Domestication Has Reshaped Gene Families, Gene Expressions and Flavonoid Metabolites in Green Jujube (Ziziphus mauritiana Lam.) Fruit
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome-Wide Identification of Genes Related to Flavonoid Biosynthetic Pathways in Green Jujube
2.2. Transcriptomic Profiles in Different Tissues and Fruits
2.3. Weighted Gene Co-Expression Network Analysis (WGCNA) and Gene Evolution Analysis
2.4. Metabolomic Profiles in Different Fruits
2.5. Statistical Analysis
3. Results
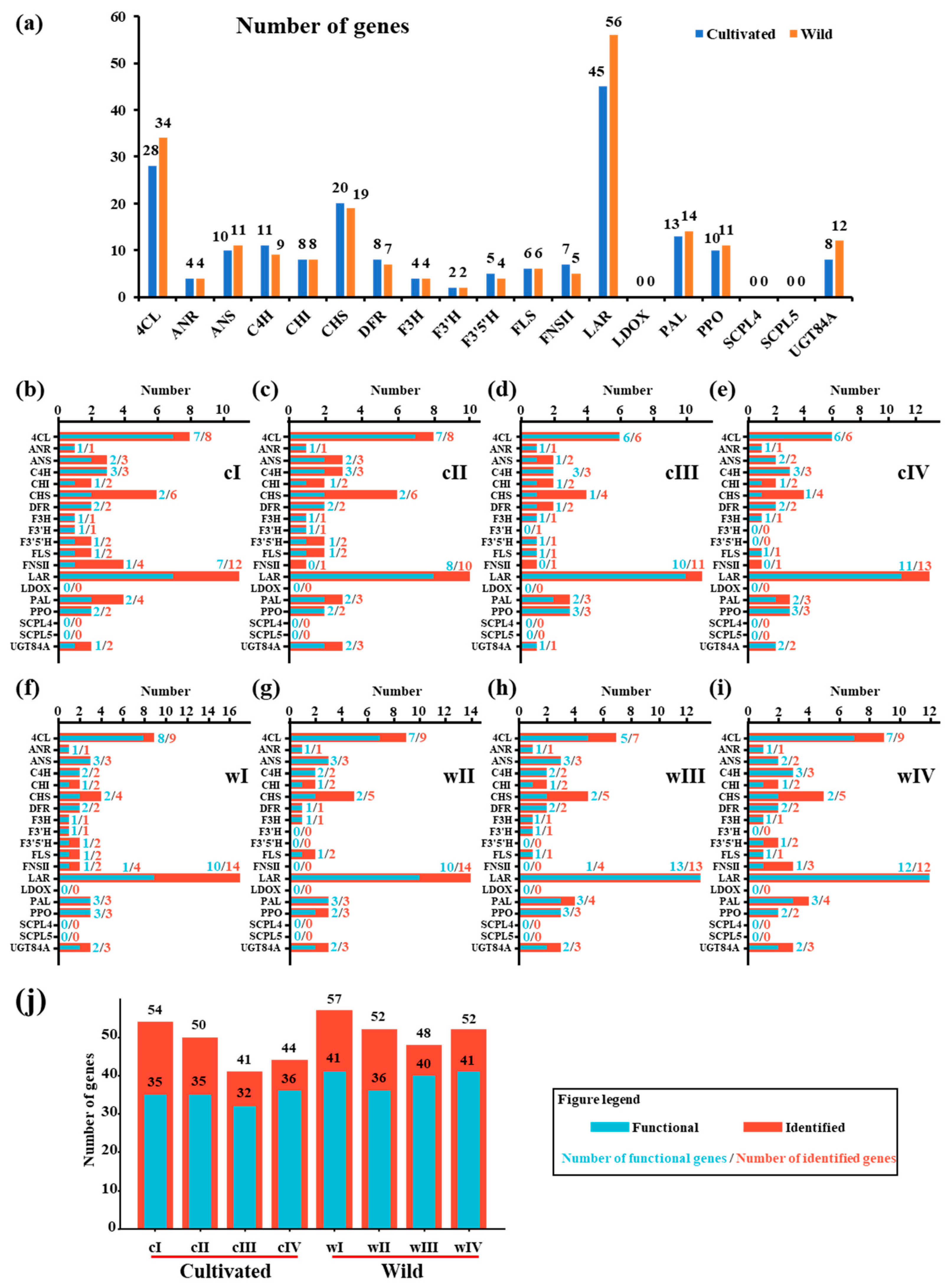
3.1. Flavonoid-Related Gene Families Differed Between Wild and Cultivated Green Jujube Genomes
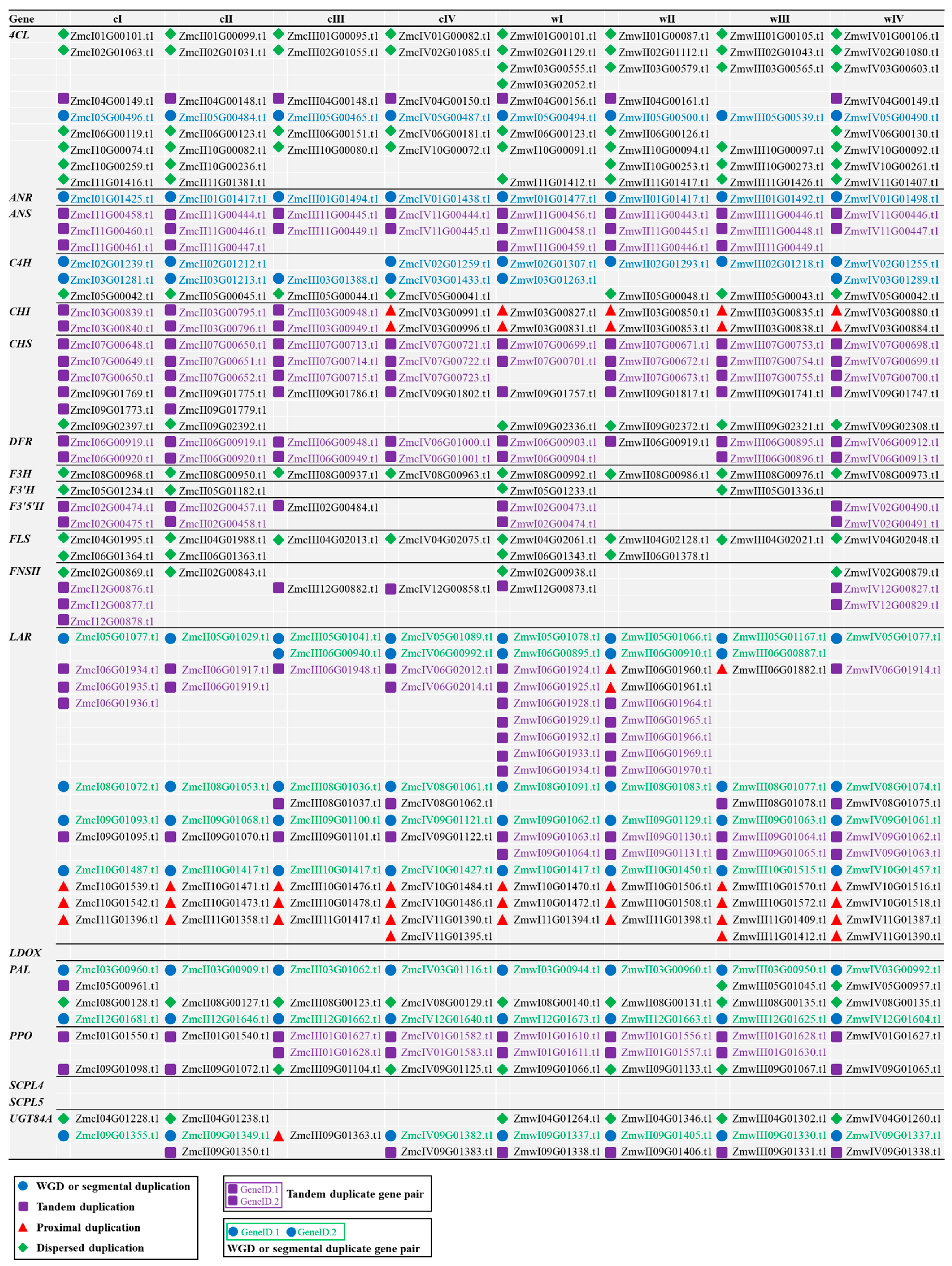
3.2. Evolutionary Analysis Revealed the Complexity of Flavonoid-Related Genes During Domestication
3.3. Gene “Loss and Gain” in Wild and Cultivated Green Jujube Genomes
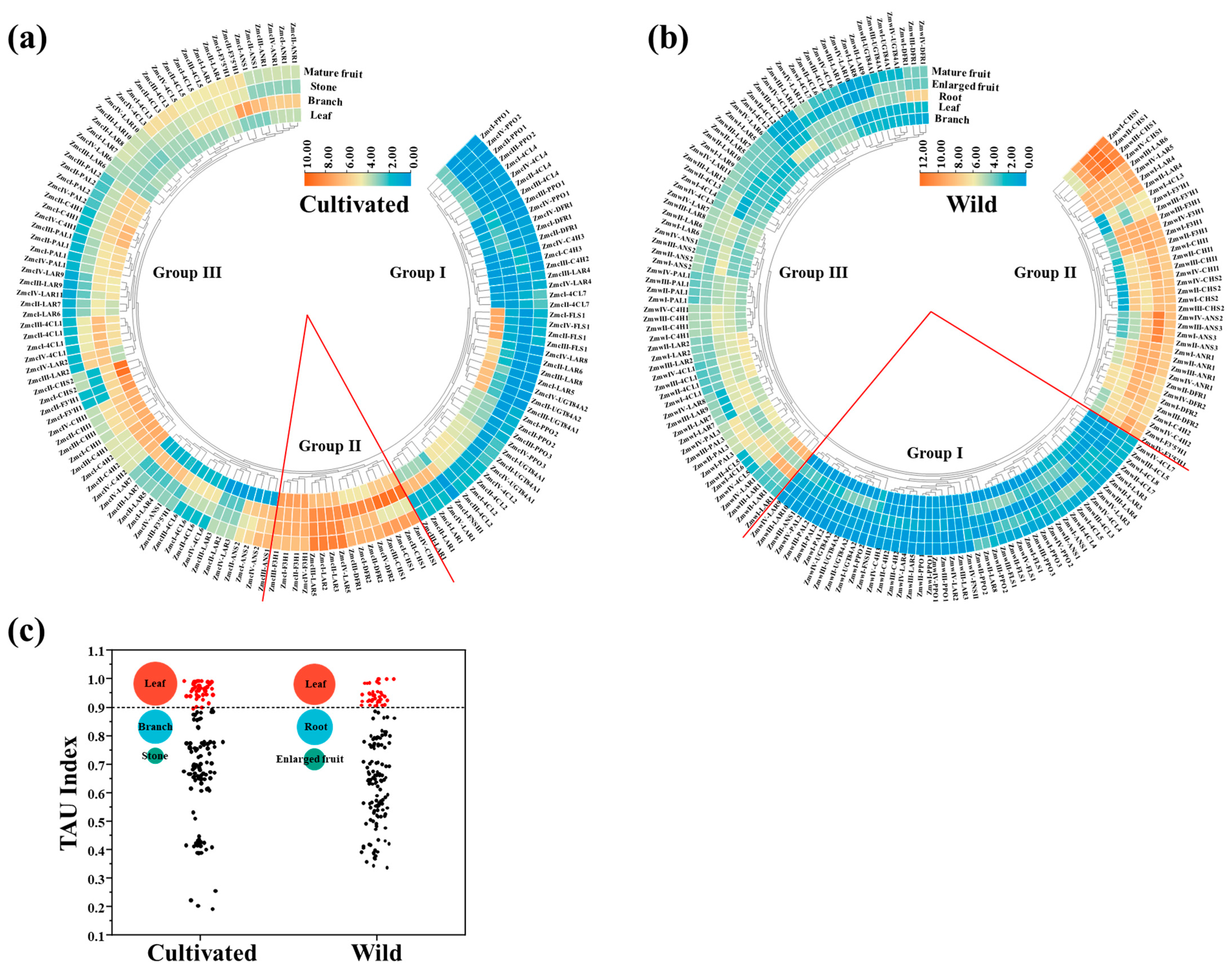
3.4. Flavonoid-Related Genes Exhibited Tissue-Specific Expression Profiles
3.5. Functional Flavonoid-Related Genes Were Screened Out in the Wild and Cultivated Green Jujube
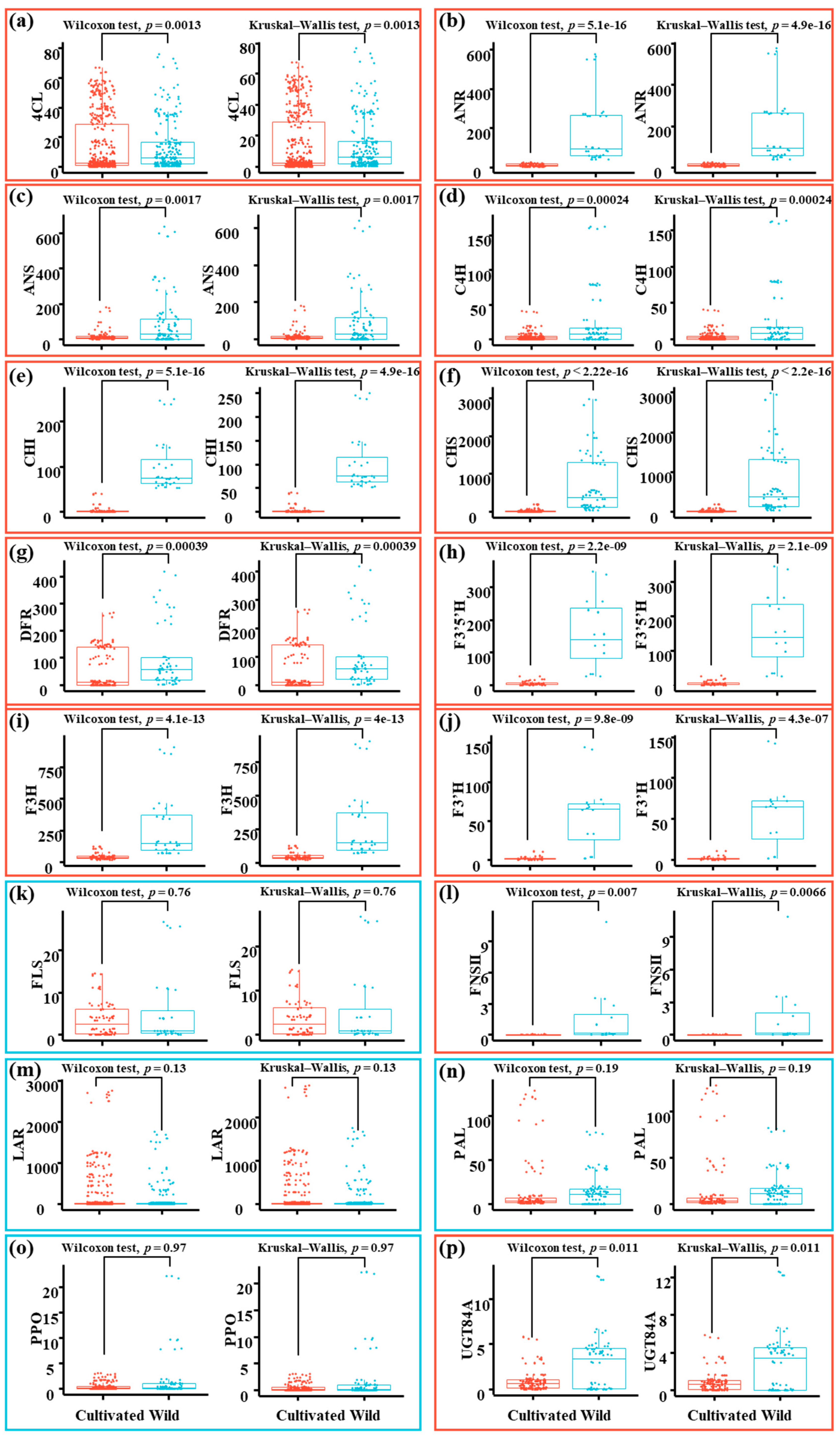
3.6. Functional Flavonoid-Related Genes Were Downregulated in Cultivated Green Jujube
3.7. The Downregulation of Functional Flavonoid-Related Genes Was Statistically Significant
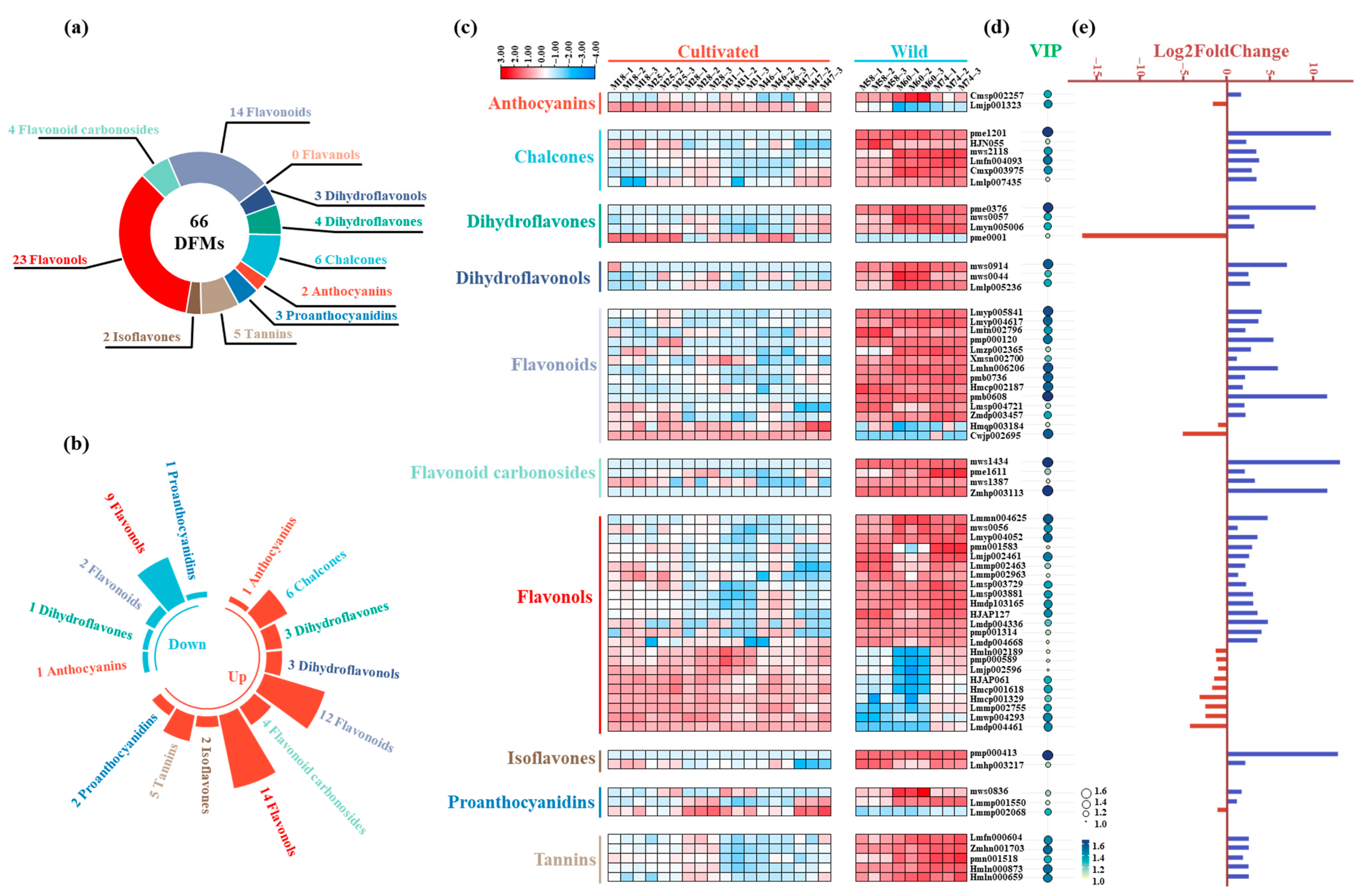
3.8. Accumulation of Flavonoid Metabolites Differed Between Wild and Cultivated Green Jujube
3.9. Wild Green Jujube Fruits Accumulated Significantly More Flavonoids
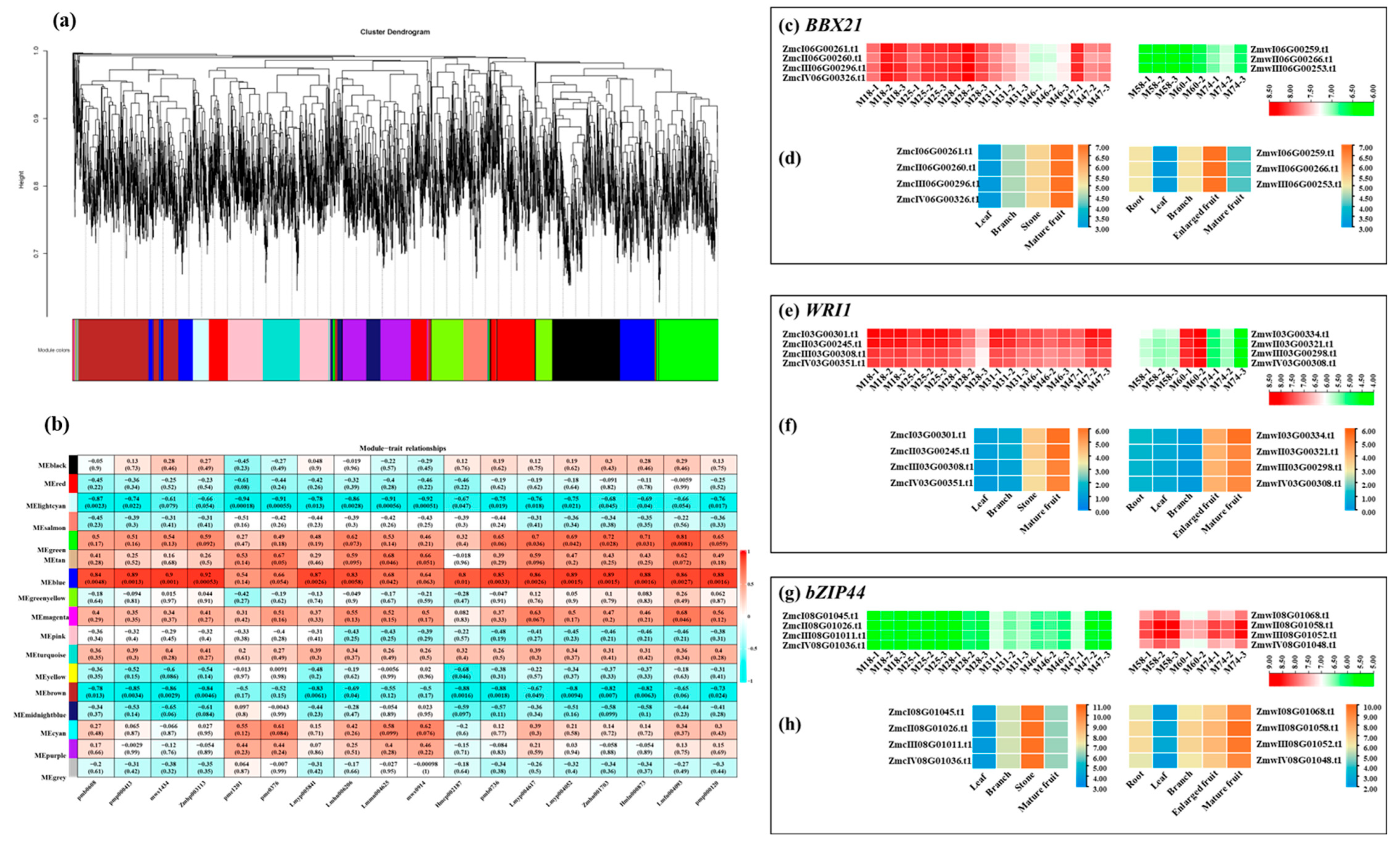
3.10. Gene Co-Expression Network Unveiled the Potential Transcription Factors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 4CL | 4-coumarate: coenzyme a ligase |
| ANR | anthocyanidin reductase |
| ANS | anthocyanidin synthase |
| C4H | trans-cinnamate 4-monooxygenase |
| CHI | chalcone isomerase |
| CHS | chalcone synthase |
| DFR | dihydroflavonol 4-reductase |
| F3H | flavanone 3-hydroxylase |
| F3′H | flavonoid 3′-hydroxylase |
| F3′5′H | flavonoid 3′5′-hydroxylase |
| FLS | flavonol synthase |
| FNSII | flavone synthase II |
| LAR | leucoanthocyanidin reductase |
| LDOX | leucoanthocyanidin dioxygenase |
| PAL | phenylalanine ammonia lyase |
| PPO | polyphenol oxidase |
| SCPL | serine carboxypeptidase-like |
| UGT84A | UDP-glycosyltransferase |
| P450 | cytochrome P450-dependent monooxygenase |
| 2-OGD | 2-oxoglutarate dependent dioxygenase |
| SDR | short-chain dehydrogenase/reductase |
| BBX | B-box |
| WRI1 | WRINKLED1 |
| bZIP | basic leucine zipper |
References
- Paauw, M.; Koes, R.; Quattrocchio, F.M. Alteration of flavonoid pigmentation patterns during domestication of food crops. J. Exp. Bot. 2019, 70, 3719–3735. [Google Scholar] [CrossRef]
- Rathore, R.S.; Jiang, W.; Sedeek, K.; Mahfouz, M. Harnessing neo-domestication of wild pigmented rice for enhanced nutrition and sustainable agriculture. Theor. Appl. Genet. 2025, 138, 108. [Google Scholar] [CrossRef]
- Kumar, S.N.; Das, U.; Immanuel, S.C. Indian jujube a potential fruit tree to improve the livelihood. Saudi J. Biol. Sci. 2023, 30, 103769. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, C.J.; Campbell, S.; Young, A.; Vogler, W.; Galea, V.J. Chinee apple (Ziziphus mauritiana): A comprehensive review of its weediness, ecological impacts and management approaches. Plants 2023, 12, 3213. [Google Scholar] [CrossRef]
- Memon, A.A.; Memon, N.; Bhanger, M.I.; Luthria, D.L. Assay of phenolic compounds from four species of ber (Ziziphus mauritiana L.) fruits: Comparison of three base hydrolysis procedure for quantification of total phenolic acids. Food Chem. 2013, 139, 496–502. [Google Scholar] [CrossRef] [PubMed]
- Zozio, S.; Servent, A.; Cazal, G.; Mbéguié-A-Mbéguié, D.; Ravion, S.; Pallet, D.; Abel, H. Changes in antioxidant activity during the ripening of jujube (Ziziphus mauritiana Lamk). Food Chem. 2014, 150, 448–456. [Google Scholar] [CrossRef]
- Wang, Z.T.; Liu, Y.P.; Ma, Y.L.; Pan, S.Y.; Li, J.K.; Shi, S.J.; Wu, Z.F.; Li, Z.; Shang, Y.F.; Wei, Z.J. Insight into the phenolics and antioxidant activity of Indian jujube (Ziziphus mauritiana Lamk) peel and pulp subjected to the simulated digestion. Heliyon 2023, 9, e16226. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Yin, X.; Liu, L.; Zhang, L.; Shen, W.; Fang, Z.; Yu, Q.; Qin, L.; Chen, L.; Jia, R.; et al. Flavonoid localization in soybean seeds: Comparative analysis of wild (Glycine soja) and cultivated (Glycine max) varieties. Food Chem. 2024, 456, 139883. [Google Scholar] [CrossRef]
- Huang, Z.; Shen, F.; Chen, Y.; Cao, K.; Wang, L. Chromosome-scale genome assembly and population genomics provide insights into the adaptation, domestication, and flavonoid metabolism of Chinese plum. Plant J. 2021, 108, 1174–1192. [Google Scholar] [CrossRef]
- Cao, K.; Wang, B.; Fang, W.; Zhu, G.; Chen, C.; Wang, X.; Li, Y.; Wu, J.; Tang, T.; Fei, Z.; et al. Combined nature and human selections reshaped peach fruit metabolome. Genome Biol. 2022, 23, 146. [Google Scholar] [CrossRef]
- Yuan, P.; Xu, C.; He, N.; Lu, X.; Zhang, X.; Shang, J.; Zhu, H.; Gong, C.; Kuang, H.; Tang, T.; et al. Watermelon domestication was shaped by stepwise selection and regulation of the metabolome. Sci. China Life Sci. 2023, 66, 579–594. [Google Scholar] [CrossRef]
- Xie, X.; Tian, J.; Wang, Y.; Shu, C.; Gui, H.; Cheng, Z.; Wang, Z.; Song, Z.; Tian, Q.; Wen, B.; et al. Comparison of the content of taste and nutrition-related non-volatile compounds in cultivated and wild blueberry varieties in Northeast China. Food Chem. X 2025, 25, 102235. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Chen, J.; Liu, X.; Wang, B.; Zhao, Y.; Liao, L.; Allan, A.C.; Sun, C.; Duan, Y.; Li, X.; et al. A metabolic perspective of selection for fruit quality related to apple domestication and improvement. Genome Biol. 2023, 24, 95. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.J.; Wu, S.; Duan, M.; Wang, L. Antioxidant metabolites in primitive, wild, and cultivated citrus and their role in stress tolerance. Molecules 2021, 26, 5801. [Google Scholar] [CrossRef]
- Li, Z.; Li, J.; Shu, Z.; Xu, M.; Zhang, Y.; Gu, J.; Chen, J.; Li, X.; Wang, M. Comparative metabolomic analysis provides insights into the metabolite profiles of wild and cultivated Dendrobium flexicaule. BMC Plant Biol. 2025, 25, 217. [Google Scholar] [CrossRef] [PubMed]
- Olennikov, D.N.; Kashchenko, N.I. Flavonoids in Cucurbitaceae herbs: Targeted HPLC-PDA-mS profiling of wild and cultivated Cucumis, Echinocystis, and Melothria species and their pancreatic lipase inhibitory potential. Nat. Prod. Res. 2025; ahead of print. [Google Scholar] [CrossRef]
- Milošević, M.; Vulić, J.; Kukrić, Z.; Lazić, B.; Četojević-Simin, D.; Čanadanović-Brunet, J. Polyphenolic composition, antioxidant and antiproliferative activity of edible and inedible parts of cultivated and wild pomegranate (Punica granatum L.). Food Technol. Biotechnol. 2023, 61, 485–493. [Google Scholar] [CrossRef]
- Tomou, E.M.; Goula, K.; Skaltsa, H.; Urmann, C. Comparative phytochemical analysis of cultivated and wild Sideritis raeseri Boiss. & Heldr. subsp. raeseri infusions. Nat. Prod. Res. 2025, 39, 2608–2613. [Google Scholar] [CrossRef]
- Ouerghemmi, I.; Rebey, I.B.; Rahali, F.Z.; Bourgou, S.; Pistelli, L.; Ksouri, R.; Marzouk, B.; Tounsi, M.S. Antioxidant and antimicrobial phenolic compounds from extracts of cultivated and wild-grown Tunisian Ruta chalepensis. J. Food Drug Anal. 2017, 25, 350–359. [Google Scholar] [CrossRef]
- Jing, D.; Liu, X.; He, Q.; Dang, J.; Hu, R.; Xia, Y.; Wu, D.; Wang, S.; Zhang, Y.; Xia, Q.; et al. Genome assembly of wild loquat (Eriobotrya japonica) and resequencing provide new insights into the genomic evolution and fruit domestication in loquat. Hortic. Res. 2022, 10, uhac265. [Google Scholar] [CrossRef]
- Zheng, J.; Wu, H.; Zhu, H.; Huang, C.; Liu, C.; Chang, Y.; Kong, Z.; Zhou, Z.; Wang, G.; Lin, Y.; et al. Determining factors, regulation system, and domestication of anthocyanin biosynthesis in rice leaves. New Phytol. 2019, 223, 705–721. [Google Scholar] [CrossRef]
- Qiao, W.; Wang, Y.; Xu, R.; Yang, Z.; Sun, Y.; Su, L.; Zhang, L.; Wang, J.; Huang, J.; Zheng, X.; et al. A functional chromogen gene C from wild rice is involved in a different anthocyanin biosynthesis pathway in indica and japonica. Theor. Appl. Genet. 2021, 134, 1531–1543. [Google Scholar] [CrossRef]
- Huang, X.; He, Y.; Zhang, K.; Shi, Y.; Zhao, H.; Lai, D.; Lin, H.; Wang, X.; Yang, Z.; Xiao, Y.; et al. Evolution and domestication of a novel biosynthetic gene cluster contributing to the flavonoid metabolism and high-altitude adaptability of plants in the Fagopyrum genus. Adv. Sci. 2024, 11, e2403603. [Google Scholar] [CrossRef]
- Guo, M.; Bi, G.; Wang, H.; Ren, H.; Chen, J.; Lian, Q.; Wang, X.; Fang, W.; Zhang, J.; Dong, Z.; et al. Genomes of autotetraploid wild and cultivated Ziziphus mauritiana reveal polyploid evolution and crop domestication. Plant Physiol. 2024, 196, 2701–2720. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Lu, C.; Wang, Z.; Zou, L.; Xiong, Y.; Zhang, Q. GFAnno: Integrated method for plant flavonoid biosynthesis pathway gene annotation. Beverage Plant Res. 2024, 4, e008. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Xie, J.; Chen, Y.; Cai, G.; Cai, R.; Hu, Z.; Wang, H. Tree Visualization By One Table (tvBOT): A web application for visualizing, modifying and annotating phylogenetic trees. Nucleic Acids Res. 2023, 51, W587–W592. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Mantica, F.; Iñiguez, L.P.; Marquez, Y.; Permanyer, J.; Torres-Mendez, A.; Cruz, J.; Franch-Marro, X.; Tulenko, F.; Burguera, D.; Bertrand, S.; et al. Evolution of tissue-specific expression of ancestral genes across vertebrates and insects. Nat. Ecol. Evol. 2024, 8, 1140–1153. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. Fast R functions for robust correlations and hierarchical clustering. J. Stat. Softw. 2012, 46, i11. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liang, S.; Yang, W.; Yu, K.; Liang, F.; Zhao, B.; Zhu, X.; Zhou, C.; Mur, L.A.J.; Roberts, J.A.; et al. MetMiner: A user-friendly pipeline for large-scale plant metabolomics data analysis. J. Integr. Plant Biol. 2024, 66, 2329–2345. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Zhang, Z. KaKs_Calculator 3.0: Calculating selective pressure on coding and non-coding sequences. Genom. Proteom. Bioinform. 2022, 20, 536–540. [Google Scholar] [CrossRef]
- Huang, Z.; Duan, W.; Song, X.; Tang, J.; Wu, P.; Zhang, B.; Hou, X. Retention, molecular evolution, and expression divergence of the auxin/indole acetic acid and auxin response factor gene families in Brassica rapa shed light on their evolution patterns in plants. Genome Biol. Evol. 2015, 8, 302–316. [Google Scholar] [CrossRef]
- Allen, M.; Poggiali, D.; Whitaker, K.; Marshall, T.R.; van Langen, J.; Kievit, R.A. Raincloud plots: A multi-platform tool for robust data visualization. Wellcome Open Res. 2021, 4, 63. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Wang, M.; Li, X.; Jiu, S.; Wang, C.; Fang, J. Genome-wide analysis of the sucrose synthase gene family in grape (Vitis vinifera): Structure, evolution, and expression profiles. Genes 2017, 8, 111. [Google Scholar] [CrossRef]
- Anjum, A.; Haram, A.; Ahmad, R.; Bashir, A. Physico-chemical attributes of fresh and dried Indian jujube (Zizyphus mauritiana) fruits. Pak. J. Agri. Sci. 2020, 57, 165–176. [Google Scholar] [CrossRef]
- Ishtiaq, A.; Muhammad, N.; Irfan, A.; Maryam, M.; Al-Khayri, M.; Yousaf, M.; Rahmatullah, Q. Fruit morphological attributes to assess genetic diversity in jujube (Ziziphus mauritiana L.) germplasm of Bahawalpur. Pure Appl. Biol. 2016, 5, 921–926. [Google Scholar] [CrossRef]
- Ngigi, P.B.; Termote, C.; Pallet, D.; Amiot, M.J. Mainstreaming traditional fruits, vegetables and pulses for nutrition, income, and sustainability in sub-Saharan Africa: The case for Kenya and Ethiopia. Front. Nutr. 2023, 10, 1197703. [Google Scholar] [CrossRef]
- Nyanga, L.K.; Gadaga, T.H.; Nout, M.J.; Smid, E.J.; Boekhout, T.; Zwietering, M.H. Nutritive value of masau (Ziziphus mauritiana) fruits from Zambezi Valley in Zimbabwe. Food Chem. 2013, 138, 168–172. [Google Scholar] [CrossRef]
- Muhammad, N.; Luo, Z.; Meng, X.; Zhao, X.; Wang, J.; Yang, M.; Liu, Z.; Liu, M. Comparative transcriptome profiling unveils the potential key genes involved in peel coloration between Ziziphus mauritiana Lam. and Z. jujuba Mill. Sci. Hortic. 2023, 319, 112175. [Google Scholar] [CrossRef]
- Deng, Y.; Li, C.; Li, H.; Lu, S. Identification and characterization of flavonoid biosynthetic enzyme genes in Salvia miltiorrhiza (Lamiaceae). Molecules 2018, 23, 1467. [Google Scholar] [CrossRef]
- Zhang, L.; Sun, X.; Wilson, I.W.; Shao, F.; Qiu, D. Identification of the genes involved in anthocyanin biosynthesis and accumulation in Taxus chinensis. Genes 2019, 10, 982. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, C.; Li, Y. Genome-wide identification and expression profiles of 13 key structural gene families involved in the biosynthesis of rice flavonoid scaffolds. Genes 2022, 13, 410. [Google Scholar] [CrossRef]
- Tong, W.; Wang, Y.; Li, F.; Zhai, F.; Su, J.; Wu, D.; Yi, L.; Gao, Q.; Wu, Q.; Xia, E. Genomic variation of 363 diverse tea accessions unveils the genetic diversity, domestication, and structural variations associated with tea adaptation. J. Integr. Plant Biol. 2024, 66, 2175–2190. [Google Scholar] [CrossRef]
- Klčová, B.; Balarynová, J.; Trněný, O.; Krejčí, P.; Cechová, M.Z.; Leonova, T.; Gorbach, D.; Frolova, N.; Kysil, E.; Orlova, A.; et al. Domestication has altered gene expression and secondary metabolites in pea seed coat. Plant J. 2024, 118, 2269–2295. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Fu, L.; Wang, B.; Ye, J.; Ou, W.; Yan, Y.; Li, M.; Zeng, L.; Dong, X.; Tie, W.; et al. Metabolic GWAS-based dissection of genetic basis underlying nutrient quality variation and domestication of cassava storage root. Genome Biol. 2023, 24, 289. [Google Scholar] [CrossRef] [PubMed]
- Borredá, C.; Perez-Roman, E.; Talon, M.; Terol, J. Comparative transcriptomics of wild and commercial Citrus during early ripening reveals how domestication shaped fruit gene expression. BMC Plant Biol. 2022, 22, 123. [Google Scholar] [CrossRef]
- Wang, X.; Chen, X.; Luo, S.; Ma, W.; Li, N.; Zhang, W.; Tikunov, Y.; Xuan, S.; Zhao, J.; Wang, Y.; et al. Discovery of a DFR gene that controls anthocyanin accumulation in the spiny Solanum group: Roles of a natural promoter variant and alternative splicing. Plant J. 2022, 111, 1096–1109. [Google Scholar] [CrossRef]
- Su, W.; Tao, R.; Liu, W.; Yu, C.; Yue, Z.; He, S.; Lavelle, D.; Zhang, W.; Zhang, L.; An, G.; et al. Characterization of four polymorphic genes controlling red leaf colour in lettuce that have undergone disruptive selection since domestication. Plant Biotechnol. J. 2020, 18, 479–490. [Google Scholar] [CrossRef]
- Crocco, C.D.; Ocampo, G.G.; Ploschuk, E.L.; Mantese, A.; Botto, J.F. Heterologous expression of AtBBX21 enhances the rate of photosynthesis and alleviates photoinhibition in Solanum tuberosum. Plant Physiol. 2018, 177, 369–380. [Google Scholar] [CrossRef]
- Cai, Y.; Liang, Y.; Shi, H.; Cui, J.; Prakash, S.; Zhang, J.; Anaokar, S.; Chai, J.; Schwender, J.; Lu, C.; et al. Creating yellow seed Camelina sativa with enhanced oil accumulation by CRISPR-mediated disruption of Transparent Testa 8. Plant Biotechnol. J. 2024, 22, 2773–2784. [Google Scholar] [CrossRef]
- Dou, F.; Phillip, F.O.; Liu, H. Combined metabolome and transcriptome analysis revealed the accumulation of anthocyanins in grape berry (Vitis vinifera L.) under high-temperature stress. Plants 2024, 13, 2394. [Google Scholar] [CrossRef] [PubMed]
- Guan, M.; Shi, X.; Chen, S.; Wan, Y.; Tang, Y.; Zhao, T.; Gao, L.; Sun, F.; Yin, N.; Zhao, H.; et al. Comparative transcriptome analysis identifies candidate genes related to seed coat color in rapeseed. Front. Plant Sci. 2023, 14, 1154208. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, J.; Li, P.; Sun, C.; Dong, W. Integrative metabolome and transcriptome analyses reveals the black fruit coloring mechanism of Crataegus maximowiczii C. K. Schneid. Plant Physiol. Biochem. 2023, 194, 111–121. [Google Scholar] [CrossRef]
- Li, C.; Pei, J.; Yan, X.; Cui, X.; Tsuruta, M.; Liu, Y.; Lian, C. A poplar B-box protein PtrBBX23 modulates the accumulation of anthocyanins and proanthocyanidins in response to high light. Plant Cell Environ. 2021, 44, 3015–3033. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, Y.; Yue, M.; Jiang, L.; Zhang, N.; Luo, Y.; Chen, Q.; Zhang, Y.; Wang, Y.; Li, M.; et al. FaMYB5 interacts with FaBBX24 to regulate anthocyanin and proanthocyanidin biosynthesis in Strawberry (Fragaria × ananassa). Int. J. Mol. Sci. 2023, 24, 12185. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Xu, Y.; Lu, Y. A regulatory mechanism on pathways: Modulating roles of MYC2 and BBX21 in the flavonoid network. Plants 2024, 13, 1156. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.; Song, Q.; Ji, K.; Gong, S.; Wang, L.; Chen, L.; Zhang, J.; Yuan, D. Full-length transcriptome from Camellia oleifera seed provides insight into the transcript variants involved in oil biosynthesis. J. Agric. Food Chem. 2020, 68, 14670–14683. [Google Scholar] [CrossRef] [PubMed]
- Shi, P.; Hua, W.; Htwe, Y.M.; Zhang, D.; Li, J.; Wang, Y. Abscisic acid improves linoleic acid accumulation possibly by promoting expression of EgFAD2 and other fatty acid biosynthesis genes in oil palm mesocarp. Front. Plant Sci. 2021, 12, 748130. [Google Scholar] [CrossRef]
- Ying, J.; Wen, S.; Cai, Y.; Ye, Y.; Li, L.; Qian, R. Decoding anthocyanin biosynthesis regulation in Asparagus officinalis peel coloration: Insights from integrated metabolomic and transcriptomic analyses. Plant Physiol. Biochem. 2024, 215, 108980. [Google Scholar] [CrossRef]
- Zhou, Y.; Xu, Y.; Zhu, G.F.; Tan, J.; Lin, J.; Huang, L.; Ye, Y.; Liu, J. Pigment diversity in leaves of Caladium × hortulanum Birdsey and transcriptomic and metabolic comparisons between red and white leaves. Int. J. Mol. Sci. 2024, 25, 605. [Google Scholar] [CrossRef] [PubMed]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, F.; Zhu, X.; Wu, M.; Chang, P.; Wu, H.; Li, H. Domestication Has Reshaped Gene Families, Gene Expressions and Flavonoid Metabolites in Green Jujube (Ziziphus mauritiana Lam.) Fruit. Horticulturae 2025, 11, 974. https://doi.org/10.3390/horticulturae11080974
Jiang F, Zhu X, Wu M, Chang P, Wu H, Li H. Domestication Has Reshaped Gene Families, Gene Expressions and Flavonoid Metabolites in Green Jujube (Ziziphus mauritiana Lam.) Fruit. Horticulturae. 2025; 11(8):974. https://doi.org/10.3390/horticulturae11080974
Chicago/Turabian StyleJiang, Fan, Xudong Zhu, Miaohong Wu, Pengyan Chang, Huini Wu, and Haiming Li. 2025. "Domestication Has Reshaped Gene Families, Gene Expressions and Flavonoid Metabolites in Green Jujube (Ziziphus mauritiana Lam.) Fruit" Horticulturae 11, no. 8: 974. https://doi.org/10.3390/horticulturae11080974
APA StyleJiang, F., Zhu, X., Wu, M., Chang, P., Wu, H., & Li, H. (2025). Domestication Has Reshaped Gene Families, Gene Expressions and Flavonoid Metabolites in Green Jujube (Ziziphus mauritiana Lam.) Fruit. Horticulturae, 11(8), 974. https://doi.org/10.3390/horticulturae11080974





