Abstract
Soil salinity severely impairs plant growth, and polyamines such as spermidine (Spd) are known to bolster stress tolerance by acting as osmoprotectants and signaling molecules. Using TiO2 enrichment, iTRAQ quantification, and bioinformatics analysis, we identified 870 proteins and 157 differentially phosphorylated proteins. Functional annotation showed that salt stress activated key components of the Salt Overly Sensitive pathway, particularly serine threonine kinases (SOS2) and Ca2+ binding sensors (SOS3). Among thirty-six SOS-associated kinases detected, eight SOS2 isoforms, four MAPKs, and two SOS3 homologs were significantly upregulated by NaCl, and Spd further increased the phosphorylation of six SOS2 proteins and one SOS3 protein under salt stress, with no detectable effect on SOS1. qRT PCR revealed enhanced expression of MAPKs and calcium-dependent protein kinases, suggesting a phosphorylation-centered model in which Spd amplifies Ca2+-mediated SOS signaling and reinforces ion homeostasis through coordinated transcriptional priming and post-translational control. Additional, proteins involved in protein synthesis and turnover (ribosomal subunits, translation initiation factors, ubiquitin–proteasome components), DNA replication and transcription, and RNA processing showed differential expression under salt or Spd treatment. Central metabolic pathways were reprogrammed, involving glycolysis, the TCA cycle, the pentose phosphate pathway, as well as ammonium transporters and amino acid biosynthetic enzymes. These findings indicate that exogenous Spd regulated phosphorylation-mediated networks involving the SOS signaling pathway, protein homeostasis, and metabolism, thereby enhancing cucumber salt tolerance.
1. Introduction
Soil salinization poses a formidable abiotic stress to global agriculture, significantly impairing the yield and quality of virtually all salt-sensitive crops such as cucumber. With more than one billion hectares of arable land affected and projections indicating up to 50% of cultivable land impacted by 2050, developing strategies to enhance crop salt tolerance has become indispensable for ensuring future food security [1]. Plants have developed important defensive responses that enable their survival and reproduction in hostile environments, which can be activated within minutes to several weeks after exposure to a high salinity environment [2], including early osmotic and ionic stress signaling, activation of the Salt Overly Sensitive (SOS) pathway [3], hormone-mediated regulation, transcriptional remodeling, and detoxification systems to restore cellular homeostasis and support survival, growth, and reproduction. In particular, calcium-dependent kinases, reactive oxygen species (ROS) signaling, and post-translational modifications converge to orchestrate a dynamic and adaptive stress response network.
Among cellular regulators, polyamines—especially spermidine (Spd) and spermine—are rapidly accumulated in many plant species in response to salt stress and are closely associated with enhanced tolerance. Spd has been shown in cucumber to activate antioxidant enzyme systems, stabilize membranes, maintain photosynthetic efficiency, and modulate osmolytes and ethylene metabolism during NaCl stress [4]. Previous proteomic studies in cucumber leaves and roots have shown that exogenous Spd elevates levels of proteins involved in energy metabolism, protein synthesis, ROS detoxification, and SAM-dependent pathways under salt stress [5,6]. Transcriptomic analyses in other plant species have similarly demonstrated Spd-induced transcriptional reprogramming under saline conditions. However, these omic approaches, focused on total protein abundance or mRNA levels, cannot reveal whether the observed regulatory effects are mediated through phosphorylation or other post-translational modifications. Mechanistically, polyamines may interact with ATP-Mg2+ complexes and influence phosphorylation events mediated by protein kinases, suggesting possible cross-talk with signaling cascades regulated via reversible protein phosphorylation [7].
Protein phosphorylation is a major type of post-translational modification observed during salinity stress in plants [8]. Phosphorylation could regulate protein function and cell signaling by triggering a change in the three-dimensional structure of the protein, which in turn influences the protein into behaving differently by activating or deactivating its catalytic function [9]. A variety of biological processes are closely related to the protein phosphorylation that play an on/off regulatory role for many biochemical functions, such as transcriptional and translational regulation, signal transduction, DNA damage repair, cell metabolism, secretion, homeostasis, and so on [10]. Protein phosphorylation also plays a critical role in ion homeostasis under salinity stress through the SOS pathway [11].
High-throughput phosphoproteomics—employing TiO2 enrichment coupled with iTRAQ or TMT quantification and tandem mass spectrometry—offers an unparalleled ability to profile global phosphorylation changes under stress treatments. This approach has proven effective in delineating phosphorylation networks involved in plant abiotic stress responses. Although previous research found cucumber seedlings treated with Spd under salt stress had higher photosynthesis efficiency, stronger ROS scavenging ability, and more protein biosynthesis activity than NaCl treatment by proteomics [5,12], these data did not distinguish whether such effects were driven by changes at the phosphorylation level. Roots are the primary interface for salt entry and osmotic and ionic signaling, and they deploy the SOS ion–homeostasis cascade at the plasma membrane, making phosphorylation control of kinases and transporters most directly observable in this tissue. Therefore, in the present study we enriched phosphopeptides from cucumber roots subjected to NaCl stress with or without exogenous Spd using TiO2 chromatography, followed by iTRAQ-based quantitative phosphoproteomics. Our goal is to reveal specific signaling components and phosphorylation-driven regulatory mechanisms through which Spd alleviates salt-induced damage at the post-translational level. The findings are expected to shed light on the phosphorylation-mediated pathways by which polyamines enhance plant salt tolerance, thus contributing to future strategies for crop improvement.
2. Materials and Methods
2.1. Plant Materials and Treatments
Cucumber (Jinchun No. 2) seeds were germinated on moist filter paper in the dark at 28 °C for 30 h. The germinated seedlings were transferred to plastic trays (41 × 41 × 5 cm) containing quartz sand and were grown in a greenhouse at Shanxi Agricultural University at 25–30 °C (day) and 15–18 °C (night) under natural light with relative humidity of 60–75%. After emergence of the third leaf, cucumber seedlings were transferred to 25 L containers in a hydroponic system containing one-half-strength Hoagland’s nutrient solution, and the solution was renewed every 3 days. The nutrient solutions were kept at 20–25 °C and were continuously aerated using an air pump at an interval of 20 min to maintain the dissolved oxygen at 8.0 ± 0.2 mg·L−1.
After 3 d of pre-culture, the cucumber seedlings were treated as follows: (1) CK, control plants were grown in Hoagland’s solution; (2) CS, plants were grown in Hoagland’s solution with 0.1 mM Spd; (3) S, plants were grown in Hoagland’s solution with 75 mM NaCl; and (4) SS, plants were grown in Hoagland’s solution containing 75 mM NaCl and 0.1 mM Spd. A total of 36 plants from each treatment were harvested after 3-day treatment, and 1 cm root tips were separated and washed with deionized water three times before immersion into liquid nitrogen. The samples with three independent biological replicates were stored at −80 °C for further analyses.
2.2. Protein Sample Preparation and Labeling
The samples were homogenized in liquid nitrogen. Two milliliters of lysis buffer (30% sucrose, 0.5 M Tris-HCl, 50 mM EDTA, 20 mM DTT, 0.1 M KCl, 2% SDS, 1 mM phenylmethanesulfonyl fluoride (PMSF), and PhosSTOP Phosphatase Inhibitor Cocktail (Roche, Basel, Switzerland)) was added into the samples. Cell lysis was performed by sonication on ice for 1 h and followed by centrifugation at 4000× g for 30 min at 4 °C. Equal volumes of Tris-saturated phenol (pH 7.5) was added and the resulting mixture was thoroughly vortexed for 30 min at 4 °C and centrifuged at 4000× g for 15 min at 4 °C for phase separation. The organic phase was collected and transferred into a new Eppendorf tube. The extraction step was repeated once. The combined organic phase was precipitated overnight using five volumes of prechilled acetone at −20 °C. The precipitate was washed twice with cold acetone and then resuspended with 1% SDS in 8 M urea.
The protein concentration was determined with the BCA assay, and 400 μg of protein from each sample was precipitated by incubation with five volumes of cold acetone at −20 °C for 1 h. Centrifuge at 12,000 rpm for 15 min at 4 °C, collect the deposit, and dry by vacuum freeze dryer. Add 50 μL dissolution buffer to the deposit and add 4 μL reducing reagent, incubate the solution at 60 °C for 1 h, and then add 2 μL cysteine-blocking reagents at room temperature for 10 min. Clean the protein solution by using a 10 KDa ultrafiltration tube to centrifuge at 12,000 rpm for 20 min. Add 100 μL dissolution buffer, centrifuge at 12,000 rpm for 15 min, and repeat this step three times. Place the column in a new tube, add 100 μL sequencing-grade trypsin (50 ng/μL), and incubate at 37 °C for 12 h. Centrifuge at 12,000 rpm for 20 min and collect the peptide. Transfer the filter units to new collection tubes and add 50 μL dissolution buffers to centrifuge the tube again. Combine the two filter solutions and the resultant peptide mixture was labeled with the iTRAQ 8Plex labeling kit (Sciex, Framingham, MA, USA), following the manufacturer’s instructions. The labeled peptide samples were then pooled and lyophilized in a vacuum concentrator.
2.3. High pH Reverse Phase Separation
The peptide mixture was redissolved in buffer A (buffer A: 20 mM ammonium formate in water, pH 10.0, adjusted with ammonium hydroxide) and then fractionated by high pH separation using an Ultimate 3000 system (ThermoFisher Scientific, Waltham, MA, USA) connected to a reverse phase column (XBridge C18 column, 4.6 mm × 250 mm, 5 μm, (Waters Corporation, Milford, MA, USA)). High pH separation was performed using a linear gradient, starting from 5% B to 45% B in 40 min (B: 20 mM ammonium formate in 80% ACN, pH 10.0, adjusted with ammonium hydroxide). The column was re-equilibrated at the initial condition for 15 min. The column flow rate was maintained at 1 mL/min, and the column temperature was maintained at 30 °C. Twelve fractions were collected; each fraction was dried in a vacuum concentrator for the next step.
2.4. Phosphopeptide Enrichment by TiO2 Kit
A Phosphopeptide Enrichment TiO2 kit (Calbiochem, San Diego, CA, USA) was used to enrich the phosphopeptides after peptide digestion, according to the manufacturer’s instruction with minor modifications. To evaluate the performance of TiO2 enrichment, we monitored the proportion of phosphopeptide spectral matches and the number of confidently localized phosphosites in each LC–MS/MS run as proxies for selectivity and yield. Nonphosphorylated peptide carryover was low, indicating effective capture. Absolute recovery was not directly quantified, but enrichment quality was further supported by consistent identification profiles across runs acquired under the same instrument settings. In brief, the dry sample was resuspended with 1 mL Nano-RPLC buffer A, with 3 mL TiO2 phosphobind buffer added. Next, TiO2 phosphobind resin was added and incubated for 30 min. After discarding the supernatant, TiO2 was rinsed three times with the wash buffer. Phosphopeptides were eluted twice with elution buffer, after which the eluates were pooled. The eluates were then dried using a Speed-Vac concentrator (Thermo Fisher Scientific, Waltham, MA, USA).
2.5. RPLC-MSMS Analysis
Samples were resuspended with Nano-RPLC buffer A (0.1% FA, 2% ACN), and analyzed online using Nano-RPLC on the Eksigent nanoLC-Ultra™2D System (Eksigent Technologies, Dublin, CA, USA). The samples were loaded on a C18 nanoLC trap column (100 µm × 3 cm, C18, 3 µm, 150 Å) and washed by Nano-RPLC Buffer A at 2 μL/min for 10 min. An elution gradient of 5–35% acetonitrile (0.1% formic acid) in a 70 min gradient was used on an analytical ChromXP C18 column (75 μm × 15 cm, C18, 3 μm, 120 Å) with a spray tip.
Data acquisition was performed with a Triple TOF 5600 System (AB SCIEX, USA) fitted with a Nanospray III source (AB SCIEX, USA) and a pulled quartz tip as the emitter (New Objectives, Woburn, MA, USA). Data were acquired using an ion spray voltage of 2.5 kV, curtain gas of 30 PSI, nebulizer gas of 5 PSI, and an interface heater temperature at 150 °C. For information-dependent acquisition (IDA), survey scans were acquired in 250 ms and as many as 35 product ion scans were collected if they exceeded a threshold of 150 counts per second (counts/s) with a 2+ to 5+ charge-state. The total cycle time was fixed at 2.5 s. A rolling collision energy setting was applied to all precursor ions for collision-induced dissociation (CID). Dynamic exclusion was set for ½ of the peak width (18 s), and the precursor was then refreshed off the exclusion list.
2.6. Protein Identification and Quantification
Data were processed using Protein Pilot Software v. 5.0 (AB SCIEX, USA) against the Cucumis sativus L. database by means of the Paragon algorithm [13]. Protein identification was performed with the search option that was described previously [12]. To control identification error rates, spectra were searched with a target decoy strategy, and peptide-spectrum matches (PSMs) together with protein identifications were filtered at a 1% false discovery rate (FDR). For quantification, the intensities of reporter ions from iTRAQ labeling were extracted and normalized across runs. Differential phosphorylation was assessed using a significance threshold of p < 0.05 in combination with a fold change greater than or equal to 1.2 or less than or equal to 0.83. Based on the identified proteins, Gene Ontology (GO) enrichment analysis was performed using the widely used databases DAVID 6.7 and QuickGO to classify and annotate GO terms and to conduct enrichment analysis of the screened differentially expressed proteins. The Kyoto Encyclopedia of Genes and Genomes (KEGG) database was used to predict the major metabolic pathways involved.
2.7. Quantitative Real-Time PCR (qRT-PCR) Analysis
Total RNA was extracted from the leaves using Tranzol (TransGen Biotech, Beijing, China). The HiScript II One Step RT-PCR Kit (Vazyme Biotech, Beijing, China) was used to perform reverse transcription with 1 μg of RNA according to the manufacturer’s instructions. According to the genome sequences of cucumber, the gene-specific primers were designed on Primer 5 software; qRT-PCR was performed using the SYBR PrimeScript TM RT-PCR Kit (Takara Bio Inc, Shiga, Japan.) on an ABI 7500 Real-Time PCR machine (Applied Biosystems, Foster City, CA, USA) with the following amplification program: 40 cycles at 94 °C for 15 s, 60 °C for 15 s, and 72 °C for 15 s. The melting curve was recorded after 40 cycles to verify the primer specificity. The relative quantization of gene expressions was calculated using the 2−ΔΔct method and normalized to actin. Three replicates were used for qRT-PCR.
2.8. Statistical Analysis
All data were statistically analyzed using SPSS 20.0 for Windows, and significance was assigned at the p < 0.05 level using Duncan’s multiple comparisons test.
3. Results
3.1. Differential Expression of Phosphorylated Protein
Previously, 5014 proteins were identified via iTRAQ in cucumber roots [14], providing a comprehensive baseline for total proteome coverage. To resolve phosphorylation dynamics under salinity with or without spermidine in cucumber, we applied iTRAQ phosphoproteomics to root tips. After three days of treatments, 870 phosphorylated proteins and 157 differentially phosphorylated proteins were identified. Using an unused score greater than 1.3, at least one unique peptide per protein, and an expression ratio threshold of at least 1.20 or at most 0.83, we quantified differential phosphorylation across the three contrasts. In salt stress versus control, 133 proteins changed in abundance, including 82 increases and 51 decreases. Relative to salt stress, Spd under salt altered 104 proteins, including 64 increases and 40 decreases. Under control conditions, Spd altered 76 proteins compared to the control, including 56 increases and 20 decreases. These distributions are shown in Figure 1A. Venn analysis indicated substantial overlap among contrasts and a core set shared by all three comparisons, as shown in Figure 1B. Previous TiO2-enriched plant phosphoproteomes studies have typically reported a serine:threonine:tyrosine ratio of approximately 90:10:<1 [15], and our dataset likewise shows many serine- and threonine-phosphorylated proteins. Consistent findings in dicots, including Arabidopsis, indicate that this pattern reflects the selectivity of TiO2 for phosphoserine and phosphothreonine and is consistent with what is known in dicotyledons.
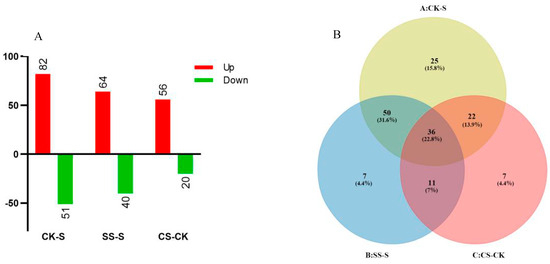
Figure 1.
Distribution of phosphorylated variant proteins in cucumber root tips induced by salt stress and Spd. (A) differentially expressed phosphoprotein count; (B) Venn analysis.
3.2. Isolation and Identification of Differential Expression Protein
The differentially expressed phosphorylated proteins were compared with the cucumber genome database to obtain annotation information, and the signals or metabolic pathways involved were preliminarily explored: (1) serine/threonine protein kinase, phosphatase, MAPK, and calcium signaling pathways; (2) protein synthesis, post-translational modification, and degradation pathways; (3) DNA replication and transcription factors; (4) cell division-associated proteins; (5) transporters; (6) metabolic pathways such as glycolysis, nitrogen metabolism, and cellulose synthesis (Table S1). Since the research species cucumber is a non-pattern species, according to the sequence similarity, the protein sequence was retrieved from the cucumber genome database, and its BLAST (version 2.12.0, NCBI, Bethesda, MD, USA) homology was mapped to the model species, Arabidopsis thaliana, which was converted into an international Uniprot ID, which contributed to analysis of biological information functions.
3.3. Analysis of Phosphorylation Differential Expression Induced by Salt Stress and Spd
The expression data of CK-S, SS-S, and CS-CK was analyzed by heat map analysis and clustering. The results showed that salt stress significantly induced the upregulation of kinase-like protein expression, among which serine/threonine protein kinase was one of the most identified proteins, and MAPK protein kinase, calcium-dependent protein kinase, and protein Phosphatase were also upregulated. In addition, exogenous Spd treatment upregulated six serine/threonine protein kinases, reducing MAPK protein expression (Figure 2A).
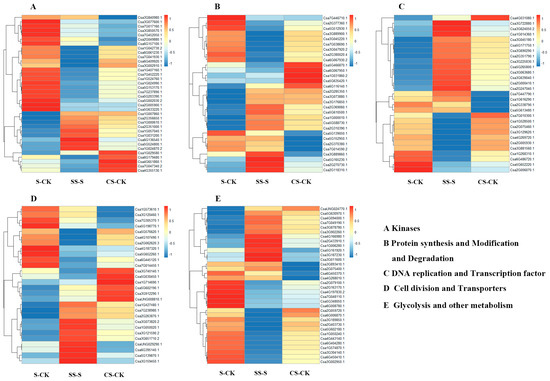
Figure 2.
Heat map expression analysis of differentially phosphorylated proteins induced by salt stress and polyamines.
The phosphorylation shifts coincide with a transient protein synthesis and translational slowdown: several ribosomal proteins and the translation regulator RPS6 were reduced under salt but restored by Spd, indicating a relief of salt induced translational bottlenecks and reactivation of protein synthesis. Concordantly, eIF4G and eIF5 decreased under salt but increased with Spd, further supporting Spd-mediated recovery of initiation. Ribosomal proteins and elongation initiations factors thus point to global repression of translation by salt and its partial reversal by Spd, while the ubiquitin proteasome pathway remains active to clear damaged protein (Figure 2B).
The DNA replication and repair of plants are inhibited under salt stress, and the expression of multiple transcription factors is significantly enhanced (Figure 2C). The application of Spd under salt stress induces upregulation of multiple transcription factors, indicating that polyamines can be regulated under salt stress. Transcription factors promote the expression of some anti-retroviral related proteins; as shown in Figure 2D, the expression of cell division-associated proteins is downregulated under salt stress, while some transmembrane transporters are upregulated, and the expression of programmed cell death proteins is upregulated after polyamine treatment. The vacuolar cation exchange protein, vacuolar protein-sorting protein, ATPase protein, and sugar transporter were significantly upregulated, indicating that polyamine can promote the transport of some molecules under salt stress. Figure 2E shows that salt stress upregulated glycolysis and nitrogen metabolism while reducing cellulose synthesis. In contrast, spermidine treatment under salt stress decreased glycolysis and increased cellulose synthesis, indicating enhanced cellulose deposition in the cell wall.
3.4. Phosphorylation Differential Protein GO Analysis
Phosphorylation differential protein GO analysis was based on David 6.7 (http://david.ncifcrf.gov/, accessed on 15 March 2021) and QuickGO (http://www.ebi.ac.uk/QuickGO/, accessed on 15 March 2021) for GO classification of screened differentially expressed proteins. As shown in Figure 3, compared with CK-S, the biological processes engaged with differential proteins involve ribosomal subunit synthesis, stimulation response, defense response, gene expression regulation, ion homeostasis, biosynthesis processes, root development, actin filament movement, cell development, etc.; enriched cellular components include plasma membrane, organelles, vesicles, respiratory chains, vacuoles, micro-organisms, metastatic complexes, etc.; and the molecular functions involved include transport activity, ion transmembrane movement, protein transport, rRNA binding, DNA binding, RNA helicase activity, isomerase, ATPase activity, etc. Compared with SS-S, the biological processes engaged with differential proteins involve stimulation response, defense response, gene expression regulation, ion homeostasis, biosynthesis, root development, actin filament movement, cell development, etc.; enriched cellular components include plasma membrane, external packaging structure, organelles, vesicles transfer complex, etc.; the molecular functions involved include DNA binding, transport activity, lipid binding, ATPase activity, ion transmembrane movement, vitamin B6 binding, ribonucleotide binding, rRNA binding, etc.
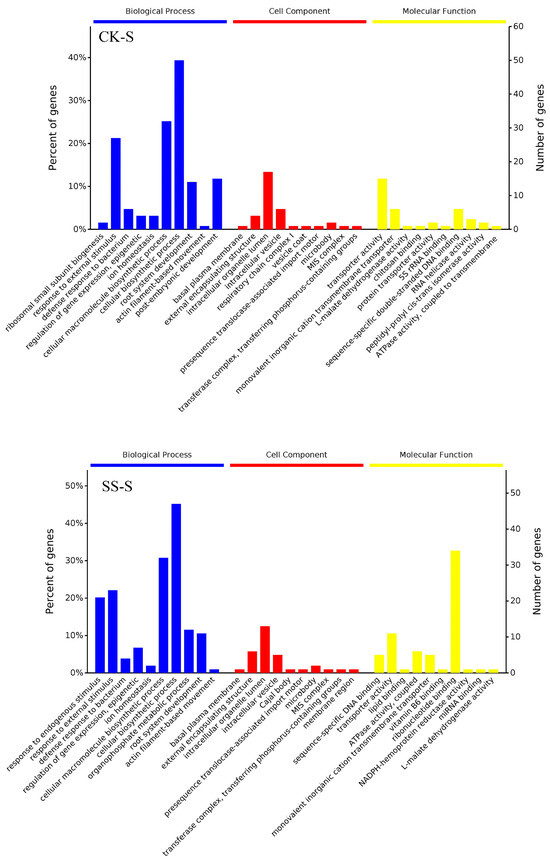
Figure 3.
GO analysis of differentially phosphorylated proteins induced by salt stress and polyamines.
3.5. Phosphorylation Differential Protein KEGG Pathway Analysis
As shown in Figure 4, the results show that compared with CK-S, the enriched pathways include sugar metabolism, energy metabolism, amino acid metabolism, transcription factors, translation, protein processing in folding, sorting, and degradation, DNA replication and repair, signal transduction, transport metabolism, environmental adaptation, etc.; compared with SS-S, the enriched metabolic pathways include sugar metabolism, energy metabolism, amino acid metabolism, coenzyme vitamins, transcription factors, translation, protein folding sequencing and degradation, DNA replication and repair, signal transduction, transport metabolism, environmental adaptation, etc.
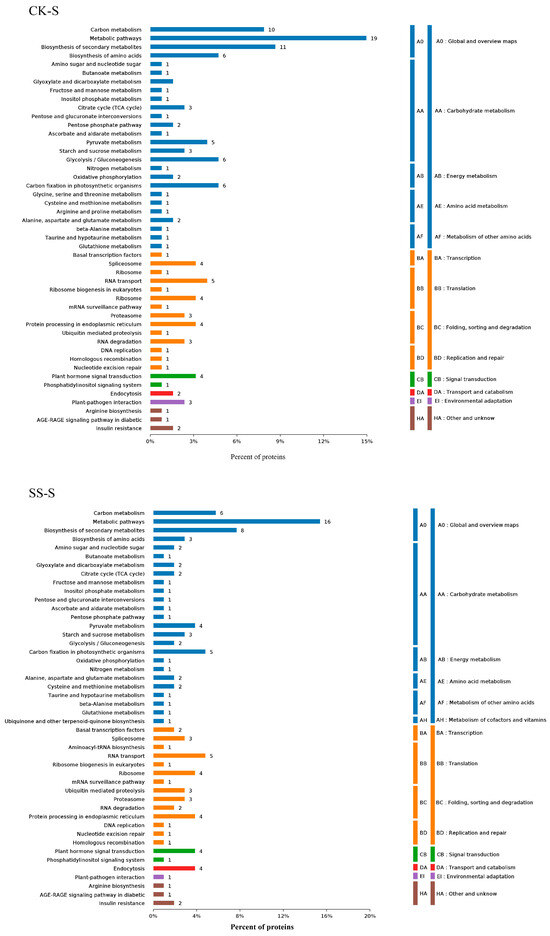
Figure 4.
KEGG and Pathway analysis of differentially phosphorylated proteins induced by salt stress and polyamines.
KEGG enrichment under Spd plus salt highlighted central carbon metabolism, consistent with phosphorylation of key enzymes and metabolite shifts that increase NADPH regeneration and temper glycolysis. Proteostasis pathways, including the ribosome and ubiquitin proteasome, were significant, together with DNA replication and repair, transcription, and RNA processing. Transport functions were enriched through phosphorylation of proton pumps, aquaporins, vacuolar exchangers, and nutrient and hormone carriers. Signaling modules mapped to calcium dependent kinases and MAPKs within the SOS framework, indicating strengthened ion homeostasis. Overall, Spd redirects phosphorylation toward energy and nitrogen metabolism, proteostasis, genome maintenance, RNA metabolism, membrane transport, and ion homeostasis, thereby reinforcing cucumber-root salt tolerance.
3.6. Phosphorylation Differential Protein MapMan Analysis
Before MapMan analysis, reporter ion intensities were normalized across iTRAQ channels and converted to protein-level fold changes, as described in Section 2.6. Fold change values were then log2 transformed and mean-centered within each comparison prior to import into MapMan. Only differentially phosphorylated proteins meeting the stated thresholds were included in the MapMan input. No additional scaling was applied within MapMan, and BIN assignments followed the default mapping for Cucumis sativus. MapMan software (version 3.6.0) was used to enrich the differentially phosphorylated proteins in metabolic pathways (Figure 5). MapMan indicated inhibition of DNA replication and protein synthesis under salt, whereas spermidine partly restored replication and tempered glycolysis, consistent with regulated reprogramming rather than irreversible failure. Concordantly, several ribosomal proteins and initiation factors decreased under salt but were restored by spermidine, supporting a transient slowdown of translation that is actively relieved by polyamine treatment. In parallel, phosphorylation of DNA replication and repair factors and the induction of multiple transcription regulators indicate genome maintenance and transcriptional reprogramming, features typical of adaptive responses.
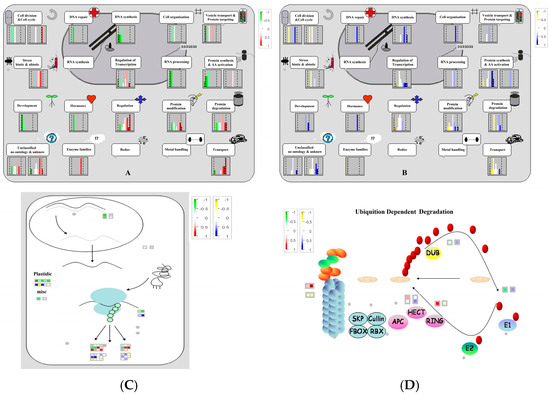
Figure 5.
MapMan analysis of differentially phosphorylated proteins involved in cellular functions and protein synthesis, modification, and degradation. (A) CK-S cell function; (B) SS-S cell function; (C) RNA–protein synthesis; (D) Ubiquition dependent degradation.
3.7. Relative Levels of Gene Expression by qRT-PCR
For qRT-PCR normalization, relative transcript levels were calculated using the 2−ΔΔCt method with actin as the internal control. As shown in Figure 6, quantitative RT-PCR analysis revealed that 13 serine/threonine protein kinase (SOS2) genes were upregulated under salt stress, most notably Csa6G409920.1 and Csa1G057040.1, which exhibited 11.9- and 10.6-fold increases, respectively. Three Calcium-dependent protein kinase (SOS3) genes (Csa3G077600.1, Csa2G062620.3, and Csa4G355130.1) were also induced, with Csa2G062620.3 showing a 9.9-fold upregulation. Additionally, three MAPK genes (Csa6G061230.1, Csa5G002030.2, and Csa2G009280.2) were upregulated 2.2-, 4.6-, and 3.2-fold under salt stress; of these, Csa6G061230.1 exhibited a pronounced 15.3-fold increase when salt stress was combined with spermidine (Spd) treatment. The expression of four protein phosphatase genes (Csa1G000610.1, Csa7G047340.2, Csa4G658620.1, and Csa7G010300.1) increased significantly under salt stress, with fold changes of 1.9, 2.7, 2.1, and 2.2, respectively. Two receptor kinase genes (Csa1G407160.1 and Csa1G031200.1) were significantly upregulated after three days of salt stress, showing 1.97- and 3.3-fold increases. The potassium ion transporter gene (Csa4G107490.1) was upregulated by salt stress. Following Spd treatment under salt stress, the induction of most genes was slightly attenuated; however, Csa5G153170.1 (SOS2) and Csa6G061230.1 (MAPK) were significantly upregulated by Spd. These changes paralleled increased phosphorylation of SOS2 and SOS3 with Spd, whereas MAPKs displayed elevated mRNA but reduced phospho peptide abundance, consistent with feedback dephosphorylation and post-transcriptional control. Concordantly, upregulation of the potassium transporter gene aligned with differential phosphorylation observed on multiple transporters in the proteomic dataset, further supporting cross-modality consistency. All of these genes are involved in the SOS pathway, indicating that Spd may modulate SOS pathway activity under saline conditions.
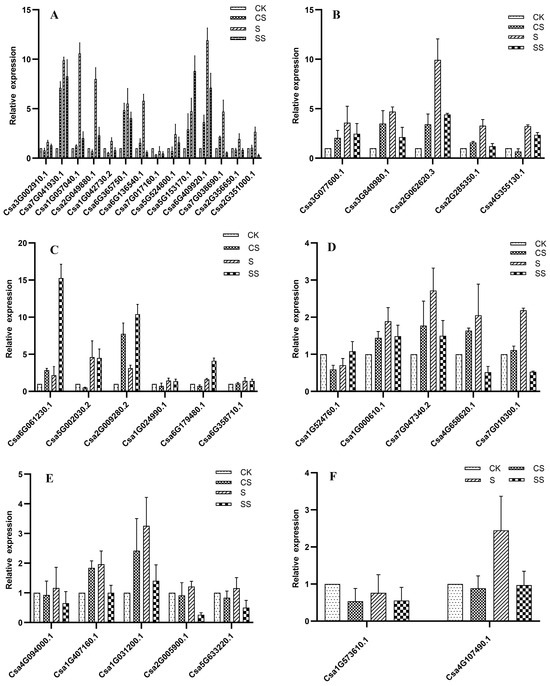
Figure 6.
Analysis of differentially phosphorylated protein gene expression under salt stress and polyamine treatment: (A) serine/threonine protein kinase; (B) calcium-dependent protein kinase; (C) MAPK; (D) protein phosphatase; (E) receptor kinase; (F) transporter.
4. Discussion
4.1. Salt Overly Sensitive (SOS) Signal Transduction Pathway
The SOS signaling cascade is central to plant adaptation under salt stress, chiefly by maintaining cellular ion homeostasis. It comprises three core components: SOS1, a plasma membrane Na+/H+ antiporter that is released upon phosphorylation to extrude Na+; SOS2, a serine/threonine kinase whose activity requires SOS3 and Ca2+ despite its capacity for autophosphorylation; and SOS3, a ubiquitous Ca2+ sensor that initiates the pathway by binding Ca2+ and activating SOS2. In response to elevated cytosolic Ca2+ under salinity, the Ca2+-SOS3 complex binds the C-terminus of SOS2, triggering its kinase activity; the resultant SOS2-SOS3 complex then phosphorylates SOS1, thereby promoting Na+ efflux [11]. Evidence from Arabidopsis demonstrates that sos2 and sos3 mutants are hypersensitive to salt and defective in Na+ and K+ homeostasis, and that SOS3 physically interacts with and activates the SOS2 kinase, establishing the core signaling module [16]. This canonical pathway has been extensively characterized in model species. However, how polyamines intersect with SOS signaling at the level of phosphorylation in crops remains insufficiently defined.
Abscisic acid (ABA)-induced polyamine catabolism yields H2O2, which directly activates plasma membrane Ca2+ channels to facilitate Ca2+ influx, and also generates second messengers such as IP3 and cADPR that release Ca2+ from internal stores [17]. The concomitant rise in Ca2+ triggers SOS pathway activation and Na+ extrusion, so polyamines might modulate this cascade by influencing cytosolic Ca2+ levels. Previous studies have reported polyamine involvement in Ca2+ signaling and MAPK cascades [16,17,18,19,20], but these were largely inferred from transcriptomic or proteomic changes without direct phosphoproteomic evidence. Our data demonstrate that salt stress robustly induces key components of the SOS signaling cascade, specifically 36 phosphorylated kinases associated with SOS signaling, including 1 SOS1, 8 SOS2 isoforms, 4 MAPKs, and 2 SOS3 proteins, while Spd further enhanced the phosphorylation of 6 SOS2 proteins and 1 SOS3 protein and reduced several MAPKs under salt stress (Table S1). The qRT-PCR analysis revealed that 13 SOS2 genes, 3 SOS3 genes, 3 MAPK genes, 4 protein phosphatase, 2 receptor kinase were upregulated under salt stress, and Spd specifically enhanced Csa5G153170.1 (SOS2) and Csa6G061230.1 (MAPK). The observed reduction in MAPK protein reflects decreased MAPK phospho-peptide abundance rather than changes in total protein. As phosphorylation occurs within minutes while mRNA accumulates over hours, this pattern suggests post-transcriptional regulation and feedback dephosphorylation that restrain MAPK over-activation while maintaining transcriptional readiness. By strengthening SOS-centered ion homeostasis and membrane transport, Spd lessens the need for sustained MAPK activation, resulting in lower phospho-MAPK levels despite elevated transcript abundance. Critically, polyamines appear to potentiate this cascade. Polyamine oxidase-mediated catabolism generates H2O2 and NO, both of which enhance serine/threonine kinase expression in diverse stress responses [18]. Polyamines also act as redox modulators under salt stress: they both scavenge reactive oxygen species (ROS) and serve as precursors of ROS via catabolism, thus helping maintain cellular redox balance. This dual role may reinforce Ca2+-mediated activation of the SOS pathway, and perhaps promote MAPK signaling and serine/threonine kinase induction more broadly. The discovery of Spd-induced phosphorylation on both SOS kinases and upstream calcium sensors expands the current model of polyamine–SOS cross-talk, suggesting that phosphorylation-dependent network shifts play a decisive role in fine-tuning ion homeostasis.
Additionally, evidence indicates certain polyamine transporters physically interact with SOS pathway components. For example, polyamine pretreatments in other species reduce Na+ accumulation, limit K+ loss, and increase the K+/Na+ ratio, a widely recognized marker of salt tolerance driven by SOS-centered ion homeostasis [21]. The polyamine transporter PUT3 has been shown to bind both SOS1 and SOS2, and SOS2 can phosphorylate PUT3, suggesting a direct mechanistic link between polyamine movement and SOS kinase activation [22]. Furthermore, ABA and polyamines may engage in reciprocal transcriptional and post-translational feedback, amplifying SOS pathway output through iterative rounds of phosphorylation and gene induction [20]. Taken together, our phosphoproteome and the cucumber data support a model in which Spd strengthens the SOS pathway by reshaping phosphorylation states across Ca2+ sensors, CIPK-like kinases, and MAPKs. This represents a phosphorylation-centric mechanism for polyamine–SOS cross-talk and provides a mechanistic bridge between polyamine signaling and ion homeostasis reported in other species. Future work could use functional genetics or pharmacological inhibitors of PAO, Ca2+ channels, sos2 and sos3 mutants, or SOS transporters to further dissect the precise routes through which polyamines augment SOS pathway activation.
4.2. Protein Synthesis, Modification, and Degradation Pathways
Protein synthesis, modification, and degradation critically regulate protein content in plants, with phosphorylation being a key post-translational modification governing protein function and activity across diverse biological processes. In rice roots, early salinity rapidly remodels phosphorylation independent of total protein levels, highlighting transport and DNA-related processes [8]. Studies in Arabidopsis and Brachypodium reported rapid phosphorylation of signaling and RNA-turnover factors and shifts in translation and defense [23]. Our phosphoproteomic analysis revealed 29 proteins responsive to salt or Spd. Three ribosomal proteins (Csa5G588730.1, Csa1G000010.1, Csa2G369060.1) were upregulated, suggesting that exogenous polyamines stabilize ribosome assembly under salt stress, thereby maintaining translational capacity [19,24,25]. Two eukaryotic translation initiation factors (Csa7G446710.1, Csa5G512930.1) were likewise hyperphosphorylated, consistent with reports that phosphorylation of initiation factors enhances translational control and stress tolerance in transgenic plants [26,27]. Concurrent upregulation of two heatshock proteins indicates a coordinated chaperone response, whereby polyamines facilitate correct protein folding during adversity [28]. In the ubiquitin–proteasome system, we identified seven differentially phosphorylated components: four E3 ligases—UPL1 (Csa3G047920.2), MARCH8 (Csa6G067930.2), UPL4 (Csa2G270730.1), and HOS1 (Csa5G165230.1) and three core proteasome subunits (Csa2G377360.1, Csa3G045220.1, Csa7G038690.1). UPL1 and MARCH8 phosphorylation likely promotes targeted degradation of regulatory proteins to enhance salt tolerance and disease resistance [29,30], whereas UPL4 and HOS1 down-phosphorylation may redirect resources toward root growth and leaf expansion under stress [31,32]. The up-phosphorylated proteasome subunits presumably accelerate clearance of oxidized proteins, mitigating cellular damage [33,34]. We detected a small ubiquitin-related modifier (Csa5G610500.1) whose phosphorylation by polyamines may modulate both protein localization and ABA-mediated signaling [35]. Previous studies emphasized transcriptional changes driven by polyamines during stress. The present results extend that view by showing that Spd directly modifies the phosphorylation status of translation factors, ribosomal proteins, and E3 ligases. This post-translational regulation provides fast and reversible control that complements gene expression changes. Reports that polyamines activate MAPKs and the unfolded-protein response further support a proteostasis-centered mode of action that is compatible with our observation of Spd-responsive phosphorylation on chaperones and UPS components [36]. Collectively, these findings portray a network shift from generalized proteome suppression under salinity to an Spd-enabled state that favors selective synthesis of protective proteins while accelerating the degradation of damaged proteins.
4.3. The Process of Genetic Central Dogma and Cell Division
The central law is a hereditary information flow proposed by Francis Crick in 1958. It mainly includes the three processes of replication, transcription, and translation [37]. Polyamine metabolites (putrescine, spermidine, spermine) interact electrostatically with nucleic acids and proteins, influencing gene expression and cell division. Our phosphoproteome analysis under salt stress identified 38 proteins across replication, transcription, RNA processing, translation, and cell division whose phosphorylation is modulated by salt stress or Spd. Four DNA replication factors (MCM2, Topoisomerases I/II, RecA homolog 3) and one ATP-dependent DNA helicase were hyperphosphorylated in response to polyamines, suggesting enhanced origin licensing, topology management, and repair under salinity [38,39,40,41,42]. Three ATP-dependent RNA helicases and nine transcription regulators exhibited divergent phosphorylation profiles: five (SPT6, bHLH80, FYVE-and CCCH-type zinc fingers) were induced by salt, while four (TIF IIF, BZIP family, CCCH zinc fingers) responded specifically to polyamines. These modifications likely fine-tune transcript elongation and stress-responsive gene expression [43,44,45,46,47,48,49]. Two RNA-binding proteins and three 3′-end processing factors were phosphorylated under salt, and then normalized by polyamines, indicating dynamic control of mRNA maturation. The decapping enhancer EDC4 and RNase P subunit p25 were differentially regulated by polyamines, suggesting targeted decay of mRNAs and tRNAs to reprogram translation. Two eIFs and two heat-shock proteins showed elevated phosphorylation with polyamines, consistent with enhanced initiation and chaperone-mediated folding during stress. Seven components of the protein-degradation machinery-including four E3 ligases (UPL1, MARCH8, UPL4, HOS1) and three proteasome subunits displayed polyamine-dependent phosphorylation, implying accelerated turnover of damaged or regulatory proteins. A SUMO modifier was also activated, potentially coordinating ABA signaling and protein localization. Six regulators of cytokinesis, microtubule dynamics, and programmed cell death were modulated by polyamines, indicating controlled proliferation and reduced metabolic loss under stress. In parallel, 14 transporters (proton, amino acid, water channels; vacuolar exchangers; auxin and sugar carriers) were differentially phosphorylated by polyamines and 16 by salt—highlighting the role of polyamines in sustaining ion balance, vesicle trafficking, and hormone transport during salt adaptation. Collectively, these data define a multilayered network in which polyamines fine-tune phosphorylation across the central dogma, protein quality control, and cellular logistics to preserve genome integrity, reprogram gene expression, and safeguard growth under saline conditions.
4.4. Carbohydrate Metabolism, Nitrogen Metabolism, and Other Metabolic Regulation
Energy metabolism, notably carbohydrate and nitrogen pathways, is fundamental to plant adaptation under salinity. These pathways sustain ATP and reducing-power supply, enabling cells to mount effective stress responses. Polyamines, especially putrescine and spermidine, play pivotal roles by modulating enzyme phosphorylation, antioxidant balance, and hormonal cross-talk, thereby stabilizing energy homeostasis in salt-stressed cucumber seedlings [50]. Our phosphoproteomic data identified key carbohydrate-metabolic enzymes whose expression responds to salt stress or Spd. Crucial enzymes such as G6PDH, fructose–bisphosphate aldolase, GAPDH, phosphoglycerate mutase, PEP carboxylase, and pyruvate dehydrogenase were hyperphosphorylated, consistent with enhanced flux through glycolysis, the pentose phosphate pathway (PPP), and the TCA cycle, thereby preserving NADPH regeneration and carbon flux. These patterns resonate with metabolomic observations showing that exogenous Spd supports PPP activity and NADPH production while alleviating salt-induced flux suppression in glycolysis and the TCA cycle [51]. Specifically, Spd treatment in cucumber roots reduced glucose-6-phosphate accumulation, increased 6-phosphogluconic acid and erythrose-4-phosphate, and enhanced G6PDH activity traits associated with strengthened antioxidant defense and energy metabolism [6].
In nitrogen metabolism, two ammonium transporters (Csa2G079100.1, Csa2G163170.1) facilitated transmembrane ammonium flux, enhancing nitrogen assimilation. Three amino acid metabolism enzymes—glutamate decarboxylase (Csa5G348050.1), glutamine synthetase (Csa3G304140.1), and pyrroline-5-carboxylate synthetase (Csa6G008780.1)—regulated glutamate conversion, organic nitrogen synthesis, and proline production. Methyltransferases showed divergent responses: Csa2G403730.1 and Csa3G002950.1 were salt-activated, while Csa3G187230.1 was inhibited, indicating context-dependent regulation beyond nitrate control. Upregulation of S-adenosylmethionine synthetase (CsaUNG024770.1) by polyamines promoted methyl donor synthesis, supporting nitrogen metabolism. Two electron transfer proteins—NADPH-cytochrome P450 reductase (Csa2G059720.1) and NADH-ubiquinone oxidoreductase (Csa3G189850.1)—sustained energy release, stabilizing metabolic pathways. Polyamines activated two ADP-ribosylation factor GTPase-activating proteins (Csa3G902260.1, Csa3G011600.1), restoring GDP synthesis and energy conversion. Finally, five cellulose synthases (Csa3G878780.1, Csa5G630970.1, Csa4G166980.1, Csa2G433910.1, Csa1G006280.1) were upregulated by polyamines, counteracting salt inhibition of cell wall development. Collectively, our phosphoproteomic evidence supports a model in which polyamines alleviate salt-induced inhibition of central metabolism by restoring phosphorylation-dependent enzymatic activities across carbohydrate and nitrogen pathways, electron transport, and cell wall synthesis. These molecular adaptations are corroborated by metabolomic data demonstrating enhanced PPP flux, restored glycolysis, elevated amino acid levels, and improved carbon allocation under exogenous spermidine treatment in cucumber roots.
5. Conclusions
Using TiO2 enrichment and iTRAQ quantification, we map a phosphorylation program by which exogenous spermidine (Spd) strengthens cucumber-root salt tolerance. Salt activates the Ca2+-dependent SOS network; Spd further boosts phosphorylation of multiple SOS2 (CIPK-like) kinases and a SOS3 sensor (with no change on SOS1). Concordant transcription and dampened MAPK phosphorylation indicate that Spd amplifies Ca2+ signaling while imposing post-transcriptional brakes on stress kinases. Beyond ion control, Spd eases translational bottlenecks by modulating ribosomal and initiation-factor phosphosites and tuning the ubiquitin proteasome system, supporting orderly protein synthesis and proteostasis. Central metabolic pathways, including glycolysis, the TCA cycle, and the pentose phosphate pathway, were reprogrammed, accompanied by differential changes in ammonium transporters and amino acid biosynthetic enzymes.These nodes offer tractable targets for breeding or biostimulant intervention against salinity in cucurbits.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae11080973/s1, Table S1: Identification of differentially phosphorylated proteins in cucumber root tips in response to salt stress and exogenous Spd by iTRAQ.
Author Contributions
Conceptualization, L.H. and B.L.; methodology B.Q. and L.R.; formal analysis, L.W.; investigation, L.H.; resources, B.L.; data curation, B.L. and D.W.; writing—original draft, D.W. and B.L.; writing—review and editing, B.L.; visualization, L.R.; supervision, L.H.; project administration, B.L.; funding acquisition, B.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Shanxi Province Key R&D Plan (202302010101003), the National Natural Science Foundation of China (31501806), and the Applied Fundamental Research Program of Shanxi Province (20210302123401).
Data Availability Statement
The data presented in this study are available upon request from the corresponding author. Data are not readily available for public release because of privacy and other issues.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Chen, C.; Yu, W.; Xu, X.; Wang, Y.; Wang, B.; Xu, S.; Lan, Q.; Wang, Y. Research Advancements in Salt Tolerance of Cucurbitaceae: From Salt Response to Molecular Mechanisms. Int. J. Mol. Sci. 2024, 25, 9051. [Google Scholar] [CrossRef]
- Gao, W.; Xu, F.-C.; Guo, D.-D.; Zhao, J.-R.; Liu, J.; Guo, Y.-W.; Singh, P.K.; Ma, X.-N.; Long, L.; Botella, J.R. Calcium-dependent protein kinases in cotton: Insights into early plant responses to salt stress. BMC Plant Biol. 2018, 18, 15. [Google Scholar] [CrossRef]
- Türkan, I.; Demiral, T. Recent developments in understanding salinity tolerance. Environ. Exp. Bot. 2009, 67, 2–9. [Google Scholar] [CrossRef]
- Yuan, Y.; Zhong, M.; Shu, S.; Du, N.; Sun, J.; Guo, S. Proteomic and Physiological Analyses Reveal Putrescine Responses in Roots of Cucumber Stressed by NaCl. Front. Plant Sci. 2016, 7, 1035. [Google Scholar] [CrossRef]
- Li, B.; He, L.; Guo, S.; Li, J.; Yang, Y.; Yan, B.; Sun, J.; Li, J. Proteomics reveal cucumber Spd-responses under normal condition and salt stress. Plant Physiol. Biochem. 2013, 67, 7–14. [Google Scholar] [CrossRef]
- Liu, B.; Peng, X.; Han, L.; Hou, L.; Li, B. Effects of Exogenous Spermidine on Root Metabolism of Cucumber Seedlings Under Salt Stress by GC-MS. Agronomy 2020, 10, 459. [Google Scholar] [CrossRef]
- Kapoor, R.T. Role of polyamines in plants under abiotic stresses: Regulation of biochemical interactions. In Plant Stress Mitigators; Academic Press: Cambridge, MA, USA, 2023; pp. 209–220. [Google Scholar] [CrossRef]
- Chitteti, B.R.; Peng, Z. Proteome and phosphoproteome differential expression under salinity stress in rice (Oryza sativa) roots. J. Proteome Res. 2007, 6, 1718–1727. [Google Scholar] [CrossRef] [PubMed]
- Al-Momani, S.; Qi, D.; Ren, Z.; Jones, A.R. Comparative qualitative phosphoproteomics analysis identifies shared phosphorylation motifs and associated biological processes in evolutionary divergent plants. J. Proteom. 2018, 181, 152–159. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-M.; Liu, W.-T.; Yang, F.; Yi, Q.-J.; Zhang, S.; Jia, H.-L. Phosphorylated proteomics analysis of human coronary artery endothelial cells stimulated by Kawasaki disease patients serum. BMC Cardiovasc. Disord. 2019, 19, 21. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Plant salt tolerance. Trends Plant Sci. 2001, 6, 66–71. [Google Scholar] [CrossRef]
- Sang, T.; Shan, X.; Li, B.; Shu, S.; Sun, J.; Guo, S. Comparative proteomic analysis reveals the positive effect of exogenous spermidine on photosynthesis and salinity tolerance in cucumber seedlings. Plant Cell Rep. 2016, 35, 1769–1782. [Google Scholar] [CrossRef]
- Shilov, I.V.; Seymour, S.L.; Patel, A.A.; Loboda, A.; Tang, W.H.; Keating, S.P.; Hunter, C.L.; Nuwaysir, L.M.; Schaeffer, D.A. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteom. 2007, 6, 1638–1655. [Google Scholar] [CrossRef]
- Lyu, J.; Wu, Y.; Jin, X.; Tang, Z.; Liao, W.; Dawuda, M.M.; Hu, L.; Xie, J.; Yu, J.; Calderón-Urrea, A. Proteomic analysis reveals key proteins involved in ethylene-induced adventitious root development in cucumber (Cucumis sativus L.). PeerJ 2021, 9, e10887. [Google Scholar] [CrossRef]
- Possemato, A.P.; Paulo, J.A.; Mulhern, D.; Guo, A.; Gygi, S.P.; Beausoleil, S.A. Multiplexed phosphoproteomic profiling using titanium dioxide and immunoaffinity enrichments reveals complementary phosphorylation events. J. Proteome Res. 2017, 16, 1506–1514. [Google Scholar] [CrossRef]
- Liu, J.; Ishitani, M.; Halfter, U.; Kim, C.-S.; Zhu, J.-K. The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad. Sci. USA 2000, 97, 3730–3734. [Google Scholar] [CrossRef] [PubMed]
- Alcázar, R.; Altabella, T.; Marco, F.; Bortolotti, C.; Reymond, M.; Koncz, C.; Carrasco, P.; Tiburcio, A.F. Polyamines: Molecules with regulatory functions in plant abiotic stress tolerance. Planta 2010, 231, 1237–1249. [Google Scholar] [CrossRef]
- Mellidou, I.; Karamanoli, K.; Beris, D.; Haralampidis, K.; Constantinidou, H.-I.A.; Roubelakis-Angelakis, K.A. Underexpression of apoplastic polyamine oxidase improves thermotolerance in Nicotiana tabacum. J. Plant Physiol. 2017, 218, 171–174. [Google Scholar] [CrossRef]
- Law, G.L.; Raney, A.; Heusner, C.; Morris, D.R. Polyamine regulation of ribosome pausing at the upstream open reading frame of S-adenosylmethionine decarboxylase. J. Biol. Chem. 2001, 276, 38036–38043. [Google Scholar] [CrossRef] [PubMed]
- Kusano, T.; Yamaguchi, K.; Berberich, T.; Takahashi, Y. The polyamine spermine rescues Arabidopsis from salinity and drought stresses. Plant Signal. Behav. 2007, 2, 251–252. [Google Scholar] [CrossRef] [PubMed]
- Cheng, B.; Hassan, M.J.; Peng, D.; Huang, T.; Peng, Y.; Li, Z. Spermidine or spermine pretreatment regulates organic metabolites and ions homeostasis in favor of white clover seed germination against salt toxicity. Plant Physiol. Biochem. 2024, 207, 108379. [Google Scholar] [CrossRef]
- Chai, H.; Guo, J.; Zhong, Y.; Hsu, C.C.; Zou, C.; Wang, P.; Zhu, J.K.; Shi, H. The plasma-membrane polyamine transporter PUT3 is regulated by the Na+/H+ antiporter SOS1 and protein kinase SOS2. New Phytol. 2020, 226, 785–797. [Google Scholar] [CrossRef]
- Stecker, K.E.; Minkoff, B.B.; Sussman, M.R. Phosphoproteomic analyses reveal early signaling events in the osmotic stress response. Plant Physiol. 2014, 165, 1171–1187. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, Y.; Peng, D.; Peng, Y.; Zhang, X.; Ma, X.; Huang, L.; Yan, Y. The inhibition of polyamine biosynthesis weakens the drought tolerance in white clover (Trifolium repens) associated with the alteration of extensive proteins. Protoplasma 2018, 255, 803–817. [Google Scholar] [CrossRef]
- Teubner, M.; Fuß, J.; Kühn, K.; Krause, K.; Schmitz-Linneweber, C. The RNA recognition motif protein CP33A is a global ligand of chloroplast mRNAs and is essential for plastid biogenesis and plant development. Plant J. 2017, 89, 472–485. [Google Scholar] [CrossRef]
- Bian, D.; Zhao, X.; Li, C.; Tian, J.; Liu, Q.; Zhou, C.; Tang, B. Molecular cloning and expression analysis of the highly conserved eukaryotic translation initiation factor 5A (eIF) from Antheraea pernyi. Entomol. Res. 2018, 48, 11–17. [Google Scholar] [CrossRef]
- Gallino, J.P.; Ruibal, C.; Casaretto, E.; Fleitas, A.L.; Bonnecarrère, V.; Borsani, O.; Vidal, S. A dehydration-induced eukaryotic translation initiation factor iso4G identified in a slow wilting soybean cultivar enhances abiotic stress tolerance in Arabidopsis. Front. Plant Sci. 2018, 9, 262. [Google Scholar] [CrossRef] [PubMed]
- Kose, S.; Imamoto, N. Nucleocytoplasmic transport under stress conditions and its role in HSP70 chaperone systems. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2014, 1840, 2953–2960. [Google Scholar] [CrossRef] [PubMed]
- Bates, P.W.; Vierstra, R.D. UPL1 and 2, two 405 kDa ubiquitin–protein ligases from Arabidopsis thaliana related to the HECT-domain protein family. Plant J. 1999, 20, 183–195. [Google Scholar] [CrossRef]
- Jahnke, M.; Trowsdale, J.; Kelly, A.P. Ubiquitination of HLA-DO by MARCH family E 3 ligases. Eur. J. Immunol. 2013, 43, 1153–1161. [Google Scholar] [CrossRef]
- Lee, K.; Seo, P.J. The E3 ubiquitin ligase HOS1 is involved in ethylene regulation of leaf expansion in Arabidopsis. Plant Signal. Behav. 2015, 10, e1003755. [Google Scholar] [CrossRef]
- Lee, K.; Seo, P.J. The Arabidopsis E3 ubiquitin ligase HOS1 contributes to auxin biosynthesis in the control of hypocotyl elongation. Plant Growth Regul. 2015, 76, 157–165. [Google Scholar] [CrossRef]
- Kovács, J.; Poór, P.; Kaschani, F.; Chandrasekar, B.; Hong, T.N.; Misas-Villamil, J.C.; Xin, B.T.; Kaiser, M.; Overkleeft, H.S.; Tari, I. Proteasome activity profiling uncovers alteration of catalytic β2 and β5 subunits of the stress-induced proteasome during salinity stress in tomato roots. Front. Plant Sci. 2017, 8, 107. [Google Scholar] [CrossRef]
- Mano, J.i.; Nagata, M.; Okamura, S.; Shiraya, T.; Mitsui, T. Identification of oxidatively modified proteins in salt-stressed Arabidopsis: A carbonyl-targeted proteomics approach. Plant Cell Physiol. 2014, 55, 1233–1244. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, G.; Xu, Z.; Li, J.; Sun, M.; Guo, J.; Ji, W. Organization and regulation of soybean SUMOylation system under abiotic stress conditions. Front. Plant Sci. 2017, 8, 1458. [Google Scholar] [CrossRef]
- Sagor, G.; Chawla, P.; Kim, D.W.; Berberich, T.; Kojima, S.; Niitsu, M.; Kusano, T. The polyamine spermine induces the unfolded protein response via the MAPK cascade in Arabidopsis. Front. Plant Sci. 2015, 6, 687. [Google Scholar] [CrossRef]
- Olby, R. Francis Crick, DNA, and the central dogma. Daedalus 1970, 99, 938–987. [Google Scholar]
- Mimura, S.; Kubota, Y.; Takisawa, H. MCM interference during licensing of DNA replication in Xenopus egg extracts-Possible Role of a C-terminal region of MCM3. Cell Cycle 2018, 17, 492–505. [Google Scholar] [CrossRef]
- Yan, H.; Tammaro, M.; Liao, S. Collision of trapped topoisomerase 2 with transcription and replication: Generation and repair of DNA double-strand breaks with 5′ adducts. Genes 2016, 7, 32. [Google Scholar] [CrossRef]
- Shafiq, S.; Chen, C.; Yang, J.; Cheng, L.; Ma, F.; Widemann, E.; Sun, Q. DNA Topoisomerase 1 prevents R-loop accumulation to modulate auxin-regulated root development in rice. Mol. Plant 2017, 10, 821–833. [Google Scholar] [CrossRef] [PubMed]
- Maslowska, K.H.; Makiela-Dzbenska, K.; Fijalkowska, I.J. The SOS system: A complex and tightly regulated response to DNA damage. Environ. Mol. Mutagen. 2019, 60, 368–384. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, Q. Far upstream element binding protein 1: A commander of transcription, translation and beyond. Oncogene 2013, 32, 2907–2916. [Google Scholar] [CrossRef]
- Franziska, B.; Baserga, S.J. The long unwinding road of RNA helicases. Mol. Cell 2007, 27, 339–352. [Google Scholar] [CrossRef]
- Zhang, X.-M.; Zhao, X.-Q.; Feng, C.-X.; Liu, N.; Feng, H.; Wang, X.-J.; Mu, X.-Q.; Huang, L.-L.; Kang, Z.-S. The cloning and characterization of a DEAD-Box RNA helicase from stress-responsive wheat. Physiol. Mol. Plant Pathol. 2014, 88, 36–42. [Google Scholar] [CrossRef]
- Wang, A.H.; Juan, A.H.; Ko, K.D.; Tsai, P.-F.; Zare, H.; Dell’Orso, S.; Sartorelli, V. The elongation factor Spt6 maintains ESC pluripotency by controlling super-enhancers and counteracting polycomb proteins. Mol. Cell 2017, 68, 398–413.e6. [Google Scholar] [CrossRef]
- Wu, J.; Jiang, Y.; Liang, Y.; Chen, L.; Chen, W.; Cheng, B. Expression of the maize MYB transcription factor ZmMYB3R enhances drought and salt stress tolerance in transgenic plants. Plant Physiol. Biochem. 2019, 137, 179–188. [Google Scholar] [CrossRef]
- Pohlmann, T.; Baumann, S.; Haag, C.; Albrecht, M.; Feldbrügge, M. A FYVE zinc finger domain protein specifically links mRNA transport to endosome trafficking. eLife 2015, 4, e06041. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Yang, Y.; Wang, W.; Guo, G.; Liu, W.; Bi, C. Overexpression of a wheat (Triticum aestivum L.) bZIP transcription factor gene, TabZIP6, decreased the freezing tolerance of transgenic Arabidopsis seedlings by down-regulating the expression of CBFs. Plant Physiol. Biochem. 2018, 124, 100–111. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Xu, Z.; Cao, S.; Chen, K.; Li, S.; Liu, X.; Gao, C.; Zhang, B.; Zhou, Y. An uncanonical CCCH-tandem zinc-finger protein represses secondary wall synthesis and controls mechanical strength in rice. Mol. Plant 2018, 11, 163–174. [Google Scholar] [CrossRef]
- Shao, J.; Huang, K.; Batool, M.; Idrees, F.; Afzal, R.; Haroon, M.; Noushahi, H.A.; Wu, W.; Hu, Q.; Lu, X.; et al. Versatile roles of polyamines in improving abiotic stress tolerance of plants. Front. Plant Sci. 2022, 13, 1003155. [Google Scholar] [CrossRef] [PubMed]
- Shan, X.; Zhou, H.; Sang, T.; Shu, S.; Sun, J.; Guo, S. Effects of exogenous spermidine on carbon and nitrogen metabolism in tomato seedlings under high temperature. J. Am. Soc. Hortic. Sci. 2016, 141, 381–388. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).