Fruit Crop Improvement with Genome Editing, In Vitro and Transgenic Approaches
Abstract
1. Introduction
2. Induced Mutagenesis
3. In Vitro Approaches
4. Genomics Insights into Fruit Quality
5. Transgenic Approaches
6. New Breeding Techniques
7. Genome Editing
| Plant | Method | Target Gene | Trait | Modification | Reference |
|---|---|---|---|---|---|
| Citrus | CRISPR/Cas9 (SDN1) | CsLOB1 | Disease susceptibility gene for citrus bacterial canker | Mutant plants exhibited improved fungal resistance | [188] |
| Grapevine (Vitis vinifera L.) | CRISPR/Cas9 | MLO-7 | Resistance to powdery mildew | Efficient targeted mutagenesis | [194] |
| Grape | CRISPR/Cas9 (SDN1) | VvWRKY52 | Disease resistance against Botrytis cinerea | Mutants plants showed higher resistance | [189] |
| Grape | CRISPR/Cas9 (SDN1) | IdnDH | Tartaric acid biosynthetic pathway | High levels of tartaric acid in mutants | [185] |
| Apple | CRISPR/Cas9 | MdPDS | Important enzyme in TA biosynthetic pathway | Albino phenotype in plants | [184,195] |
| CRISPR/Cas9 | MdDIPM4 | Fire blight disease susceptibility protein | Reduced susceptibility to the pathogen, Erwinia amylovora | [190] | |
| Banana | CRISPR/Cas9 | RAS-PDS genes (RAS-PDS1 and RAS-PDS2) | Complete albino and variegated phenotypein the plantlets | [196] | |
| CRISPR/Cas9 | PDS | 100% mutation rate and triallelic deletions or insertions among the plants | [197] | ||
| CRISPR/Cas9 | MA-ACO1 | A key component of the ethylene biosynthetic pathway | Plants were characterized by extendedshelf-time | [198] | |
| Musa dmr6 | Banana Xanthomonas wilt | Mutants showed enhanced resistance to important disease, BX | [199] |
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. World Food and Agriculture—Statistical Yearbook 2021; FAO: Rome, Italy, 2021. [Google Scholar] [CrossRef]
- Cornara, L.; Xiao, J.; Smeriglio, A.; Trombetta, D.; Burlando, B. Emerging Exotic Fruits: New Functional Foods in the European Market. Food 2020, 1, 126–139. [Google Scholar] [CrossRef]
- Rai, M.K.; Shekhawat, N.S. Recent advances in genetic engineering for improvement of fruit crops. Plant Cell Tissue Organ Cult. 2014, 116, 1–15. [Google Scholar] [CrossRef]
- Sabbadini, S.; Capocasa, F.; Battino, M.; Mazzoni, L.; Mezzetti, B. Improved nutritional quality in fruit tree species through traditional and biotechnological approaches. Trends Food Sci. Technol. 2021, 117, 125–138. [Google Scholar] [CrossRef]
- Sun-Waterhouse, D. The development of fruit-based functional foods targeting the health and wellness market: A review. Int. J. Food Sci. Technol. 2011, 46, 899–920. [Google Scholar] [CrossRef]
- Litz, R.E.; Padilla, G. Genetic transformation of fruit trees. In Genomics of Tree Crops; Springer International Publishing: New York City, NY, USA, 2012; pp. 117–154. [Google Scholar]
- Dhawan, O.; Lavania, U. Enhancing the productivity of secondary metabolites via induced polyploidy: A review. Euphytica 1996, 87, 81–89. [Google Scholar] [CrossRef]
- Blasco, M.; Badenes, M.L.; Naval, M.M. Colchicine-induced polyploidy in loquat (Eriobotrya japonica (Thunb.) Lindl.). Plant Cell Tissue Organ 2015, 120, 453–461. [Google Scholar] [CrossRef]
- Sattler, M.C.; Carvalho, C.R.; Clarindo, W.R. The polyploidy and its key role in plant breeding. Planta 2016, 243, 281–296. [Google Scholar] [CrossRef]
- Paterson, A.H. Polyploidy, evolutionary opportunity, and crop adaptation. Genetica 2005, 123, 191–196. [Google Scholar] [CrossRef]
- Wu, J.-H.; Ferguson, A.; Murray, B.; Duffy, A.M.; Jia, Y.; Cheng, C.; Martin, P. Fruit Quality in Induced Polyploids of Actinidia chinensis. HortScience 2013, 48, 701–707. [Google Scholar] [CrossRef]
- Wu, J.H.; Ferguson, A.R.; Murray, B.G.; Jia, Y.; Datson, P.M.; Zhang, J. Induced polyploidy dramatically increases the size and alters the shape of fruit in Actinidia chinensis. Ann. Bot. 2012, 109, 169–179. [Google Scholar] [CrossRef]
- Wu, J.H.; Ferguson, A.R.; Murray, B.G. Manipulation of ploidy for kiwifruit breeding: In vitro chromosome doubling in diploid Actinidia chinensis Planch. Plant Cell Tissue Organ Cult. 2011, 106, 503–511. [Google Scholar] [CrossRef]
- Predieri, S. Mutation induction and tissue culture in improving fruits. Plant Cell Tissue Organ 2001, 64, 185–210. [Google Scholar] [CrossRef]
- Liu, P.; Zhao, Z.H.; Dai, L.; Liu, X.Y.; Peng, J.Y.; Peng, S.Q.; Zhou, Z.J. Genetic variations of Ziziphus cultivar ‘Zanhuangdazao’ by using RAPD technique. Acta Hortic. 2009, 840, 149–154. [Google Scholar] [CrossRef]
- Tan, F.Q.; Zhang, M.; Xie, K.D.; Fan, Y.J.; Song, X.; Wang, R.; Wu, X.M.; Zhang, H.Y.; Guo, W.W. Polyploidy remodels fruit metabolism by modifying carbon source utilization and metabolic flux in Ponkan mandarin (Citrus reticulata Blanco). Plant Sci. 2019, 289, 110276. [Google Scholar] [CrossRef] [PubMed]
- Touchell, D.H.; Palmer, I.E.; Ranney, T. In vitro Ploidy Manipulation for Crop Improvement. Front. Plant Sci. 2020, 11, 722. [Google Scholar] [CrossRef] [PubMed]
- Suprasanna, P.; Mirajkar, S.J.; Bhagwat, S.G. Induced mutations and crop improvement. In Plant Biology And Biotechnology; Bahadur, B., Rajam, M.V., Sahijram, L., Krishnamurthy, K.V., Eds.; Springer: Berlin/Heidelberg, Germany, 2015; pp. 593–618. [Google Scholar]
- Sarsu, F. Contribution of induced mutation in crops to global food security. ACI Av. Cienc. Ing. 2020, 12, 10. [Google Scholar] [CrossRef]
- Soriano, J.M. Molecular Marker Technology for Crop Improvement. Agronomy 2020, 10, 1462. [Google Scholar] [CrossRef]
- Zhang, C.; Hao, Y.-J. Advances in Genomic, Transcriptomic, and Metabolomic Analyses of Fruit Quality in Fruit Crops. Hortic. Plant J. 2020, 6, 361–371. [Google Scholar] [CrossRef]
- Lobato-Gómez, M.; Hewitt, S.; Capell TChristou, P.; Dhingra, A.; Girón-Calva, P.S. Transgenic and genome-edited fruits: Background, constraints, benefits, and commercial opportunities. Hortic. Res. 2021, 8, 166. [Google Scholar] [CrossRef]
- Bapat, V.A.; Jagtap, U.B.; Ghag, S.B.; Ganapathi, T.R. Molecular Approaches for the Improvement of Under-Researched Tropical Fruit Trees: Jackfruit, Guava, and Custard Apple. Int. J. Fruit Sci. 2020, 20, 233–281. [Google Scholar] [CrossRef]
- Mathiazhagan, M.; Chidambara, B.; Hunashikatti, L.R.; Ravishankar, K.V. Genomic Approaches for Improvement of Tropical Fruits: Fruit Quality, Shelf Life and Nutrient Content. Genes 2021, 12, 1881. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.M. Major mutation-assisted plant breeding programs supported by FAO/IAEA. Plant Cell Tissue Organ Cult. 2005, 82, 113–123. [Google Scholar] [CrossRef]
- Jain, S.M.; Suprasanna, P. Induced Mutations for Enhancing Nutrition and Food Production. Gene Conserve 2011, 10, 201–215. [Google Scholar]
- Kodym, A.; Afza, R. Physical and chemical mutagenesis. Methods Mol. Biol. 2003, 236, 189–204. [Google Scholar] [CrossRef] [PubMed]
- Mba, C.; Afza, R.; Bado, S.; Jain, S.M. Induced mutagenesis in plants using physical and chemical agents. In Plant Cell Culture: Essential Methods; Davey, M.R., Anthony, P., Eds.; John Wiley & Sons: New York, NY, USA, 2010; pp. 111–130. [Google Scholar]
- Maluszynski, M.; Szarejko, I.; Bhatia, C.R.; Nichterlein, K.; Lagoda, P.J.L. Methodologies for generating variability. Part 4: Mutation techniques. In Plant Breeding and Farmers Participation; Ceccarelli, S., Guimarães, E.P., Weltzien, E., Eds.; Food and Agriculture Organization of the United Nations: Rome, Italy, 2009; pp. 159–194. [Google Scholar]
- Matsukura, C.; Yamaguchi, I.; Inamura, M.; Ban, Y.; Kobayashi, Y.; Yin, Y.-G.; Saito, T.; Kuwata, C.; Imanishi, S.; Nishimura, S. Generation of gamma irradiation-induced mutant lines of the miniature tomato (Solanum lycopersicum L.) cultivar ‘Micro-Tom’. Plant Biotechnol. 2007, 24, 39–44. [Google Scholar] [CrossRef]
- Kharkwal, M.C.; Shu, Q.Y. The role of induced mutations in world food security. In Induced Plant Mutations in the Genomics Era, Proceedings of the International Joint FAO/IAEA Symp IAEA, Vienna, Austria, 27–31 August 2018; Food and Agriculture Organization of the United Nations: Rome, Italy, 2009; pp. 33–38. [Google Scholar]
- Jain, S.M.; Till, B.; Suprasanna, P.; Roux, N. Mutations and cultivar development of banana. In Banana Breeding-Progress and Challenges; Pillay, M., Tenkouano, A., Eds.; CRC Press: Boca Raton, FL, USA, 2011; pp. 203–217. [Google Scholar]
- Badigannavar, A.M.; Mondal, S.; Suprasanna, P. Role of induced mutagenesis and improved crop varieties in food security: Impact in the Indian context. CABI Rev. 2022. [CrossRef]
- IAEA. 2022. Available online: https://www.iaea.org/resources/databases/mutant-varieties-database (accessed on 13 October 2022).
- Geier, T. Chimeras: Properties and dissociation in vegetatively propagated plants. In Plant Mutation Breeding and Biotechnology; CABI: Wallingford, UK, 2012; pp. 191–201. [Google Scholar]
- Sattar, M.N.; Iqbal, Z.; Al-Khayri, J.M.; Jain, S.M. Induced Genetic Variations in Fruit Trees Using New Breeding Tools: Food Security and Climate Resilience. Plants 2021, 10, 1347. [Google Scholar] [CrossRef]
- Lamo, K.; Bhat, D.; Kour, K.; Solanki, S. Mutation Studies in Fruit Crops: A Review. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 3620–3633. [Google Scholar] [CrossRef]
- Jain, S.M. A review of induction of mutations in fruits of tropical and subtropical regions. Acta Hortic. 2002, 575, 295–302. [Google Scholar] [CrossRef]
- Jain, S.M. Mutagenesis in crop improvement under the climate change. Rom. Biotechnol. Lett. 2010, 15, 88–106. [Google Scholar]
- Jain, S.M. Date palm biotechnology: Current status and prospective—An overview. Emir. J. Food Agric. 2012, 24, 386–399. [Google Scholar]
- Jain, S.M. Radiation-Induced Mutations for Date Palm Improvement. In Date Palm Biotechnology; Jain, S., Al-Khayri, J., Johnson, D., Eds.; Springer: Dordrecht, The Netherlands, 2011; pp. 271–286. [Google Scholar]
- Lokko, Y.; Amoatey, H. Improvement of Pineapple Using in vitro and Mutation Breeding Techniques; IAEA-TECDOC-1227; IAEA: Vienna, Austria, 2001; pp. 25–29. [Google Scholar]
- Prasanna, S.; Jain, S.M. Mutant Resources and Mutagenomics in Crop Plants. Emir. J. Food Agric. 2017, 29, 651–657. [Google Scholar] [CrossRef]
- Till, B.J.; Datta, S.; Jankowicz-Cieslak, J. TILLING, The Next Generation. Adv. Biochem. Eng. Biotechnol. 2018, 164, 139–160. [Google Scholar] [PubMed]
- Jankowicz-Cieslak, J.; Ingelbrecht, I.L.; Till, B.J. Mutation Detection in Gamma-Irradiated Banana Using Low Coverage Copy Number Variation. In Efficient Screening Techniques to Identify Mutants with TR4 Resistance in Banana; Jankowicz-Cieslak, J., Ingelbrecht, I.L., Eds.; Springer: Berlin/Heidelberg, Germany, 2022. [Google Scholar] [CrossRef]
- Abe, A.; Kosugi, S.; Yoshida, K.; Natsume, S.; Takagi, H.; Kanzaki, H.; Matsumura, H.; Yoshida, K.; Mitsuoka, C.; Tamiru, M.; et al. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 2012, 30, 174. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Martín, J.; Steuernagel, B.; Ghosh, S.; Herren, G.; Hurni, S.; Adamski, N.; Vrána, J.; Kubaláková, M.; Krattinger, S.G.; Wicker, T. Rapid gene isolation in barley and wheat by mutant chromosome sequencing. Genome Biol. 2016, 17, 221. [Google Scholar] [CrossRef]
- Takagi, H.; Tamiru, M.; Abe, A.; Yoshida, K.; Uemura, A.; Yaegashi, H.; Obara, T.; Oikawa, K.; Utsushi, H.; Kanzaki, E. MutMap accelerates breeding of a salt-tolerant rice cultivar. Nat. Biotechnol. 2015, 33, 445. [Google Scholar] [CrossRef]
- Chaudhary, J.; Deshmukh, R.; Sonah, H. Mutagenesis Approaches and Their Role in Crop Improvement. Plants 2019, 8, 467. [Google Scholar] [CrossRef]
- Fekih, R.; Takagi, H.; Tamiru, M.; Abe, A.; Natsume, S.; Yaegashi, H.; Sharma, S.; Sharma, S.; Kanzaki, H.; Matsumura, H.; et al. MutMap+: Genetic mapping and mutant identification without crossing in rice. PLoS ONE 2013, 8, e68529. [Google Scholar] [CrossRef]
- Tribhuvan, K.U.; Sandhya; Kumar, K.; Sevanthi, A.M.; Gaikwad, K. MutMap: A versatile tool for identification of mutant loci and mapping of genes. Ind. J. Plant Physiol. 2018, 23, 612–621. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, K.; Bai, J.; Lu, J.; Lu, X.; Hu, J.; Pan, C.; He, S.; Yuan, J.; Zhang, Y.; et al. All-flesh fruit in tomato is controlled by reduced expression dosage of AFF through a structural variant mutation in the promoter. J. Exp. Bot. 2022, 73, 123–138. [Google Scholar] [CrossRef]
- Karmarkar, V.M.; Kulkarni, V.M.; Suprasanna, P.; Bapat, V.A.; Rao, P.S. Radio-sensitivity of in vivo and in vitro cultures of banana cv. Basrai (AAA). Fruits 2001, 56, 67–74. [Google Scholar] [CrossRef]
- Wang, X.; Wang, A.; Li, Y.; Xu, Y.; Wei, Q.; Wang, J.; Lin, F.; Gong, D.; Liu, F.; Wang, Y.; et al. A Novel Banana Mutant “RF 1” (Musa spp. ABB, Pisang Awak Subgroup) for Improved Agronomic Traits and Enhanced Cold Tolerance and Disease Resistance. Front. Plant Sci. 2021, 12, 730718. [Google Scholar] [CrossRef]
- Egertsdotter, U.; Ahmad, I.; Clapham, D. Automation and Scale Up of Somatic Embryogenesis for Commercial Plant Production, With Emphasis on Conifers. Front. Plant Sci. 2019, 10, 109. [Google Scholar] [CrossRef]
- Kulkarni, V.M.; Ganapathi, T.R.; Suprasanna, P.; Bapat, V.A. In vitro mutagenesis in banana (Musa spp.) using gamma irradiation. In Protocols for Micropropagation of Woody Trees and Fruits; Springer: Dordrecht, The Netherlands, 2007; pp. 543–559. [Google Scholar]
- Jain, S.M. In vitro mutagenesis in banana (Musa spp.) Improvement. Acta Hortic. 2010, 879, 605–614. [Google Scholar] [CrossRef]
- Saraswathi, M.S.; Kannan, G.; Uma, S.; Kalaiponman, K. Improvement in Banana through Mutation Breeding: Status and Prospect. In Bananas and Plantains: Leading-Edge Research and Development. Vol. 1: Diversity, Improvement and Protection; Uma, S., Vaganan, M.M., Agrawal, A., Eds.; ICAR-National Research Centre for Banana: Tiruchirappalli, India, 2020; pp. 288–308. [Google Scholar]
- Ganapathi, T.R.; Srinivas, L.; Suprasanna, P.; Bapat, V.A. Regeneration of plants from alginate-encapsulated somatic embryos of banana cv. Rasthali (Musa SPP. AAB Group). In vitro Cell. Dev. Biol.-Plant 2001, 37, 178–181. [Google Scholar] [CrossRef]
- Gómez Kosky, R.; de Feria Silva, M.; Posada Pérez, L.; Gilliard, T.; Martínez, F.B.; Vega, M.R.; Milian, M.C.; Mendoza, E.Q. Somatic embryogenesis of the banana hybrid cultivar FHIA-18 (AAAB) in liquid medium and scaled-up in a bioreactor. Plant Cell Tissue Organ Cult. 2002, 68, 21–26. [Google Scholar] [CrossRef]
- Jain, S.M.; Swennen, R. (Eds.) Banana Improvement: Cellular, Molecular and Mutagenesis Approaches; Science Publisher: NH, USA, 2004. [Google Scholar]
- Suprasanna, P.; Ghag, S.; Ganapathi, T.R.; Jain, S.M. Induced genetic diversity in banana. In Genetic Diversity in Horticultural Plants; Nandwani, D., Ed.; Sustainable Development and Biodiversity; Springer: Heidelberg, Germany, 2019; Volume 22, pp. 273–297. [Google Scholar]
- Ganapathi, T.R.; Negi, S.; Tak, H.; Bapat, V.A. Transgenic Banana: Current Status, Opportunities and Challenges. In Genetically Modified Crops; Kavi Kishor, P.B., Rajam, M.V., Pullaiah, T., Eds.; Springer: Singapore, 2021; pp. 111–128. [Google Scholar]
- García-Gonzáles, R.; Quiroz, K.; Carrasco, B.; Caligari, P.D.S. Plant tissue culture: Current status, opportunities and challenges. Cienc. Investig. Agrar. 2010, 37, 5–30. [Google Scholar] [CrossRef]
- Anuradha, S.; Malik, A. Biotechnology a Modern Tool for Fruits Production—A Review. Int. J. Curr. Microbiol. App. Sci. 2017, 6, 1902–1912. [Google Scholar]
- Kumaravel, M.; Backiyarani, S.; Marimuthu, S.; Kumar, A.; Uma, S. Induction of somatic embryogenesis (SE) in recalcitrant Musa spp. by media manipulation based on SE’s molecular mechanism. Acta Hortic. 2020, 1272, 119–127. [Google Scholar] [CrossRef]
- Uma, S.; Kumaravel, M.; Backiyarani, S.; Saraswathi, M.S.; Durai, P.; Karthic, R. Somatic embryogenesis as a tool for reproduction of genetically stable plants in banana and confirmatory field trials. Plant Cell Tissue Organ Cult. 2021, 147, 181–188. [Google Scholar] [CrossRef]
- Ghosh, A.; Ganapathi, T.R.; Nath, P.; Bapat, V.A. Establishment of embryogenic cell suspension cultures and Agrobacterium-mediated transformation in an important Cavendish banana cv. Robusta (AAA). Plant Cell Tissue Organ Cult. 2009, 97, 131–139. [Google Scholar] [CrossRef]
- Sahijram, L. Soma clonal Variation in Micropropagated Plants. In Plant Biology and Biotechnology; Bahadur, B., Venkat Rajam, M., Sahijram, L., Krishnamurthy, K., Eds.; Springer: New Delhi, India, 2015; pp. 407–416. [Google Scholar]
- Sahijram, L.; Soneji, J.R.; Bollamma, K.T. Analyzing soma clonal variation in micropropagated bananas (Musa spp.). Vitr. Cell. Dev. Biol.-Plant 2003, 39, 551–556. [Google Scholar] [CrossRef]
- Smith, M.K. A review of factors influencing the genetic stability of micropropagated bananas. Fruits 1988, 43, 219–223. [Google Scholar]
- Larkin, P.; Scowcroft, W. Somaclonal variation—A novel source of variability from cell cultures for plant improvement. Theor. Appl. Genet. 1981, 60, 197–214. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.M.; Brar, D.S.; Ahluwalia, B.S. (Eds.) Somaclonal Variation and Induced Mutations in Crop Improvement; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1998. [Google Scholar]
- Krishna, H.; Alizadeh, M.; Singh, D.; Singh, U.; Chauhan, N.; Eftekhari, M.; Sadh, R.K. Somaclonal variations and their applications in horticultural crops improvement. 3 Biotech 2016, 6, 54. [Google Scholar] [CrossRef]
- Jain, S.M. Tissue culture-derived variation in crop improvement. Euphytica 2001, 118, 153–166. [Google Scholar] [CrossRef]
- Rai, M.; Kalia, R.; Singh, R.; Gangola, M.P.; Dhawan, A. Developing stress tolerant plants through in vitro selection—An overview of the recent progress. Environ. Exp. Bot. 2011, 71, 89–98. [Google Scholar] [CrossRef]
- Yoo, C.M.; Dalid, C.; Moyer, C.; Whitaker, V.; Lee, S. Improving Strawberry Varieties by Somaclonal Variation. UF-IFS Extension. 2022, 2022, 1–5. [Google Scholar] [CrossRef]
- Rai, M.K. Somaclonal Variation in Improvement of Agricultural Crops: Recent Progress. In Agricultural Biotechnology: Latest Research, and Trends; Kumar Srivastava, D., Kumar Thakur, A., Kumar, P., Eds.; Springer Nature Pvt Ltd.: Singapore, 2021; pp. 129–146. [Google Scholar] [CrossRef]
- Tang, C.Y.; Liu, C.C.; Hwang, S.C. Improvement of the horticultural traits of Cavendish banana (Musa spp., AAA group I). Selection and evaluation of a semi-dwarf clone resistant to Fusarium wilt. Chin. Soc. Hortic. Sci. 2000, 46, 173–182. [Google Scholar]
- Hwang, S.C.; Ko, W.H. Cavendish banana cultivars resistant to Fusarium wilt acquired through soma clonal variation in Taiwan. Plant Dis. 2004, 88, 580–588. [Google Scholar] [CrossRef]
- Giménez, C.; De Garcia, E.; De Enrech, N.X.; Blanca, I. Somaclonal variation in banana: Cytogenetic and molecular characterization of the soma clonal variant CIEN BTA-03. Vitr. Cell. Dev. Biol. Plant 2001, 37, 217–222. [Google Scholar] [CrossRef]
- Martin, K.; Pachathundikandi, S.; Zhang, C.; Slater, A.; Madassery, J. RAPD analysis of a variant of banana (Musa sp.) cv. grande naine and its propagation via shoot tip culture. Vitr. Cell. Dev. Biol. Plant 2006, 42, 188–192. [Google Scholar] [CrossRef]
- Lee, S.Y.; Su, Y.U.; Chou, C.S.; Liu, C.C.; Chen, C.C.; Chao, C.P. Selection of a new somaclone cultivar ‘Tai-Chiao No. 5′ (AAA, Cavendish) with resistance to Fusarium wilt of banana in Chinese Taipei. Acta Hortic. 2011, 897, 391–397. [Google Scholar] [CrossRef]
- Hall, H.K.; Skirvin, R.M.; Braam, W.F. Germplasm release of ‘Lincoln Logan’, a tissue culture-derived genetic thornless ‘Loganberry’. Fruit Var. J. 1986, 40, 134–135. [Google Scholar]
- Ge, H.; Li, Y.; Fu, H.; Long, G.; Luo, L.; Li, R.; Deng, Z. Production of sweet orange somaclones tolerant to citrus canker disease by in vitro mutagenesis with EMS. Plant Cell Tissue Organ Cult. 2015, 123, 29–38. [Google Scholar] [CrossRef]
- Germana, M.A.; Aleza, P.; Grosser, J.W.; Dutt, M.; Wang, N.; Cuenca, J.; Kaur, P. Citrus biotechnology. In The Genus Citrus; Talon, M., Caruso, M., Gmitter, F.G., Eds.; Woodhead Publishing: Cambridge, UK, 2020; pp. 171–192. [Google Scholar]
- Pérez, G.; Mbogholi, A.; Sagarra, F.; Aragón, C.; González, J.; Isidrón, M.; Lorenzo, J.C. Morphological and physiological characterization of two new pineapple somaclones derived from in vitro culture. Vitr. Cell. Dev. Biol. Plant 2011, 47, 428–433. [Google Scholar] [CrossRef]
- Pérez, G.; Yanez, E.; Mbogholi, A.; Valle, B.; Sagarra, F.; Yabor, L.; Aragón, C.; González, J.; Isidrón, M.; Lorenzo, J.C. New pineapple soma clonal variants: P3R5 and dwarf. Am. J. Plant Sci. 2012, 3, 16625. [Google Scholar] [CrossRef]
- Evans, D.A. Somaclonal variation—Genetic basis and breeding applications. Trend Genet. 1989, 5, 46–50. [Google Scholar] [CrossRef]
- Chen, F.; Song, Y.; Li, X.; Chen, J.; Mo, L.; Zhang, X.; Lin, Z.; Zhang, L. Genome sequences of horticultural plants: Past, present, and future. Hortic. Res. 2019, 6, 112. [Google Scholar] [CrossRef]
- Sharma, N.; Singh, S.K.; Lal, S.; Singh, N.K. Genome Sequence Information in Fruit Crops: Current Status. Transcriptomics 2016, 4, 130. [Google Scholar] [CrossRef]
- Allan, A.; Espley, R. MYBs Drive Novel Consumer Traits in Fruits and Vegetables. Trends Plant Sci. 2018, 23, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Can’ani, A.; Spitzer-Rimon, B.; Ravid, J.; Farhi, M.; Masci, T.; Aravena-Calvo, J.; Ovadis, M.; Vainstein, A. Two showy traits, scent emission and pigmentation, are finely coregulated by the MYB transcription factor PH4 in petunia flowers. New Phytol. 2015, 208, 708–714. [Google Scholar]
- Espley, R.V.; Butts, C.A.; Laing, W.A.; Martell, S.; Smith, H.; McGhie, T.K.; Zhang, J.; Paturi, G.; Hedderley, D.; Bovy, A.; et al. Dietary Flavonoids from Modified Apple Reduce Inflammation Markers and Modulate Gut Microbiota in Mice. J. Nutr. 2014, 144, 146–154. [Google Scholar] [CrossRef]
- Xu, Q.; Chen, L.L.; Ruan, X.; Chen, D.; Zhu, A.; Chen, C.; Bertrand, D.; Jiao, W.B.; Hao, B.H.; Lyon, M.P.; et al. The draft genome of sweet orange (Citrus sinensis). Nat. Genet. 2013, 45, 59–66. [Google Scholar] [CrossRef]
- Huang, J.; Zhang, C.; Zhao, X.; Fei, Z.; Wan, K.; Zhang, Z.; Pang, X.; Yin, X.; Bai, Y.; Sun, X.; et al. The Jujube Genome Provides Insights into Genome Evolution and the Domestication of Sweetness/Acidity Taste in Fruit Trees. PLoS Genet. 2016, 12, e1006433. [Google Scholar] [CrossRef]
- Wu, J.; Wang, Z.W.; Shi, Z.B.; Zhang, S.; Ming, R.; Zhu, S.L.; Khan, M.A.; Tao, S.T.; Korban, S.S.; Wang, H.; et al. The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res. 2013, 23, 396–408. [Google Scholar] [CrossRef]
- Qiao, X.; Yin, H.; Li, L.; Wang, R.; Wu, J.; Zhang, S. Different modes of gene duplication show divergent evolutionary patterns and contribute differently to the expansion of gene families involved in important fruit traits in pear (Pyrus bretschneideri). Front. Plant Sci. 2018, 9, 161. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Hu, J.; Han, X.; Li, J.; Gao, Y.; Richards, C.M.; Zhang, C.; Tian, Y.; Liu, G.; Gul, H.; et al. A high-quality apple genome assembly reveals the association of a retrotransposon and red fruit colour. Nat. Commun. 2019, 10, 1494. [Google Scholar] [CrossRef]
- Zhang, T.Q.; Xu, Z.G.; Shang, G.D.; Wang, J.W. A single-cell RNA sequencing profiles the developmental landscape of Arabidopsis root. Mol. Plant 2019, 12, 648–660. [Google Scholar] [CrossRef]
- Yao, J.; Dong, Y.; Morris, B. Parthenocarpic apple fruit production conferred by transposon insertion mutations in a MADS-box transcription factor. Proc. Nat. Acad. Sci. USA 2001, 98, 1306–1311. [Google Scholar] [CrossRef]
- Kobayashi, S.; Goto-Yamamoto, N.; Hirochika, H. Retrotransposon-induced mutations in grape skin color. Science 2004, 304, 982. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.R.; Lee, E.; Mcdavid, D.A.J.; Thomas, M.R.; Robinson, S.P. White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J. 2007, 49, 772–785. [Google Scholar] [CrossRef] [PubMed]
- Butelli, E.; Licciardello, C.; Zhang, Y.; Liu, J.; Mackay, S.; Bailey, P.; Reforgiato-Recupero, G.; Martin, C. Retrotransposons Control Fruit-Specific, Cold-Dependent Accumulation of Anthocyanins in Blood Oranges. Plant Cell 2012, 24, 1242–1255. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Jiang, N.; Schaffner, E. A retrotransposon mediated gene duplication underlies morphological variation of tomato fruit. Science 2008, 319, 1527–1530. [Google Scholar] [CrossRef] [PubMed]
- Tatsuki, M.; Soeno, K.; Shimada, Y.; Sawamura, Y.; Suesada, Y.; Yaegaki, H.; Sato, A.; Kakei, Y.; Nakamura, A.; Bai, S.L.; et al. Insertion of a transposon-like sequence in the 5 -flanking region of the YUCCA gene causes the stony hard phenotype. Plant J. 2018, 96, 815–827. [Google Scholar] [CrossRef]
- van Nocker, S.; Gardiner, S.E. Breeding better cultivars, faster: Applications of new technologies for the rapid deployment of superior horticultural tree crops. Hortic. Res. 2014, 1, 14022. [Google Scholar] [CrossRef]
- Savadi, S.; Mangalassery, S.; Sandesh, M.S. Advances in genomics and genome editing for breeding next generation of fruit and nut crops. Genomics 2021, 113, 3718–3734. [Google Scholar] [CrossRef]
- Khan, M.A.; Korban, S.S. Association mapping in forest trees and fruit crops. J. Exp. Bot. 2012, 63, 4045–4060. [Google Scholar] [CrossRef]
- Kumar, S.; Chagné, D.; Bink, M.C.A.M.; Volz, R.K.; Whitworth, C.; Carlisle, C. Genomic selection for fruit quality traits in apple (Malus × domestica Borkh.). PLoS ONE 2012, 7, e36674. [Google Scholar] [CrossRef]
- Luby, J.J.; Shaw, D.V. Does marker-assisted selection make dollars and sense in a fruit breeding program? HortScience 2001, 36, 872–879. [Google Scholar] [CrossRef]
- Ru, S.; Main, D.; Evans, K.; Peace, C. Current applications, challenges, and perspectives of marker-assisted seedling selection in Rosaceae tree fruit breeding. Tree Genet. Genomes 2015, 11, 8. [Google Scholar] [CrossRef]
- Kunihisa, M.; Moriya, S.; Abe, K.; Okada, K.; Haji, T.; Hayashi, T.; Kim, H.; Nishitani, C.; Terakami, S.; Yamamoto, T. Identification of QTLs for fruit quality traits in Japanese apples: QTLs for early ripening are tightly related to preharvest fruit drop. Breed. Sci. 2014, 64, 240–251. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Terakami, S.; Takada, N.; Nishio, S.; Onoue, N.; Nishitani, C.; Kunihisa, M.; Inoue, E.; Iwata, H.; Hayashi, T.; et al. Identification of QTLs controlling harvest time and fruit skin color in Japanese pear (Pyrus pyrifolia Nakai). Breed. Sci. 2014, 64, 351–361. [Google Scholar] [CrossRef]
- Eduardo, I.; Pacheco, I.; Chietera, G.; Bassi, D.; Pozzi, C.; Vecchietti, A.; Rossini, L. QTL analysis of fruit quality traits in two peach intraspecific populations and importance of maturity date pleiotropic effect. Tree Genet. Genomes 2011, 7, 323–335. [Google Scholar] [CrossRef]
- Martínez-García, P.J.; Parfitt, D.E.; Ogundiwin, E.A.; Fass, J.; Chan, H.M.; Ahmad, R.; Lurie, S.; Dandekar, A.; Gradziel, T.M.; Crisosto, C.H. High density SNP mapping and QTL analysis for fruit quality characteristics in peach (Prunus persica L.). Tree Genet. Genomes 2013, 9, 19–36. [Google Scholar] [CrossRef]
- Kuhn, D.N.; Bally, I.S.E.; Dillon, N.L.; Innes, D.; Groh, A.M.; Rahaman, J.; Ophir, R.; Cohen, Y.; Sherman, A. Genetic map of mango: A tool for mango breeding. Front. Plant Sci. 2017, 8, 577. [Google Scholar] [CrossRef]
- Nantawan, U.; Kanchana-udomkan, C.; Bar, I.; Ford, R. Linkage mapping and quantitative trait loci analysis of sweetness and other fruit quality traits in papaya. BMC Plant Biol. 2019, 2019, 449. [Google Scholar] [CrossRef]
- Kuhn, D.N.; Livingstone, D.S.; Richards, J.H.; Manosalva, P.; Van den Berg, N.; Chambers, A.H. Application of genomic tools to avocado (Persea americana) breeding: SNP discovery for genotyping and germplasm characterization. Sci. Hortic. 2019, 246, 1–11. [Google Scholar] [CrossRef]
- Lasserre-Zuber, P.; Caffier, V.; Stievenard, R.; Lemarquand, A.; Le Cam, B.; Durel, C.E. Pyramiding Quantitative Resistance with a Major Resistance Gene in Apple: From Ephemeral to Enduring Effectiveness in Controlling Scab. Plant Dis. 2018, 102, 2220–2223. [Google Scholar] [CrossRef]
- Pierantoni, L.; Dondini, L.; Cho, K.-H.; Shin, I.-S.; Gennari, F.; Chiodini, R.; Tartarini, S.; Kang, S.-J.; Sansavini, S. Pear scab resistance QTLs via a European pear (Pyrus communis) linkage map. Tree Genet. Genome 2007, 3, 311–317. [Google Scholar] [CrossRef]
- Soriano, J.M.; Vera-Ruiz, E.M.; Vilanova, S.; Martínez-Calvo, J.; Llácer, G.; Badenes, M.L.; Romero, C. Identification and mapping of a locus conferring plum pox virus resistance in two apricot-improved linkage maps. Tree Genet. Genomes 2008, 4, 391–402. [Google Scholar] [CrossRef][Green Version]
- Pacheco, I.; Bassi, D.; Eduardo, I.; Ciacciulli, A.; Pirona, R.; Rossini, L.; Vecchietti, A. QTL mapping for brown rot (Monilinia fructigena) resistance in an intraspecific peach (Prunus persica L. batsch) F1 progeny. Tree Genet. Genomes 2014, 10, 1223–1242. [Google Scholar] [CrossRef]
- Riaz, S.; Tenscher, A.C.; Ramming, D.W.; Walker, M.A. Using a limited mapping strategy to identify major QTLs for resistance to grapevine powdery mildew (Erysiphe necator) and their use in marker-assisted breeding. Theor. Appl. Genet. 2011, 122, 1059–1073. [Google Scholar] [CrossRef] [PubMed]
- van Heerden, C.J.; Burger, P.; Vermeulen, A.; Prins, R. Detection of downy and powdery mildew resistance QTL in a ‘Regent’ × ‘RedGlobe’ population. Euphytica 2014, 200, 281–295. [Google Scholar] [CrossRef]
- Aranzana, M.J.; Decroocq, V.; Dirlewanger, E.; Eduardo, I.; Gao, Z.S.; Gasic, K.; Iezzoni, A.; Jung, S.; Peace, C.; Prieto, H.; et al. Prunus genetics and applications after de novo genome sequencing: Achievements and prospects. Hortic. Res. 2019, 6, 58. [Google Scholar] [CrossRef]
- García-Gómez, B.E.; Salazar, J.A.; Nicolás-Almansa, M.; Razi, M.; Rubio, M.; Ruiz, D.; Martínez-Gómez, P. Molecular Bases of Fruit Quality in Prunus Species: An Integrated Genomic, Transcriptomic and Metabolic Review with a Breeding Perspective. Int. J. Mol. Sci. 2021, 22, 333. [Google Scholar] [CrossRef]
- Minamikawa, M.F.; Takada, N.; Terakami, S.; Saito, T.; Onogi, A. Genome-wide association study and genomic prediction using parental and breeding populations of Japanese pear (Pyrus pyrifolia Nakai). Sci. Rep. 2018, 8, 11994. [Google Scholar] [CrossRef]
- Font, C.; Guajardo, V.; Chin-wo, S.R. Association mapping analysis for fruit quality traits in Prunus persica using SNP markers. Front. Plant Sci. 2019, 9, 2005. [Google Scholar] [CrossRef]
- Salazar, J.A.; Pacheco, I.; Shinya, P.; Zapata, P.; Silva, C.; Aradhya, M.; Velasco, D.; Ruiz, D.; Martínez-gómez, P. Genotyping by sequencing for SNP-based linkage analysis and identification of QTLs linked to fruit quality traits in japanese plum (Prunus salicina Lindl.). Front. Plant Sci. 2017, 8, 476. [Google Scholar] [CrossRef]
- Pandey, A.; Alok, A.; Lakhwani, D.; Singh, J.; Asif, M.H. Genome-wide expression analysis and metabolite profiling elucidate transcriptional regulation of flavonoid biosynthesis and modulation under abiotic stresses in banana. Nat. Publ. Gr. 2016, 2016, 31361. [Google Scholar] [CrossRef]
- Cao, K.; Wang, L.; Zhu, G.; Fang, W.; Chen, C.; Luo, J. Genetic diversity, linkage disequilibrium, and association mapping analyses of peach (Prunus persica) landraces in China. Tree Genet. Genomes 2012, 8, 975–990. [Google Scholar] [CrossRef]
- Iwata, H.; Minamikawa, M.F.; Kajiya-Kanegae, H.; Ishimori, M.; Hayashi, T. Genomics-assisted breeding in fruit trees. Breed. Sci. 2016, 66, 100–115. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Garrick, D.J.; Bink, M.C.; Whitworth, C.; Chagné, D.; Volz, R.K. Novel genomic approaches unravel genetic architecture of complex traits in apple. BMC Genom. 2013, 14, 393. [Google Scholar] [CrossRef]
- Gantait, S.; Mukherjee, E.; Jogam, P.; Babu, K.H.; Jain, S.M.; Suprasanna, P. Improving crops through transgenic breeding—Technological advances and prospects. In Advances in Plant Tissue Culture; Rai, A.C., Kumar, A., Modi, A., Singh, M., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 295–324. [Google Scholar]
- Gambino, G.; Gribaudo, I. Genetic transformation of fruit trees: Current status and remaining challenges. Transgenic Res. 2012, 21, 1163–1181. [Google Scholar] [CrossRef] [PubMed]
- Kramer, M.G.; Redenbaugh, K. Commercialization of a tomato with an antisense polygalacturonase gene: The FLAVR SAVR™ tomato story. Euphytica 1994, 79, 293–297. [Google Scholar] [CrossRef]
- Pasquali, G.; Biricolti, S.; Locatelli, F.; Baldoni, E.; Mattana, M. Osmyb4 expression improves adaptive responses to drought and cold stress in transgenic apples. Plant Cell Rep. 2008, 27, 1677–1686. [Google Scholar] [CrossRef] [PubMed]
- Shekhawat, U.K.S.; Ganapathi, T.R.; Srinivas, L. Cloning and Characterization of a novel stress-responsive WRKY transcription factor gene (MusaWRKY71) from Musa spp. cv. Karibale Monthan (ABB group) using transformed banana cells. Mol. Biol. Rep. 2011, 38, 4023–4035. [Google Scholar] [CrossRef]
- Shekhawat, U.K.S.; Srinivas, L.; Ganapathi, T.R. MusaDHN-1, a novel multiple stress-inducible SK3-type dehydrin gene, contributes affirmatively to drought- and salt-stress tolerance in banana. Planta 2011, 234, 915–932. [Google Scholar] [CrossRef]
- Dhekney, S.A.; Litz, R.E.; Moraga, D.A.; Yadav, A.K. Potential for introducing cold tolerance into papaya by transformation with C-repeat binding factor (CBF) genes. In vitro Cell. Dev. Biol. Plant 2007, 43, 195–202. [Google Scholar] [CrossRef]
- Tian, N.; Wang, J.; Xu, Z.Q. Overexpression of Na+/H+ antiporter gene AtNHX1 from Arabidopsis thaliana improves the salt tolerance of kiwifruit (Actinidia deliciosa). S. Afr. J. Bot. 2011, 77, 160–169. [Google Scholar] [CrossRef]
- Namukwaya, B.; Tripathi, L.; Tripathi, J.N.; Arinaitwe, G.; Mukasa, S.B.; Tushemereirwe, W.K. Transgenic banana expressing Pflp gene confers enhanced resistance to Xanthomonas wilt disease. Transgenic Res. 2012, 21, 855–865. [Google Scholar] [CrossRef] [PubMed]
- Vishnevetsky, J.; White, T.L.; Palmateer, A.J.; Flaishman, M.; Cohen, Y.; Elad, Y.; Velcheva, M.; Hanania, U.; Sahar, N.; Dgani, O. Improved tolerance toward fungal diseases in transgenic cavendish banana (Musa spp. AAA Group) Cv. Grand Nain. Transgenic Res. 2011, 20, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Paul, J.; Khanna, H.; Kleidon, J.; Hoang, P.; Geijskes, J.; Daniells, J.; Zaplin, E.; Rosenberg, Y.; James, A.; Mlalazi, B. Golden bananas in the field: Elevated fruit pro-vitamin A from the expression of a single banana transgene. Plant Biotechnol. J. 2017, 15, 520–553. [Google Scholar] [CrossRef] [PubMed]
- Kumar, G.B.S.; Srinivas, L.; Ganapathi, T.R. Iron fortification of banana by the expression of soybean ferritin. Biol. Trace Elem. Res. 2010, 142, 232–241. [Google Scholar] [CrossRef]
- Brookes, G.; Barfoot, P. GM crop technology use 1996–2018: Farm income and production impacts. GM Crops Food 2020, 11, 242–261. [Google Scholar] [CrossRef] [PubMed]
- Limera, C.; Sabbadini, S.; Sweet, J.B.; Mezzetti, B. New Biotechnological Tools for the Genetic Improvement of Major Woody Fruit Species. Front. Plant Sci. 2017, 8, 1418. [Google Scholar] [CrossRef]
- Suprasanna, P.; Saddhe, A.; Ghuge, S.A.; Ingle, K.P. New and novel genetic tools for improving crops. CAB Rev. Perspect. Agric. Vet. Sci. Nutr. Nat. Resour. 2021, 16, 1–15. [Google Scholar] [CrossRef]
- Carvalho, R.F.; Folta, K.M. Assessment of promoters and a selectable marker for development of strawberry intragenic vectors. Plant Cell Tissue Organ Cult. 2017, 128, 259–271. [Google Scholar] [CrossRef]
- Schouten, H.J.; Krens, F.A.; Jacobsen, E. Cisgenic plants are similar to traditionally bred plants. EMBO Rep. 2006, 7, 750–753. [Google Scholar] [CrossRef]
- Mezzetti, B.; Smagghe, G.; Arpaia, S.; Christiaens, O.; Dietz-Pfeilstetter, A.; Jones, H.; Kostov, K.; Sabbadini, S.; Opsahl-Sorteberg, H.-G.; Ventura, V.; et al. RNAi: What is its position in agriculture? J. Pest Sci. 2020, 93, 1125–1130. [Google Scholar] [CrossRef]
- Ricci, A.; Sabbadini, S.; Prieto, H.; Padilla, I.M.; Dardick, C.; Li, Z.; Scorza, R.; Limera, C.; Mezzetti, B.; Perez-Jimenez, M.; et al. Genetic transformation in peach (Prunus persica L.): Challenges and ways forward. Plants 2020, 9, 971. [Google Scholar] [CrossRef] [PubMed]
- Sabbadini, S.; Capriotti, L.; Molesini, B.; Pandolfini, T.; Navacchi, O.; Limera, C.; Ricci, A.; Mezzetti, B. Comparison of regeneration capacity and Agrobacterium-mediated cell transformation efficiency of different cultivars and rootstocks of Vitis spp. via organogenesis. Sci. Rep. 2019, 9, 582. [Google Scholar] [CrossRef] [PubMed]
- Kanchiswamy, C.N.; Sargent, D.J.; Velasco, R.; Maffei, M.E.; Malnoy, M. Looking forward to genetically edited fruit crops. Trends Biotechnol. 2015, 33, 62–64. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S.G.; Schaart, J.G.; Groenwold, R.; Jacobsen, E.; Schouten, H.J.; Krens, F.A. Functional analysis and expression profiling of HcrVf1 and HcrVf2 for development of scab resistant cisgenic and intragenic apples. Plant Mol. Biol. 2011, 75, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Vanblaere, T.; Szankowski, I.; Schaart, J.; Schouten, H.; Flachowsky, H.; Broggini, G.A.; Gessler, C. The development of a cisgenic apple plant. J. Biotechnol. 2011, 154, 304–311. [Google Scholar] [CrossRef]
- Vanblaere, T.; Flachowsky, H.; Gessler, C.; Broggini, G.A. Molecular characterization of cisgenic lines of apple ‘Gala’ carrying the Rvi6 scab resistance gene. Plant Biotechnol. J. 2014, 12, 2–9. [Google Scholar] [CrossRef]
- Krens, F.A.; Schaart, J.G.; Van der Burgh, A.M.; Tinnenbroek-Capel, I.E.; Groenwold, M.R.; Kodde, L.P.; Broggini, G.A.L.; Gessler, C.; Schouten, H.J. Cisgenic apple trees; development, characterization, and performance. Front. Plant Sci. 2015, 6, 286. [Google Scholar] [CrossRef][Green Version]
- Wurdig, J.; Flachowsky, H.; Sab, A.; Peil, A.; Hanke, M. Improving resistance of different apple cultivars using the Rvi6 scab resistance gene in a cisgenic approach based on the FLP/FRT recombinase system. Mol. Breed. 2015, 35, 95. [Google Scholar] [CrossRef]
- Gessler, C.; Vanblaere, T.; Parravicini, G.; Broggini GA, L. Cisgenic ‘Gala’ containing the scab resistance gene from Malus floribunda 821 and the fire blight resistance genes from M. ‘Evereste’. Acta Hortic. 2014, 1048, 43–50. [Google Scholar] [CrossRef]
- Kost, T.D.; Gessler, C.; Jänsch, M.; Flachowsky, H.; Patocchi, A.; Broggini, G.A.L. Development of the first cisgenic apple with increased resistance to fire blight. PLoS ONE 2015, 10, e0143980. [Google Scholar] [CrossRef]
- Pessina, S.; Lenzi, L.; Perazzolli, M.; Campa, M.; Dalla Costa, L.; Urso, S.; Valè, G.; Salamini, F.; Velasco, R.; Malnoy, M. Knockdown of MLO genes reduce susceptibility to powdery mildew in grapevine. Hortic. Res. 2016, 3, 16016. [Google Scholar] [CrossRef] [PubMed]
- Viss, W.J.; Pitrak, J.; Humann, J.; Cook, M.; Driver, J.; Ream, W. Crown-gall-resistant transgenic apple trees that silence Agrobacterium tumefaciens oncogenes. Mol. Breed. 2003, 12, 283–295. [Google Scholar] [CrossRef]
- Freiman, A.; Shlizerman, L.; Golobovitch, S.; Yablovitz, Z.; Korchinsky, R.; Cohen, Y.; Samach, A.; Chevreau, E.; le Roux, P.; Patocchi, A.; et al. Development of a transgenic early flowering pear (Pyrus communis L.) genotype by RNAi silencing of PcTFL1-1 and PcTFL1-2. Planta 2012, 235, 1239–1251. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Zhang, F.; Yang, Y.; Ma, Y.; Liu, Y.; Li, H.; Dai, H.; Zhang, Z. Modification of plant height via RNAi suppression of MdGA20-ox gene expression in apple. J. Am. Soc. Hortic. Sci. 2016, 141, 242–248. [Google Scholar] [CrossRef]
- Klocko, A.L.; Borejsza-Wysocka, E.; Brunner, A.M.; Shevchenko, O.; Aldwinckle, H.; Strauss, S.H. Transgenic suppression of AGAMOUS genes in apple reduces fertility and increases floral attractiveness. PLoS ONE 2016, 11, e0159421. [Google Scholar] [CrossRef]
- Atkinson, R.G.; Sutherland, P.W.; Johnston, S.L.; Gunaseelan, K.; Hallett, I.C.; Mitra, D.; Brummell, D.A.; Schröder, R.; Johnston, J.W.; Schaffer, R.J. Down-regulation of POLYGALACTURONASE1 alters firmness, tensile strength and water loss in apple (Malus x domestica) fruit. BMC Plant Biol. 2012, 12, 129. [Google Scholar] [CrossRef]
- Dhekney, S.A.; Li, Z.T.; Gray, D.J. Grapevines engineered to express cisgenic Vitis vinifera thaumatin-like protein exhibit fungal disease resistance in vitro cell. Dev. Biol. Plant 2011, 47, 458–466. [Google Scholar] [CrossRef]
- Gonsalves, D. Control of papaya ringspot virus in papaya: A case study. Annu. Rev. Phytopathol. 1998, 36, 415–437. [Google Scholar] [CrossRef]
- Gonsalves, D. Transgenic papaya: Development, release, impact and challenges. Adv. Virus Res. 2006, 67, 317–354. [Google Scholar] [CrossRef]
- Scorza, R.; Ravelonandro, M.; Callahan, A.M.; Cordts, J.M.; Fuchs, M.; Dunez, J. Transgenic plums (Prunus domestica L.) express the plum pox virus coat protein gene. Plant Cell Rep. 1994, 14, 18–22. [Google Scholar] [CrossRef]
- Scorza, R.; Callahan, A.; Levy, L.; Damsteegt, V.; Webb, K.; Ravelonandro, M. Post-transcriptional gene silencing in plum pox virus resistant transgenic European plum containing the plum pox poty virus coat protein gene. Transgenic Res. 2001, 10, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Scorza, R.; Callahan, A.; Dardick, C.; Ravelonandro, M.; Polak, J.; Malinowski, T.; Zagrai, I.; Cambra, M.; Kamenova, I. Genetic engineering of Plum pox virus resistance:‘ Honey Sweet ’plum—From concept to product. Plant Cell Tissue Organ Cult. 2013, 115, 1–12. [Google Scholar] [CrossRef]
- Reyes, C.A.; Francesco, A.D.; Pena, E.J.; Costa, N.; Plata, M.I.; Sendin, L.; Castagnaro, A.P.; García, M.L. Resistance to Citrus psorosis virus in transgenic sweet orange plants is triggered by coat protein–RNA silencing. J. Biotechnol. 2011, 151, 151–158. [Google Scholar] [CrossRef]
- Febres, V.J.; Lee, R.F.; Moore, G.A. Transgenic resistance to Citrus tristeza virus in grapefruit. Plant Cell Rep. 2008, 27, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Shelake, R.M.; Kadam, U.S.; Kumar, K.; Pramanik, D.; Singh, A.K.; Kim, J.Y. Engineering drought and salinity tolerance traits in crops through CRISPR-mediated genome editing: Targets, tools, challenges, and perspectives. Plant Commun. 2022, 3, 100417. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, Z.; Unver, T.; Zhang, B. CRISPR/Cas: A powerful tool for gene function study and crop improvement. J. Adv. Res. 2021, 29, 207–221. [Google Scholar] [CrossRef]
- Puchta, H.; Jiang, J.; Wang, K.; Zhao, Y. Updates on gene editing and its applications. Plant Physiol. Vol. 2022, 188, 1725–1730. [Google Scholar] [CrossRef]
- Wada, N.; Osakabe, K.; Osakabe, Y. Expanding the plant genome editing toolbox with recently developed CRISPR-Cas systems. Plant Physiol. 2022, 188, 1825–1837. [Google Scholar] [CrossRef]
- Kim, H.; Kim, S.T.; Ryu, J.; Kang, B.C.; Kim, J.S.; Kim, S.G. CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat. Commun. 2017, 8, 14406. [Google Scholar] [CrossRef]
- Ito, Y.; Nishizawa-Yokoi, A.; Endo, M.; Mikami, M.; Toki, S. CRISPR/Cas9-mediated mutagenesis of the RIN locus that regulates tomato fruit ripening. Biochem. Biophys. Res. Commun. 2015, 467, 76–82. [Google Scholar] [CrossRef]
- Nishitani, C.; Hirai, N.; Komori, S.; Wada, M.; Okada, K.; Osakabe, K.; Yamamoto, T.; Osakabe, Y. Efficient Genome Editing in Apple Using a CRISPR/Cas9 system. Sci. Rep. 2016, 6, 31481. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.; Liu, X.; Zhang, Z.; Wang, Y.; Duan, W.; Li, S.; Liang, Z. CRISPR/Cas9-Mediated Efficient Targeted Mutagenesis in Chardonnay (Vitis Vinifera L.). Sci. Rep. 2016, 6, 32289. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Xu, J.; Orbović, V.; Zhang, Y.; Wang, N. Editing citrus genome via SaCas9/sgRNA system. Front. Plant Sci. 2017, 8, 2135. [Google Scholar] [CrossRef] [PubMed]
- Song, G.Q.; Sink, K.C.; Walworth, A.E.; Cook, M.A.; Allison, R.F.; Lang, G.A. Engineering cherry rootstocks with resistance to Prunus necrotic ring spot virus through RNAi-mediated silencing. Plant Biotechnol. J. 2013, 11, 702–708. [Google Scholar] [CrossRef] [PubMed]
- Peng, A.; Chen, S.; Lei, T.; Xu, L.; He, Y.; Wu, L.; Yao, L.; Zou, X. Engineering canker resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotech. J. 2017, 15, 1509–1519. [Google Scholar] [CrossRef]
- Wang, X.; Tu, M.; Wang, D.; Liu, J.; Li, Y.; Li, Z.; Wang, Y.; Wang, X. CRISPR/Cas9-mediated efficient targeted mutagenesis in grape in the first generation. Plant Biotechnol. J. 2018, 16, 844–885. [Google Scholar] [CrossRef]
- Pompili, V.; Dalla Costa, L.; Piazza, S.; Pindo, M.; Malnoy, M. Reduced fire blight susceptibility in apple cultivars using a high-efficiency CRISPR/Cas9-FLP/FRT-based gene editing system. Plant Biotechnol. J. 2020, 18, 845–858. [Google Scholar] [CrossRef]
- Tripathi, J.N.; Ntui, V.O.; Ron, M.; Muiruri, S.K.; Britt, A.; Tripathi, L. CRISPR/Cas9 editing of endogenous banana streak virus in the B genome of Musa spp. overcomes a major challenge in banana breeding. Commun. Biol. 2019, 2, 46. [Google Scholar] [CrossRef]
- Chen, J.; Xie, J.; Duan, Y.; Hu, H.; Hu, Y.; Li, W. Genome-wide identification and expression profiling reveal tissue-specific expression and differentially-regulated genes involved in gibberellin metabolism between Williams banana and its dwarf mutant. BMC Plant Biol. 2016, 16, 123. [Google Scholar] [CrossRef]
- Shao, X.; Wu, S.; Dou, T.; Zhu, H.; Hu, C.; Huo, H.; He, W.; Deng, G.; Sheng, O.; Bi, F. Using CRISPR/Cas9 genome editing system to create MaGA20ox2 gene-modified semi-dwarf banana. Plant Biotechnol. J. 2019, 18, 17–19. [Google Scholar] [CrossRef]
- Malnoy, M.; Viola, R.; Jung, M.-H.; Koo, O.-J.; Kim, S.; Kim, J.-S.; Velasco, R.; Kanchiswamy, C.N. DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front. Plant Sci. 2016, 7, 1904. [Google Scholar] [CrossRef] [PubMed]
- Malabarba, J.; Chevreau, E.; Dousset, N.; Veillet, F.; Moizan, J.; Vergne, E. New Strategies to Overcome Present CRISPR/Cas9 Limitations in Apple and Pear: Efficient Dechimerization and Base Editing. Int. J. Mol. Sci. 2021, 22, 319. [Google Scholar] [CrossRef]
- Kaur, N.; Alok, A.; Shivani Kaur, N.; Pandey, P.; Awasthi, P.; Tiwari, S. CRISPR/Cas9-mediated efficient editing in phytoene desaturase (PDS) demonstrates precise manipulation in banana cv. Rasthali genome. Funct. Integr. Genom. 2018, 18, 89–99. [Google Scholar] [CrossRef]
- Naim, F.; Dugdale, B.; Kleidon, J. Gene editing the phytoene desaturase alleles of Cavendish banana using CRISPR/Cas9. Transgenic Res. 2018, 27, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Sheng, O.; Deng, G.; He, W.; Dong, T.; Yang, Q.; Dou, T.; Li, C.; Gao, H.; Liu, S.; et al. CRISPR/Cas9-mediated genome editing of MaACO1 (aminocyclopropane-1-carboxylate oxidase 1) promotes the shelf life of banana fruit. Plant Biotechnol. J. 2021, 19, 654–656. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, J.N.; Ntui, V.O.; Shah, T.; Tripathi, L. CRISPR/Cas9-mediated editing of DMR6 orthologue in banana (Musa spp.) confers enhanced resistance to bacterial diseases. Plant Biotechnol. J. 2021, 19, 1291–1293. [Google Scholar] [CrossRef] [PubMed]
- Charrier, A.; Vergne, E.; Dousset, N.J.-P.; Richer, A.; Petiteau, A.; Chevreau, E. Efficient targeted mutagenesis in apple and first-time edition of pear using the CRISPR-Cas9 system. Front. Plant Sci. 2019, 10, 40. [Google Scholar] [CrossRef]
- Flachowsky, H.; Szankowski, I.; Waidmann, S.; Peil, A.; Tränkner, C.; Hanke, V.-M. The MdTFL1 gene of apple (Malus × domestica Borkh.) reduces vegetative growth and generation time. Tree Physiol. 2012, 32, 1288–1301. [Google Scholar] [CrossRef]
- Wenzel, S.; Flachowsky, H.; Hanke, M.V. The fast-track breeding approach can be improved by heat-induced expression of the FLOWERING LOCUS T genes from poplar (Populus trichocarpa) in apple (Malus × domestica Borkh.). Plant Cell Tissue Organ Cult. 2013, 115, 127–137. [Google Scholar] [CrossRef]
- Gao, C. Genome engineering for crop improvement and future agriculture. Cell 2021, 184, 1621–1635. [Google Scholar] [CrossRef]
- Wolter, F.; Puchta, H. In planta gene targeting can be enhanced by the use of CRISPR/Cas12a. Plant J. 2019, 100, 1083–1094. [Google Scholar] [CrossRef] [PubMed]
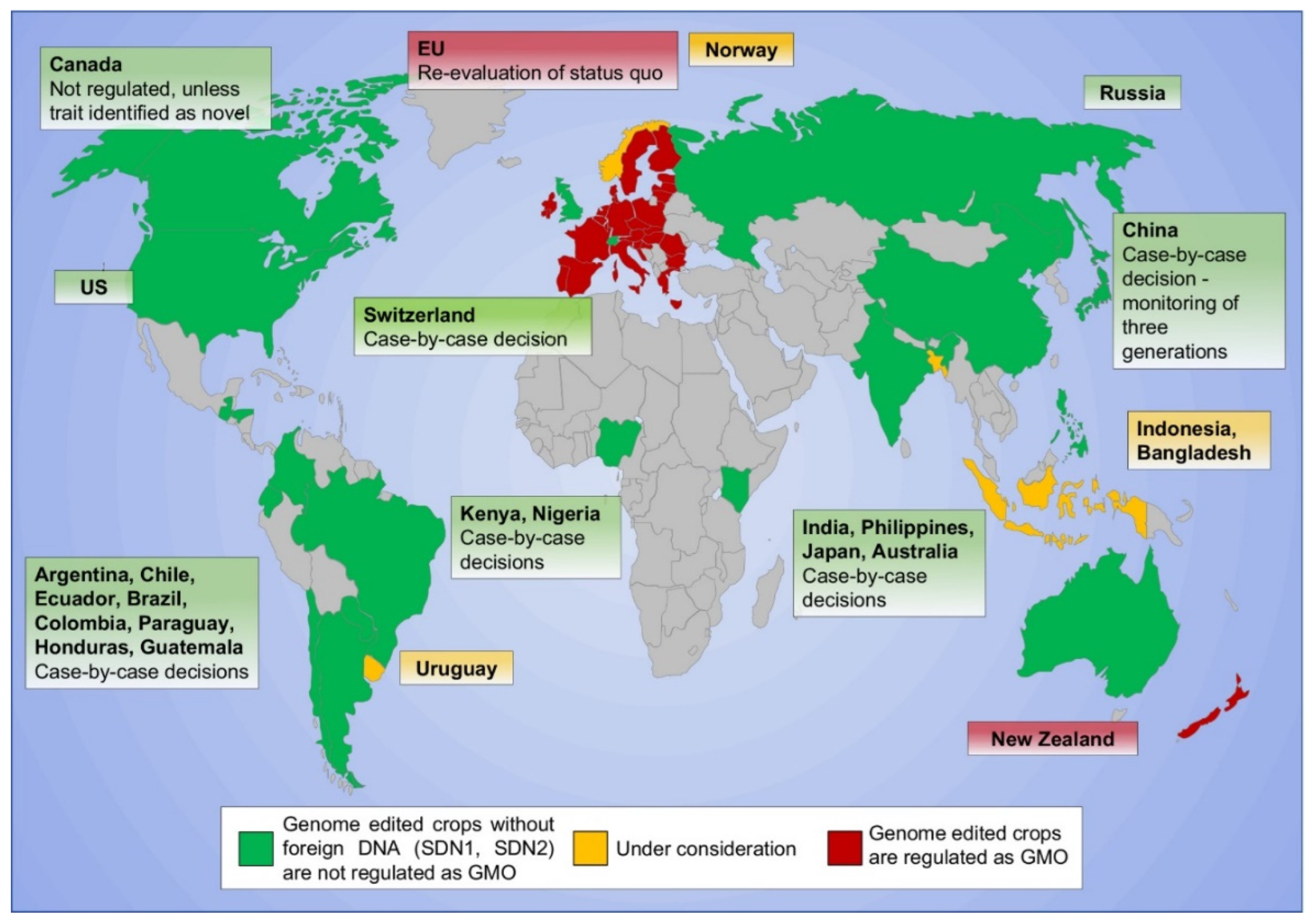
- Buchholzer, M.; Frommer, W.B. An increasing number of countries regulate genome editing in crops. New Phytol. 2022, 237, 12–15. [Google Scholar] [CrossRef] [PubMed]
- Menz, J.; Modrzejewski, D.; Hartung, F.; Wilhelm, R.; Sprink, T. Genome Edited Crops Touch the Market: A View on the Global Development and Regulatory Environment. Front. Plant Sci. 2020, 11, 586027. [Google Scholar] [CrossRef]
- Ntui, V.O.; Tripathi, J.N.; Tripathi, L. Robust CRISPR/Cas9 mediated genome editing tool for banana and plantain (Musa spp.). Curr. Plant Biol. 2020, 21, 100128. [Google Scholar] [CrossRef]
- Schaart, J.G.; van de Wiel, C.C.M.; Smulders, M.J.M. Genome editing of polyploid crops: Prospects, achievements and bottlenecks. Transgenic Res. 2021, 30, 337–351. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Zhang, H.; Zhu, H. CRISPR technology is revolutionizing the improvement of tomato and other fruit crops. Hortic. Res. 2019, 6, 77. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Torres, F.; Ghogare, R.; Stowe, E.; Cerdá-Bennasser, P.; Lobato-Gómez, M.; Williamson-Benavides, B.A.; Giron-Calva, P.S.; Hewitt, S.; Christou, P.; Dhingra, A. Genome editing in fruit, ornamental, and industrial crops. Transgenic Res. 2021, 30, 499–528. [Google Scholar] [CrossRef]
- Zhou, J.; Li, D.; Wang, G.; Wang, F.; Kunjal, M.; Joldersma, D.; Liu, Z. Application and future perspective of CRISPR/Cas9 genome editing in fruit crops. J. Integr. Plant Biol. 2020, 62, 269–286. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace Genome Editing without Double-Strand Breaks or Donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
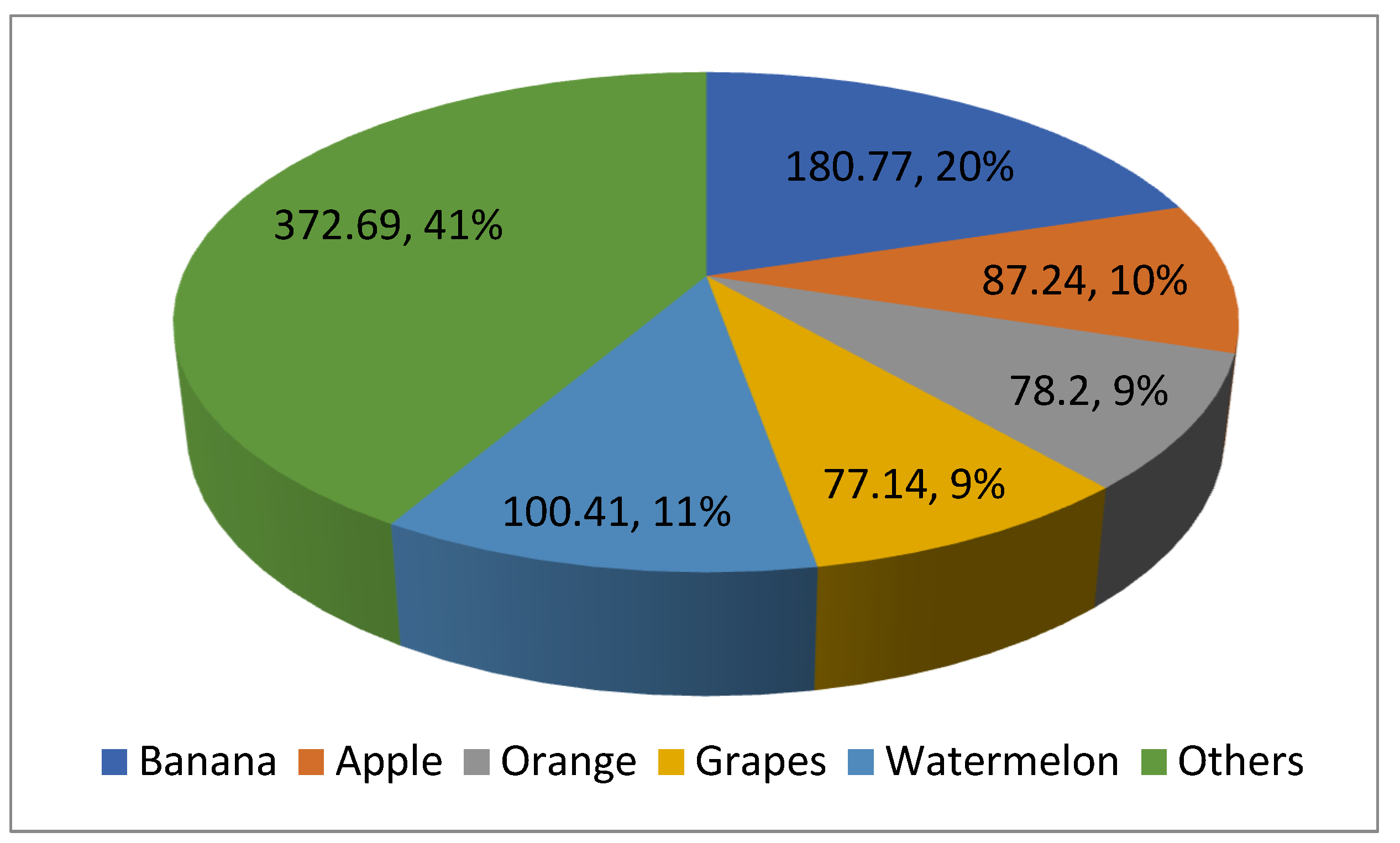
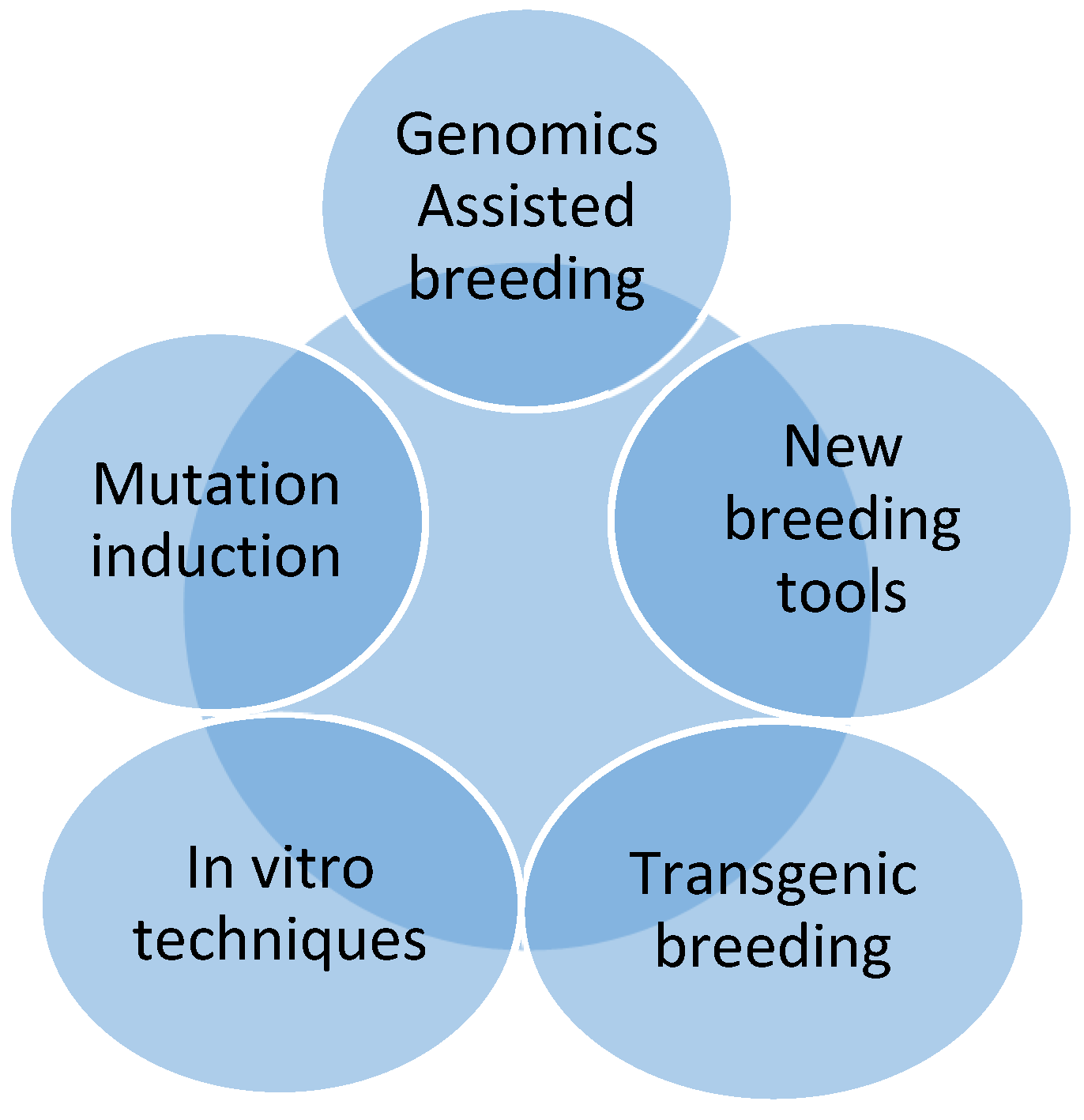
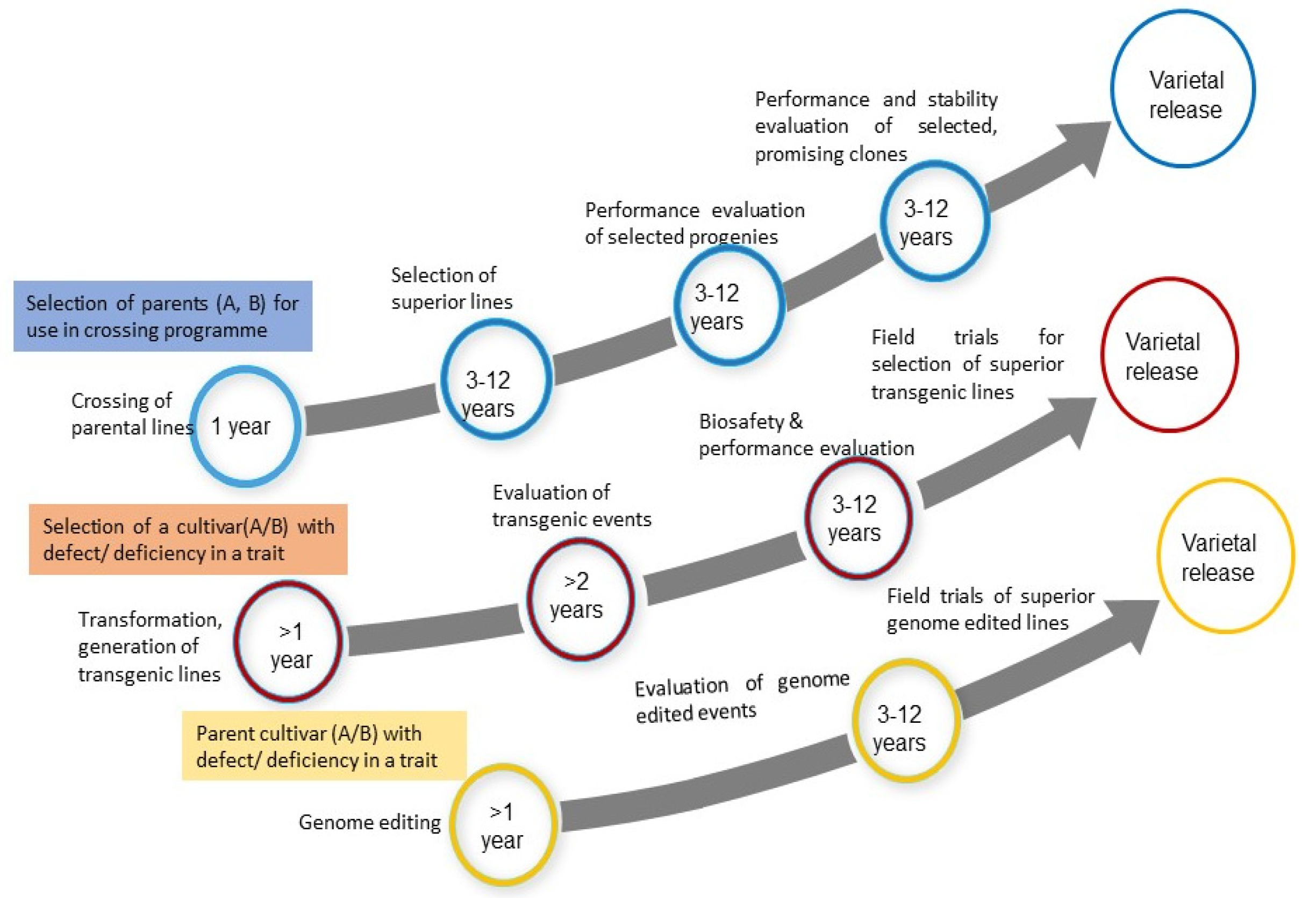
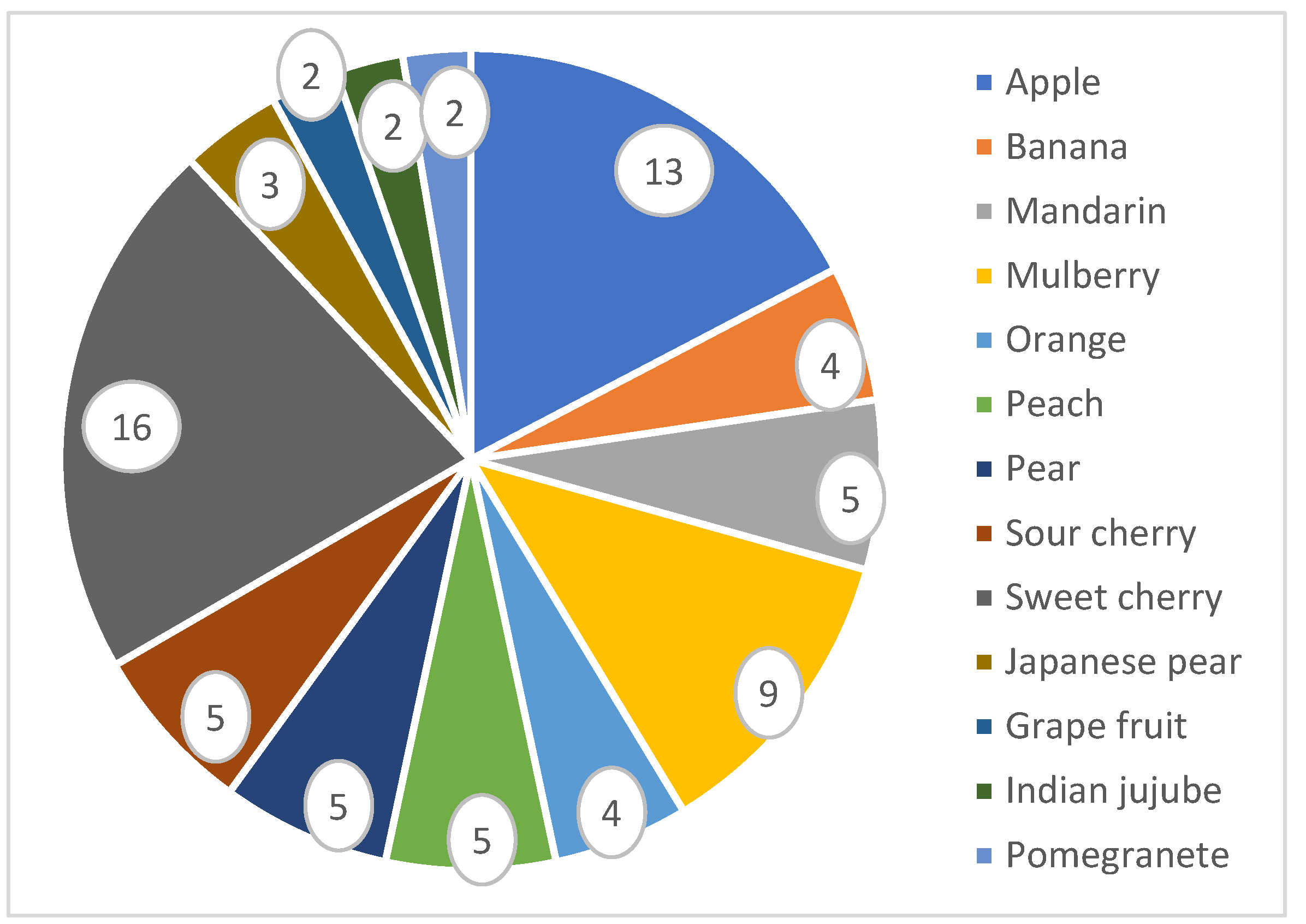

| Plant | Released Variety | Improved Traits | Reference |
|---|---|---|---|
| Banana | TC1-229 | Semi-dwarf and resistant to Fusarium wilt | [79] |
| TC2-425 | Larger bunch size, resistant to Fusarium oxysporum f. sp. cubense (Foc) race 4; high yield | [80] | |
| CIEN-BTA-03, | Resistant to yellow Sigatoka | [81] | |
| CUDBT-B1 | Reduced height and early flowering | [82] | |
| Tai-Chiao No. 5 | Superior horticultural traits and resistance to Fusarium wilt | [83] | |
| Blackberry | var. ‘Lincoln Logan’ | Thornless | [84] |
| Sweet orange (Citrus sinensis) | DG-2 | Tolerant to citrus canker disease | [85] |
| Sweet orange (Citrus sinensis) | EV1, EV2, N7-3, N13-32, OLL-4, OLL-8, Valquarius, SF14W-62, UF 111-24 | Better yield and fruit quality | [86] |
| Pineapple (Ananas comosus L., Merr.) | Cvs. P3R5 and Dwarf, | Variation in fruit color, growth habit, fruit size, and length of plant generation cycle | [87,88] |
| Tomato (Lycopersicon esculentum L.) | DNAP9 | High solid contents | [89] |
| Improved Trait | Gene | Method | Achievement | References |
|---|---|---|---|---|
| Apple | ||||
| Resistance to Apple scab (Venturia inaequalis) | HcrVf2 | Cisgenesis | Plant exhibited reduction in fungal infection | [157,158,159] |
| Resistance to Apple scab (Venturia inaequalis) | Rvi6 | Cisgenesis | Plants had similar resistance to the M. floribunda control | [160] |
| Resistance to Apple scab (Venturia inaequalis) strain 104 (Race 1) | Rvi6 | Cisgenesis | Plants showed resistance to Venturia inaequalis strain 104 (Race 1) | [161] |
| Resistance to Apple Rvi6 scab | HcrVf2 | Cisgenesis | Cisgenic lines containing the HcrVf2gene | [162] |
| Resistance to fire blight (Erwinia amylovora) | FB_MR5 | Cisgenesis | Plants expressed lower disease symptoms | [163] |
| Resistance to powdery mildew (Podosphaera leucotricha) | MdMLO19 | RNA interference | Transgenic apple lines resistant to powdery mildew | [164] |
| Resistance to crown gall formation | iaaM and ipt | RNA interference | Transgenic apple lines resistant to crown gall formation on tree roots | [165] |
| Early flowering induction | MdTFL1 | RNA interference | Silencing of PcTFL1-1 and PcTFL1-2 genes in transgenic pear with consequent early flowering phenotype | [166] |
| Dwarf plant type | MdGA20-ox | RNA interference | Transgenic apple lines with reduced height, shorter internode length, and higher number of nodes | [167] |
| The reduction of fertility and the increase of floral attractiveness | MdAG-like genes: MdMADS15 and MdMADS22 | RNA interference | Trees with polypetalous flowers. Reduced male and female fertility of flowers | [168] |
| Improve post-harvest fruit quality | Endo-polygalacturonase1 PG1) | RNA interference | Increased post-harvest fruit quality | [169] |
| Grapevine (Vitis vinifera L.): Resistance to Powdery mildew (Erysiphe necator) | VVTL-1 | Cisgenesis | Plants showed delayed disease development and decreased severity of black rot (Guignardia bidwellii) | [170] |
| Papaya (Carica papaya): Papaya ringspot virus (PRSV) | PRSV-CP | RNA interference | Resistance to PRSV Transgenic papaya resistant to Papaya ringspot virus (PRSV) | [171,172] |
| Plum (Prunus domestica L.): plum pox virus (PPV) | PPV-CP | Resistance to Sharka (PPV) Transgenic plum clone Honeysweet resistant to sharka disease | [173,174,175] | |
| Sweet orange (Citrus sinensis): Citrus psorosis virus (CPsV) | CPsV-CP | RNA interference | Resistance to CPsV Transgenic sweet orange plants resistant to CPsV | [176] |
| Grapefruit (Citrus paradisi): Citrus tristeza virus (CTV) | CTV | RNA interference | Resistance to CTV Transgenic grapefruit lines resistant to CTV | [177] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Penna, S.; Jain, S.M. Fruit Crop Improvement with Genome Editing, In Vitro and Transgenic Approaches. Horticulturae 2023, 9, 58. https://doi.org/10.3390/horticulturae9010058
Penna S, Jain SM. Fruit Crop Improvement with Genome Editing, In Vitro and Transgenic Approaches. Horticulturae. 2023; 9(1):58. https://doi.org/10.3390/horticulturae9010058
Chicago/Turabian StylePenna, Suprasanna, and Shri Mohan Jain. 2023. "Fruit Crop Improvement with Genome Editing, In Vitro and Transgenic Approaches" Horticulturae 9, no. 1: 58. https://doi.org/10.3390/horticulturae9010058
APA StylePenna, S., & Jain, S. M. (2023). Fruit Crop Improvement with Genome Editing, In Vitro and Transgenic Approaches. Horticulturae, 9(1), 58. https://doi.org/10.3390/horticulturae9010058







