V.B.; Piven, N.M.; Gleba, Y.Y. Somaclonal variation and in vitro induced mutagenesis in grapevine. Plant. Cell Tiss. Organ. Cult. 1997, 49, 17–27. [Google Scholar] [CrossRef]
- Palombi, M.A.; Damiano, C. Comparison between RAPD and SSR molecular markers in detecting genetic variation in kiwifruit (Actinidia deliciosa A. Chev). Plant. Cell Rep. 2002, 20, 1061–1066. [Google Scholar] [CrossRef]
- Kaeppler, S.M.; Phillips, R.L. DNA methylation and tissue culture-induced variation in plants. Vitr. Cell Dev. Biol. Plant. 1993, 29, 125–130. [Google Scholar] [CrossRef]
- Nivas, S.K.D.; Souza, L. Genetic fidelity in micropropagated plantlets of Anacardium occidentale L. (Cashew) an important fruit tree. Int. J. Sci. Res. 2014, 3, 2142–2146. [Google Scholar]
- Smulders, M.; de Klerk, G. Epigenetics in plant tissue culture. Plant. Growth Regul. 2011, 63, 137–146. [Google Scholar] [CrossRef]
- Prado, M.J.; Rodriguez, E.; Rey, L.; González, M.V.; Santos, C.; Rey, M. Detection of somaclonal variants in somatic embryogenesis-regenerated plants of Vitis vinifera by flow cytometry and microsatellite markers. Plant. Cell Tiss. Organ. Cult. 2010, 103, 49–59. [Google Scholar] [CrossRef]
- Marcotrigiano, M.; Bernatzky, R. Arrangement of cell layers in the shoot apical meristems of periclinal chimeras influences cell fate. Plant. J. 1995, 7, 193–202. [Google Scholar] [CrossRef]
- Burge, G.K.; Morgan, E.R.; Seelye, J.F. Opportunities for synthetic plant chimeral breeding: Past and future. Plant. Cell Tiss. Organ. Cult. 2002, 70, 13–21. [Google Scholar] [CrossRef]
- Franks, T.; Botta, R.; Thomas, M.R.; Franks, J. Chimerism in grapevines: Implications for cultivar identity, ancestry and genetic improvement. Theor. Appl. Genet. 2002, 104, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Bertsch, C.; Kieffer, F.; Maillot, P.; Farine, S.; Butterlin, G.; Merdinoglu, D.; Walter, B. Genetic chimerism of Vitis vinifera cv. Chardonnay 96 is maintained through organogenesis but not somatic embryogenesis. BMC Plant. Biol. 2005, 5, 20. [Google Scholar] [CrossRef]
- Riaz, S.; Garrison, K.E.; Dangl, G.S.; Boursiquot, J.M.; Meredith, C.P. Genetic divergence and chimerism within ancient asexually propagated winegrape cultivars. J. Amer. Soc. Hort. Sci. 2002, 127, 508–514. [Google Scholar] [CrossRef][Green Version]
- Quackenbush, J.; Liang, F.; Holt, I.; Pertea, G.; Upton, J. The TIGR gene indices: Reconstruction and representation of expressed gene sequences. Nucleic Acids Res. 2000, 28, 141–145. [Google Scholar] [CrossRef][Green Version]
- Siemann, E.; Creasy, L. Concentration of the phytoalexin resveratrol in wine. Am. J. Enol. Viticult. 1992, 43, 49–52. [Google Scholar]
- Sotheeswaran, S.; Pasupathy, V. Distribution of resveratrol oligomers in plants. Photochemistry 1993, 32, 1083–1092. [Google Scholar] [CrossRef]
- Kikkert, J.K.; Ali, G.S.; Wallace, P.G.; Reisch, B.; Reustle, G.M. Expression of fungal chitinase in Vitis vinifera L. ‘Merlot’ and ‘Chardonnay’ plants produced by biolistic information. Acta Hortic. 2000, 528, 297–303. [Google Scholar] [CrossRef]
- Tamaoki, M.; Imai, H.; Takahashi, H.; Toda, Y.; Niwa, Y.; Nakajima, N.; Aono, M.; Kubo, A.; Saji, H. Development of visible markers for transgenic plants and their availability for environmental risk assessment. Z. Für Nat. 2006, 61, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Braidot, E.; Zancani, M.; Petrussa, E.; Peresson, C.; Bertolini, A.; Patui, S.; Macri, F.; Vianello, A. Transport and accumulation of flavonoids in grapevine (Vitis vinifera L.). Plant. Signal. Behav. 2008, 3, 626–632. [Google Scholar] [CrossRef] [PubMed]
- Basha, S.M.; Musingo, M.; Colova, V.S. Compositional differences in the phenolics compounds of muscadine and bunch grape wines. Afr. J. Biotechnol. 2004, 10, 523–528. [Google Scholar] [CrossRef]
- INRA-CNRGV The French Plant Genomic Resource Center. Available online: http://cnrgv.toulouse.inra.fr (accessed on 10 October 2022).
- Huang, X.; Wei, X.; Sang, T.; Zhao, Q.; Feng, Q.; Zhao, Y.; Li, C.; Zhy, C.; Lu, T.; Zhang, Z.; et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 2010, 42, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Meuwissen, T.H.E.; Hayes, B.J.; Goddard, M.E. Prediction of Total Genetic Value Using Genome-Wide Dense Marker Maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef]
- Jannink, J.-L.; Lorenz, A.J.; Iwata, H. Genomic selection in plant breeding: From theory to practice. Brief. Funct. Genom. 2010, 9, 166–177. [Google Scholar] [CrossRef]
- Nakaya, A.; Isobe, S.N. Will genomic selection be a practical method for plant breeding? Ann. Bot. 2012, 110, 1303–1316. [Google Scholar] [CrossRef]
- Fodor, A.; Segura, V.; Denis, M.; Neuenschwander, S.; Fournier-Level, A.; Chatelet, P.; Homa, F.A.; Lacombe, T.; This, P.; Le Cunff, L. Genome-wide prediction methods in highly diverse and heterozygous species: Proof-of-concept through simulation in grapevine. PLoS ONE 2014, 9, e110436. [Google Scholar] [CrossRef]
- Wang, M.; Jiang, N.; Jia, T.; Leach, L.; Cockram, J.; Waugh, R.; Ramsay, L.; Thomas, B.; Luo, Z. Genome-wide association mapping of agronomic and morphologic traits in highly structured populations of barley cultivars. Theor. Appl. Genet. 2012, 124, 233–246. [Google Scholar] [CrossRef]
- Laucou, V.; Lacombe, T.; Dechesne, F.; Siret, R.; Bruno, J.-P.; Dessup, M.; Dessup, T.; Ortigosa, P.; Parra, P.; Roux, C. High throughput analysis of grape genetic diversity as a tool for germplasm collection management. Theor. Appl. Genet. 2011, 122, 1233–1245. [Google Scholar] [CrossRef] [PubMed]
- Myles, S.; Chia, J.-M.; Hurwitz, B.; Simon, C.; Zhong, G.Y.; Buckler, E.; Ware, D. Rapid Genomic Characterization of the Genus Vitis. PLoS ONE 2010, 5, e8219. [Google Scholar] [CrossRef] [PubMed]
- Bowers, J.; Boursiquot, J.M.; This, P.; Chu, K.; Johansson, H.; Meredith, C. Historical Genetics: The Parentage of Chardonnay, Gamay, and Other Wine Grapes of Northeastern France. Science 1999, 285, 1562–1565. [Google Scholar] [CrossRef]
- Boursiquot, J.-M.; Lacombe, T.; Laucou, V.; Julliard, S.; Perrin, F.-X.; Lanier, N.; Legrand, D.; Meredith, C.; This, P. Parentage of Merlot and related winegrape cultivars of southwestern France: Discovery of the missing link. Aust. J. Grape Wine Res. 2009, 15, 144–155. [Google Scholar] [CrossRef]
- Myles, S.; Boyko, A.R.; Owens, C.L.; Brown, P.J.; Grassi, F.; Aradhya, M.K.; Prins, B.; Reynolds, A.; Chia, I.-M.; Ware, D.; et al. Genetic Structure and Domestication History of the Grape. Proc. Natl. Acad. Sci. USA 2011, 108, 3530–3535. [Google Scholar] [CrossRef] [PubMed]
- Lacombe, T.; Boursiquot, J.-M.; Laucou, V.; Di Vecchi-Staraz, M.; Péros, J.-P.; This, P. Large-scale parentage analysis in an extended set of grapevine cultivars (Vitis vinifera L.). Theor. Appl. Genet. 2012, 126, 1–14. [Google Scholar] [CrossRef]
- Hannah, L.; Roehrdanz, P.R.; Ikegami, M.; Shepard, A.V.; Shaw, M.R.; Tabor, G.; Zhi, L.; Marquet, P.A.; Hijmans, R.J. Climate change, wine, and conservation. Proc. Natl. Acad. Sci. USA 2013, 110, 6907–6912. [Google Scholar] [CrossRef] [PubMed]
- Moriondo, M.; Jones, G.V.; Bois, B.; Dibari, C.; Ferrise, R.; Trombi, G.; Bindi, M. Projected shifts of wine regions in response to climate change. Clim. Change 2013, 119, 825–839. [Google Scholar] [CrossRef]
- Gray, D.J.; Jayasankar, S.; Li, Z.T. Vitaceae (Grape Vitis spp.). In Biotechnology of Fruit and Nut Crops. Biotechnology in Agriculture Series; No. 29; Litz, R.E., Ed.; CAB International: Wallingford, UK, 2005; Volume Chapter 22, pp. 672–706. [Google Scholar]
- Maraš, V.; Tello, J.; Gazivoda, A.; Mugoša, M.; Perišić, M.; Raičević, J.; Štajner, N.; Ocete, R.; Božović, V.; Popović, T.; et al. Population genetic analysis in old Montenegrin vineyards reveals ancient ways currently active to generate diversity in Vitis vinifera. Sci. Rep. 2020, 10, 15000. [Google Scholar] [CrossRef] [PubMed]
- Čuš, F.; Cesnik, H.B.; Bolta, S.V.; Gregorcic, A. Pesticide residues and microbio-logical quality of bottled wines. Food Control. 2010, 21, 150–154. [Google Scholar] [CrossRef]
- Pretorius, I.S.; Høj, P.B. Grape and wine biotechnology: Challenges, opportunities and potential benefits. Aust. J. Grape Wine Res. 2006, 11, 83–108. [Google Scholar] [CrossRef]
- Gray, D.J.; Li, Z.T.; Dhekney, S.A. Precision breeding of grapevine (Vitis vinifera L.) for improved traits. Plant. Sci. 2014, 228, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.T.; Dhekney, S.A.; Dutt, M.; Van Aman, M.; Tattersall, J.; Kelley, K.T.; Gray, D.J. Optimizing Agrobacterium-mediated transformation of grapevine. Vitr. Cell. Dev. Biol. Plant. 2006, 42, 220–227. [Google Scholar] [CrossRef]
- Li, Z.T.; Dhekney, S.A.; Dutt, M.; Gray, D.J. An improved protocol for Agrobacterium-mediated transformation of grapevine (Vitis vinifera L.). Plant. Cell Tiss. Organ. Cult. 2008, 93, 311–321. [Google Scholar] [CrossRef]
- Torregrosa, L.; Rienth, M.; Luchaire, N.; Novelli, F.; Bigard, A.; Chatbanyong, R.; Lopez, G.; Farnos, M.; Roux, C.; Adivèze, A.; et al. The Microvine, A Biological Model, Very Versatile and Efficient to Boost Grapevine Research in Physiology and Genetics. In Proceedings of the 39th OIV Meeting, Bento Gonzalvez, Brazil, 24–28 October 2016. [Google Scholar]
- Dalla Costa, L.; Piazza, S.; Campa, M.; Flachowsky, H.; Hanke, M.V.; Malnoy, M. Efficient heat-shock removal of the selectable marker gene in genetically modified grapevine. Plant. Cell Tiss. Organ. Cult. 2016, 124, 471–481. [Google Scholar] [CrossRef]
- Muruganantham, M.; Moskovitz, Y.; Haviv, S.; Horesh, T.; Fenigstein, A.; Preez, J.D.; Stephan, D.; Burger, J.T.; Mawassi, M. Grapevine virusA-mediated gene silencing in Nicotiana benthamiana and Vitis vinifera. J. Virol. Methods 2009, 155, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Kurth, E.G.; Peremyslov, V.V.; Prokhnevsky, A.I.; Kasschau, K.D.; Miller, M.; Carrington, J.C.; Dolja, V.V. Virus-derived gene expression and RNA interference vector for grapevine. J. Virol. 2012, 86, 6002–6009. [Google Scholar] [CrossRef] [PubMed]
- Meng, B.; Venkataraman, S.; Li, C.; Wang, W.; Dayan-Glick, C.; Mawassi, M. Construction and biological activities of the first infectious cDNA clones of the genus Foveavirus. Virology 2013, 435, 453–462. [Google Scholar] [CrossRef]
- Wood, K. Marker proteins for gene expression. Curr. Opin. Biotechnol. 1995, 6, 50–58. [Google Scholar] [CrossRef]
- Rosellini, D. Selectable markers and reporter genes. A well furnished tool-box for plant science and genetic engineering. Crit. Rev. Plant. Sci. 2012, 31, 401–453. [Google Scholar] [CrossRef]
- Dutt, M.; Li, Z.T.; Dhekney, S.A.; Gray, D.J. A co-transformation system to produce transgenic grapevines free of marker genes. Plant. Sci. 2008, 175, 423–430. [Google Scholar] [CrossRef]
- Li, Z.T.; Dhekney, S.A.; Gray, D.J. Use of the VvMybA1 gene for non-destructive quantification of promoter activity via color histogram analysis in grapevine (Vitis vinifera) and tobacco. Transgenic Res. 2011, 20, 1087–1097. [Google Scholar] [CrossRef]
- Dutt, M.; Li, Z.T.; Gray, D.J.; Gmitter, F.; Grosser, J.W. Purple citrus? Utilization of a Myb-related transcription factor gene for Anthocyanin production. AMER Soc. Hortic. Sci. 2013, 48, 10–11. [Google Scholar]
- Li, Z.T.; Jayasankar, S.; Gray, D.J. Expression of a bifunctional green fluorescent protein (GFP) fusion marker under the control of three constitutive promoters and enhanced derivatives in transgenic grape (Vitis vinifera). Plant. Sci. 2001, 160, 877–887. [Google Scholar] [CrossRef] [PubMed]
- Conn, S.; Zhang, W.; Franco, C. Anthocyanic vacuolar inclusions (AVIs) selectively bind acylated anthocyanins in Vitis vinifera L. (grapevine) suspension culture. Biotechnol. Lett. 2003, 25, 835–839. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.T.; Kim, K.-H.; Jasinski, J.R.; Creech, M.R.; Gray, D.J. Large-scale characterization of promoters from grapevine (Vitis spp.) using quantitative anthocyanin and GUS assay systems. Plant. Sci. 2012, 196, 132–142. [Google Scholar] [CrossRef]
- Pilati, S.; Perazzolli, M.; Malossini, A.; Cestaro, A.; Damattè, L.; Fontana, P.; Dal Ri, A.; Viola, R.; Velasco, R.; Moser, C. Genome-wide transcriptional analysis of grapevine berry ripening reveals a set of genes similarly modulated during three seasons and the occurrence of an oxidative burst at veraison. BMC Genom. 2007, 8, 428. [Google Scholar] [CrossRef]
- Mezzetti, B.; Pandolfini, T.; Navacchi, O.; Landi, L. Genetic transformation of Vitis vinifera via organogenesis. BMC Biotechnol. 2002, 2, 18. [Google Scholar] [CrossRef]
- Xie, X.; Agüero, C.B.; Wang, Y.; Andrew Walker, M. Genetic transformation of grape varieties and rootstocks via organogenesis. Plant. Cell Tiss. Organ. Cult. 2016, 126, 541–552. [Google Scholar] [CrossRef]
- Sabbadini, S.; Capriotti, L.; Molesini, B.; Pandolfini, T.; Navacchi, O.; Limera, C.; Ricci, A.; Mezzetti, B. Comparison of regeneration capacity and Agrobacterium-me.ediated cell transformation efficiency of different cultivars and rootstocks of Vitis spp. via organogenesis. Sci. Rep. 2019, 9, 582. [Google Scholar] [CrossRef]
- de Carvalho, D.C.; da Silva, A.L.L.; Schuck, M.R.; Purcino, M.; Tanno, G.N.; Biasi, L.A. Fox grape cv. Bordô (Vitis labrusca L.) and grapevine cv. Chardonnay (Vitis vinifera L.) cultivated in vitro under different carbohydrates, amino acids and 6-benzylaminopurine levels. Brazil. Arch. Biol. Technol. 2013, 56, 191–201. [Google Scholar] [CrossRef]
- Colby, S.M.; Juncosa, A.M.; Meredith, C.P. Cellular differences in Agrobacterium susceptibility and regenerative capacity restrict the development of transgenic grapevines. J. Amer. Soc. Hort. Sci. 1991, 116, 356–361. [Google Scholar] [CrossRef]
- Maillot, P.; Deglène-Benbrahim, L.; Walter, B. Efficient somatic embryogenesis from meristematic explants in grapevine (Vitis vinifera L.) cv. Chardonnay: An improved protocol. Trees 2016, 30, 1377–1387. [Google Scholar] [CrossRef]
- Zhou, Q.; Dai, L.; Cheng, S.; He, J.; Wang, D.; Zhang, J.; Wang, Y. A circulatory system useful both for long-term somatic embryogenesis and genetic transformation in Vitis vinifera L. cv. Thompson Seedless. Plant. Cell Tiss. Organ. Cult. 2014, 118, 157–168. [Google Scholar] [CrossRef]
- Saporta, R.; San Pedro, T.; Gisbert, C. Attempts at grapevine (Vitis vinifera L.) breeding through genetic transformation: The main limiting factors. Vitis 2016, 55, 173–186. [Google Scholar] [CrossRef]
- Wang, X.; Tu, M.; Wang, D.; Liu, J.; Li, Y.; Li, Z.; Wang, Y.; Wang, X. CRISPR/Cas9-mediated efficient targeted mutagenesis in grape in the first generation. Plant. Biotechnol. J. 2018, 16, 844–855. [Google Scholar] [CrossRef]
- Carra, A.; Sajeva, M.; Abbate, L.; Siragusa, M.; Pathirana, R.; Carimi, F. Factors affecting somatic embryogenesis in eight Italian grapevine cultivars and the genetic stability of embryo-derived regenerants as assessed by molecular markers. Sci. Hort. 2016, 204, 123–127. [Google Scholar] [CrossRef]
- de Carvalho, D.C.; da Silva, A.L.L.; Tanno, G.N.; Purcino, M.; Biasi, L.A. Organogenesis from leaf segments and internodes of grapevine cv. Merlot. Ciência Agrotecnol. 2011, 35, 108–114. [Google Scholar] [CrossRef][Green Version]
- Perrin, M.; Martin, D.; Joly, D.; Demangeat, G.; This, P.; Masson, J.E. Medium-dependent response of grapevine somatic embryogenic cells. Plant. Sci. 2001, 161, 107–116. [Google Scholar] [CrossRef]
- Jelly, N.S.; Valat, L.; Walter, B.; Maillot, P. Transient expression assays in grapevine: A step towards genetic improvement. Plant. Biotechnol. J. 2014, 12, 1231–1245. [Google Scholar] [CrossRef]
- Torregrosa, L.; Verries, C.; Tesniere, C. Grapevine (Vitis vinifera L.) promoter analysis by biolistic-mediated transient transformation of cell suspensions. Vitis 2002, 41, 27–32. [Google Scholar] [CrossRef]
- Verries, C.; Pradal, M.; Chatelet, P.; Torregrosa, L.; Tesniere, C. Isolation and analysis of the promoter of VvAdh2, a grapevine Vitis vinifera L. ripening-related gene. Plant. Sci. 2004, 167, 1067–1074. [Google Scholar] [CrossRef]
- Harris, N.N.; Luczo, J.M.; Robinson, S.P.; Walker, A.R. Transcriptional regulation of the three grapevine chalcone synthase genes and their role in flavonoid synthesis in Shiraz. Aus. J. Grape Wine Res. 2013, 19, 221–229. [Google Scholar] [CrossRef]
- Deluc, L.; Bogs, J.; Walker, A.R.; Ferrier, T.; Decendit, A.; Merillon, J.M.; Robinson, S.P.; Barrieu, F. The transcription factor VvMYB5b contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis in developing grape berries. Plant. Physiol. 2008, 147, 2041–2053. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.R.; Lee, E.; Bogs, J.; McDavid, D.A.; Thomas, M.R.; Robinson, S.P. White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant. J. 2007, 49, 772–785. [Google Scholar] [CrossRef]
- Czemmel, S.; Stracke, R.; Weisshaar, B.; Cordon, N.; Harris, N.N.; Walker, A.R.; Robinson, S.P.; Bogs, J. The grapevine R2R3-MYB transcription factor VvMYBF1 regulates flavonol synthesis in developing grape berries. Plant. Physiol. 2009, 151, 1513–1530. [Google Scholar] [CrossRef]
- Bogs, J.; Jaffe, F.W.; Takos, A.M.; Walker, A.R.; Robinson, S.P. The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant. Physiol. 2007, 143, 1347–1361. [Google Scholar] [CrossRef]
- Hichri, I.; Heppel, S.C.; Pillet, J.; Leon, C.; Czemmel, S.; Delrot, S.; Lauvergeat, V.; Bogs, J. The basic helix-loop-helix transcription factor MYC1 is involved in the regulation of the flavonoid biosynthesis pathway in grapevine. Mol. Plant. 2010, 3, 509–523. [Google Scholar] [CrossRef]
- Höll, J.; Vannozzi, A.; Czemmel, S.; D’Onofrio, C.; Walker, A.R.; Rausch, T.; Lucchin, M.; Boss, P.K.; Dry, I.B.; Bogs, J. The R2R3-MYB transcription factors MYB14 and MYB15 regulate stilbene biosynthesis in Vitis vinifera. Plant. Cell 2013, 25, 4135–4414. [Google Scholar] [CrossRef]
- Joubert, D.A.; de Lorenzo, G.; Vivier, M.A. Regulation of the grapevine polygalacturonase-inhibiting protein encoding gene: Expression pattern, induction profile and promoter analysis. J. Plant. Res. 2013, 126, 267–281. [Google Scholar] [CrossRef]
- Saumonneau, A.; Laloi, M.; Lallemand, M.; Rabot, A.; Atanassova, R. Dissection of the transcriptional regulation of grape ASR and response to glucose and abscisic acid. J. Exp. Bot. 2012, 63, 1495–1510. [Google Scholar] [CrossRef] [PubMed]
- Marchive, C.; Leon, C.; Kappel, C.; Coutos-Thevenot, P.; Corio-Costet, M.F.; Delrot, S.; Lauvergeat, V. Over-expression of VvWRKY1 in grapevines induces expression of jasmonic acid pathway-related genes and confers higher tolerance to the downy mildew. PLoS ONE 2013, 8, e54185. [Google Scholar] [CrossRef]
- Saumonneau, A.; Agasse, A.; Bidoyen, M.T.; Lallemand, M.; Cantereau, A.; Medici, A.; Laloi, M.; Atanassova, R. Interaction of grape ASR proteins with a DREB transcription factor in the nucleus. FEBS Lett. 2008, 582, 3281–3287. [Google Scholar] [CrossRef] [PubMed]
- Urso, S.; Zottini, M.; Ruberti, C.; Schiavo, F.L.; Stanca, A.M.; Cattivelli, L.; Vale, G. An Agrobacterium tumefaciens-mediated gene silencing system for functional analysis in grapevine. Plant. Cell Tiss. Organ. Cult. 2013, 114, 49–60. [Google Scholar] [CrossRef]
- Visser, M.; Stephan, D.; Jaynes, J.M.; Burger, J.T. A transient expression assay for the in planta efficacy screening of an antimicrobial peptide against grapevine bacterial pathogens. Lett. Appl. Microbiol. 2012, 54, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Bertazzon, N.; Raiola, A.; Castiglioni, C.; Gardiman, M.; Angelini, E.; Borgo, M.; Ferrari, S. Transient silencing of the grapevine gene VvPGIP1 by agroinfiltration with a construct for RNA interference. Plant. Cell Rep. 2012, 31, 133–143. [Google Scholar] [CrossRef]
- Santos-Rosa, M.; Poutaraud, A.; Merdinoglu, D.; Mestre, P. Development of a transient expression system in grapevine via agro-infiltration. Plant. Cell Rep. 2008, 27, 1053–1063. [Google Scholar] [CrossRef]
- Guan, X.; Zhao, H.; Xu, Y.; Wang, Y. Transient expression of glyoxal oxidase from the Chinese wild grape Vitis pseudoreticulata can suppress powdery mildew in a susceptible genotype. Protoplasma 2011, 248, 415–423. [Google Scholar] [CrossRef]
- He, M.; Xu, Y.; Cao, J.; Zhu, Z.; Jiao, Y.; Wang, Y.; Guan, X.; Yang, Y.; Xu, W.; Fu, Z. Subcellular localization and functional analyses of a PR10 protein gene from Vitis pseudoreticulata in response to Plasmopara viticola infection. Protoplasma 2013, 250, 129–140. [Google Scholar] [CrossRef]
- Xu, W.; Yu, Y.; Ding, J.; Hua, Z.; Wang, Y. Characterization of a novel stilbene synthase promoter involved in pathogen- and stress-inducible expression from Chinese wild Vitis pseudoreticulata. Planta 2010, 231, 475–487. [Google Scholar] [CrossRef]
- Xu, T.F.; Zhao, X.C.; Jiao, Y.T.; Wei, J.Y.; Wang, L.; Xu, Y. A pathogenesis related protein, VpPR-10.1, from Vitis pseudoreticulata: An insight of its mode of antifungal activity. PLoS ONE 2014, 9, e95102. [Google Scholar] [CrossRef] [PubMed]
- Le Henanff, G.; Heitz, T.; Mestre, P.; Mutterer, J.; Walter, B.; Chong, J. Characterization of Vitis vinifera NPR1 homologs involved in the regulation of pathogenesis-related gene expression. BMC Plant. Biol. 2009, 9, 54. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.P.; Peremyslov, V.V.; Medina, V.; Dolja, V.V. Tandem leader proteases of grapevine leafroll-associated virus-2: Host-specific functions in the infection cycle. Virology 2009, 383, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Gollop, R.; Even, S.; Colova-Tsolova, V.; Perl, A. Expression of the grape dihydroflavonol reductase gene and analysis of its promoter region. J. Exp. Bot. 2002, 53, 1397–1409. [Google Scholar] [CrossRef]
- Jelly, N.S.; Schellenbaum, P.; Walter, B.; Maillot, P. Transient expression of artificial microRNAs targeting Grapevine fanleaf virus and evidence for RNA silencing in grapevine somatic embryos. Transgenic Res. 2012, 21, 1319–1327. [Google Scholar] [CrossRef]
- Li, Z.T.; Jayasankar, S.; Gray, D.J. Bi-directional duplex promoters with duplicated enhancers significantly increase transgene expression in grape and Tobacco. Transgenic Res. 2004, 13, 143–154. [Google Scholar] [CrossRef]
- Gutoranov, G.P.; Tsvetkov, I.J.; Colova-Tsolova, V.M.; Atanassov., A.I. Genetically engineered grapevines carrying GFLV coat protein and antifreeze genes. Agricult. Conspect. Sci. 2001, 66, 71–76. [Google Scholar]
- Mulwa, R.M.S.; Norton, M.A.; Farrand, S.K.; Skirvin, R.M. Agrobacterium-mediated transformation and regeneration of transgenic ‘Chancellor’ wine grape plants expressing the tfdA gene. Vitis 2007, 46, 110–115. [Google Scholar]
- Nirala, N.K.; Das, D.K.; Srivastava, P.S.; Sopory, S.K.; Upadhyaya, K.C. Expression of a rice chitinase gene enhances antifungal potential in transgenic grapevine (Vitis vinifera L.). Vitis 2010, 49, 181–187. [Google Scholar] [CrossRef]
- Dhekney, S.A.; Li, Z.T.; Gray, D.J. Grapevines engineered to express cisgenic Vitis vinifera thaumatin-like protein exhibit fungal disease resistance. Vitr. Cell. Dev. Biol. Plant. 2011, 47, 458–466. [Google Scholar] [CrossRef]
- Nookaraju, A.; Agrawal, D.C. Enhanced tolerance of transgenic grapevines expressing chitinase and β-1,3-glucanase genes to downy mildew. Plant. Cell Tiss. Organ. Cult. 2012, 111, 15–28. [Google Scholar] [CrossRef]
- Dai, L.; Zhou, Q.; Li, R.; Du, Y.; He, J.; Wang, D.; Cheng, S.; Zhang, J.; Wang, Y. Establishment of a picloram-induced somatic embryogenesis system in Vitis vinifera cv. Chardonnay and genetic transformation of a stilbene synthase gene from wild-growing Vitis species. Plant. Cell Tiss. Organ. Cult. 2015, 121, 397–412. [Google Scholar] [CrossRef]
- Li, Z.T.; Hopkins, D.L.; Gray, D.J. Overexpression of antimicrobial lytic peptides protects grapevine from Pierce’s disease under greenhouse but not field conditions. Transgenic Res. 2015, 24, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Xie, X.; Xu, Y.; Zhang, C.; Wang, X.; Zhang, J.; Wang, Y. Genetic transformation of a fruit-specific, highly expressed stilbene synthase gene from Chinese wild Vitis quinquangularis. Planta 2016, 243, 1041–1053. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Wang, D.; Xie, X.; Zhang, C.; Wang, X.; Xu, Y.; Wang, Y.; Zhang, J. The novel gene VpPR4-1 from Vitis pseudoreticulata increases powdery mildew resistance in transgenic Vitis vinifera L. Front. Plant. Sci. 2016, 7, 695. [Google Scholar] [CrossRef] [PubMed]
- Pessina, S.; Lenzi, L.; Perazzolli, M.; Campa, M.; Dalla Costa, L.; Urso, S.; Valè, G.; Salamini, F.; Velasco, R.; Malnoy, M. Knockdown of MLO genes reduces susceptibility to powdery mildew in grapevine. Hortic. Res. 2016, 3, 16016. [Google Scholar] [CrossRef]
- He, R.; Wu, J.; Zhang, Y.; Agüero, C.B.; Li, X.; Liu, S.; Wang, C.; Andrew Walker, M.; Lu, J. Overexpression of a thaumatin-like protein gene from Vitis amurensis improves downy mildew resistance in Vitis vinifera grapevine. Protoplasma 2017, 254, 1579–1589. [Google Scholar] [CrossRef]
- Jiao, L.; Zhang, Y.; Lu, J. Overexpression of a stressresponsive U-box protein gene VaPUB affects the accumulation of resistance related proteins in Vitis vinifera ‘Thompson Seedless’. Plant. Physiol. Biochem. 2017, 112, 53–63. [Google Scholar] [CrossRef]
- Ma, H.; Xiang, G.; Li, Z.; Wang, Y.; Dou, M.; Su, L.; Yin, X.; Liu, R.; Wang, Y.; Xu, Y. Grapevine VpPR10.1 functions in resistance to Plasmopara viticola through triggering a cell death-like defense response by interacting with VpVDAC3. Plant. Biotechnol. J. 2018, 16, 1488–1501. [Google Scholar] [CrossRef]
- Su, H.; Jiao, Y.T.; Wang, F.F.; Liu, Y.E.; Niu, W.L.; Liu, G.T.; Xu, Y. Overexpression of VpPR10.1 by an efficient transformation method enhances downy mildew resistance in V. vinifera. Plant. Cell Rep. 2018, 37, 819–832. [Google Scholar] [CrossRef]
- Soliman, H.I.A. Production of genetically modified grape (Vitis vinifera L.) plants. Int. J. Hortic. Agric. Food Sci. 2018, 2, 111–120. [Google Scholar] [CrossRef]
- Jiang, J.; Xi, H.; Dai, Z.; Lecourieux, F.; Yuan, L.; Liu, X.; Patra, B.; Wei, Y.; Li, S.; Wang, L. VvWRKY8 represses stilbene synthase genes through direct interaction with VvMYB14 to control resveratrol biosynthesis in grapevine. J. Exp. Bot. 2019, 70, 715–729. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Ma, F.; Li, R.; Zhou, Q.; Yao, W.; Jiao, Y.; Zhang, C.; Zhang, J.; Wang, X.; Xu, Y.; et al. VpSTS29/STS2 enhances fungal tolerance in grapevine through a positive feedback loop. Plant. Cell Environ. 2019, 42, 2979–2998. [Google Scholar] [CrossRef] [PubMed]
- Tu, M.; Wang, X.; Yin, W.; Wang, Y.; Li, Y.; Zhang, G.; Li, Z.; Song, J.; Wang, X. Grapevine VlbZIP30 improves drought resistance by directly activating VvNAC17 and promoting lignin biosynthesis through the regulation of three peroxidase genes. Hortic. Res. 2020, 7, 150. [Google Scholar] [CrossRef]
- Carroll, D. Genome engineering with zinc-finger nucleases. Genetics 2011, 188, 773–782. [Google Scholar] [CrossRef]
- Mahfouz, M.M.; Li, L.; Shamimuzzaman, M.; Wibowo, A.; Fang, X.; Zhu, J.-K. De novo-engineered transcription activator-like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double-strand breaks. Proc. Nat. Acad. Sci. USA 2011, 108, 2623–2628. [Google Scholar] [CrossRef]
- Li, L.; Piatek, M.J.; Atef, A.; Piatek, A.; Wibowo, A.; Fang, X.; Sabir, J.S.M.; Zhu, J.-K.; Mahfouz, M.M. Rapid and highly efficient construction of TALE-based transcriptional regulators and nucleases for genome modification. Plant. Molec. Biol. 2012, 78, 407–416. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. Programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Kaul, T.; Sony, S.K.; Verma, R.; Motelb, K.F.A.; Prakash, A.T.; Eswaran, M.; Bharti, J.; Nehra, J.; Kaul, R. Revisiting CRISPR/Cas-mediated crop improvement: Special focus on nutrition. J. Biosci. 2020, 45, 1–37. [Google Scholar] [CrossRef]
- van der Oost, J.; Westra, E.R.; Jackson, R.N.; Wiedenheft, B. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat. Rev. Microbiol. 2014, 12, 479–492. [Google Scholar] [CrossRef]
- Xie, K.; Zhang, J.; Yang, Y. Genome-wide prediction of highly specific guide RNA spacers for CRISPR-Cas9-mediated genome editing in model plants and major crops. Mol. Plant. 2014, 7, 923–926. [Google Scholar] [CrossRef] [PubMed]
- Bortesi, L.; Fischer, R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 2015, 33, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Butiuc-Keul, A.; Farkas, A.; Carpa, R.; Dobrota, C.T.; Iordache, D. Development of smart fruit crops by genome editing. Turk. J. Agric. For. 2022, 46, 129–140. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O.; et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef]
- Carpa, R.; Remizovschi, A.; Culda, C.A.; Butiuc-Keul, A.L. Inherent and composite hydrogels as promising materials to limit antimicrobial resistance. Gels 2022, 8, 70. [Google Scholar] [CrossRef]
- Ali, Z.; Abulfaraj, A.; Idris, A.; Ali, S.; Tashkandi, M.; Mahfouz, M.M. CRISPR/ Cas9-mediated viral interference in plants. Genome Biol. 2015, 16, 238. [Google Scholar] [CrossRef]
- Malnoy, M.; Viola, R.; Jung, M.-H.; Koo, O.-J.; Kim, J.-S.; Velasco, R.; Kanchiswamy, C.N. DNAfree genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front. Plant. Sci. 2016, 7, 1904. [Google Scholar] [CrossRef]
- Osakabe, Y.; Liang, Z.; Ren, C.; Nishitani, C.; Osakabe, K.; Wada, M.; Komori, S.; Malnoy, M.; Velasco, R.; Poli, M.; et al. CRISPR-Cas9-mediated genome editing in apple and grapevine. Nat. Prot. 2018, 13, 2844–2863. [Google Scholar] [CrossRef]
- Li, M.Y.; Jiao, Y.T.; Wang, Y.T.; Zhang, N.; Wang, B.B.; Liu, R.Q.; Yin, X.; Xu, Y.; Liu, G.T. CRISPR/Cas9-mediated VvPR4b editing decreases downy mildew resistance in grapevine (Vitis vinifera L.). Hortic. Res. 2020, 7, 149. [Google Scholar] [CrossRef]
- Wan, D.Y.; Guo, Y.; Cheng, Y.; Hu, Y.; Xiao, S.Y.; Wang, Y.J.; Wen, Y.Q. CRISPR/Cas9-mediated mutagenesis of VvMLO3 results in enhanced resistance to powdery mildew in grapevine (Vitis vinifera). Hortic. Res. 2020, 7, 116. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.; Liu, X.; Zhang, Z.; Wang, Y.; Duan, W.; Li, S.; Liang, Z. CRISPR/Cas9-mediated efficient targeted mutagenesis in Chardonnay (Vitis vinifera L.). Sci. Rep. 2016, 6, 32289. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, I.; Ban, Y.; Azuma, A.; Onoue, N.; Moriguchi, T.; Yamamoto, T.; Toki, S.; Endo, M. CRISPR/Cas9-mediated targeted mutagenesis in grape. PLoS ONE 2017, 12, e0177966. [Google Scholar] [CrossRef]
- Ren, F.; Ren, C.; Zhang, Z.; Duan, W.; Lecourieux, D.; Li, S.; Liang, Z. Efficiency Optimization of CRISPR/Cas9-Mediated Targeted Mutagenesis in Grape. Front. Plant. Sci. 2019, 10, 612. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).