Abstract
Salinity is a major constraint limiting the yield of tomatoes. However, grafting strategies may help to overcome the salt toxicity of this important horticultural species if appropriate rootstocks are identified. The present study aimed to test a new rootstock, JUPAFORT1, obtained by crossing the glycophyte Solanum lycopersicum (cv. Poncho Negro) with the halophyte wild-related species Solanum chilense to improve the salinity tolerance of the Chilean tomato landrace Old Limachino Tomato (OLT). Intact OLT plants were exposed to 0, 80, or 160 mM of NaCl for 21 days at the vegetative stage and compared with self-grafted (L/L) and Limachino plants grafted on JUPAFORT1 rootstock (L/R) under a completely randomized design. JUPAFORT1 increased OLT scion vigor in the absence of salt but did not significantly increase fresh weight under stress conditions. However, JUPAFORT1 confers to the scion an anisohydric behavior contrasting with the isohydric behavior of L and L/L plants as indicated by measurements of stomatal conductance; L/R plants were able to maintain their metabolic status despite a slight decrease in the leaf’s relative water content. JUPAFORT1 rootstock also enabled the maintenance of photosynthetic pigment concentrations in the scion in contrast to L and L/L plants, which exhibited a decrease in photosynthetic pigments under stress conditions. L/R plants encountered oxidative stress at the highest stress intensity (160 mM of NaCl) only, while L and L/L plants suffered from oxidative damage at a lower dose (80 mM of NaCl). L/R plants behaved as includer plants and did not sequester Na+ in the root system, in contrast to L and L/L, which behaved as excluder plants retaining Na+ in the root system to avoid its translocation to the shoots. The expression of genes coding for ion transporters (HKT1.1, HKT1.2, LKT1, SKOR, SOS2, and SOS3) in the root system was not modified by salinity in L/R. In contrast, their expression varied in response to salinity in L and L/L. Overall, L/R plants exhibited higher physiological stability than L/L or L plants in response to an increasing NaCl dose and did not require additional energy investment to trigger an adaptative response to salinity. This suggests that the constitutive salinity tolerance of the halophyte S. chilense was maintained in the interspecific rootstock. JUPAFORT1 issued from S. lycopersicum x S. chilense may thus improve salt-stress resilience in OLT tomatoes. Additional studies are required to identify the molecular components involved in the root-to-shoot signaling pathway in this promising material.
1. Introduction
Salinity is one of the main factors limiting plant growth and productivity in cultivated species. Tomato is a glycophyte species considered to be highly salt sensitive. Further, salinity affects almost all aspects of plant physiology and biochemistry, such as plant growth, plant water and oxidative status, mineral nutrition, photosynthesis, and hormonal profile [1,2]. Improving the salinity tolerance of crops thus requires selecting an adequate combination of several physiological traits that influence the plant’s adaptive responses to the environment at all stages of development and ultimately contribute to yield maintenance [1,2]. It is, however, likely that some genes controlling these physiological traits are no longer present in modern commercial cultivars of domesticated glycophytes, which were selected in the past for their high yield potential under non-saline conditions [3,4].
In tomatoes grown under salinity stress, long-term damage has been linked to the low water potential of the external medium hampering water absorption by the plant and the excessive accumulation of Na+ and Cl− within the leaves. Photosynthesis is one of the most sensitive processes affected by saline stress. Indeed, salinity reduced the net photosynthetic rate as a consequence of a decrease in stomatal conductance (gs) and alteration of the intercellular CO2 concentration (Ci) of plants [5]. Moreover, salinity reduces PSII efficiency by altering the photosystem II (PSII) complex, causing primary charge separation in PSII, damage pigment–protein complexes of chloroplast thylakoid membranes, and a decrease in PSII electron transport quantum yield [1,3]. Because salinity severely limits photosynthetic CO2 fixation, the rate of light energy absorption by photosynthetic pigments frequently exceeds the rate of its consumption in the chloroplast. Excess energy can accelerate PSII damage through the generation of reactive oxygen species (ROS) such as superoxide ions (*O2−, hydrogen peroxide (H2O2), and hydroxyl radicals (*OH−) [6]. These highly toxic and reactive compounds can damage DNA, proteins, chlorophyll, and membrane structures [1,2].
Some plants have selected adaptative strategies to cope with high salt levels in soil. Among them, the limitation of Na+ absorption by the roots and translocation to the shoot on the one hand, as well as antioxidative mechanisms in photosynthetic organs on the other hand, are undoubtedly the most efficient strategies [1]. Because they share a similar ionic radius, Na+ absorption by K+ transporters often leads to K+ deficiencies, and the maintenance of K+ versus Na+ selectivity is paramount for salt-exposed plants [7,8].
Membrane transporters for K+ flux identified in plants are generally classified into three channels and three transporter families based on phylogenetic analyses. The first group includes shaker-type K+ channels and two-pore K+ channels, such as non-selective cation channels (NSCCs) [9]. The second group consists of the KUP/HAK/KT (potassium uptake permease, high-affinity K+ transporters, and K+ transporters, respectively) family of transporters, and the third group consists of the HKT (high-affinity K+ transport) family of transporters and K+/H+ antiporters [10]. The SOS (Salt Overly Sensitive) pathway is also associated with two important processes that lead to salinity tolerance. One is a root cell-based mechanism that relies on the flux of Na+ back into the soil solution. The second is the control of Na+ unloading in the xylem, which has been confirmed in Arabidopsis, tomato, rice, and barley [10,11]. Three distinct proteins are involved in the SOS pathway: SOS1 is a plasma membrane Na+/H+ antiporter, SOS2 is a serine/threonine protein kinase, and SOS3 is a calcium-binding protein regulating SOS1 activity [10,12,13].
As far as tolerance to secondary oxidative stress is concerned, plants rely on a large set of endogenous non-enzymatic and enzymatic antioxidants. Ascorbate and glutathione assume key functions in the Asada cycle and contribute to limiting oxidative damage [14]. Ascorbate peroxidase (APX; EC 1.11.1.11), superoxide dismutase (SOD; EC 1.15.1.1), catalase (CAT; EC 1.11.1.6), glutathione reductase (GR; EC 1.6.4.2), glutathione-S-transferase (GST; 2.5.1.18), dehydroascorbate reductase (DHAR; EC 1.8.5.1), and monodehydroascorbate reductase (MDHAR) are important enzymes directly involved in ROS detoxification [1,15,16].
Most studies on salt’s impact on cultivated tomatoes deal with modern commercial cultivars. However, some neglected local varieties now receive considerable attention concerning the high quality of produced fruits from both organoleptic and nutritional points of view. This is especially the case of the promising “OLT Limachino tomato” (OLT) [17], which has been recently rediscovered in the Limache basin of Central Chile and exhibits attractive sensory and health properties. The plant is well adapted to the local environment, does not require high amounts of pesticides or fertilizers, and has great potential for sustainable agriculture. The consumers’ demand for this high-quality product is ever-increasing. However, OLT is unfortunately sensitive to NaCl [18], and there is therefore considerable interest in increasing its mean level of salinity tolerance.
Since roots are the first organs in contact with salinity, using adapted rootstock is a convenient strategy to limit salt accumulation and improve Na+ tolerance in tomatoes [6,19,20]. He et al. [6] stated that alleviating salinity-induced growth inhibition in tomato plants grafted on a Chinese commercial rootstock would be associated with improved scion antioxidant capacity and a concomitant improvement in photosynthesis. On the other hand, some authors investigated the mechanisms involved in the effects of grafting on yield in tomatoes and reported that the salinity tolerance conferred by some grafting combinations was related to the ability of the rootstock to reduce the rates of salt ion uptake and transport to the shoot [19,20]. The selection of the rootstock is nevertheless a challenging task and requires a combination of numerous root-related characteristics. Local salt-tolerant varieties and wild-related halophyte plant species might be used for this purpose. The tomato cv. Poncho Negro was ancestrally developed and established in the extreme conditions of Northern Chile (Arica and Parinacota region). It is tolerant to mineral toxicities (salinity and boron) [21,22] and biotic stress, specifically to Pseudomona syrengae pv tomato attack [23]. On the other hand, numerous data are available for some close-related wild halophyte species [24,25,26,27] and interspecific crosses with cultivated S. lycopersicum [19,28]. Introgression lines from numerous wild species have been successfully performed. Paradoxically, only a few data are available for the wild halophyte Solanum chilense. This species is native to the Atacama Desert and can grow at salinity levels similar to those found in seawater. The fascinating ability of S. chilense to adapt and cope with saline environments has been underexploited, mainly due to the self-incompatibility inducing cross-pollination of this wild species, the small size of produced fruits, and other traits that are undesirable to incorporate into breeding programs [24,29,30,31,32,33,34,35,36]. These authors, however, also demonstrated that the halophyte S. chilense behaves as an includer and does not restrict Na+ translocation from the roots to the shoots, while the salt-resistant glycophyte cultivars of cultivated S. lycopersicum behave as excluder, sequestering Na+ in the roots and thus avoiding its translocation to the shoot. Previous studies [24,29,30,31,32,33,34,35,36] have confirmed that the wild relative tomato S. chilense is tolerant to salinity and can cope with high endogenous Na+ levels. In this case, hormonal signals issued from the roots may trigger efficient salt-tolerance mechanisms in the shoots [37,38]. However, to the best of our knowledge, no data are available on the use of interspecific hybrids issued from Solanum lycopersicum x Solanum chilense for rootstock selection, especially considering that this cross is difficult to conduct considering the self-incompatible nature of the wild parent.
Interspecific crosses successfully performed between Solanum lycopersicum cv. Poncho Negro (accession SLY0001) and wild halophyte Solanum chilense (donor accession LA 4170) allowed us to recently select, for the first time to the best of our knowledge, a promising F1 rootstock material (JUPAFORT1; INIA), which was cloned in vitro [39]. The purpose of the present study was to assess the Na+ distribution in the OLT/JUPAFOR scion/rootstock combination in relation to mineral status, the management of oxidative stress and photosynthetically related parameters in the shoots, and the expression of genes coding for the Na+/K+ transporter in the roots.
2. Material and Methods
The experiment was conducted in greenhouses at the INIA-La Cruz Regional Center located in La Cruz, Region of Valparaíso (32° 59′ LS; 71° 10′ LW, 105 m.a.s.l.). The trials were conducted during the vegetative stage of plant development in the spring–summer season of 2021 and 2022.
2.1. Plant Material and Growing Conditions
The tomato genotype (Solanum lycopersicum Mill.) cv. known, in English, as Old Limachino tomato (OLT, accession SLY074) [40,41], was used as a scion, while the rootstock (R) used is a F1 hybrid clonal material (INIA JUPAFORT1). This rootstock material was previously created at the INIA’ premises by crossing a cultivated tomato Solanum lycopersicum (salinity-resistant tomato genotype cv. called Poncho Negro, accession SLY0001) with a wild halophytic tomato Solanum chilense (Solanum chilense Dun., donor accession LA 4170). It was propagated in vitro through a protocol developed at INIA that optimizes the micropropagation process for grafting [39]. One F1 plant (JUPAFORT1) was selected and clonally propagated in vitro so that all the rootstocks were genetically identical. Clonal rootstock seedlings (R) maintained in vitro were then acclimatized in greenhouse conditions to perform the grafting process with the OLT plants at the three true leaves stage. OLT seeds were sown and seedlings were allowed to grow inside a growth chamber (Plant Growth Incubator 487 Liter, Laboratory Instruments-MRC) in four plumavit containers filled with a perlitevermiculite mixture (1: 3 v/v), regularly moistened for three weeks with half-strength modified Hoagland solution containing 5 mM of KNO3, 1 mM of NH4H2PO4, 0.5 mM of MgSO4, and 5.5 mM of Ca(NO3)2; 25 μM of KCl, 10 μM of H3BO3, 1 μM of MnSO4, 1 μM of ZnSO4, 0.25 μM of CuSO4, and 10 μM of Na2MoO4; and 50 mg L−1 of Fe-EDDHA [40].
In this study, three groups of OLT plants were considered: (1) ungrafted plants (L), (2) self-grafted (L/L), and (3) plants grafted with the clonal rootstock (L/R). When the OLT plants were used for grafting, the scion contained three true leaves. The grafted plants (L/L and L/R) were obtained using a manual splice-grafting procedure and were secured with a silicone clip. All plants were then transplanted into 1.5 L pots when they had three to four fully unfolded true leaves. Seven days after stress application (scions exhibiting 4–5 true leaves), a nutrient solution without NaCl was used as a control treatment (EC = 3 dS m−1), while solutions containing 80 (EC = 9 dS m−1) and 160 (EC = 14 dS m−1) mM of NaCl were applied every 3 days until the end of the experiment, 21 days after treatment (DAT). The first experimental factor was the rootstock (R), while the second factor was the salinity level. Each treatment included six replicates (six plants) arranged in a completely randomized design. The natural light intensity at the top of the canopy averaged 500 μmol m−2 s−1 (photosynthetic photon flux density, PPFD). The air temperature ranged between 25 °C and 32 °C during the day and approximately 18 °C at night. The relative humidity was maintained at 85 ± 5% at night and 45 ± 5% during the day. The environmental conditions were permanently recorded using a Hobo MX2301A temperature/RH Data Logger; data were transferred to the HOBO connect Monitoring Application.
2.2. Measurements
2.2.1. Plant Growth and Water Status
The fresh (FW) and dry weight (DW) of leaves, stems, and roots were estimated at 21 DAT (days after stress imposition) on six plants per treatment. The dry weight was considered after 48 h in an oven at 80 °C. To evaluate vegetative growth, the plant height (length of the dominant axis), basal diameter, and number of leaves were also measured at 1, 7, 14, and 21 DAT. The number of leaves was counted by recording all leaves on the main stem with blades longer than 1 cm.
The leaf’s water potential was measured before sunrise (Ψwp) and at midday (Ψwn) using a Scholander-type pressure chamber (PMS Instrument Co., Orlando, FL, USA) [29] on the sixth fully expanded leaf (counted from the base). The ylem pressure potential (Ψwx) was quantified using the same chamber after conditioning leaves for two hours in dark bags to eliminate leaf transpiration [42]. The relative water content (RWC) evaluated before sunrise (RWCp) and at midday (RWCn) was determined according to Martínez et al. [42] and expressed in %:
Relative Water Content at predawn ()
Relative Water Content at noon ()
where and are the fresh weight at predawn and noom, respectively, and are the turgid weight at predawn and noom, respectively measured after 24 h of saturation (when leaf weight reached a plateau) on deionized water at 4 °C in the dark, and DW is the dry weight determined after 48 h in an oven at 80 °C.
The leaf’s water content on dry matter before dawn (WCDWp, g/g) and at midday (WCDWn, g/g) were determined using the method developed by Martínez et al. [29]. These measurements were performed on six plants per plant group and treatment at 21 DAT.
2.2.2. Leaf Stomatal Conductance, Chlorophyll Fluorescence, and Pigment Concentrations
The leaf stomatal conductance (gs) was measured at midday on the sixth fully expanded leaf using a porometer (Decagon devices, SC1, NE Hopkins Ct., Pullman, WA, USA). The chlorophyll content index was estimated using a portable chlorophyll meter (Chlorophyll Content Meter Opti-Sciences Model CCM-200 Plus GPS) on the same leaf. The basal (F0) and maximal fluorescence (Fm) and maximum PSII yield (Fv/Fm) of chlorophyll fluorescence were determined as previously detailed [43] using a chlorophyll a fluorescence monitoring system (FMS2, Hansatech, Norfolk, UK) on the same leaf. These measurements were performed on six plants per plant group and treatment at 6, 13, and 20 DAT.
The chlorophyll a and b and carotenoid concentrations were determined using extracts of 100 mg of pre-cooled mortar-ground fresh leaf material in 8 mL of 80% v/v cold acetone. The absorbance of the extracts was read at 663.2, 646.8, and 470 nm using a Shimadzu UV-1800 spectrophotometer (Shimadzu, Kyoto, Japan), and the concentrations of chlorophyll a, chlorophyll b, and carotenoids were calculated according to Lichtenthaler [44]. These measurements were performed on the sixth fully expanded leaf of six plants per plant group and treatment at 21 DAT.
2.2.3. Oxidative Damage and Total Antioxidant Capacity
The malondialdehyde (MDA) content was measured according to Heath and Paker [45] using a Shimadzu UV-1800 Spectrophotometer (Shimadzu, Kyoto, Japan)–Metler Toledo, Zaventem, Belgium. The antioxidant capacity at the leaf level was measured using two methods: (i) ferric-reducing antioxidant power (FRAP) and (ii) a DPPH (2,2-diphenyl 1-picrylhydrazyle) assay. The FRAP was measured based on the methodology of Benzie and Strain [46] with slight modifications [40,41]. The DPPH was determined using the colorimetric method based on work by Brand-Williams et al. [47]. These measurements were performed on the sixth fully expanded leaf of six plants per plant group and treatment at 21 DAT.
2.2.4. Non-Enzymatic Antioxidant Compounds
Non-enzymatic antioxidant compound concentrations were measured on the sixth fully expanded leaf of six plants per plant group and treatment at 21 DAT.
Total polyphenols were measured spectrophotometrically using the adapted colorimetric assay with a Folin–Ciocalteu reagent and reading absorbance at 765 nm [48] in reference to a standard curve established with gallic acid.
Ascorbate quantification was performed according to Kampfenkel [49]. A 0.25 g leaf tissue sample was homogenized with 4 mL of trichloroacetic acid and left for 15 min on ice. The samples were then centrifuged for 5 min at 10,000× g at 4 °C, and the supernatant was recovered. To determine the total ascorbate, oxidized ascorbate (DHA) was reduced to AsA with DL-ditriotreitol (DTT), and excess DTT was removed with NEM. For the determination of reduced ascorbate, 0.2 mL of the supernatant, 0.4 mL of 0.2 M phosphate buffer with a pH of 7.4, 0.4 mL of Milli-Q water, 1 mL of 10% w/v TCA, 0.8 mL of 85% H3PO4, 0.8 mL of 4% dipyridyl, and 0.4 mL of 3% FeCl3 were used. The solutions to determine the reduced ascorbate and total ascorbate were incubated for 1 h at 37 °C, and the absorbance was read in a spectrophotometer at a wavelength of 525 nm. The determination of oxidized ascorbate (DHA) was estimated by subtracting the total minus reduced ascorbate values.
Oxidized Ascorbate (DHA) = total ascorbate (ASAT) − Reduced Ascorbate (AsA)
To determine reduced glutathione (GSH) and total glutathione (total GSH), 0.2 g of frozen leaf tissue was extracted and derivatized using ortho-phthalaldehyde (OPA) [50]. The extraction was performed as previously detailed [51]. The derivatives were separated on a reverse-phase HPLC column with a fluorescence-detected acetonitrile gradient system. A volume of 5 µL of the sample was injected into a Shimadzu HPLC system coupled with a Piramide C18 column (125 × 4.6 mm internal diameter; 5 μm particle size) (Macherey-Nagel, Düren, Germany). The derivatives were eluted in acetonitrile gradient in 50 mM of sodium acetate buffer (pH 6.2) at 30 °C at a flow rate of 0.7 mL min−1. Detection was performed with a Shimadzu RF-20 fluorescence detector at an emission wavelength of 420 nm and an excitation wavelength of 340 nm. GSH was quantified using a calibration curve with standard solutions from 0.0625 to 50 µM. Oxidized glutathione (GSSG) was determined by subtracting the GSH from the Total GSH content.
2.2.5. Antioxidant Enzyme Activity
Approximately 0.5 g of fresh leaf tissue (from the sixth fully expanded leaf of six plants per plant group and treatment at 21 DAT) was ground in liquid nitrogen and then homogenized with 4 mL of 50 mM phosphate buffer containing 0.1 mM EDTA, 1% w/v polyvinylpyrrolidone (PVP), 1 mM of ascorbate, 5% w/v glycerol, and 1 M NaCl. Subsequently, the samples were centrifuged at 10,000× g for 10 min at 4 °C. After the supernatant was recovered, the samples were stored at −80 °C until the determination of enzyme activities and protein quantification. The activity of glutathione reductase (GR; EC 1.6.4.2) was quantified by NADPH consumption (ε = 6.22, mM−1 cm−1) at 340 nm for 5 min [52]. The ascorbate peroxidase activity (APX; EC 1.11.1.11) was determined by the ascorbate consumption at 265 nm (ε = 13.7 mM−1 cm−1) for 30 s [53]. Enzyme activity was expressed as mmol AsA min−1 mg−1 protein. Catalase activity (EC1.11.1.6) was determined by the consumption of H2O2 at 240 nm for 5 min [53]. Dehydroascorbate reductase activity (DHAR; EC 1.8.5.1) was measured at 265 nm for 3 min following the absorbance change resulting from ascorbic acid formation [53]. Glutathion-S-transferase activity (EC 2.5.1.18) was measured by the increase in oxidized glutathione (ε = 9.6, mM−1 cm−1) via the absorbance change at 340 nm for 5 min [53]. Monodehydroascorbate reductase (MDHAR; EC 1.6.5.4) activity was measured using the method developed by Miyake and Asada [54]. The total superoxide dismutase (SOD; EC 1.15.1.1) activity was measured according to Beyer and Fridovitch [55], modified by Yu et al. [56]. Each enzyme activity was analyzed in triplicate. Protein quantification was performed spectrophotometrically at 595 nm according to the Bradford method [57] with Bio-Rad (Hercules, CA, USA) assay protein BIORAD using BSA for calibration.
2.2.6. Ion Concentration
For Na+ and K+ quantification, tissues from six plants (root, stem, and leaf) per plant group and treatment at 21 DAT were considered. For each sample, c.a. 50 mg of dry matter was digested in 4 mL of 0.5% nitric acid at 80 °C. After complete evaporation, residues were dissolved with HNO3 (68%) + HClcc (1:3 v/v) and incubated under gentle agitation of 100 rpm or 48 h. The solution was then filtered with Whatman 70 mm Grade 1, and the filtrate was used to determine the Na+ and K+ concentrations via flame emission with an atomic absorption spectrometer (Shimadzu AA-680 atomic absorption spectrometer (Shimadzu Ltd., Kyoto, Japan)).
2.2.7. Differential Gene Expression in Roots
The expression of seven genes involved in Na+ and K+ transport mechanisms associated with tomato salinity tolerance was analyzed in the root system by means of qRT-PCR in three plants per plant type and treatment. For this purpose, specific primers suitable for qPCR were designed for the following genes: [1] putative Na+ transporters HKT1.1 (HKT1.1) and [2] HKT1.2 (HKT1.2), [3] potassium channel SKOR-like (SKOR), [4] potassium channel (LKT1), [5] plasmalemma Na+/H+ antiporter (SOS1), [6] calcineurin B-like interacting protein kinase (SOS2), and [7] calcium sensor calcineurin B-like (SOS3) (Supplementary Table S1). Since the clonal rootstock used in this study was an interspecific hybrid between S. lycopersicum and S. chilense, highly transferable primers were designed between the two species. The efficiency of amplifying a single amplicon between 64 and 199 bp was tested on a root transcriptome of the S. lycopersicum x S. chilense hybrid using in silico PCR. Roots from treated and control plants were collected 21 days after initiation of the salt treatment (21 DAT), quickly washed, and immediately frozen in liquid nitrogen and stored at −80 °C. The total RNA from each sample was then obtained using the RNeasy Plant Mini Kit from Qiagen [58]. The qPCR assay was performed with the Gotaq@Probe qPCR Master Mix Kit using an Agilent Real Time PCR kit (AriaMX Base Unit Agilent Technologies). The genes coding for elongation factor 1-alpha (EF1aF) and Actin (actin) were used as references. The method described by Pfaffl [59] was used to perform all relative expression calculations. Samples were run in triplicate, including three inserts as negative. Data are presented as the mean of the three replicates ± standard deviation.
2.3. Statistical Analysis
Physiological, biochemical, gene expression, and biomass data were analyzed using a two-way analysis of variance (ANOVA), and the multiple range test was used for mean comparisons at a significance level of p < 0.05. The statistical model was defined based on fixed effects and hierarchical ranking criteria. The grafting type and salinity concentration (NaCl concentration) were considered as the main effects, as well as their interactions. When the ANOVA was significant at p ≤ 0.05, Duncan’s multiple range test was used to compare the means of each variable under study. All analyses were performed in RStudio version 3.0.2 (R Development Core Team, 2012; available online at http://www.R-project.org; accessed on 2 October 2022; the software was obtained from INIA, Chile). The plotting program was generated using GraphPad Prism (version 8.0 GraphPad software, San Diego, CA, USA).
3. Results
3.1. Plant Growth
In the absence of NaCl, the leaf FW was higher in L/R plants compared to L and L/L plants (Figure 1A). In response to salinity, it significantly decreased only in L/R plants in response to 160 mM. Leaf DW exhibited the lowest value in L/L and remained unaffected by salinity. Similarly, the stem FW (Figure 1C) was higher in L and L/R than in L/L in control conditions. Salinity had no significant effect on the stem FW or stem DW (Figure 1D), whatever the considered grafting type. The root FW (Figure 1E) of L/L exhibited a low value compared to other grafting types in the absence of stress. The salinity increased the root FW in self-grafted plants L/L, but did not impact the root DW, which remained similar in all treatments.
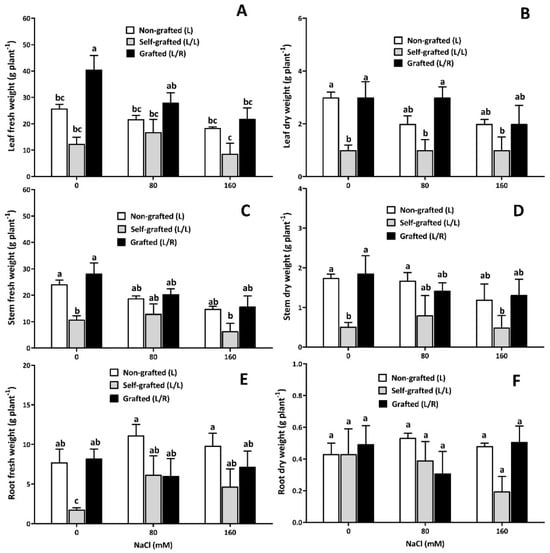
Figure 1.
Plant growth-related properties as a function of NaCl dose. Fresh and dry weights of leaves (A,B), stems (C,D), and roots (E,F) (g.plant−1) for plants corresponding to treatments L (ungrafted Old Limachino tomato), L/L (self-grafted Old Limachino tomato), and L/R (grafted Old Limachino tomato on INIA JUPAFORT1 rootstock) grown under three salinity treatments: 0 mM of NaCl; 80 mM of NaCl, and 160 mM of NaCl for 21 days (DAT). Each value is the mean of six replicates, and vertical bars are S.E. Lowercase letters represent differences (p ≤ 0.05 Duncan’s test) between treatments and grafting types.
The plant height was significantly reduced in L plants 7 days with 160 mM of NaCl and after 21 days with 80 mM of NaCl (Figure 2A). L/L were smaller than L plants (Figure 2B), and 80 mM of NaCl unexpectedly increased the plant height, while 160 mM of NaCl decreased it compared to 0 mM of NaCl. The plant height was only slightly affected by salinity in L/R plants (Figure 2C). No effect of grafting type or salinity was observed regarding the stem diameter (Figure 2D–F). The number of leaves in L plants was reduced by salinity after 14 days (Figure 2G). In L/L, 80 mM of NaCl increased the number of leaves starting on day 7 (Figure 2H), while plants exposed to 160 mM of NaCl produced the lowest number of leaves. The leaf production was not affected by NaCl in L/R plants, except for a slight decrease in the number of leaves recorded for L/R plants exposed to 160 mM on day 7 and day 21 (Figure 2I).
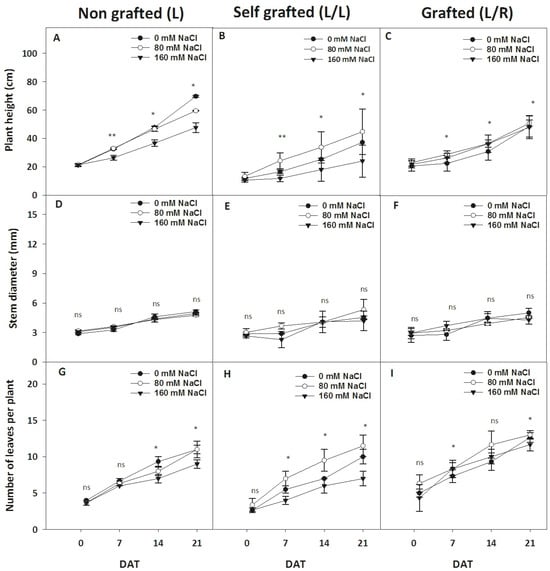
Figure 2.
The evolution of plant morphological parameters during exposure to different NaCl doses. The plant height (cm; A–C), basal stem diameter (mm; D–F), and number of leaves per plant (G–I) for plants corresponding to treatment L (ungrafted Old Limachino tomato), L/L (self-grafted Old Limachino tomato), and L/R (grafted Old Limachino tomato on INIA JUPAFORT1 rootstock) grown under three salinity treatments: 0 mM of NaCl; 80 mM of NaCl, and 160 mM of NaCl for 1, 7, 14, and 21 days after beginning treatment (DAT). Each value is the mean of six replicates, and vertical bars are S.E. Multifactorial ANOVA test: (*) p ≤ 0.05, and p ≤ 0.01 (**), by date; ns: non significant.
3.2. Plant Water Status and Photosynthesis-Related Parameters
The plant water status-related parameters are presented in Table 1. Ψwp and Ψwx remained unaffected by salinity in the three plant groups. ΨWn increased in response to 160 mM of NaCl in L/L and L/R. ΨWn was globally higher in L/R than in L/L plants, regardless of the salt treatment. RWCp decreased in response to 160 mM of NaCl in the three plant groups, and self-grafted L/L was the only combination affected by 80 mM of NaCl. RWCn decreased in response to 80 and 160 mM of NaCl in L plants, while it decreased only at the highest dose in L/L and L/R plants. WCDWp and WCDWn remained unaffected by salinity and were similar for the three plant groups.

Table 1.
The leaf water potential at predawn (Ψwp, MPa), the leaf water potential at noon (ΨWn, MPa), the xylem pressure potential (ΨWx, MPa), the leaf relative water content at predawn (RWCp, %), the leaf relative water content at noon (RWCn, %), the leaf water content on dry matter at predawn (WCDWp, g/g), and the leaf water content on dry matter at noon (WCDWn, g/g) in non-grafted (L), self-grafted (L/L), and grafted on INIA JUPAFOR1 rootstock (L/R) in shoots of Old Limachino tomato plants grown under 0, 80, or 160 mM of NaCl for 21 days. Each value is the mean of six replicates ± S.E. Lowercase letters represent differences (p ≤ 0.05 Duncan’s test) between treatments and grafting types.
The stomatal conductance (gs) and chlorophyll index are shown in Figure 3. In L plants, gs increased up to 14 DAT and thereafter decreased up to 21 DAT at 80 mM and 0 mM of NaCl, while such a pattern was less pronounced at 160 mM. In contrast, gs decreased at 14 DAT in L/L plants and then increased during the following days up to 21 DAT at 0 and 160 mM of NaCl. At 80 mM of NaCl, gs remained constant until 7 DAT, increased between 7 and 14 DAT, and decreased between 14 and 21 DAT (Figure 3B). For the L/R treatment, gs remained constant regardless of the NaCl dose until 14 DAT, but a significant increase was recorded at 21 DAT for plants exposed to 160 mM of NaCl (Figure 3C). Figure 3D–F show that the chlorophyll index of L and L/R plants increased over time. It was also observed that the chlorophyll index increased progressively starting at 14 DAT for 80 mM of NaCl for L and L/L plants. In the case of L/R plants, a significant increase in the presence of 160 mM of NaCl was recorded at 21 DAT.
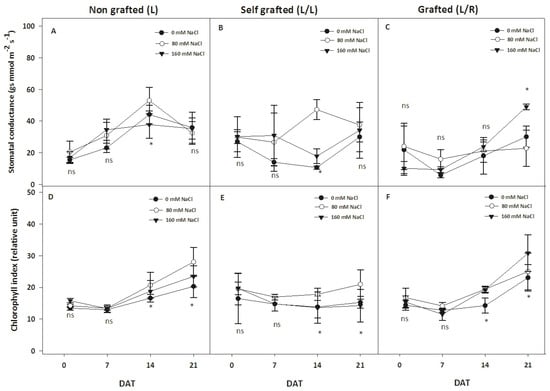
Figure 3.
The evolution of the stomatal conductance and chlorophyll content index during exposure to different NaCl doses. Stomatal conductance (gs, mmol m−2 s−1 A–C) and chlorophyll index (%; D–F) for plants corresponding to treatment L (ungrafted Old Limachino tomato), L/L (self-grafted Old Limachino tomato), and L/R (grafted Old Limachino tomato on INIA JUPAFORT1 rootstock) grown under three salinity treatments: 0 mM of NaCl; 80 mM of NaCl, and 160 mM of NaCl at 1, 7, 14, and 21 days after beginning treatment (DAT). Each value is the mean of six replicates, and vertical bars are S.E. Multifactorial ANOVA test: (*) p ≤ 0.05, by date. ns: non significant.
All fluorescence-related parameters showed high stability and remained unaffected by the type of plants or salinity level, except at 20 DAT, where a reduction in Fm and Fv/Fm was observed, especially in L/L plants at 160 mM of NaCl (Figure 4E). In control conditions, the Chl a content was the highest in L/R plants (Figure 5). It slightly increased in response to 80 mM of NaCl in L/L plants and in response to 160 mM of NaCl in L plants (Figure 5A). The three plant groups had similar Chl b content in control conditions (Figure 5B) and 80 mM of NaCl, but it decreased in response to 160 mM of NaCl in L/L only. The Chl a/b ratio was the lowest in L/L plants in the control treatment, and it increased for all three plant groups at 80 and 160 mM of NaCl (Figure 5C). The carotenoid content was higher in L/L plants compared to L/R plants under control conditions. Salinity increased the carotenoid content in all plant groups at 80 mM of NaCl (Figure 5D). It then remained constant at 160 mM of NaCl, except for the carotenoid content of L/L plants, which slightly decreased at 160 mM of NaCl compared to 80 mM of NaCl.
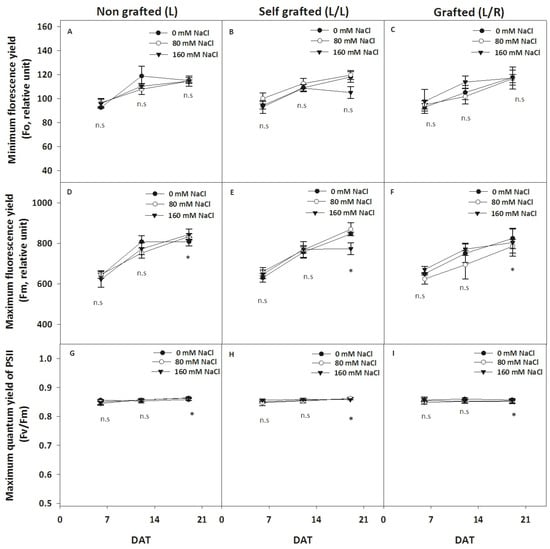
Figure 4.
The evolution of chlorophyll fluorescence-related parameters during exposure to different NaCl doses. The basal (F0, relative unit; A–C) and maximum (Fm, relative unit; D–F) fluorescence and maximum PSII yield (Fv/Fm, G–I) in leaves for plants corresponding to treatment L (ungrafted Old Limachino tomato), L/L (self-grafted Old Limachino tomato), and L/R (grafted Old Limachino tomato on INIA JUPAFORT1 rootstock) grown under three salinity treatments: 0 mM of NaCl; 80 mM of NaCl, and 160 mM of NaCl at 6, 13, and 20 days after the beginning of the treatment (DAT). Each value is the mean of six replicates, and vertical bars are S.E. Multifactorial ANOVA test: (*) p ≤ 0.05, n.s, non significant.
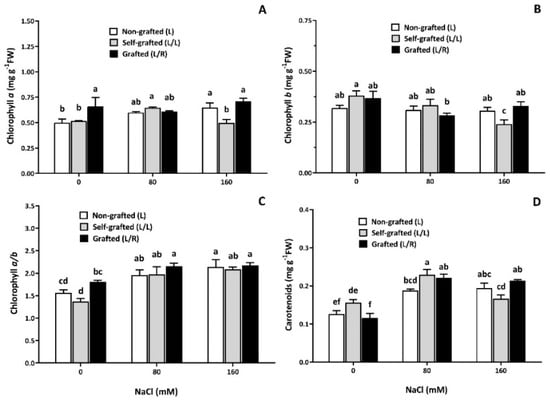
Figure 5.
The pigment concentration as a function of the NaCl dose. Chlorophyll (Chl) a (A) and b (B) contents, chlorophyll a/b ratio (C), and carotenoids (D) for plants corresponding to treatment L (ungrafted Old Limachino tomato), L/L (self-grafted Old Limachino tomato), and L/R (grafted Old Limachino tomato on INIA JUPAFORT1 rootstock) grown for 21 days under three salinity treatments: 0 mM of NaCl; 80 mM of NaCl, and 160 mM of NaCl. Each value is the mean of six replicates, and vertical bars are S.E. Lowercase letters represent differences (p ≤ 0.05 Duncan’s test) between treatments and grafting types.
Under control conditions, the total protein content was higher in L/L and L/R than in L plants (Figure 6A). Under salinity, this variable remained unaffected in L/R plants, while the total protein content increased in both L and L/L plants, especially at the highest NaCl concentration (160 mM) (Figure 6A).
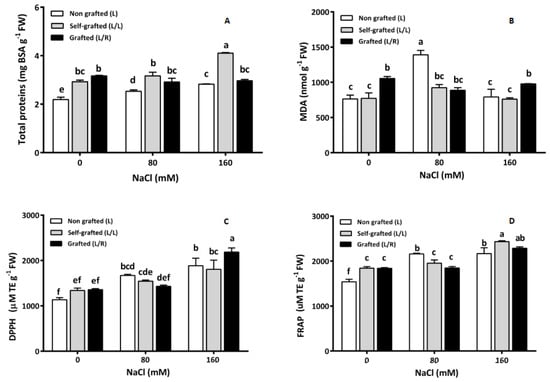
Figure 6.
The total proteins (mg g−1 FW; (A)), malondialdehyde (MDA; nmol g−1 FW, (B)), and antioxidant capacity: DPPH (2,2-diphenyl 1-picrylhydrazyle, (C)) and FRAP (ferric-reducing antioxidant power (D)) (mM Trolox equivalent TE.g−1 FW) in leaves for plants corresponding to treatment L (ungrafted Old Limachino tomato), L/L (self-grafted Old Limachino tomato), and L/R (grafted Old Limachino tomato on INIA JUPAFORT1 rootstock) grown for 21 days under three salinity treatments: 0 mM of NaCl; 80 mM of NaCl, and 160 mM of NaCl. Each value is the mean of six replicates, and vertical bars are S.E. Lowercase letters represent differences (p ≤ 0.05 Duncan’s test) between treatments and grafting types.
3.3. Management of Oxidative Stress
In the absence of NaCl, the MDA (Figure 6B) content was the highest in L/R plants and remained constant in the presence of salt. MDA also remained unaffected by salt in L/L, while it exhibited a maximal value in response to 80 mM of NaCl in L. The antioxidant capacity, assessed by DPPH and FRAP, is shown in Figure 6C and Figure 6D, respectively. Salinity increased DPPH in all plant groups, with a more pronounced effect at 160 mM of NaCl. At this dose, the highest value was recorded for L/R plants, while activity did not differ among plant groups at 0 and 80 mM of NaCl. L plants showed the lowest FRAP value in the control treatment; 80 mM of NaCl increased FRAP values in L plants only, while the highest salt concentration (160 mM of NaCl) also increased it in L/L and L/R.
The polyphenol concentration did not vary with the plant groups at 0 and 80 mM of NaCl. When the plants were exposed to salinity stress, L and L/L plants progressively increased their polyphenol concentrations. They exhibited the highest value at 160 mM of NaCl, whereas this concentration remained stable in L/R plants (Figure 7).
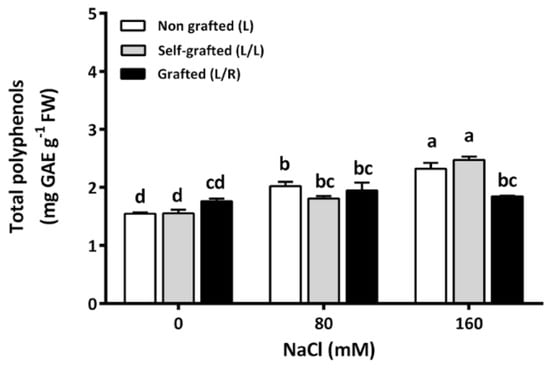
Figure 7.
The leaf total polyphenols concentration (mg GAE (gallic acid equivalent) g−1 FW) for plants corresponding to treatment L (ungrafted Old Limachino tomato), L/L (self-grafted Old Limachino tomato), and L/R (grafted Old Limachino tomato on INIA JUPAFORT1 rootstock) grown for 21 days under three salinity treatments: 0 of NaCl; 80 of NaCl, and 160 of NaCl). Each value is the mean of six replicates, and vertical bars are S.E. Lowercase letters represent differences (p ≤ 0.05 Duncan’s test) between treatments and grafting types.
The ascorbic acid (AsA) concentration (Figure 8A) was higher in the non-saline treatment than in the saline treatments. In control conditions, L/R had a lower AsA concentration than L plants, while no differences among plant groups were recorded under salinity. L plants showed the lowest DHA (Figure 8B) concentration compared to the two other plant groups under control conditions. At 80 of NaCl, no differences were observed among plant groups. At high salinity (160 of NaCl), L/R exhibited higher values than L plants (Figure 8B). Moreover, DHA increased between 80 and 160 of NaCl in L/R plants, while it remained constant in L and L/L plants (Figure 8B). Figure 8C shows that L plants under non-saline conditions had the highest AsA/DHA ratio, which strongly decreased in response to 80 and 160 mM of NaCl. For the other plant groups, the AsA/DHA ratio remained constant, whatever the salinity level. For all plant groups, the total ascorbic acid content decreased between 0 and 80 of NaCl and then remained stable up to 160 mM of NaCl (Figure 8D).
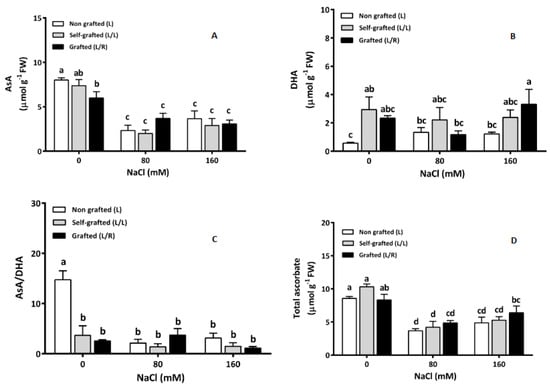
Figure 8.
Reduced ascorbic acid (AsA; μmol g−1 FW; (A)) and dehydroascorbate (DHA; μmol g−1 FW; (B)), AsA/DHA (C) and total ascorbate (μmol g−1 FW; (D)) in leaves for plants corresponding to treatment L (ungrafted Old Limachino tomato), L/L (self-grafted Old Limachino tomato), and L/R (grafted Old Limachino tomato on INIA JUPAFORT1 rootstock) grown for 21 days under three salinity treatments: 0 mM of NaCl; 80 of NaCl, and 160 of NaCl. Each value is the mean of six replicates, and vertical bars are S.E. Lowercase letters represent differences (p ≤ 0.05 Duncan’s test) between treatments and grafting types.
The reduced glutathione (GSH) concentration was similar for all plant groups under control conditions (Figure 9A). The GSH increased in L/R plants exposed to 80 of NaCl, while it remained constant in L and L/L. At high salinity (160 mM), GSH increased in all plants and was higher in L/L and L/R than in L plants. The concentration of oxidized glutathione (GSSG) (Figure 9B) was lower in L/L plants than in other plants in the absence of NaCl. In response to salinity, GSSG increased in L/L and L/R plants and exhibited the highest value in L/R plants exposed to 160 of NaCl. The highest GSH/GSSG ratio (Figure 9C) was recorded in L/L plants in the absence of salt. Under salinity, an increase in this ratio was observed in L plants at 160 of NaCl. On the contrary, a significant reduction in GSH/GSSG was observed in L/L plants at 80 and 160 mM of NaCl. The GSH/GSSG ratio remained constant in L/R plants, regardless of the salinity level. At 0 of NaCl, the total glutathione concentration (Figure 9D) was similar in all plant groups. Under salinity, the total GSH increased in L/R plants at 80 mM of NaCl and 160 mM of NaCl. In L and L/L plants, the total GSH increased at 160 mM only (Figure 9D).
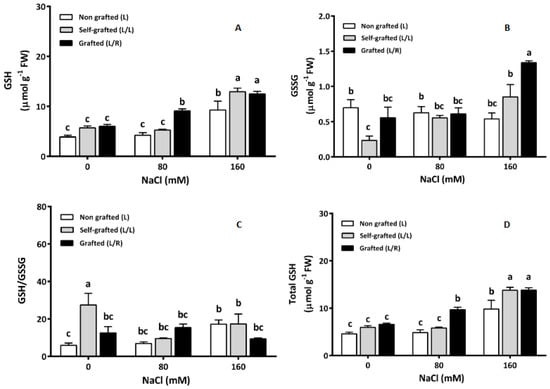
Figure 9.
Reduced glutathione (GSH; μmol g−1 FW; (A)) and oxidized glutathione (GSSG; μmol g−1 FW; (B)) concentrations, GSH/GSSG (C), and total GSH concentrations (μmol g−1 FW; (D)) in the leaves of plants corresponding to treatment L (ungrafted Old Limachino tomato), L/L (self-grafted Old Limachino tomato), and L/R (grafted Old Limachino tomato on INIA JUPAFORT1 rootstock) grown for 21 days under three salinity treatments: 0 of NaCl; 80 of NaCl, and 160 of NaCl. Each value is the mean of six replicates, and vertical bars are S.E. Lowercase letters represent differences (p ≤ 0.05 Duncan’s test) between treatments and grafting types.
The GR activity (Figure 10A) was higher in L than in L/L and L/R under control and 80 mM NaCl conditions. All plant groups exhibited similar GR activities at 160 of NaCl. L/R plants were the only ones for which the GR enzyme activity was not reduced by salinity. The APX activity (Figure 10B) under control conditions was higher in L/L and L/R plants, and it slightly increased at 80 of NaCl for L plants only. APX activities still increased at 160 of NaCl for L and L/R plants, while it remained constant in L/L plants at all NaCl doses. In the absence of salt, the CAT activity (Figure 10C) was similar for all plant groups. It increased in L and L/L plants at 80 mM and 160 mM of NaCl but only increased in L/R plants at 160 mM of NaCl.
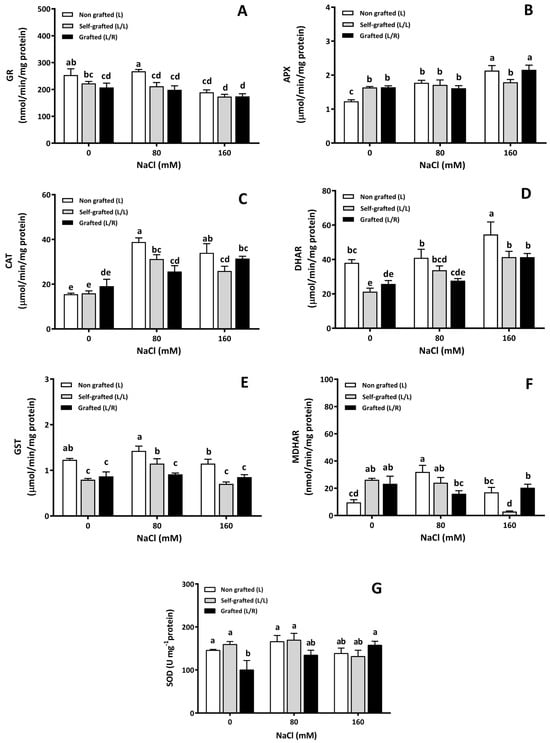
Figure 10.
The antioxidant enzyme activity as a function of different NaCl doses. The enzymatic activity of antioxidant enzymes (glutathione reductase (GR; (A)), ascorbate peroxidase (APX; (B)), catalase (CAT; (C)), dehydro-ascorbate reductase (DHAR; (D)), glutathione-S-transferase (GST; (E)), monodehydroascorbate reductase (MDHAR; (F)), and superoxide dismutase (SOD; (G))) in leaves of plants corresponding to treatment L (ungrafted Old Limachino tomato), L/L (self-grafted Old Limachino tomato), and L/R (grafted Old Limachino tomato on INIA JUPAFORT1 rootstock) grown for 21 days under three salinity treatments: 0 of NaCl; 80 of NaCl, and 160 of NaCl. Each value is the mean of six replicates, and vertical bars are S.E. Lowercase letters represent differences (p ≤ 0.05 Duncan’s test) between treatments and grafting types.
The DHAR activity under control conditions and at 160 of NaCl was higher in L plants compared to the other plant groups (Figure 10D). At 80 mM, DHAR was higher in L plants than in L/R plants (Figure 10D). Increasing salinity caused an increase in DHAR in all plant groups, reaching higher values in L plants. The GST enzyme activity for the control treatment and the two salinities was higher in the L plants compared to the L/L and L/R grafted plants (Figure 10E). The GST activity increased at 80 of NaCl for L/L plants, while it remained stable in L/R plants at all salinity levels. The MDHAR activity (Figure 10F) was lower in L plants under control conditions than in the other plant groups. At 80 of NaCl, the MDHAR activity was the lowest in L/R plants and the highest in L plants, while the lowest value was found in L/L plants at 160 of NaCl (Figure 10F). Once again, the MDHAR activity in L/R remained constant at all salinity levels. Under control conditions, the SOD enzyme activity was higher in L and L/L than in L/R plants (Figure 10G). Under salinity, an increase in SOD activity was only observed in L/R plants at 160 of NaCl (Figure 10G).
3.4. Na+ and K+ Concentrations
The effects of the plant group and salt stress on mineral content (Na+ and K+) in roots, stems, and leaves are presented in Table 2. Under salinity, all plant groups showed significantly higher Na+ levels compared to the control treatment, with the highest Na+ concentrations being observed in L/L plants. The Na+ content was slightly higher in leaves and stems of L/R compared to L plants, while an inverse trend was recorded for roots at 160 of NaCl. This trend was consistently observed across independent experiments but remained insignificant from a statistical point of view. Salinity induced a decrease in the K+ concentration in leaves and stems, but as far as L plants are concerned, such a decrease was insignificant for leaves. L/L and L/R plants exhibited similar K+ concentrations at 160 of NaCl. In leaves, the highest Na+/K+ ratio was observed in L/L plants exposed to 160 of NaCl. In L and L/R plants, the leaf Na+/K+ ratio increased between 0 and 80 of NaCl and then remained stable between 80 and 160 of NaCl. In the stem, the Na+/K+ ratio increased at 80 of NaCl for all plant groups and then remained stable up to 160 of NaCl. In the roots, the Na+/K+ ratio increased at 80 of NaCl and then remained stable up to 160 of NaCl in L and L/L plants, while it still increased in L/R plants, reaching the maximal value for L/R roots exposed to 160 of NaCl.

Table 2.
Potassium (mmol K+ g−1 DW) and sodium (mmol Na+ g−1 DW) concentration and sodium-to-potassium ratios (Na+/K+) in the leaf, stem, and root of non-grafted (L), self-grafted (L/L), and grafted on INIA JUPAFOR1 rootstock (L/R)) Old Limachino tomato plants grown under 0, 80, or 160 of NaCl for 21 days. Each value is the mean of six replicates ± S.E. Lowercase letters represent differences (p ≤ 0.05 Duncan’s test) between treatments and grafting types.
3.5. Gene Expression
The gene expression in roots was considered for six of the seven transporters selected in silico: the Na+ transporter HKT1.1 (HKT1.1), Na+ transporter HKT1.2 (HKT1.2), potassium channel SKOR-like (SKOR), potassium channel (LKT1), calcineurin B-like interacting protein kinase (SOS2), and calcium sensor calcineurin B-like (SOS3) (Figure 11). The expression of SOS1 could not be reliably detected due to its very low threshold cycle. The expression of gene coding for the Na+ transporter HKT1.1 (Figure 11A) was significantly reduced by salinity in L/R plants compared to L/L plants, which did not show statistically significant changes in response to salinity. Similarly, HKT1;1 expression was not significantly affected by salinity in L plants. The expression of HKT1.2 (Figure 11B) was increased by salinity only in L/L plants, with the highest value reported at 160 of NaCl. The relative expression of the SOS2 gene showed a significant increase in L plants at 80 mM of NaCl but was unaffected by salinity in L/L and L/R (Figure 11E).
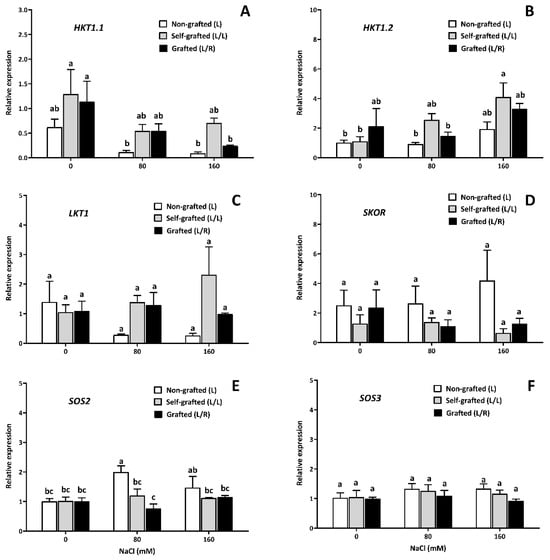
Figure 11.
The expression of gene coding for the ion transporter as a function of the NaCl dose. The gene expression in the roots (relative units) of the Na+ transporter HKT1.1 (HKT1.1, (A)), Na+ transporter HKT1.2 (HKT1. 2, (B)), potassium channel SKOR-like (SKOR, (D)); potassium channel (LKT1, (C)), calcineurin B-like interacting protein kinase (SOS2, (E)), and calcium sensor calcineurin B-like (SOS3, (F)) for plants corresponding to treatment L (ungrafted Old Limachino tomato), L/L (self-grafted Old Limachino tomato), and L/R (grafted Old Limachino tomato on INIA JUPAFORT1 rootstock) grown under three salinity treatments: 0 of NaCl, 80 of NaCl, and 160 of NaCl at 21 days after beginning treatment (DAT). Each value is the mean of three replicates, and vertical bars are S.E. Lowercase letters represent differences (p ≤ 0.05 Duncan’s test) between treatments and types of graftings.
4. Discussion
4.1. Effect of JUPAFORT1 Rootstock on Growth and Plant Water Status under Salinity
Since the fresh biomass in leaves and stems was significantly higher in L/R plants under control conditions compared to L and L/L plants (Figure 1A), the interspecific rootstock JUPAFORT1 was able to induce a strong growth stimulation under these conditions. On the other hand, an opposite effect was observed in L/L plants, which showed the lowest growth in the control treatment and at the extreme salinity level (Figure 1A). These different responses in the absence of salt were similar to those previously found on the effects of the Durinta F1 cultivar grafted on Beaufort rootstock (interspecific, De Ruiter) [60]. Our results suggest that JUPAFORT1 is a vigorous clonal rootstock for OLT plants under non-saline conditions.
The poor response of L/L plants in terms of their growing capacity suggests a strong weakening of their vigor due to the effect of cutting in OLT plants. The leaf, stem, and root FW were indeed lower in L/L than in L plants (Figure 1). It is known that mechanical damage such as cutting may induce a transient burst of ethylene, which could inhibit growth. However, our data indicate that the JUPAFORT1 rootstock in L/R treatment was able to overcome such inhibition in the absence of salt. Salinity, however, reduced the leaf weight in L/R plants, while stems and roots were hardly affected. These results indicate that the vigorous vegetative growth obtained under control conditions (e.g., commercial F1 hybrids) using rootstocks is not necessarily maintained under saline conditions. This contrasts with data obtained in other studies for other rootstocks, which demonstrated that the good performances obtained using vigorous rootstocks under control conditions may be transferable to saline conditions. This deleterious impact on leaf growth in L/R plants was not recorded for root growth, which remained unaffected by salinity. The JUPAFORT1 rootstock is issued from a cross between a salt-tolerant cultivar of S. lycopersicum (Poncho Negro) and the wild halophyte relative S. chilense. Such a decrease in the shoot/root ratio is frequently reported in halophytes [2,4] and might be regarded as an attempt to reduce evaporative surface under water stress conditions and to save energy for triggering salt-adaptative mechanisms [61,62].
In terms of the OLT plant height, our results indicate that the JUPAFORT1 rootstock tended to reduce it (Figure 2A,C) under both control and saline conditions. This indicates that JUPAFORT1 may reduce the internode elongation of OLT plants independently of salinity. Solanum chilense is known to have a bushy growing behavior with a higher number of ramifications than its cultivated counterpart. It is also well established that internode elongation depends on the hormonal profile of the plant, and S. chilense was reported to strongly differ from S. lycopersicum [31,32,33], suggesting that root phytohormones translocated from the root to the shoot may drastically modify the plant’s growing habit. Similarly, this property of JUPAFORT1 could be translated into a higher production potential per axis in L/R plants. In contrast, the stem diameter was similar in all groups and remained unaffected by salinity. Solanum chilense exhibits a narrow stem that hampers its direct use as rootstock and justifies preliminary crosses with S. lycopericum. The present work demonstrates that JUPAFOR1 exhibited a larger stem than its wild parent, allowing for efficient grafting processes.
L/R plants were mainly affected by two water status variables under salt stress conditions, namely, an increase in the midday water potential (Ψwn) and a decrease in the midday relative leaf water content (RWCn). The first hypothesis to explain such a striking rise of Ψwn in L/R plants under extreme-salinity conditions could be an increase in the osmotic potential (Ψs) resulting from increased solute dilution by increased water entry into the cells as a consequence of succulence adaptative processes. A second hypothesis could be a change in the turgor potential due to greater cell wall elasticity, which contributes to salinity tolerance [1,63]. It has to be mentioned that the relative content (RWC) was affected by salinity but that the actual water content (WC) estimated on a dry weight basis was not, which supports the second hypothesis. The maintenance of metabolic activity despite a decrease in the relative water content is relevant from a “tolerance strategy” rather than an “avoidance strategy”, which would require expensive osmotic adjustment processes or precocious stomatal closure compromising photosynthetic activity [1,64]. The increase in Ψwn at 160 mM compared to 80 mM in R/L was not associated with a significant decrease in leaf and stem fresh or dry weights, suggesting that R/L tolerated the water component of salt stress. Regarding the effect of JUPAFORT1 on the stomatal conductance of OLT plants, the results suggest that this variable was affected only at the end of the cycle (21 DAT) and under extreme-salinity conditions. This stability of stomatal conductance observed in L/R plants suggests that these plants were less affected by salt stress than the other two treatments (Figure 3A–C), which aligns with previous results [65]. It is also noteworthy that the highest stress intensity (160 of NaCl) at 21 DAT increased rather than decreased gs value in L/R plants, suggesting that those plants displayed a typical anisohydric behavior and maintained their capacity of CO2 fixation at high salinities. At the same time, L and L/L still behaved as isohydric plants [66].
4.2. Effect of JUPAFORT1 on the Content of Photosynthetic Pigments, Protein, and MDA in the Plant under Saline Conditions
The basal fluorescence (F0) of Chl a in the leaves of L/R plants was not altered by the salinity stress. In contrast, the maximal fluorescence (Fm) was only marginally modified at the end of the experiment for the highest NaCl dose. Since a similar behavior was recorded for L and L/L plants, it may be assumed that the JUPAFORT1 rootstock did not afford specific advantages regarding the light phase of photosynthesis. Using other rootstock issued from S. lycopersicum, other authors observed that PSII efficiency in dark-adapted leaves at 50 DAT, measured as the Fv/Fm ratio, was 5% higher in high-vigor grafting combinations. In contrast, in low-vigorousness grafting combinations, the values were similar to those of control plants under 75 of NaCl [27]. Such a difference in the Fv/Fm ratio was not recorded in the present work.
At the highest stress intensities, Chla, Chlb, and carotenoid concentrations were higher in L/R plants than in L/L. Although the higher pigment content might be regarded as a consequence of leaf growth inhibition, leading to an “accumulating” effect, a positive impact of R rootstock on pigment stability could not be ruled out. The JUPAFORT1 rootstock may help stressed plants avoid or overcome salt-induced senescence, which could also result from the specific hormonal status conferred by the halophyte parents in relation to ethylene, salicylic acid, and polyamine synthesis [31,32,33]. Ethylene is a well-known stress-induced senescing hormone, and a recent study using Poncho Negro as rootstock for Limachino scion reported great erethylene production in L/R plants than in L/L plants [23]. The fact that the pigment concentration remained higher in our L/R plants than in L/L plants provides a different picture and suggests that under high-salt-stress conditions, S. chilense used in the present work in combination with Poncho Negro may confer specific properties to JUPAFORT1. This might be related to the high level of salicylic acid synthesis reported for S. chilense [31,32,33] since salicylic acid is frequently reported as an antagonist of ethylene action [37]. This hypothesis is reinforced by the fact that in terms of the protein and MDA content, L/R plants showed no variation under salinity, while L plants exhibited a strong increase in MDA in response to 80 of NaCl. This suggests that JUPAFORT1 confers a degree of protection to OLT plants against salt stress, preventing damages caused by ionic toxicity at the level of proteins and cell membranes [14].
4.3. Effect of JUPAFORT1 on Plant Antioxidant Capacity and Activity under Salinity Conditions
Salt-tolerant glycophytes may cope with transient or episodic high doses of salt and can trigger adaptative processes to cope with NaCl. Most of these processes are induced by stress, and such induction has an energetic cost [52,62]. In contrast, halophyte species are often permanently exposed to salinity, as is the case for S. chilense in the Atacama desert, and their high level of tolerance relies on constitutive properties that do not imply additional energetical investment in the presence of NaCl [2,4,12]. It is noteworthy that numerous properties related to the management of secondary oxidative stress remained constant in L/R plants at all salinity levels (MDA and polyphenol concentrations, AsA/DHA and GSH/GSSG ratios, GR, GST, and MDHAR activities), while those properties fluctuated depending on the NaCl dose in L and L/L plants. This suggests that the JUPAFORT1 rootstock was able to confer to OLT scion the ability to display constitutive regulation of properties such as the Asada cycle [16]. It is of special interest to notice that, in our case, the halophytic properties should involve the roots since S. chilense was used for rootstock obtention. At the same time, the scion is still issued from a glycophyte species. However, the homeostasis of the redox status was clearly observed in the shoots, and this suggests that some messages sent by the roots may directly influence the scion behavior.
DPPH and FRAP antioxidant capacities in L/R plants increased under extreme-salinity conditions, while they already increased in response to moderate salinity (80 of NaCl) in L plants. This suggests that L/R plants mainly encountered oxidative stress requiring antioxidant mechanisms only at very high stress intensities. This hypothesis is supported by MDA concentrations, which strongly increased at 80 of NaCl in L but not in L/R plants. As the polyphenol (antioxidant compound) content of L/R plants remained constant under salinity conditions, this variable would not explain the antioxidant capacity of these plants at 160 mM. It is known that the main non-enzymatic antioxidant molecules involved in the ascorbate–glutathione cycle are ascorbate and glutathione [14,16,67]. Ascorbate (vitamin C, AsA) is one of the most important non-enzymatic antioxidant compounds and has been detected in most plant cells, organelles, and apoplasts. The AsA content in L/R plants exposed to salinity was lower than that in the control condition. Similarly, the total ascorbate in L/R plants decreased at 80 of NaCl and even to a lower extent at 160 of NaCl. This suggests that ascorbate was not the major contributor to the above-mentioned increase in antioxidant capacity in stressed plants.
GSH is a tripeptide (alpha-glutamyl cysteinyl glycine) found in cellular compartments such as the cytosol, chloroplasts, endoplasmic reticulum, vacuoles, and mitochondria. It has been described to act directly as an antioxidant in reducing most stress-generated free radicals [16]. In our study, all plant groups showed the same GSH content under control conditions (Figure 9A), but NaCl increased the GSH content in L/R plants at both 80 of NaCl and 160 of NaCl, indicating a positive correlation between salinity and the GSH content in these plants (Figure 9A). L/R plants also exhibited the highest GSSG concentration in response to the highest NaCl dose, indicating that GSH oxidation is directly involved in managing the stress situation that total GSH may contribute to the highest antioxidant capacity recorded by FRAP and DPPH activity in salt-treated L/R plants. In addition, the change in the GSH/GSSG ratio during H2O2 degradation is important for signaling processes through the redox pathway [68]. It has been suggested that the GSH/GSSG ratio indicates cellular redox balance and may be involved in ROS sensing [16,67,68,69]. L/R plants showed no change in this ratio under salinity conditions, while the ratio was modified (although in the opposite direction) in L and L/L plants. This, once again, suggests that the JUPAFORT1 rootstock helps the scion to maintain the oxidation state of glutathione under both control and salinity conditions.
Under salinity, SOD activity was only increased in response to extreme salinity, suggesting that JUPAFORT1 activates a protective mechanism against leaf oxidation in the OLT scion under salt stress by reducing O2*. This type of increase in SOD activity in response to salinity was also observed in the tomato landraces Hail 747 and Hail1072 and is considered a crucial mechanism against secondary oxidative stress [70]. Similarly, CAT activity was increased in L/R plants under extreme-salinity conditions, in contrast to L and L/L plants, in which CAT activity increased at 80 of NaCl (Figure 10C). The lower CAT activity in L/R plants could indicate that this type of transplant is less altered in its redox state (GSH/GSSG), as the tissues show lower oxidation with stable MDA levels and slight changes in PSII Chl a fluorescence. This might also be related to the anisohydric behavior of the scion in this combination, which reduces the risk of photorespiration resulting from stomatal closure.
GST enzyme activity can be induced by various environmental stimuli. For example, increased GST levels maintain cellular redox homeostasis and protect organisms against oxidative stress [15,66,71]. Although many GST genes have been cloned and analyzed in various plant species, including tomatoes, the effect of tomato rootstocks on GST enzyme activity has rarely been investigated. Our results showed that this activity was lower in both L/L and L/R plants than in L plants (Figure 10E). Under salinity, this activity remained unaffected in L/R plants. These results suggest once again that JUPAFORT1 helps to maintain homeostatic conditions in stressed plants, thus conferring tolerance to salinity through the stabilization of antioxidative enzymes.
Within the AsA-GSH glutathione cycle, the activities of the four enzymes involved in this cycle (APX, MDHAR, DHAR, and GR) were measured. The enzyme ascorbate peroxidase (APX) is part of the isoenzyme family of hem peroxidases found in different plant organs [66,72]. Under control conditions, APX activity was higher in both L/L and L/R plants. Under salinity, this activity increased in plants at high salinity, in contrast to L plants, in which APX increased at intermediate salinities (Figure 10B). A similar trend was observed for DHAR (Figure 10D). This once again corroborates our hypothesis that oxidative stress triggering APX activation occurred at lower salinity in L than in L/R and that JUPAFORT1 thus efficiently contributes to reducing secondary oxidative stress in L scion.
4.4. Effect of JUPAFORT1 on Plant Na+ and K+ Levels under Salinity Conditions and Expression of Genes Coding for Ion Transporters
JUPAFORT1 was obtained after crossing the wild halophyte S. chilense, which behaves as an includer, [31,32,33,35,36], and the salt-resistant cv. Poncho Negro from the halophyte S. lycopersicum, which behaves as an excluder [21]. No significant difference was recorded for the shoot Na+ concentration between the three groups of plants at both 80 and 160 mM. However, the Na+ in root/Na+ leaf ratios were always lower for L/R plants than other groups, suggesting that L/R plants were less efficient in root Na+ sequestration. Such “including” behavior is often reported for halophyte plant species, especially those that can sequester Na+ in salt glands [73]. The specificity of our grafting system is that the root system is influenced by the halophytic origin of the rootstock. At the same time, Na+ accumulation occurs in the aerial part of the glycophyte scion.
Further, plant salinity resistance is not only a function of the Na+ total concentration but also depends on the Na+ distribution between cell compartments. Na+ may contribute to osmotic adjustment if it is correctly sequestered in vacuoles through the NHX transporter. It may also be less detrimental if correctly extruded by the SOS system in the apoplasm, thus avoiding accumulation in the cytosol [1]. Na+ transport is known to be, at least partly, mediated by HKT transporter proteins that contribute to salinity tolerance in several species, including tomatoes [7,74]. These proteins are localized in the xylem and phloem of roots and shoots and play a role in Na+ exclusion from the xylem [8]. Some reports question the assumption that Na+ exclusion leads to improved salinity tolerance, while others reported the opposite [75].
In our investigation, the expression level of the HKT1.1 gene was the same in all plant groups under non-saline conditions. However, this level was reduced in the roots of L/R plants under high salinity (Figure 11A). On the other hand, the HKT1.2 level was also the same in the three plant groups under control conditions. Under salinity, this level was not modified in L/R plants, but it was particularly modified in L/L plants at 160 of NaCl (Figure 11B). These results suggest that JUPAFORT1 maintained Na+ flux to the top of the plant, confirming its includer behavior. However, this increase in toxicity may be mitigated by the ability of JUPAFORT1 to produce greater succulence at the leaf level. Since the level of HKT1.2 was increased under salinity in L/L plants, they transported a higher level of Na+ into stems and leaves, thus increasing their toxicity. These results are similar to those reported in wild tomato species under salinity conditions [74]. On the other hand, L and L/R plants did not show changes in the gene expression of this transporter (HKT1.2) in roots (Figure 11B), as previously reported in roots of S. lycopersicum plants under saline conditions [35,36].
On the other hand, the SOS pathway is involved in Na+ extrusion in plants [13,76], andSOS2 plays a role in Na+ and K+ fluxes to the aerial part of the plant [77]. In the case of the SOS2 gene, our research provided evidence that salinity only increased gene expression in the roots of L plants, which could be related to the low level of the Na+/K+ ratio in the plant (leaves and stems) compared to L/L plants (Figure 11E). Although SOS1 expression was not detected in our study, SOS2 upregulation may suggest that Na+ was excluded from the roots of L plants by the SOS pathway, which could contribute to the excluder properties of the domesticated S. lycopersicum.
However, despite the increased gene expression of the SOS pathway in the roots of L plants, the Na+ content in this organ increased at the highest salinity, suggesting that the exclusion process ensured by SOS2 may be saturated in L plants, as previously observed for SOS1 in tomato under salinity [78]. Concerning the SOS3-SOS2 complex, no changes in these two genes were observed in the roots of L/R plants under salinity, suggesting that JUPAFORT1 does not activate the ion-exclusion system in the roots of these plants through the plasma membrane Na+/H+ antiporter [79,80]. In this regard, several studies have observed that tomato SOS3 is mainly expressed in roots [80] and that the SOS3-SOS2 complex can activate the Na+/H+ antiporter of Arabidopsis AtSOS1 [80,81]. It is noteworthy that in L/R plants, gene expression was characterized in the root system for all six genes studied (HKT1.1; HKT1.2; LKT1; SKOR; SOS2, and SOS3), and all of them, except HKT1.1, showed no expression changes under salinity. This suggests, once again, that the expression of the vast majority of these genes is constitutive in these L/R plants and that there is no need for them to activate additional transcription that will require energy input [61,62]. Therefore, the gene expression results at the root level suggest that JUPAFORT1confers increased salinity tolerance to OLT plants. However, we should keep in mind that JUPAFORT1 is a hybrid resulting from the cross between a salt-tolerant glycophyte (S. lycopersicum cv. Poncho Negro) and the halophyte S. chilense. Hence, regulation of the SOS pathway in this type of material should be rather complex since the physiological importance of this pathway is not the same for excluding glycophytes and including halophytes.
5. Conclusions
Our study shows that the clonal rootstock JUPAFORT1, derived from an interspecific cross between wild halophyte Solanum chilense and the landrace Poncho Negro of cultivated glycophyte Solanum lycopersicum, confer to the Old Limachino scion some typical halophyte properties helping it to cope with salinity. These properties could be related to regulating the plant water status and the constitutive maintenance of the oxidative stress defense system at the foliar level, including non-enzymatic antioxidants (mainly glutathione) and enzymatic antioxidants, which exhibited higher stability when the L scion was grafted on the rootstock. The expression of genes coding for ion transporters and the extrusion system (HKT1.1, SOS2, and SOS3 genes) was not modified in the root of JUPAFORT1 rootstock, which is in line with the behavior of the includer halophyte parent, S. chilense. This is the first time that such an interspecific hybrid rootstock issued from S. lycopersicum x S. chilense was tested, and our work demonstrates that JUPAFORT1 may improve the behavior of salt-affected scion. Further works are required to identify the molecular and biochemical components of the root-to-shoot signaling pathway. This information is key for future breeding programs on salinity-tolerant rootstocks. These innovative results at the vegetative level also motivate the study of the influence of JUPAFORT1 at the reproductive stage in terms of flowering, fruit formation, and yield-related parameters. Finally, a comparison with commercial rootstock is required to validate the interest of JUPAFORT1 for salt tolerance.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae10080780/s1, Table S1. Selected primers for genes involved in Na+ and K+ transport mechanisms associated with salinity tolerance in plants to be analyzed by qPCR.
Author Contributions
Conceptualization, J.-P.M., R.F. and S.L.; methodology, C.R., D.B., J.F.A.-Q. and J.-P.M.; experiments validation, J.-P.M., B.S., C.L. and J.F.A.-Q.; statistical analysis, J.-P.M., R.F., J.F.A.-Q. and M.Q.; investigation, J.-P.M., J.F.A.-Q., D.B., C.R., CL., B.S., M.Q. and S.L.; resources, J.-P.M., M.Q. and S.L.; data curation, J.F.A.-Q., D.B., C.R., F.C., B.S. and J.-P.M.; writing—original draft preparation, J.-P.M., R.F., C.R., D.B., J.F.A.-Q., C.L., B.S., M.Q. and S.L.; writing—review and editing, J.-P.M., R.F., M.Q. and S.L.; supervision, J.-P.M. and R.F.; project administration, J.-P.M.; funding acquisition, J.-P.M., M.Q. and S.L. All authors have contributed substantially to the work reported. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Development and Research National Agency (ANID) of the Ministry of Science and Technology of Chile (FONDECYT project No. 1220909) and the Bilateral Cooperation project between Chile (AGCI) and Belgium (WBI-2023) Nº RI 13.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Acknowledgments
Juan Pablo Martínez and Boris Sagredo acknowledge the contribution of the FONDECYT project No. 1220909, funded by the Development and Research National Agency (ANID) of the Ministry of Science and Technology of Chile. Juan Pablo Martínez, Raúl Fuentes, Muriel Quinet, and Stanley Lutts also acknowledge the support of the Bilateral Cooperation project between Chile (AGCI) and Belgium (WBI-2023) Nº RI 13.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Fu, H.; Yang, Y. How plant tolerate salt stress. Curr. Issues Mol. Biol. 2023, 45, 5914–5934. [Google Scholar] [CrossRef]
- Rahman, M.M.; Mostofa, M.G.; Keya, S.S.; Siddiqui, M.N.; Ansary, M.E.U.; Das, A.K.; Rahman, M.A.; Tran, L.S.P. Adapatative mechanisms of halophyte and their potential in improving salinity tolerance in plants. Int. J. Mol. Sci. 2021, 22, 10733. [Google Scholar] [CrossRef]
- Atta, K.; Mondal, S.; Gorai, S.; Singh, A.P.; Kumari, A.; Ghosh, T.; Roy, A.; Hembram, S.; Gaiwad, D.J.; Mondal, S.; et al. Impacts of salinity stress on crop plants: Improving salt tolerance through genetic and molecular dissection. Front. Plant Sci. 2023, 14, 1241736. [Google Scholar] [CrossRef]
- Mann, A.; Lata, C.; Kumar, N.; Kumar, A.; Kumar, A.; Sheoran, P. Halophyte as new model plant species for salt tolerance strategies. Front. Plant Sci. 2023, 14, 1137211. [Google Scholar] [CrossRef]
- Marsic, N.K.; Vodnik, D.; Mikulic-Prtkovesk, M.; Veberic, R.; Sircelj, H. Photosynthetic traits of plants and biochemical profile of tomato fruits are influenced by grafting, salinity stress and growing season. J. Agric. Food Chem. 2018, 66, 5439–5450. [Google Scholar] [CrossRef]
- He, Y.; Zhu, Z.; Yang, J.; Ni, X.; Zhu, B. Grafting increases the salt tolerance of tomato by improvement of photosynthesis and enhancement of antioxidant enzymes activity. Environ. Exp. Bot. 2009, 66, 270–278. [Google Scholar] [CrossRef]
- Asins, M.J.; Villalta, A.M.M.; Olías, R.; Alvarez, D.E.; Morales, P.; Huertas, R.; Li, J.; Jaime-Pérez, N.; Haro, R.; Raga, V.; et al. Two closely linked tomato HKT coding genes are positional candidates for the major tomato QTL involved in Na+/K+ homeostasis. Plant Cell Environ. 2013, 36, 1171–1191. [Google Scholar] [CrossRef]
- Adams, E.; Shin, R. Transport, signaling, and homeostasis of potassium and sodium in plants. J. Integr. Plant Biol. 2014, 56, 231–249. [Google Scholar] [CrossRef]
- Coskun, D.; Britto, D.T.; Kronzucker, H.J. The physiology of channel-mediated K+ acquisition in roots of higher plants. Physiol. Plant. 2014, 151, 305–312. [Google Scholar] [CrossRef]
- Ali, A.; Raddatz, N.; Pardo, J.M.; Yun, D.J. HKT sodium and potassium transporters in Arabidopsis thaliana and related halophyte species. Physiol. Plant. 2021, 171, 546–558. [Google Scholar] [CrossRef]
- Ishikawa, T.; Shabala, S. Control of xylem Na+ loading and transport to the shoot in rice and barley as a determinant of differential salinity stress tolerance. Physiol. Plant. 2019, 165, 619–631. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Ann. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Xie, Q.; Zhou, Y.; Jiang, X. Structure, function and regulation of the plasma membrane Na+/H+ antiporter Salt Overly Sensitive 1 in plants. Front. Plant Sci. 2022, 13, 866265. [Google Scholar] [CrossRef]
- Parvin, K.; Hasanuzzaman, M.; Bhuyan, M.H.; Nahar, K.; Mohsin, S.M.; Fujita, M. Comparative physiological and biochemical changes in Tomato (Solanum lycopersicum L.) under salt stress and recovery: Role of antioxidant defense and glyoxalase systems. Antioxidants 2019, 8, 350. [Google Scholar] [CrossRef]
- Chen, J.H.; Jiang, H.W.; Hsieh, E.J.; Chen, H.Y.; Chien, C.T.; Hsieh, H.L.; Lin, T.P. Drought and salt stress tolerance of an Arabidopsis glutathione S-transferase U17 knockout mutant are attributed to the combined effect of glutathione and abscisic acid. Plant Physiol. 2012, 158, 340–351. [Google Scholar] [CrossRef]
- Kunert, K.J.; Foyer, C.H. The ascorbate/glutathione cycle. Adv. Bot. Res. 2023, 105, 77–112. [Google Scholar]
- Martínez, J.P.; Jana, C.; Muena, V.; Salazar, E.; Rico, J.J.; Calabrese, N.; Hernandez, J.E.; Lutts, S.; Ruentes, F. The Recovery of the Old Limachino Tomato: History, Findings, Lessons, Challenges and Perspectives. In Agriculture Value Chain—Challenges and Trends in Academia and Industry; Hernández, J., Kacprzyk, J., Eds.; Studies in Systems, Decision and Control; Springer: Cham, Switzerland, 2021; Volume 280. [Google Scholar] [CrossRef]
- Jara, C. Efecto de la Fertilizacion (Convencional y Compost) Sobre la Productividad y Calidad de Tomate Limachino Antiguo y Larga Vida Bajo Adero en la Cuenca de Limache. Master’s Thesis, Universidad Técnica Federico Santa María, Santiago, Chile, 2016; 70p. [Google Scholar]
- Singh, H.; Kumar, P.; Kyriacou, M.C.; Colla, G.; Rouphael, Y. Grafting tomato as a tool to improve salt tolerance. Agronomy 2020, 10, 263. [Google Scholar] [CrossRef]
- Di Gioia, F.; Signore, A.; Serio, F.; Santamaria, P. Grafting improves tomato salinity tolerance through sodium partitioning within the shoot. HortScience 2013, 48, 855–862. [Google Scholar] [CrossRef]
- Contreras, C.; Montoya, A.; Pacheco, P.; Martínez-Ballesta, M.C.; Carvajal, M.; Bastías, E. The effects of the combination of salinity and excess boron on the water relations of tolerant tomato (Solanum lycopersicum L.) cv. Poncho Negro, in relation to aquaporin functionality. Span. J. Agric. Res. 2011, 9, 494–503. [Google Scholar] [CrossRef]
- Huanca-Mamani, W.; Cárdenas-Ninasivincha, S.; Acosta-García, G.; Alache, K.; Bastías, E. Expression analysis of three stress-related genes in response to excess of boron in Solanum lycopersicum cv. Poncho Negro. Idesia 2018, 36, 35–40. [Google Scholar] [CrossRef]
- Alfaro-Quezada, J.F.; Martínez, J.P.; Molinett, S.; Valenzuela, M.; Montenegro, I.; Ramírez, I.; Dorta, F.; Ávila-Valdés, A.; Gharbi, E.; Zhou, M.; et al. Rootstock increases the physiological defence of tomato plants against Pseudomonas syringae pv. tomato infection. J. Exp. Bot. 2023, 74, 2891–2911. [Google Scholar] [CrossRef]
- Tapia, G.; Mendez, J.; Inostroza, L. Different combinations of morpho-physiological traits are responsible for tolerance to drought in wild tomatoes Solanum chilense and Solanum peruvianum. Plant Biol. 2016, 18, 406–416. [Google Scholar] [CrossRef]
- Gálvez, F.J.; Baghour, M.; Hao, G.; Cagnac, O.; Rodríguez-Rosales, M.P.; Venema, K. Expression of LeNHX isoforms in response to salt stress in salt sensitive and salt tolerant tomato species. Plant Physiol. Biochem. 2012, 51, 109–115. [Google Scholar] [CrossRef]
- Aydin, A. The growth, leaf antioxidant enzymes and amino acid content of tomato as affected by grafting in wild tomato rootstocks (S. pimpinellifolium and S. habrochaites) under salt stress. Sci. Hortic. 2024, 325, 112679. [Google Scholar] [CrossRef]
- Albacete, A.; Martínez-Andújar, C.; Ghanem, M.E.; Acosta, M.; Sánchez-Bravo, J.; Asins, M.J.; Cuartero, J.; Lutts, S.; Dodd, I.C.; Pérez-Alfocea, F. Rootstock-mediated changes in xylem ionic and hormonal status are correlated with delayed leaf senescence and increased leaf area and crop productivity in salinized tomato. Plant Cell Environ. 2009, 32, 928–938. [Google Scholar] [CrossRef]
- Zeist, A.C.; Henschel, J.M.; Silva Júnior, A.D.; Almeida Oliveira, G.J.; Neto, J.G.; Beauboeuf, C.M.; Parthasarathi, T.; Vilela de Resende, J.T. Responses of rootstock viability to toleate salinity in tomato. S. Afr. J. Bot. 2023, 153, 280–289. [Google Scholar] [CrossRef]
- Martínez, J.P.; Antúnez, A.; Pertuzé, R.; Acosta, M.P.; Palma, X.; Fuentes, L.; Ayala, A.; Araya, H.; Lutts, S. Effects of saline water on water status, yield and fruit quality of wild (Solanum chilense) and domesticated (Solanum lycopersicum var. cerasiforme) tomatoes. Exp. Agric. 2012, 48, 573–586. [Google Scholar] [CrossRef]
- Martínez, J.P.; Antúnez, A.; Araya, H.; Pertuzé, R.; Fuentes, L.; Lizana, X.C.; Lutts, S. Salt stress differently affects growth, water status and antioxidant enzyme activities in Solanum lycopersicum and its wild relative Solanum chilense. Aust. J. Bot. 2014, 62, 359. [Google Scholar] [CrossRef]
- Gharbi, E.; Martínez, J.P.; Benahmed, H.; Fauconnier, M.L.; Lutts, S.; Quinet, M. Salicylic acid differently impacts ethylene and polyamine synthesis in the glycophyte Solanum lycopersicum and the wild-related halophyte Solanum chilense exposed to mild salt stress. Physiol. Plant. 2016, 158, 152–167. [Google Scholar] [CrossRef]
- Gharbi, E.; Martínez, J.P.; Benahmed, H.; Lepoint, G.; Vanpee, B.; Quinet, M.; Lutts, S. Inhibition of ethylene synthesis reduces salt-tolerance in tomato wild relative species Solanum chilense. J. Plant Physiol. 2017, 210, 24–37. [Google Scholar] [CrossRef]
- Gharbi, E.; Martínez, J.P.; Benahmed, H.; Dailly, H.; Quinet, M.; Lutts, S. The salicylic acid analog 2,6-dichloroisonicotinic acid has specific impact on the response of the halophyte plant species Solanum chilense to salinity. Plant Growth Regul. 2017, 82, 517–525. [Google Scholar] [CrossRef]
- Bigot, S.; Pongrac, P.; Šala, M.; van Elteren, J.T.; Martínez, J.P.; Lutts, S.; Quinet, M. The halophyte species Solanum chilense Dun. maintains its reproduction despite sodium accumulation in its floral organs. Plant 2022, 11, 672. [Google Scholar] [CrossRef]
- Bigot, S.; Fuksová, M.; Martínez, J.P.; Lutts, S.; Quinet, M. Sodium and chloride accumulation and repartition differed between the cultivated tomato (Solanum lycopersicum) and its wild halophyte relative Solanum chilense under salt stress. Sci. Hortic. 2023, 321, 112324. [Google Scholar] [CrossRef]
- Bigot, S.; Leclef, C.; Rosales, C.; Martínez, J.P.; Lutts, S.; Quinet, M. Comparison of the salt resistance of Solanum lycopersicum x Solanum chilense hybrids and their parents. Front. Hortic. 2023, 2, 1130702. [Google Scholar] [CrossRef]
- Ghanem, M.E.; Hichri, I.; Smigocki, A.C.; Albacete, A.; Fauconnier, M.L.; Diatloff, E.; Martinez-Andujar, C.; Lutts, S.; Dodd, I.C.; Pérez-Alfocea, F. Root-targeted biotechnology to mediate hormonal signalling and improve crop stress tolerance. Plant Cell Rep. 2011, 30, 807–823. [Google Scholar] [CrossRef]
- Martínez-Andújar, C.; Martínez-Pérez, A.; Albacete, A.; Martínez Melgarejo, P.A.; Dodd, I.A.; Thompson, A.J.; Mohareb, F.; Estelles-Lopez, L.; Kevei, Z.; Ferrández-Ayela, A.; et al. Overproduction of ABA in rootstocks alleviates salinity stress in tomato shoots. Plant Cell Environ. 2021, 44, 2966–2986. [Google Scholar] [CrossRef]
- Martínez, J.P.; Rosales, C.; Salinas, L.; Farías, K.; Lizana, C.; Quinet, M.; Lutts, S.; Tan, M.L. Micropropagación de un portainjerto INIA tolerante a la salinidad creado a partir del cruce entre Solanum lycopersicum y Solanum chilense. In Proceedings of the 70° Congreso Agronómico Formación Agronómica para el 2030 Tradición y Futuro, Santiago, Chile, 7–9 January 2020. [Google Scholar]
- Martínez, J.P.; Fuentes, R.; Farías, K.; Lizana, C.; Alfaro, J.F.; Fuentes, L.; Calabrese, N.; Bigot, S.; Quinet, M.; Lutts, S. Effects of saline stress on fruit antioxidant capacity of wild (Solanum chilense) and domesticated (Solanum lycopersicum var. cerasiforme) tomatoes. Agronomy 2020, 10, 1481. [Google Scholar] [CrossRef]
- Martínez, J.P.; Fuentes, R.; Farías, K.; Loyola, N.; Freixas, A.; Stange, C.; Sagredo, B.; Quinet, M.; Lutts, S. Effects of a local tomato rootstock on the agronomic, functional and sensory quality of the fruit of a recovered local tomato (Solanum lycopersicum L.) named “Tomate Limachino Antiguo”. Agronomy 2022, 12, 2178. [Google Scholar] [CrossRef]
- Martínez, J.P.; Lutts, S.; Schanck, A.; Bajji, M.; Kinet, J.M. Is osmotic adjustment required for water stress resistance in the Mediterranean shrub Atriplex halimus L.? J. Plant Physiol. 2004, 161, 1041–1051. [Google Scholar] [CrossRef]
- Swoczyna, T.; Kalaji, H.M.; Bussotti, F.; Mojski, J.; Pllastrini, M. Environmental stress—What can we learn from chlorophyll a fluorescence analysis in woody plants? A review. Front. Plant Sci. 2022, 13, 1048582. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K. Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 1987, 148, 350–382. [Google Scholar]
- Heath, R.L.; Packer, L. Photoperoxidation in isolated chloroplasts: I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 1968, 125, 189–198. [Google Scholar] [CrossRef]
- Benzie, I.; Strain, J.J. The ferric reducing ability of plasma (FRAP) as a measure of “antioxidant power”: The FRAP assay. Anal. Biochem. 1996, 239, 70–76. [Google Scholar] [CrossRef]
- Brand-Williams, W.; Cuvelier, M.E.; Berset, C. Use of freee radical method to evaluate antioxidant activity. Food Sci. Technol. 1995, 28, 25–30. [Google Scholar]
- Singleton, V.; Rossi, J. Colorimetry of total phenolics with phosphomolybdic and phosphotungstic acid reagents. Am. J. Enol. Vitic. 1965, 16, 144–148. [Google Scholar] [CrossRef]
- Kampfenkel, K.; Vanmontagu, M.; Inze, D. Extraction and determination of ascorbate and dehydroascorbate from plant tissue. Anal. Biochem. 1995, 225, 165–167. [Google Scholar] [CrossRef]
- Cereser, C.; Guichard, J.; Drai, J.; Bannier, E.; Garcia, I.; Boget, S.; Parvaz, P.; Revol, A. Quantitation of reduced and total glutathione at the femtomole level by high-performance liquid chromatography with fluorescence detection: Application to red blood cells and cultured fibroblasts. J. Chromatogr. B Biomed. Sci. Appl. 2001, 752, 123–132. [Google Scholar] [CrossRef]
- Zhou, M.X.; Dailly, H.; Renard, M.E.; Han, R.M.; Lutts, S. NaCl impact on Kosteletzkya pentacarpos seedlings simultaneously exposed to cadmium and zinc toxicities. Environ. Sci. Pollut. Res. 2018, 25, 17444–17456. [Google Scholar] [CrossRef]
- Gupta, A.S.; Webb, R.P.; Holaday, A.S.; Allen, R.D. Overexpression of superoxide dismutase protects plants from oxidative stress (induction of ascorbate peroxidase in superoxide dismutase-overexpressing plants). Plant Physiol. 1993, 103, 1067–1073. [Google Scholar] [CrossRef]
- Nakano, Y.; Asada, K. Hydrogen peroxide scavenged by ascorbate specific peroxidase in spinach chloroplast. Plant Cell Physiol. 1981, 22, 867–880. [Google Scholar] [CrossRef]
- Miyake, C.; Asada, K. Thylakoid bound ascorbate peroxidase in spinach chloroplasts and photoreduction of its primary oxidation product monodehydroascorbate radicals in thylakoids. Plant Cell Physiol. 1992, 33, 541–553. [Google Scholar] [CrossRef]
- Beyer, W.F.; Fridovitch, I. Assaying for superoxide dismutase activity: Some large consequences of minor changes in conditions. Anal. Biochem. 1987, 161, 559–566. [Google Scholar] [CrossRef]
- Yu, Q.; Osborne, L.; Rengel, Z. Micronutrient deficiency changes activities of superoxide dismutase and ascorbate peroxidase in tobacco plants. J. Plant Nutr. 1998, 21, 1427–1437. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid sensitive method for the quantification of microquantities of protein utilizing the principle of protein dye binding. Anal. Biochem. 1976, 161, 559–566. [Google Scholar]
- López-Gómez, R.; Gómez-Lim, M.A. A method for extracting intact RNA from fruits rich in polysaccharides using ripe mango mesocarp. HortScience 1992, 27, 440–442. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Öztekin, G.B.; Giuffrida, F.; Tüzel, Y.; Leonardi, C. Is the vigour of grafted tomato plants related to root characteristics. J. Agric. Food Environ. 2009, 7, 364–368. [Google Scholar]
- Munns, R.; Gilliham, M. Salinity tolerance of crops—What is the cost? New Phytol. 2015, 208, 668–673. [Google Scholar] [CrossRef]
- Munns, R.; Day, D.A.; Fricke, W.; Watt, M.; Arsova, B.; Barkla, B.J.; Bose, J.; Byrt, C.S.; Chen, Z.-H.; Foster, K.J.; et al. Energy costs of salt tolerance in cop plants. New Phytol. 2020, 225, 1072–1090. [Google Scholar] [CrossRef]
- Martínez, J.P.; Silva, H.; Ledent, J.F.; Pinto, M. Effect of drought stress on the osmotic adjustment, cell wall elasticity and cell volume of six cultivars of common beans (Phaseolus vulgaris L.). Eur. J. Agron. 2007, 26, 30–38. [Google Scholar] [CrossRef]
- Hussain, H.A.; Hussain, S.; Khaliq, A.; Ashraf, U.; Anjum, S.A.; Men, S.; Wang, L. Chilling and drought stresses in crop plants: Implication, cross talk, and potential management opportunities. Front. Plant Sci. 2018, 9, 393. [Google Scholar] [CrossRef] [PubMed]
- Coban, A.; Akhoundnejad, Y.; Dere, S.; Yildiz Dasgan, H. Impact of salt-tolerant rootstock on the enhancement of sensitive tomato plant responses to salinity. HortScience 2020, 55, 35–39. [Google Scholar] [CrossRef]
- Hochberg, U.; Rockwell, F.E.; Holbrook, N.M.; Cochard, H. Iso/Anisohydry: A plant-environment interaction rather tan a simple hydraulic traiy. Trends Plant Sci. 2018, 23, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Martínez, J.P.; Araya, H. Ascorbate–Glutathione Cycle: Enzymatic and Non-enzymatic Integrated Mechanisms and Its Biomolecular Regulation. In Ascorbate-Glutathione Pathway and Stress Tolerance in Plants; Anjum, N., Chan, M.T., Umar, S., Eds.; Springer: Dordrecht, The Netherland, 2010. [Google Scholar] [CrossRef]
- Considine, M.J.; Foyer, C.H. Oxygen and reactive oxygen species-dependent regulation of plant growth and development. Plant Physiol 2021, 186, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Considine, M.J.; Foyer, C.H. Stress effects on the reacyiove oxygen species-dependent regulation of plant growth and development. J. Exp. Bot. 2021, 72, 5795–5806. [Google Scholar] [CrossRef] [PubMed]
- Alzahib, R.H.; Migdadi, H.M.; Al Ghamdi, A.A.; Alwahibi, M.S.; Ibrahim, A.A.; Al-Selwey, W.A. Assessment of morpho-physiological, biochemical and antioxidant responses of tomato landraces to salinity stress. Plants 2021, 10, 696. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Xing, X.J.; Tian, Y.S.; Peng, R.H.; Xue, Y.; Zhao, W.; Yao, Q.H. Transgenic Arabidopsis plants expressing tomato Glutathione S-Transferase showed enhanced resistance to salt and drought stress. PLoS ONE 2015, 10, e0136960. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Bhuyan, M.H.M.B.; Anee, T.I.; Parvin, K.; Nahar, K.; Mahmud, J.A.; Fujita, M. Regulation of ascorbate-glutathione pathway in mitigating oxidative damage in plants under abiotic stress. Antioxidants 2019, 8, 384. [Google Scholar] [CrossRef] [PubMed]
- Litalien, A.; Zeed, B. Curing the earth: A review of anthropogenic soil salinization and plant-based strategy for sustainable mitigation. Sci. Total Environ. 2020, 698, 134235. [Google Scholar] [CrossRef] [PubMed]
- Almeida, P.; de Boer, G.-J.; de Boer, A.H. Differences in shoot Na+ accumulation between two tomato species are due to differences in ion affinity of HKT1;2. J. Plant Physiol. 2014, 171, 438–447. [Google Scholar] [CrossRef]
- Venkatarama, G.; Shabal, S.; Véry, A.A.; Hariharan, G.N.; Somasundaram, S.; Pulipati, S.; Sellamuthu, G.; Harikrishnan, M.; Kumari, K.; Shabala, L.; et al. To exclude or accumulate? Revealing the role of sodium HKT1;5 transporter in plant adaptative responses to varying soil salinity. Plant Physiol. Biochem. 2021, 169, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Pardo, J.M.; Batelli, G.; Van Oosten, M.J.; Bressan, R.A.; Li, X. The Salt Overly Sensitive (SOS) pathway: Established and emerging roles. Mol. Plant 2013, 6, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Belver, A.; Olías, R.; Huertas, R.; Rodríguez-Rosales, M.P. Involvement of SlSOS2 in tomato salt tolerance. Bioengineered 2012, 3, 298–302. [Google Scholar] [CrossRef] [PubMed]
- Nieves-Cordones, M.; Al Shiblawi, F.R.; Sentenac, H. Roles and transport of sodium and potassium in plants. In The Alkali Metal Ions: Their Role For Life; Sigel, A., Sigel, H., Sigel, R.K.O., Eds.; Metal Ions in Life Sciences; Springer International Publishing: Cham, Switzerland, 2016; Volume 16, pp. 291–324. [Google Scholar]
- Yang, Y.; Guo, Y. Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 2018, 60, 796–804. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Wang, X.S.; Guo, H.D.; Bai, S.Y.; Khan, A.; Wang, X.M.; Gao, Y.M.; Li, J.S. Tomato salt tolerance mechanisms and their potential applications for fighting salinity: A review. Front. Plant Sci. 2022, 13, 949541. [Google Scholar] [CrossRef]
- Huertas, R.; Olias, R.; Eljakaoui, Z.; Gálvez, F.J.; Li, J.U.N.; De Morales, P.A.; Belver, A.; Rodríguez-Rosales, M.P. Overexpression of SlSOS2 (SlCIPK24) confers salt tolerance to transgenic tomato. Plant Cell Environ. 2012, 35, 1467–1482. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).