BnA.JAZ5 Attenuates Drought Tolerance in Rapeseed through Mediation of ABA–JA Crosstalk
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Plasmid Construction and Transformation
2.3. RNA Extraction and Real-Time PCR
2.4. MeJA and ABA Treatments
2.5. Stress Treatment at Seedling Stage
2.6. Dehydration Treatments
2.7. Measurement of Leaf Stomatal Density
2.8. Water Loss Assay
2.9. Measurement of Relative Water Content
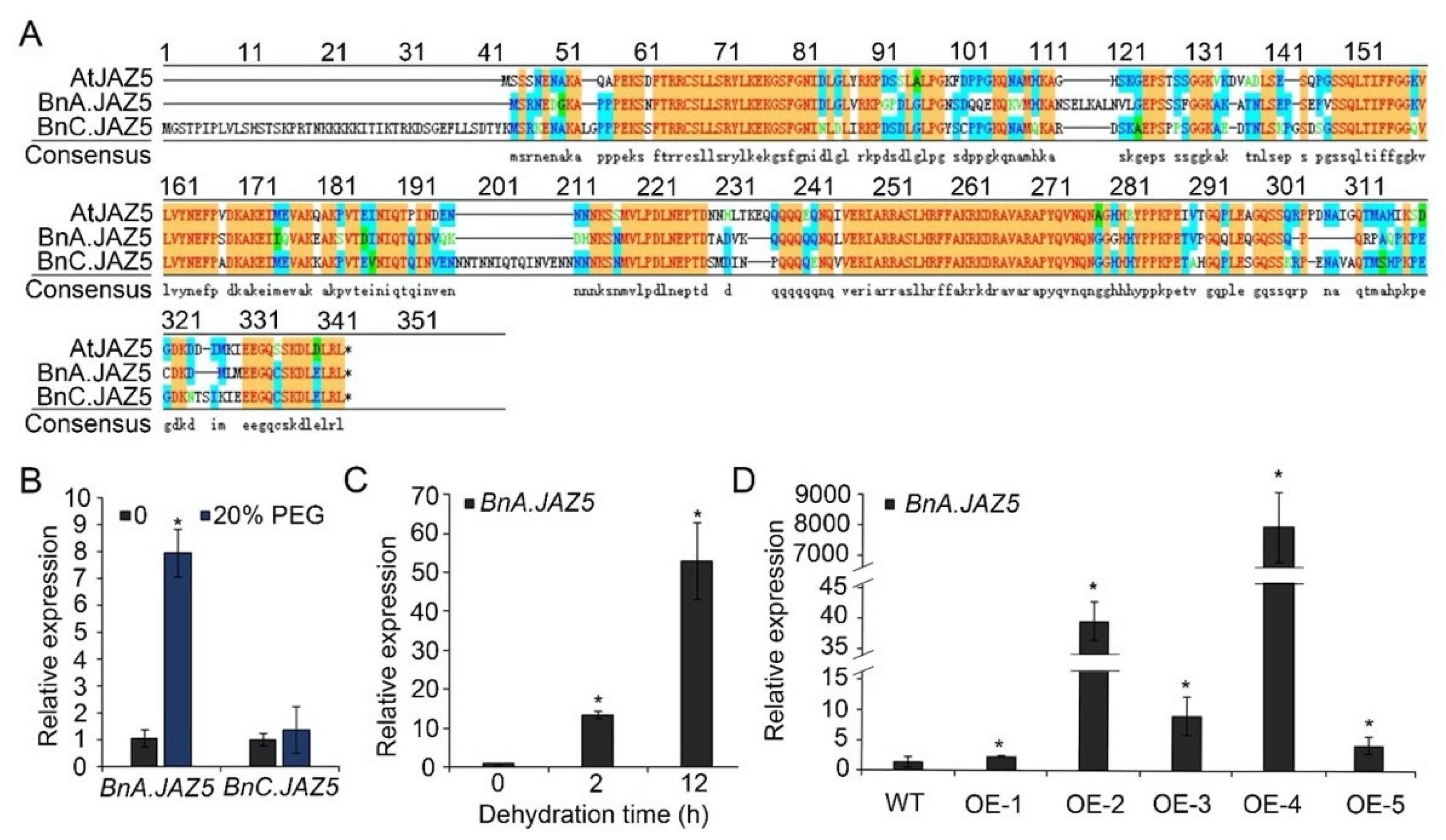
2.10. Sequence Alignment and Phylogenetic Analysis
2.11. Statistical Analysis
3. Results
3.1. Expression Patterns of JA Signaling Regulator BnA.JAZ5 Revealed Its Potential Function in Rapeseed Response to Drought Stress
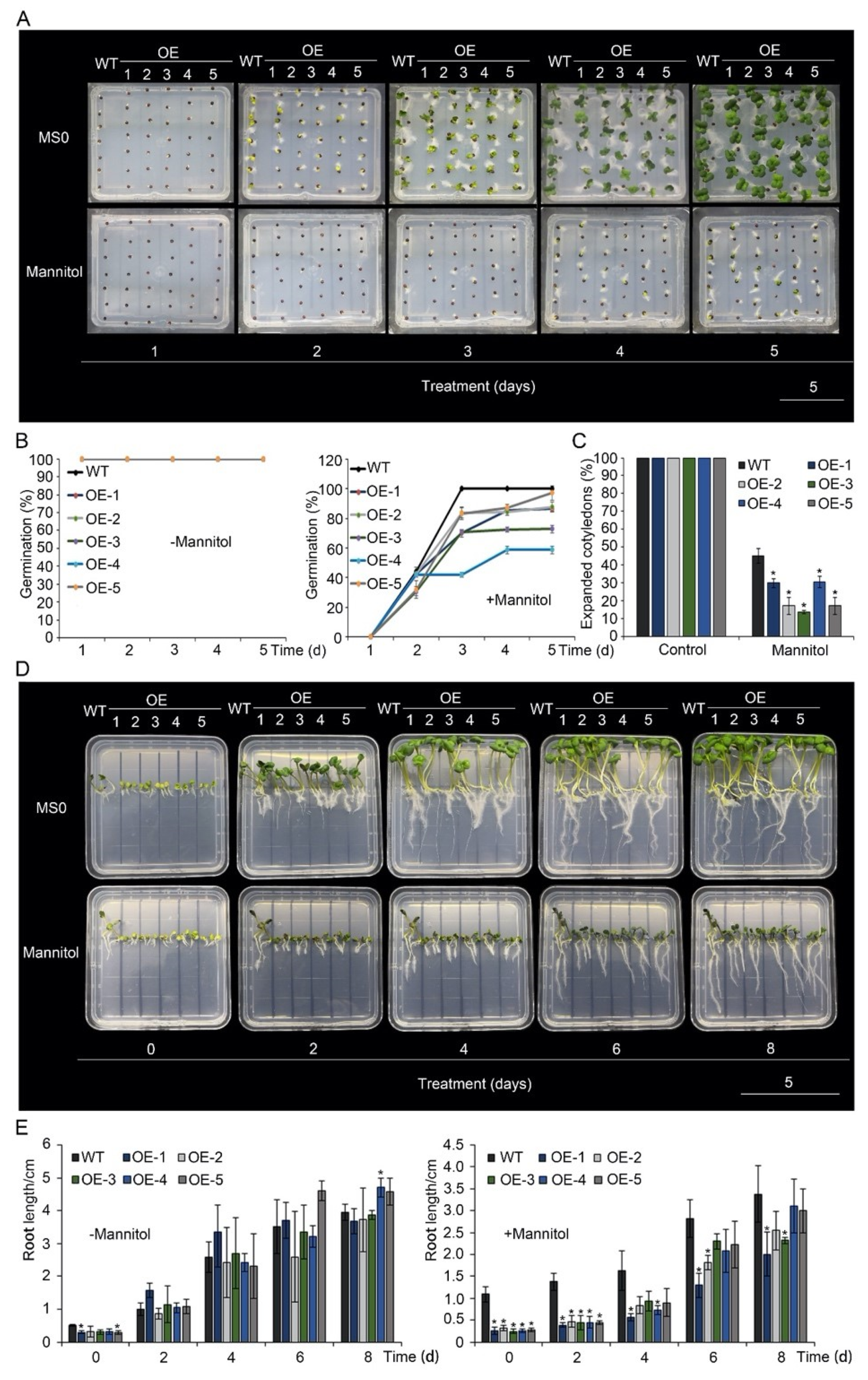
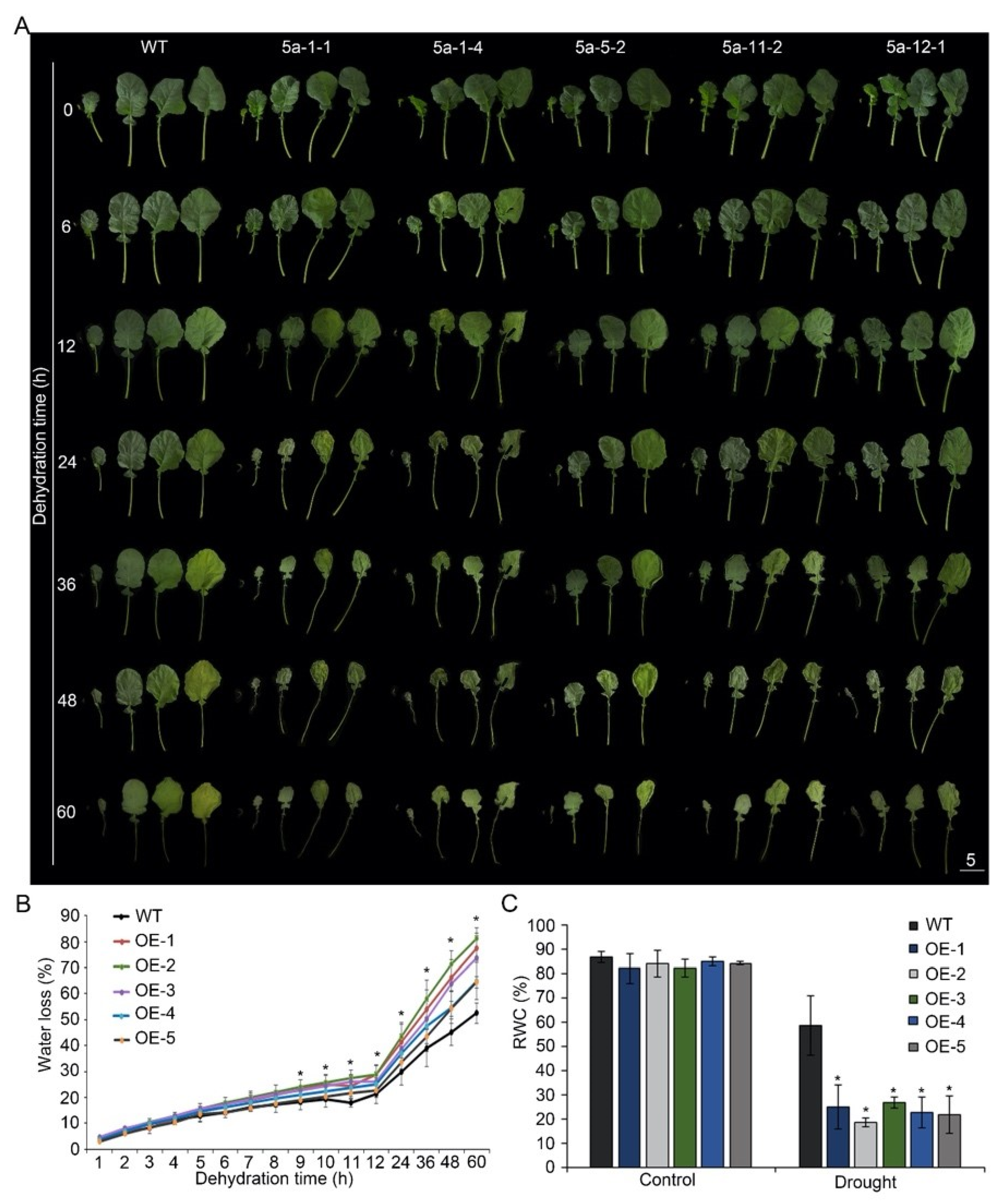
3.2. Overexpression of BnA.JAZ5 Reduced Drought Tolerance
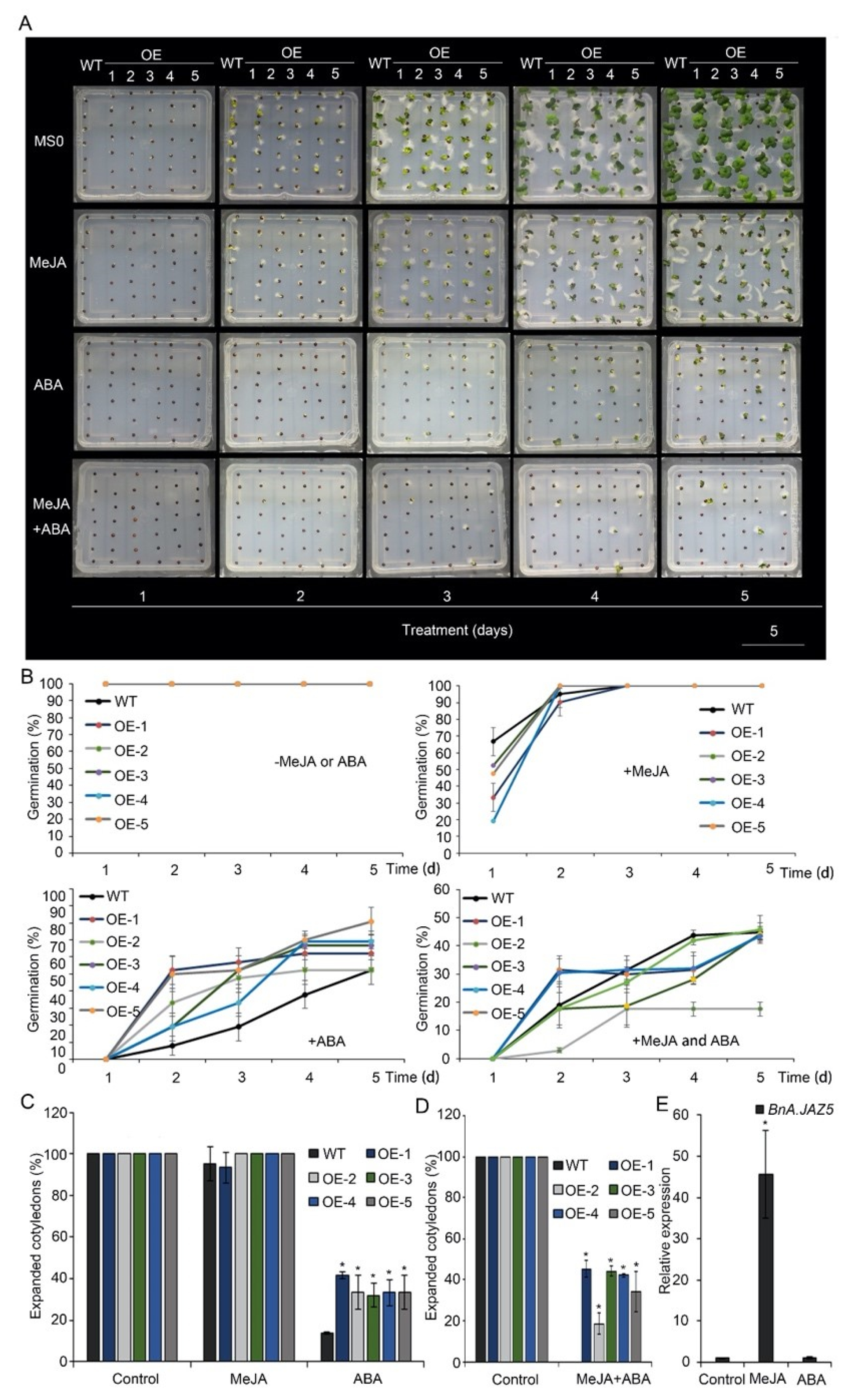
3.3. BnA.JAZ5 Overexpression Altered Plant Responses to JA and ABA
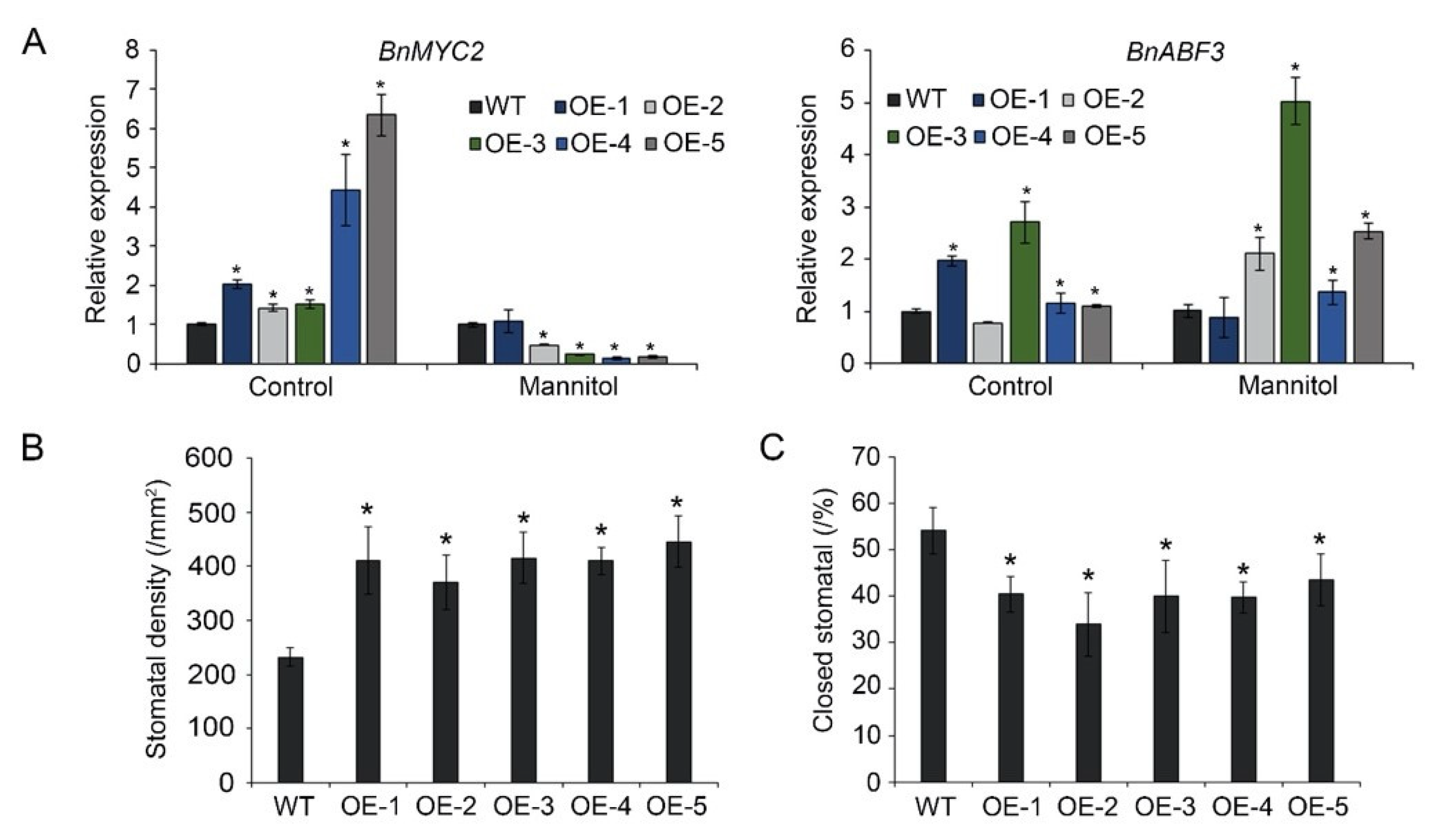
3.4. BnA.JAZ5 Regulates ABA-Dependent Stress-Responsive Genes
3.5. BnA.JAZ5 Overexpression Increased Stomatal Density and Reduced Stomatal Closure
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cook, E.R.; Seager, R.; Cane, M.A.; Stahle, D.W. North American drought: Reconstructions, causes, and consequences. Earth-Sci. Rev. 2007, 81, 93–134. [Google Scholar] [CrossRef]
- Tester, M.; Langridge, P. Breeding technologies to increase crop production in a changing world. Science 2010, 327, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Allender, C.J.; King, G.J. Origins of the amphiploid species Brassica napus L. investigated by chloroplast and nuclear molecular markers. BMC Plant Biol. 2010, 10, 54. [Google Scholar] [CrossRef] [Green Version]
- Wan, J.X.; Griffiths, R.; Ying, J.F.; McCourt, P.; Huang, Y.F. Development of drought-tolerant Canola (Brassica napus L.) through genetic modulation of ABA-mediated stomatal responses. Crop Sci. 2009, 49, 1539–1554. [Google Scholar] [CrossRef]
- Jeong, J.S.; Kim, Y.S.; Baek, K.H.; Jung, H.; Ha, S.H.; Do Choi, Y.; Kim, M.; Reuzeau, C.; Kim, J.K. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 2010, 153, 185–197. [Google Scholar] [CrossRef] [Green Version]
- Yoo, C.Y.; Pence, H.E.; Jin, J.B.; Miura, K.; Gosney, M.J.; Hasegawa, P.M.; Mickelbart, M.V. The Arabidopsis GTL1 transcription factor regulates water use efficiency and drought tolerance by modulating stomatal density via transrepression of SDD1. Plant Cell 2010, 22, 4128–4141. [Google Scholar] [CrossRef] [Green Version]
- Masle, J.; Gilmore, S.R.; Farquhar, G.D. The ERECTA gene regulates plant transpiration efficiency in Arabidopsis. Nature 2005, 436, 866–870. [Google Scholar] [CrossRef]
- Berger, D.; Altmann, T. A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes Dev. 2000, 14, 1119–1131. [Google Scholar] [CrossRef]
- Shpak, E.D.; McAbee, J.M.; Pillitteri, L.J.; Torii, K.U. Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 2005, 309, 290–293. [Google Scholar] [CrossRef]
- Ouyang, S.Q.; Liu, Y.F.; Liu, P.; Lei, G.; He, S.J.; Ma, B.; Zhang, W.K.; Zhang, J.S.; Chen, S.Y. Receptor-like kinase OsSIK1 improves drought and salt stress tolerance in rice (Oryza sativa) plants. Plant J. 2010, 62, 316–329. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Vinocur, B.; Altman, A. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta 2003, 218, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Zhu, J.K. Molecular and genetic aspects of plant responses to osmotic stress. Plant Cell Environ. 2002, 25, 131–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshida, T.; Mogami, J.; Yamaguchi-Shinozaki, K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 2014, 21, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol. Gen. Genet. 1993, 236, 331–340. [Google Scholar] [CrossRef]
- Kishor, P.; Hong, Z.; Miao, G.H.; Hu, C.; Verma, D. Overexpression of Δ1-Pyrroline-5-Carboxylate Synthetase lncreases Proline Production and Confers Osmotolerance in Transgenic Plants. Plant Physiol. 1995, 108, 1387–1394. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; van Wijk, R.; Zarza, X.; Shahbaz, M.; van Hooren, M.; Guardia, A.; Scuffi, D.; Garcia-Mata, C.; Van den Ende, W.; Hoffmann-Benning, S.; et al. Knock-down of Arabidopsis PLC5 reduces primary root growth and secondary root formation while overexpression improves drought tolerance and causes stunted root hair growth. Plant Cell Physiol. 2018, 59, 2004–2019. [Google Scholar] [CrossRef]
- Devoto, A.; Turner, J.G. Jasmonate-regulated Arabidopsis stress signalling network. Physiol. Plant 2005, 123, 161–172. [Google Scholar] [CrossRef]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef]
- Fernandez-Calvo, P.; Chini, A.; Fernandez-Barbero, G.; Chico, J.M.; Gimenez-Ibanez, S.; Geerinck, J.; Eeckhout, D.; Schweizer, F.; Godoy, M.; Franco-Zorrilla, J.M.; et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 2011, 23, 701–715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorenzo, O.; Chico, J.M.; Sanchez-Serrano, J.J.; Solano, R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 2004, 16, 1938–1950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, Y.; Figueroa, P.; Browse, J. Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J. Exp. Bot. 2011, 62, 2143–2154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, C.; Li, L.; Zhai, Q.; You, Y.; Deng, L.; Wu, F.; Chen, R.; Jiang, H.; Wang, H.; Chen, Q.; et al. Mediator subunit MED25 links the jasmonate receptor to transcriptionally active chromatin. Proc. Natl. Acad. Sci. USA 2017, 114, E8930–E8939. [Google Scholar] [CrossRef] [Green Version]
- Pauwels, L.; Barbero, G.F.; Geerinck, J.; Tilleman, S.; Grunewald, W.; Perez, A.C.; Chico, J.M.; Bossche, R.V.; Sewell, J.; Gil, E.; et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 2010, 464, 788–791. [Google Scholar] [CrossRef] [Green Version]
- Shyu, C.; Figueroa, P.; Depew, C.L.; Cooke, T.F.; Sheard, L.B.; Moreno, J.E.; Katsir, L.; Zheng, N.; Browse, J.; Howe, G.A. JAZ8 lacks a canonical degron and has an EAR motif that mediates transcriptional repression of jasmonate responses in Arabidopsis. Plant Cell 2012, 24, 536–550. [Google Scholar] [CrossRef] [Green Version]
- Katsir, L.; Schilmiller, A.L.; Staswick, P.E.; He, S.Y.; Howe, G.A. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. USA 2008, 105, 7100–7105. [Google Scholar] [CrossRef] [Green Version]
- Yan, J.; Yao, R.; Chen, L.; Li, S.; Gu, M.; Nan, F.; Xie, D. Dynamic perception of jasmonates by the F-Box protein COI1. Mol. Plant 2018, 11, 1237–1247. [Google Scholar] [CrossRef] [Green Version]
- Qi, T.; Song, S.; Ren, Q.; Wu, D.; Huang, H.; Chen, Y.; Fan, M.; Peng, W.; Ren, C.; Xie, D. The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 2011, 23, 1795–1814. [Google Scholar] [CrossRef] [Green Version]
- Chini, A.; Fonseca, S.; Fernandez, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; Garcia-Casado, G.; Lopez-Vidriero, I.; Lozano, F.M.; Ponce, M.R.; et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef]
- Song, S.; Qi, T.; Huang, H.; Ren, Q.; Wu, D.; Chang, C.; Peng, W.; Liu, Y.; Peng, J.; Xie, D. The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect Jasmonate-regulated stamen development in Arabidopsis. Plant Cell 2011, 23, 1000–1013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Wang, X.; Zhang, W.; Yu, F.; Tian, J.; Li, D.; Guo, A. Functional analysis of the two Brassica AP3 genes involved in apetalous and stamen carpelloid phenotypes. PLoS ONE 2011, 6, e20930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Li, X.; Yang, J.; He, Y. Natural antisense transcripts of MIR398 genes suppress microR398 processing and attenuate plant thermotolerance. Nat. Commun. 2020, 11, 5351. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Wu, F.; Bai, J.; Li, X.; Yang, X.; Xue, W.; Liu, H.; He, Y. BcpLH organizes a specific subset of microRNAs to form a leafy head in Chinese cabbage (Brassica rapa ssp. pekinensis). Hortic. Res. 2020, 7, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verma, S.S.; Yajima, W.R.; Rahman, M.H.; Shah, S.; Liu, J.J.; Ekramoddoullah, A.K.; Kav, N.N. A cysteine-rich antimicrobial peptide from Pinus monticola (PmAMP1) confers resistance to multiple fungal pathogens in canola (Brassica napus). Plant Mol. Biol. 2012, 79, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Moloney, M.M.; Walker, J.M.; Sharma, K.K. High-efficiency transformation of Brassica napus using Agrobacterium vectors. Plant Cell Rep. 1989, 8, 238–242. [Google Scholar] [CrossRef]
- Lv, J.; Huang, Q.; Sun, Y.; Qu, G.; Guo, Y.; Zhang, X.; Zhao, H.; Hu, S. Male sterility of an AHAS-mutant induced by tribenuron-methyl solution correlated with the decrease of AHAS activity in Brassica napus L. Front. Plant Sci. 2018, 9, 1014. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zhu, Z.; Sun, B.; Xu, X.; Chen, H.; Zou, L.; Chen, G.; Cao, B.; Chen, C.; Lei, J. Overexpression of AtEDT1/HDG11 in Chinese Kale (Brassica oleracea var. alboglabra) enhances drought and osmotic stress tolerance. Front. Plant Sci. 2016, 7, 1285. [Google Scholar] [CrossRef] [Green Version]
- Ma, Q.; Xia, Z.; Cai, Z.; Li, L.; Cheng, Y.; Liu, J.; Nian, H. GmWRKY16 enhances drought and salt tolerance through an ABA-mediated pathway in Arabidopsis thaliana. Front. Plant Sci. 2018, 9, 1979. [Google Scholar] [CrossRef] [Green Version]
- Pan, J.; Hu, Y.; Wang, H.; Guo, Q.; Chen, Y.; Howe, G.A.; Yu, D. Molecular mechanism underlying the synergetic effect of jasmonate on abscisic acid signaling during seed germination in Arabidopsis. Plant Cell 2020, 32, 3846–3865. [Google Scholar] [CrossRef] [PubMed]
- Dave, A.; Hernandez, M.L.; He, Z.; Andriotis, V.M.; Vaistij, F.E.; Larson, T.R.; Graham, I.A. 12-oxo-phytodienoic acid accumulation during seed development represses seed germination in Arabidopsis. Plant Cell 2011, 23, 583–599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piskurewicz, U.; Lopez-Molina, L. Basic techniques to assess seed germination responses to abiotic stress in Arabidopsis thaliana. Methods Mol. Biol. 2016, 1398, 183–196. [Google Scholar]
- Verslues, P.E.; Agarwal, M.; Katiyar-Agarwal, S.; Zhu, J.; Zhu, J.K. Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 2006, 45, 523–539. [Google Scholar] [CrossRef] [PubMed]
- Van der Weele, C.M.; Spollen, W.G.; Sharp, R.E.; Baskin, T.I. Growth of Arabidopsis thaliana seedlings under water deficit studied by control of water potential in nutrient-agar media. J. Exp. Bot. 2000, 51, 1555–1562. [Google Scholar] [CrossRef] [Green Version]
- Sato, H.; Suzuki, T.; Takahashi, F.; Shinozaki, K.; Yamaguchi-Shinozaki, K. NF-YB2 and NF-YB3 have functionally diverged and differentially induce drought and heat stress-specific genes. Plant Physiol. 2019, 180, 1677–1690. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Nolan, T.M.; Ye, H.; Zhang, M.; Tong, H.; Xin, P.; Chu, J.; Chu, C.; Li, Z.; Yin, Y. Arabidopsis WRKY46, WRKY54, and WRKY70 transcription factors are involved in Brassinosteroid-regulated plant growth and drought responses. Plant Cell 2017, 29, 1425–1439. [Google Scholar] [CrossRef]
- Cao, W.; Cao, B.; Wang, X.; Bai, J.; Xu, Y.Z.; Zhao, J.; Li, X.; He, Y.; Hu, S. Alternatively spliced BobCAL transcripts alter curd morphotypes in a collection of Chinese cauliflower accessions. Hortic. Res. 2020, 7, 160. [Google Scholar] [CrossRef]
- Kumar, D.; Yusuf, M.A.; Singh, P.; Sardar, M.; Sarin, N.B. Modulation of antioxidant machinery in α-tocopherol-enriched transgenic Brassica juncea plants tolerant to abiotic stress conditions. Protoplasma 2013, 250, 1079–1089. [Google Scholar] [CrossRef]
- Danisman, S.; van Dijk, A.D.; Bimbo, A.; van der Wal, F.; Hennig, L.; de Folter, S.; Angenent, G.C.; Immink, R.G. Analysis of functional redundancies within the Arabidopsis TCP transcription factor family. J. Exp. Bot. 2013, 64, 5673–5685. [Google Scholar] [CrossRef] [Green Version]
- Schopfer, P.; Plachy, C. Control of seed germination by abscisic acid. II. Effect on embryo water uptake in Brassica napus L. Plant Physiol. 1984, 76, 155–160. [Google Scholar] [CrossRef] [Green Version]
- Fu, J.; Wu, H.; Ma, S.; Xiang, D.; Liu, R.; Xiong, L. OsJAZ1 attenuates drought resistance by regulating JA and ABA signaling in rice. Front. Plant Sci. 2017, 8, 2108. [Google Scholar] [CrossRef] [Green Version]
- Ju, L.; Jing, Y.; Shi, P.; Liu, J.; Chen, J.; Yan, J.; Chu, J.; Chen, K.M.; Sun, J. JAZ proteins modulate seed germination through interaction with ABI5 in bread wheat and Arabidopsis. New Phytol. 2019, 223, 246–260. [Google Scholar] [CrossRef]
- Xue, Y.; Warburton, M.L.; Sawkins, M.; Zhang, X.; Setter, T.; Xu, Y.; Grudloyma, P.; Gethi, J.; Ribaut, J.M.; Li, W.; et al. Genome-wide association analysis for nine agronomic traits in maize under well-watered and water-stressed conditions. Theor. Appl. Genet. 2013, 126, 2587–2596. [Google Scholar] [CrossRef]
- Seo, J.S.; Joo, J.; Kim, M.J.; Kim, Y.K.; Nahm, B.H.; Song, S.I.; Cheong, J.J.; Lee, J.S.; Kim, J.K.; Choi, Y.D. OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 2011, 65, 907–921. [Google Scholar] [CrossRef]
- Wu, H.; Ye, H.; Yao, R.; Zhang, T.; Xiong, L. OsJAZ9 acts as a transcriptional regulator in jasmonate signaling and modulates salt stress tolerance in rice. Plant Sci. 2015, 232, 1–12. [Google Scholar] [CrossRef]
- Lv, Y.; Yang, M.; Hu, D.; Yang, Z.; Ma, S.; Li, X.; Xiong, L. The OsMYB30 transcription factor suppresses cold tolerance by interacting with a JAZ protein and suppressing β-amylase expression. Plant Physiol. 2017, 173, 1475–1491. [Google Scholar] [CrossRef] [Green Version]
- Zhu, D.; Cai, H.; Luo, X.; Bai, X.; Deyholos, M.K.; Chen, Q.; Chen, C.; Ji, W.; Zhu, Y. Over-expression of a novel JAZ family gene from Glycine soja, increases salt and alkali stress tolerance. Biochem. Biophys. Res. Commun. 2012, 426, 273–279. [Google Scholar] [CrossRef]
- Kim, T.H.; Bohmer, M.; Hu, H.; Nishimura, N.; Schroeder, J.I. Guard cell signal transduction network: Advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu. Rev. Plant Biol. 2010, 61, 561–591. [Google Scholar] [CrossRef] [Green Version]
- Yu, H.; Chen, X.; Hong, Y.Y.; Wang, Y.; Xu, P.; Ke, S.D.; Liu, H.Y.; Zhu, J.K.; Oliver, D.J.; Xiang, C.B. Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. Plant Cell 2008, 20, 1134–1151. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Zhou, G. Responses of leaf stomatal density to water status and its relationship with photosynthesis in a grass. J. Exp. Bot. 2008, 59, 3317–3325. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez-Guzman, M.; Pizzio, G.A.; Antoni, R.; Vera-Sirera, F.; Merilo, E.; Bassel, G.W.; Fernandez, M.A.; Holdsworth, M.J.; Perez-Amador, M.A.; Kollist, H.; et al. Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 2012, 24, 2483–2496. [Google Scholar] [CrossRef] [Green Version]
- Leung, J.; Giraudat, J. Aabscisic acid signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 199–222. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.Y.; Chao, D.Y.; Gao, J.P.; Zhu, M.Z.; Shi, M.; Lin, H.X. A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev. 2009, 23, 1805–1817. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, B.; Bai, J.; Wang, X.; Zhang, Y.; Yu, X.; Hu, S.; He, Y. BnA.JAZ5 Attenuates Drought Tolerance in Rapeseed through Mediation of ABA–JA Crosstalk. Horticulturae 2022, 8, 131. https://doi.org/10.3390/horticulturae8020131
Cao B, Bai J, Wang X, Zhang Y, Yu X, Hu S, He Y. BnA.JAZ5 Attenuates Drought Tolerance in Rapeseed through Mediation of ABA–JA Crosstalk. Horticulturae. 2022; 8(2):131. https://doi.org/10.3390/horticulturae8020131
Chicago/Turabian StyleCao, Biting, Jinjuan Bai, Xuan Wang, Yanfeng Zhang, Xiang Yu, Shengwu Hu, and Yuke He. 2022. "BnA.JAZ5 Attenuates Drought Tolerance in Rapeseed through Mediation of ABA–JA Crosstalk" Horticulturae 8, no. 2: 131. https://doi.org/10.3390/horticulturae8020131
APA StyleCao, B., Bai, J., Wang, X., Zhang, Y., Yu, X., Hu, S., & He, Y. (2022). BnA.JAZ5 Attenuates Drought Tolerance in Rapeseed through Mediation of ABA–JA Crosstalk. Horticulturae, 8(2), 131. https://doi.org/10.3390/horticulturae8020131






