Penicillium roqueforti Secondary Metabolites: Biosynthetic Pathways, Gene Clusters, and Bioactivities
Abstract
:1. Introduction
2. Secondary Metabolites of P. roqueforti
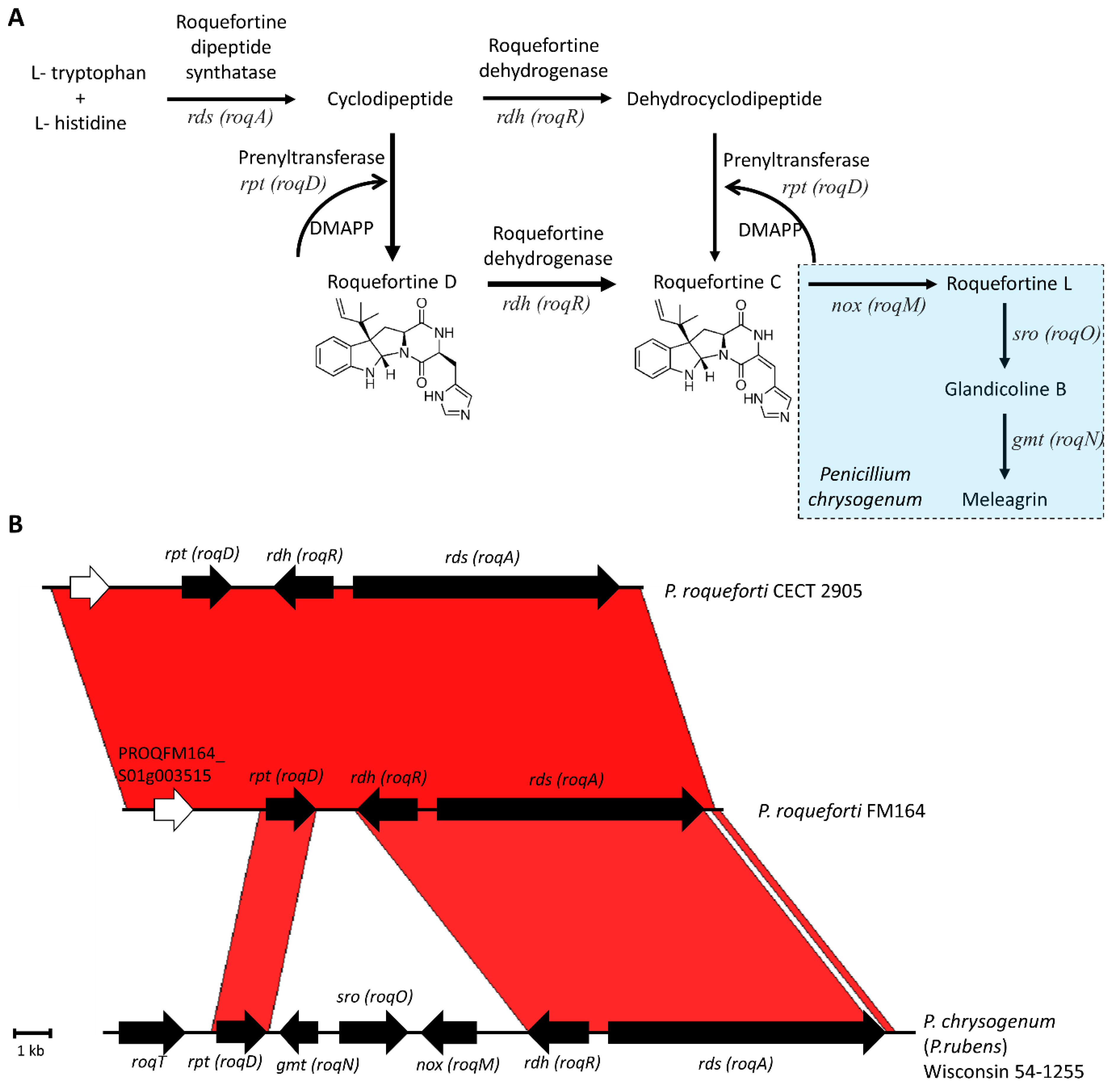
2.1. Roquefortine C
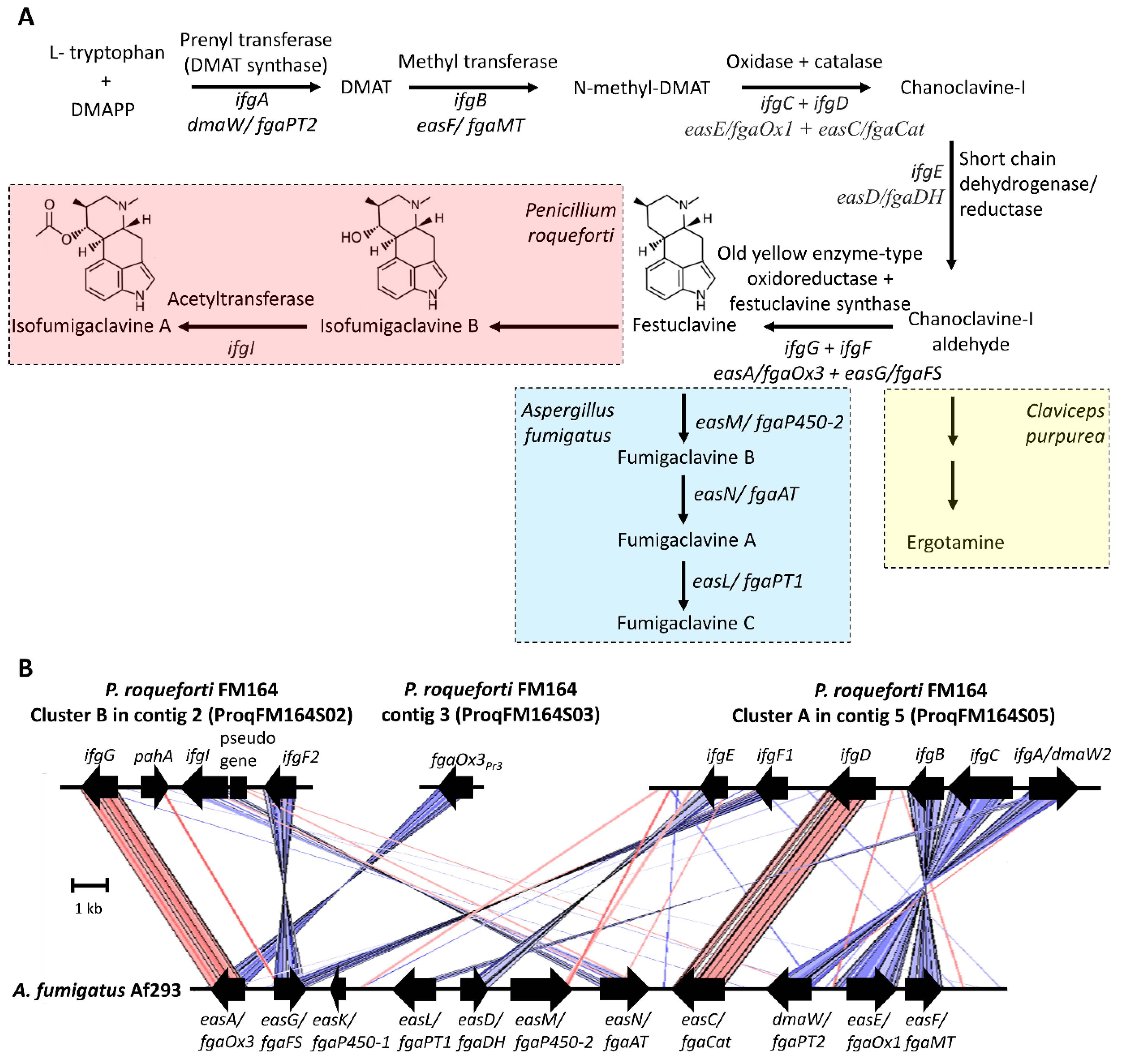
2.2. Clavine Alkaloids
2.3. Mycophenolic Acid
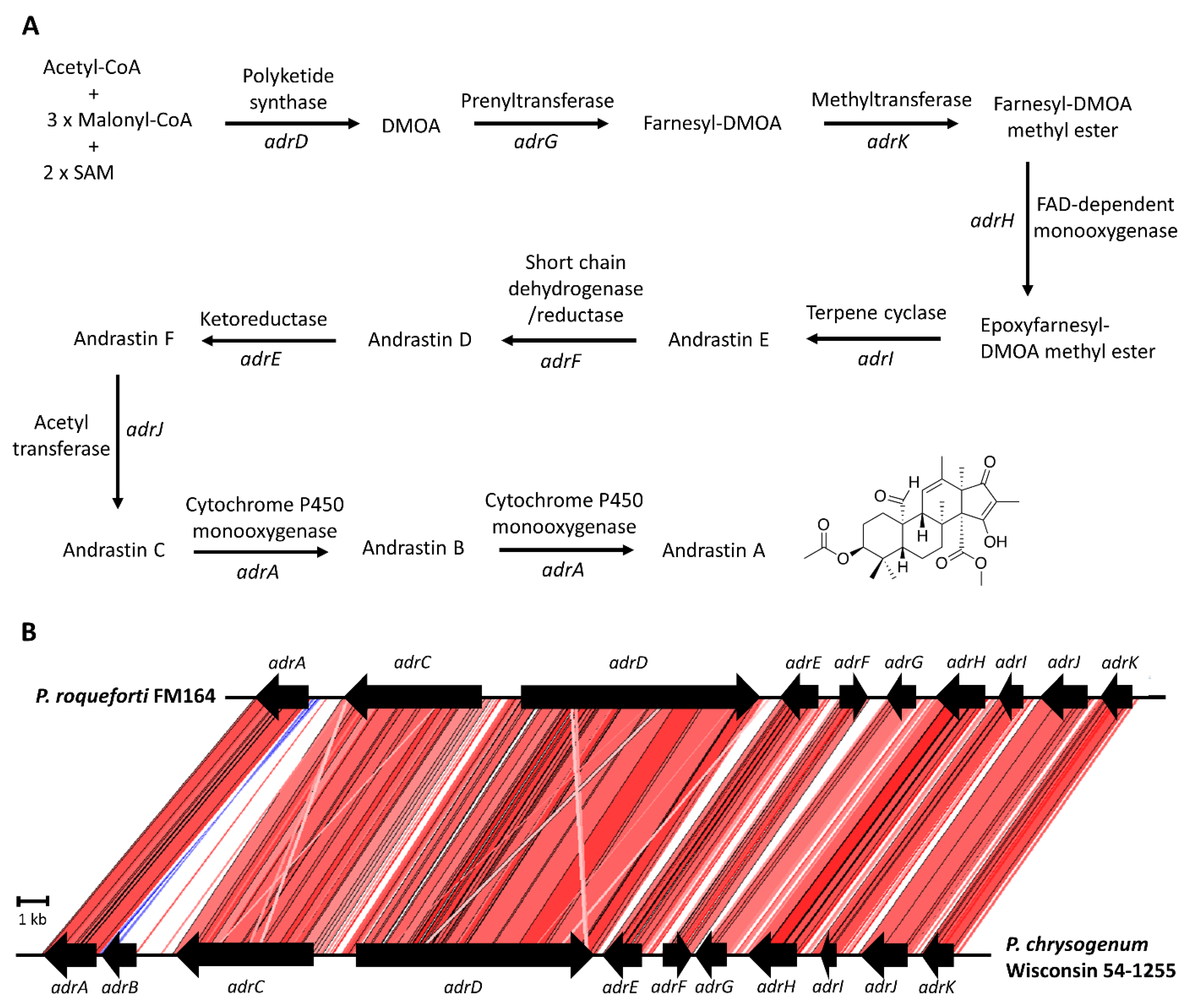
2.4. Andrastin A
2.5. PR-Toxin
2.6. Other Secondary Metabolites of P. roqueforti
2.6.1. Citreoisocoumarin
2.6.2. Orsellinic Acid
2.6.3. Sesterterpenoids: Peniroquesines A–C and 1–7, and Roquefornine A
2.6.4. Tetrapeptides D-Phe-L-Val-D-Val-L-Phe and D-Phe-L-Val-D-Val-L-Tyr
2.6.5. Cis-bis(methylthio)silvatin
2.6.6. Scytalone and Melanin
3. Secondary Metabolite Potential of P. roqueforti: Yet Undiscovered Metabolites and/or Silent Gene Clusters
4. Conclusions and Future Directions
Supplementary Materials
Funding
Data Availability Statement
Conflicts of Interest
References
- Brakhage, A.A. Regulation of Fungal Secondary Metabolism. Nat. Rev. Microbiol. 2013, 11, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Keller, N.P. Fungal Secondary Metabolism: Regulation, Function and Drug Discovery. Nat. Rev. Microbiol. 2019, 17, 167–180. [Google Scholar] [CrossRef]
- Keller, N.P.; Turner, G.; Bennett, J.W. Fungal Secondary Metabolism—From Biochemistry to Genomics. Nat. Rev. Microbiol. 2005, 3, 937–947. [Google Scholar] [CrossRef] [PubMed]
- Conrado, R.; Gomes, T.C.; Roque, G.S.; De Souza, A.O. Overview of Bioactive Fungal Secondary Metabolites: Cytotoxic and Antimicrobial Compounds. Antibiotics 2022, 11, 1604. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liang, X.; Li, Y.; Wang, P.; Keller, N.P. Genetic Regulation of Mycotoxin Biosynthesis. J. Fungi 2023, 9, 21. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, M.C.; Jenkins, F.P.; Philp, J.M. Toxicity Associated with Certain Samples of Groundnuts. Nature 1961, 192, 1095–1096. [Google Scholar] [CrossRef]
- Nesbitt, B.F.; O’Kelly, J.; Sargeant, K.; Sheridan, A. Aspergillus flavus and Turkey X Disease: Toxic Metabolites of Aspergillus flavus. Nature 1962, 195, 1062–1063. [Google Scholar] [CrossRef]
- Copetti, M.V. Fungi as industrial producers of food ingredients. Curr. Opin. Food Sci. 2019, 25, 52–56. [Google Scholar] [CrossRef]
- Macheleidt, J.; Mattern, D.J.; Fischer, J.; Netzker, T.; Weber, J.; Schroeckh, V.; Valiante, V.; Brakhage, A.A. Regulation and Role of Fungal Secondary Metabolites. Annu. Rev. Genet. 2016, 50, 371–392. [Google Scholar] [CrossRef]
- Demain, A.L.; Fang, A. The Natural Functions of Secondary Metabolites. In History of Modern Biotechnology I; Fiechter, A., Ed.; Springer: Berlin/Heidelberg, Germany, 2000; pp. 1–39. ISBN 978-3-540-44964-5. [Google Scholar]
- Skellam, E. Subcellular Localization of Fungal Specialized Metabolites. Fungal Biol. Biotechnol. 2022, 9, 11. [Google Scholar] [CrossRef]
- Pfannenstiel, B.T.; Keller, N.P. On Top of Biosynthetic Gene Clusters: How Epigenetic Machinery Influences Secondary Metabolism in Fungi. Microb. Eng. Biotechnol. 2019, 37, 107345. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Pei, R.; Zhou, J.; Zeng, B.; Tu, Y.; He, B. Molecular Regulation of Fungal Secondary Metabolism. World J. Microbiol. Biotechnol. 2023, 39, 204. [Google Scholar] [CrossRef] [PubMed]
- Metin, B. Filamentous Fungi in Cheese Production. In Microbial Cultures and Enzymes in Dairy Technology; Öztürkoğlu Budak, Ş., Akal, H.C., Eds.; IGI Global: Hershey, PA, USA, 2018; pp. 257–275. ISBN 978-1-522-55363-2. [Google Scholar] [CrossRef]
- Morales, M.B.L.; Ardö, Y.; Berthier, F.; Karatzas, K.-A.G.; Bintsis, T. Blue-Veined Cheeses. In Global Cheesemaking Technology: Cheese Quality and Characteristics; Papademas, P., Bintsis, T., Eds.; John Wiley & Sons: Chichester, UK, 2017; pp. 415–435. ISBN 978-1-119-04616-5. [Google Scholar] [CrossRef]
- Cantor, M.D.; van den Tempel, T.; Hansen, T.K.; Ardö, Y. Blue Cheese. In Cheese: Chemistry, Physics and Microbiology; Fox, P.F., McSweeney, P.L.H., Cogan, T.M., Guinee, T.P., Eds.; Academic Press: Cambridge, MA, USA, 2004; Volume 2, pp. 175–198. ISBN 978-0-122-63653-0. [Google Scholar] [CrossRef]
- Gripon, J.C. Mould-Ripened Cheeses. In Cheese: Chemistry, Physics and Microbiology; Springer: Boston, MA, USA, 1993; pp. 111–136. ISBN 978-1-4613-6137-4. [Google Scholar] [CrossRef]
- Martín, J.F.; Coton, M. Chapter 12—Blue Cheese: Microbiota and Fungal Metabolites. In Fermented Foods in Health and Disease Prevention; Frias, J., Martinez-Villaluenga, C., Peñas, E., Eds.; Academic Press: Boston, MA, USA, 2017; pp. 275–303. ISBN 978-0-12-802309-9. [Google Scholar] [CrossRef]
- Frisvad, J.C.; Samson, R.A. Polyphasic Taxonomy of Penicillium Subgenus Penicillium. A Guide to Identification of Food and Air-Borne Terverticillate Penicillia and Their Mycotoxins. Stud. Mycol. 2004, 49, C174. [Google Scholar]
- Sumarah, M.W.; Miller, J.D.; Blackwell, B.A. Isolation and Metabolite Production by Penicillium roqueforti, P. paneum and P. crustosum Isolated in Canada. Mycopathologia 2005, 159, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Mioso, R.; Toledo Marante, F.J.; Herrera Bravo de Laguna, I. Penicillium roqueforti: A Multifunctional Cell Factory of High Value-added Molecules. J. Appl. Microbiol. 2015, 118, 781–791. [Google Scholar] [CrossRef]
- Frisvad, J.C.; Smedsgaard, J.; Larsen, T.O.; Samson, R.A. Mycotoxins, Drugs and Other Extrolites Produced by Species in Penicillium Subgenus Penicillium. Stud. Mycol. 2004, 49, 201–241. [Google Scholar]
- O’Brien, M.; Nielsen, K.F.; O’Kiely, P.; Forristal, P.D.; Fuller, H.T.; Frisvad, J.C. Mycotoxins and Other Secondary Metabolites Produced in Vitro by Penicillium paneum Frisvad and Penicillium roqueforti Thom Isolated from Baled Grass Silage in Ireland. J. Agric. Food Chem. 2006, 54, 9268–9276. [Google Scholar] [CrossRef]
- Nielsen, K.F.; Sumarah, M.W.; Frisvad, J.C.; Miller, J.D. Production of Metabolites from the Penicillium roqueforti Complex. J. Agric. Food Chem. 2006, 54, 3756–3763. [Google Scholar] [CrossRef]
- Hymery, N.; Vasseur, V.; Coton, M.; Mounier, J.; Jany, J.L.; Barbier, G.; Coton, E. Filamentous Fungi and Mycotoxins in Cheese: A Review. Compr. Rev. Food Sci. Food Saf. 2014, 13, 437–456. [Google Scholar] [CrossRef]
- Martín, J.F.; Liras, P. Secondary Metabolites in Cheese Fungi. In Fungal Metabolites; Mérillon, J.-M., Ramawat, K.G., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 293–315. ISBN 978-3-319-25001-4. [Google Scholar]
- García-Estrada, C.; Martín, J.-F. Biosynthetic Gene Clusters for Relevant Secondary Metabolites Produced by Penicillium roqueforti in Blue Cheeses. Appl. Microbiol. Biotechnol. 2016, 100, 8303–8313. [Google Scholar] [CrossRef]
- Chávez, R.; Vaca, I.; García-Estrada, C. Secondary Metabolites Produced by the Blue-Cheese Ripening Mold Penicillium roqueforti; Biosynthesis and Regulation Mechanisms. J. Fungi 2023, 9, 459. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-P.; Yu, J.; Shu, Y.; Shi, Y.-X.; Luo, P.; Cai, L.; Ding, Z.-T. Peniroquesines A–C: Sesterterpenoids Possessing a 5-6-5-6-5-Fused Pentacyclic Ring System from Penicillium roqueforti YJ-14. Org. Lett. 2018, 20, 5853–5856. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-P.; Shu, Y.; Hu, J.-T.; Liu, R.; Cai, X.-Y.; Sun, C.-T.; Gan, D.; Zhou, D.-J.; Mei, R.-F.; Ding, H.; et al. Roquefornine A, a Sesterterpenoid with a 5/6/5/5/6-Fused Ring System from the Fungus Penicillium roqueforti YJ-1411. Org. Chem. Front. 2020, 7, 1463–1468. [Google Scholar] [CrossRef]
- Wang, J.-P.; Shu, Y.; Liu, R.; Gan, J.-L.; Deng, S.-P.; Cai, X.-Y.; Hu, J.-T.; Cai, L.; Ding, Z.-T. Bioactive Sesterterpenoids from the Fungus Penicillium roqueforti YJ-14. Phytochemistry 2021, 187, 112762. [Google Scholar] [CrossRef] [PubMed]
- Hammerl, R.; Frank, O.; Schmittnägel, T.; Ehrmann, M.A.; Hofmann, T. Functional Metabolome Analysis of Penicillium roqueforti by Means of Differential Off-Line LC–NMR. J. Agric. Food Chem. 2019, 67, 5135–5146. [Google Scholar] [CrossRef]
- Hammerl, R.; Frank, O.; Dietz, M.; Hirschmann, J.; Hofmann, T. Tyrosine Induced Metabolome Alterations of Penicillium roqueforti and Quantitation of Secondary Key Metabolites in Blue-Mold Cheese. J. Agric. Food Chem. 2019, 67, 8500–8509. [Google Scholar] [CrossRef]
- Zhang, W.; Du, L.; Qu, Z.; Zhang, X.; Li, F.; Li, Z.; Qi, F.; Wang, X.; Jiang, Y.; Men, P.; et al. Compartmentalized Biosynthesis of Mycophenolic Acid. Proc. Natl. Acad. Sci. USA 2019, 116, 13305–13310. [Google Scholar] [CrossRef]
- Kosalková, K.; Domínguez-Santos, R.; Coton, M.; Coton, E.; García-Estrada, C.; Liras, P.; Martín, J.F. A Natural Short Pathway Synthesizes Roquefortine C but Not Meleagrin in Three Different Penicillium roqueforti Strains. Appl. Microbiol. Biotechnol. 2015, 99, 7601–7612. [Google Scholar] [CrossRef]
- Ohmomo, S.; Sato, T.; Utagawa, T.; Abe, M. Isolation of Festuclavine and Three New Indole Alkaloids, Roquefortine A, B and C from the Cultures of Penicillium roqueforti. Agric. Biol. Chem. 1975, 39, 1333–1334. [Google Scholar] [CrossRef]
- Scott, P.M.; Merrien, M.-A.; Polonsky, J. Roquefortine and Isofumigaclavine A, Metabolites From Penicillium roqueforti. Experientia 1976, 32, 140–142. [Google Scholar] [CrossRef]
- Martín, J.F.; Liras, P. Evolutionary Formation of Gene Clusters by Reorganization: The Meleagrin/Roquefortine Paradigm in Different Fungi. Appl. Microbiol. Biotechnol. 2016, 100, 1579–1587. [Google Scholar] [CrossRef] [PubMed]
- Martín, J.-F.; Liras, P.; García-Estrada, C. Roquefortine C and Related Prenylated Indole Alkaloids. In Biosynthesis and Molecular Genetics of Fungal Secondary Metabolites; Martín, J.-F., García-Estrada, C., Zeilinger, S., Eds.; Springer New York: New York, NY, USA, 2014; pp. 111–128. ISBN 978-1-4939-1191-2. [Google Scholar]
- Wagener, R.E.; Davis, N.D.; Diener, U.L. Penitrem A and Roquefortine Production by Penicillium commune. Appl. Environ. Microbiol. 1980, 39, 882–887. [Google Scholar] [CrossRef] [PubMed]
- Lowes, N.; Smith, R.; Beck, B. Roquefortine in the Stomach Contents of Dogs Suspected of Strychnine Poisoning in Alberta. Can. Vet. J. Rev. Vét. Can. 1992, 33, 535–538. [Google Scholar]
- Häggblom, P. Isolation of Roquefortine C from Feed Grain. Appl. Environ. Microbiol. 1990, 56, 2924–2926. [Google Scholar] [CrossRef]
- Arnold, D.L.; Scott, P.M.; McGuire, P.F.; Harwig, J.; Nera, E.A. Acute Toxicity Studies on Roquefortine and PR Toxin, Metabolites of Penicillium roqueforti, in the Mouse. Food Cosmet. Toxicol. 1978, 16, 369–371. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, R.R.; Rasmussen, P.H.; Larsen, T.O.; Bladt, T.T.; Binderup, M.L. In Vitro Cytotoxicity of Fungi Spoiling Maize Silage. Food Chem. Toxicol. 2011, 49, 31–44. [Google Scholar] [CrossRef]
- Hymery, N.; Mounier, J.; Coton, E. Effect of Penicillium roqueforti Mycotoxins on Caco-2 Cells: Acute and Chronic Exposure. Toxicol. In Vitro 2018, 48, 188–194. [Google Scholar] [CrossRef]
- Aninat, C.; Hayashi, Y.; André, F.; Delaforge, M. Molecular Requirements for Inhibition of Cytochrome P450 Activities by Roquefortine. Chem. Res. Toxicol. 2001, 14, 1259–1265. [Google Scholar] [CrossRef]
- Kopp, B.; Rehm, H.J. Antimicrobial Action of Roquefortine. Eur. J. Appl. Microbiol. Biotechnol. 1979, 6, 397–401. [Google Scholar] [CrossRef]
- Kopp, B.; Rehm, H.J. Studies on the Inhibition of Bacterial Macromolecule Synthesis by Roquefortine, a Mycotoxin from Penicillium roqueforti. Eur. J. Appl. Microbiol. Biotechnol. 1981, 13, 232–235. [Google Scholar] [CrossRef]
- Scott, P.M.; Kennedy, B.P.; Harwig, J.; Blanchfield, B.J. Study of Conditions of Production of Roquefortine and Other Metabolites of Penicillium roqueforti. Appl. Environ. Microbiol. 1977, 33, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Frisvad, J.C.; Filtenborg, O. Terverticillate Penicillia: Chemotaxonomy and Mycotoxin Production. Mycologia 1989, 81, 837–861. [Google Scholar] [CrossRef]
- Fontaine, K.; Passeró, E.; Vallone, L.; Hymery, N.; Coton, M.; Jany, J.-L.; Mounier, J.; Coton, E. Occurrence of Roquefortine C, Mycophenolic Acid and Aflatoxin M1 Mycotoxins in Blue-Veined Cheeses. Food Control 2015, 47, 634–640. [Google Scholar] [CrossRef]
- García-Estrada, C.; Ullán, R.V.; Albillos, S.M.; Fernández-Bodega, M.Á.; Durek, P.; von Döhren, H.; Martín, J.F. A Single Cluster of Coregulated Genes Encodes the Biosynthesis of the Mycotoxins Roquefortine C and Meleagrin in Penicillium chrysogenum. Chem. Biol. 2011, 18, 1499–1512. [Google Scholar] [CrossRef]
- Ali, H.; Ries, M.I.; Nijland, J.G.; Lankhorst, P.P.; Hankemeier, T.; Bovenberg, R.A.L.; Vreeken, R.J.; Driessen, A.J.M. A Branched Biosynthetic Pathway Is Involved in Production of Roquefortine and Related Compounds in Penicillium chrysogenum. PLoS ONE 2013, 8, e65328. [Google Scholar] [CrossRef] [PubMed]
- Carver, T.J.; Rutherford, K.M.; Berriman, M.; Rajandream, M.-A.; Barrell, B.G.; Parkhill, J. ACT: The Artemis Comparison Tool. Bioinformatics 2005, 21, 3422–3423. [Google Scholar] [CrossRef]
- Scott, P.M.; Kennedy, B.P.C. Analysis of Blue Cheese for Roquefortine and Other Alkaloids from Penicillium roqueforti. J. Agric. Food Chem. 1976, 24, 865–868. [Google Scholar] [CrossRef]
- Boysen, M.; Skouboe, P.; Frisvad, J.; Rossen, L. Reclassification of the Penicillium roqueforti Group into Three Species on the Basis of Molecular Genetic and Biochemical Profiles. Microbiology 1996, 142, 541–549. [Google Scholar] [CrossRef]
- Hong, S.L.; Robbers, J.E. Genetics of Ergoline Alkaloid Formation in Penicillium roqueforti. Appl. Environ. Microbiol. 1985, 50, 558–561. [Google Scholar] [CrossRef]
- Vinokurova, N.; Boichenko, D.; Baskunov, B.; Zelenkova, N.; Vepritskaya, I.; Arinbasarov, M.; Reshetilova, T. Minor Alkaloids of the Fungus Penicillium roqueforti Thom 1906. Appl. Biochem. Microbiol. 2001, 37, 184–187. [Google Scholar] [CrossRef]
- McCabe, S.R.; Wipf, P. Total Synthesis, Biosynthesis and Biological Profiles of Clavine Alkaloids. Org. Biomol. Chem. 2016, 14, 5894–5913. [Google Scholar] [CrossRef] [PubMed]
- Starec, M.; Fiserova, A.; Rosina, J.; Malek, J.; Krsiak, M. Effect of Agroclavine on NK Activity In Vivo under Normal and Stress Conditions in Rats. Physiol. Res. Acad. Sci. Bohemoslov. 2001, 50, 513–519. [Google Scholar]
- Gerhards, N.; Li, S.-M. A Bifunctional Old Yellow Enzyme from Penicillium roqueforti Is Involved in Ergot Alkaloid Biosynthesis. Org. Biomol. Chem. 2017, 15, 8059–8071. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Bodega, Á.; Álvarez-Álvarez, R.; Liras, P.; Martín, J.F. Silencing of a Second Dimethylallyltryptophan Synthase of Penicillium roqueforti Reveals a Novel Clavine Alkaloid Gene Cluster. Appl. Microbiol. Biotechnol. 2017, 101, 6111–6121. [Google Scholar] [CrossRef] [PubMed]
- Bilovol, Y.; Panaccione, D.G. Functional Analysis of the Gene Controlling Hydroxylation of Festuclavine in the Ergot Alkaloid Pathway of Neosartorya fumigata. Curr. Genet. 2016, 62, 853–860. [Google Scholar] [CrossRef]
- Wallwey, C.; Matuschek, M.; Xie, X.-L.; Li, S.-M. Ergot Alkaloid Biosynthesis in Aspergillus fumigatus: Conversion of Chanoclavine-I Aldehyde to Festuclavine by the Festuclavine Synthase FgaFS in the Presence of the Old Yellow Enzyme FgaOx3. Org. Biomol. Chem. 2010, 8, 3500–3508. [Google Scholar] [CrossRef]
- Martín, J.F.; Álvarez-Álvarez, R.; Liras, P. Clavine Alkaloids Gene Clusters of Penicillium and Related Fungi: Evolutionary Combination of Prenyltransferases, Monooxygenases and Dioxygenases. Genes 2017, 8, 342. [Google Scholar] [CrossRef]
- Unsöld, I.A.; Li, S.-M. Overproduction, Purification and Characterization of FgaPT2, a Dimethylallyltryptophan Synthase from Aspergillus fumigatus. Microbiology 2005, 151, 1499–1505. [Google Scholar] [CrossRef]
- Rigbers, O.; Li, S.-M. Ergot Alkaloid Biosynthesis in Aspergillus fumigatus: Overproduction and Biochemical Characterization of a 4-Dimethylallyltryptophan N-Methyltransferase. J. Biol. Chem. 2008, 283, 26859–26868. [Google Scholar] [CrossRef]
- Ryan, K.L.; Moore, C.T.; Panaccione, D.G. Partial Reconstruction of the Ergot Alkaloid Pathway by Heterologous Gene Expression in Aspergillus nidulans. Toxins 2013, 5, 445–455. [Google Scholar] [CrossRef]
- Goetz, K.E.; Coyle, C.M.; Cheng, J.Z.; O’Connor, S.E.; Panaccione, D.G. Ergot Cluster-Encoded Catalase Is Required for Synthesis of Chanoclavine-I in Aspergillus fumigatus. Curr. Genet. 2011, 57, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Wallwey, C.; Matuschek, M.; Li, S.-M. Ergot Alkaloid Biosynthesis in Aspergillus fumigatus: Conversion of Chanoclavine-I to Chanoclavine-I Aldehyde Catalyzed by a Short-Chain Alcohol Dehydrogenase FgaDH. Arch. Microbiol. 2010, 192, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Coyle, C.M.; Cheng, J.Z.; O’Connor, S.E.; Panaccione, D.G. An Old Yellow Enzyme Gene Controls the Branch Point between Aspergillus fumigatus and Claviceps purpurea Ergot Alkaloid Pathways. Appl. Environ. Microbiol. 2010, 76, 3898–3903. [Google Scholar] [CrossRef]
- Liu, X.; Wang, L.; Steffan, N.; Yin, W.-B.; Li, S.-M. Ergot Alkaloid Biosynthesis in Aspergillus fumigatus: FgaAT Catalyses the Acetylation of Fumigaclavine B. ChemBioChem 2009, 10, 2325–2328. [Google Scholar] [CrossRef] [PubMed]
- Unsöld, I.A.; Li, S.-M. Reverse Prenyltransferase in the Biosynthesis of Fumigaclavine C in Aspergillus fumigatus: Gene Expression, Purification, and Characterization of Fumigaclavine C Synthase FGAPT1. ChemBioChem 2006, 7, 158–164. [Google Scholar] [CrossRef]
- Steiner, U.; Ahimsa-Müller, M.A.; Markert, A.; Kucht, S.; Groß, J.; Kauf, N.; Kuzma, M.; Zych, M.; Lamshöft, M.; Furmanowa, M.; et al. Molecular Characterization of a Seed Transmitted Clavicipitaceous Fungus Occurring on Dicotyledoneous Plants (Convolvulaceae). Planta 2006, 224, 533–544. [Google Scholar] [CrossRef]
- Havemann, J.; Vogel, D.; Loll, B.; Keller, U. Cyclolization of D-Lysergic Acid Alkaloid Peptides. Chem. Biol. 2014, 21, 146–155. [Google Scholar] [CrossRef]
- Young, C.A.; Schardl, C.L.; Panaccione, D.G.; Florea, S.; Takach, J.E.; Charlton, N.D.; Moore, N.; Webb, J.S.; Jaromczyk, J. Genetics, Genomics and Evolution of Ergot Alkaloid Diversity. Toxins 2015, 7, 1273–1302. [Google Scholar] [CrossRef]
- Gosio, B. Sperimentate Su Culture Pure Di Bacilli Del Carbonchio Demonstrarano Notevole Potere Antisettica. CR Acad. Med. Torino 1893, 61, 484. [Google Scholar]
- Bentley, R. Bartolomeo Gosio, 1863–1944: An Appreciation. In Advances in Applied Microbiology; Academic Press: Cambridge, MA, USA, 2001; Volume 48, pp. 229–250. ISBN 0065-2164. [Google Scholar]
- Bentley, R. Mycophenolic Acid: A One Hundred Year Odyssey from Antibiotic to Immunosuppressant. Chem. Rev. 2000, 100, 3801–3826. [Google Scholar] [CrossRef]
- Alsberg, C.; Black, O.F. Contributions to the Study of Maize Deterioration: Biochemical and Toxicological Investigations of Penicillium puberulum and Penicillium stoloniferum; US Government Printing Office: Washington, DC, USA, 1913.
- Clutterbuck, P.W.; Oxford, A.E.; Raistrick, H.; Smith, G. Studies in the Biochemistry of Micro-Organisms: The Metabolic Products of the Penicillium brevi-compactum Series. Biochem. J. 1932, 26, 1441. [Google Scholar] [CrossRef] [PubMed]
- Florey, H.W.; Jennings, M.A.; Gilliver, K.; Sanders, A.G. Mycophenolic Acid an Antibiotic from Penicillium brevicompactum Dierckx. Lancet 1946, 247, 46–49. [Google Scholar] [CrossRef] [PubMed]
- Abraham, E.P. The Effect of Mycophenolic Acid on the Growth of Staphylococcus aureus in Heart Broth. Biochem. J. 1945, 39, 398–408. [Google Scholar] [CrossRef]
- Kinoshita, H.; Wongsuntornpoj, S.; Ihara, F.; Nihira, T. Anti-Rhodotorula Activity of Mycophenolic Acid Enhanced in the Presence of Polyene Antibiotic Nystatin. Lett. Appl. Microbiol. 2017, 64, 144–149. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.W.; Chan, K.-H.; Kao, R.Y.T.; To, K.K.W.; Zheng, B.-J.; Li, C.P.Y.; Li, P.T.W.; Dai, J.; Mok, F.K.Y.; Chen, H.; et al. Broad-Spectrum Antivirals for the Emerging Middle East Respiratory Syndrome Coronavirus. J. Infect. 2013, 67, 606–616. [Google Scholar] [CrossRef]
- Silverman Kitchin, J.E.; Pomeranz, M.K.; Pak, G.; Washenik, K.; Shupack, J.L. Rediscovering Mycophenolic Acid: A Review of Its Mechanism, Side Effects, and Potential Uses. J. Am. Acad. Dermatol. 1997, 37, 445–449. [Google Scholar] [CrossRef]
- Jones, E.L.; Epinette, W.W.; Hackney, V.C.; Menendez, L.; Frost, P. Treatment of Psoriasis With Oral Mycophenolic Acid. J. Investig. Dermatol. 1975, 65, 537–542. [Google Scholar] [CrossRef]
- Domhan, S.; Muschal, S.; Schwager, C.; Morath, C.; Wirkner, U.; Ansorge, W.; Maercker, C.; Zeier, M.; Huber, P.E.; Abdollahi, A. Molecular Mechanisms of the Antiangiogenic and Antitumor Effects of Mycophenolic Acid. Mol. Cancer Ther. 2008, 7, 1656–1668. [Google Scholar] [CrossRef]
- Siebert, A.; Cholewiński, G.; Trzonkowski, P.; Rachon, J. Immunosuppressive Properties of Amino Acid and Peptide Derivatives of Mycophenolic Acid. Eur. J. Med. Chem. 2020, 189, 112091. [Google Scholar] [CrossRef]
- Allison, A.C.; Eugui, E.M. Mycophenolate Mofetil and Its Mechanisms of Action. Immunopharmacology 2000, 47, 85–118. [Google Scholar] [CrossRef]
- Adams, E.; Todd, G.; Gibson, W. Long-Term Toxicity Study of Mycophenolic Acid in Rabbits. Toxicol. Appl. Pharmacol. 1975, 34, 509–512. [Google Scholar] [CrossRef] [PubMed]
- Wilson, B.J. Miscellanous Penicillium Toxins. In Microbial Toxins: A Comprehensive Treatise; Ciegler, A., Kadis, S., Ajl, S.J., Eds.; Academic Press: New York, NY, USA; London, UK, 1971; pp. 460–517. [Google Scholar]
- Heischmann, S.; Dzieciatkowska, M.; Hansen, K.; Leibfritz, D.; Christians, U. The Immunosuppressant Mycophenolic Acid Alters Nucleotide and Lipid Metabolism in an Intestinal Cell Model. Sci. Rep. 2017, 7, 45088. [Google Scholar] [CrossRef] [PubMed]
- Wehner, F.C.; Thiel, P.G.; van Rensburg, S.J.; Demasius, I.P.C. Mutagenicity to Salmonella Typhimurium of Some Aspergillus and Penicillium Mycotoxins. Mutat. Res. Toxicol. 1978, 58, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Umeda, M.; Tsutsui, T.; Saito, M. Mutagenicity and Inducibility of DNA Single-Strand Breaks and Chromosome Aberrations by Various Mycotoxins. GANN Jpn. J. Cancer Res. 1977, 68, 619–625. [Google Scholar] [CrossRef]
- Cornforth, J. Terpenoid Biosynthesis. Chem. Br. 1968, 4, 102–106. [Google Scholar] [PubMed]
- McNaught, A.D.; Wilkinson, A. IUPAC. Compendium of Chemical Terminology, 2nd ed.; Blackwell Scientific Publications: Oxford, UK, 1997. [Google Scholar]
- Geris, R.; Simpson, T.J. Meroterpenoids Produced by Fungi. Nat. Prod. Rep. 2009, 26, 1063–1094. [Google Scholar] [CrossRef]
- Chen, X.; Wang, L.; Zhang, J.; Jiang, T.; Hu, C.; Li, D.; Zou, Y. Immunosuppressant Mycophenolic Acid Biosynthesis Employs a New Globin-like Enzyme for Prenyl Side Chain Cleavage. Acta Pharm. Sin. B 2019, 9, 1253–1258. [Google Scholar] [CrossRef]
- Del-Cid, A.; Gil-Durán, C.; Vaca, I.; Rojas-Aedo, J.F.; García-Rico, R.O.; Levicán, G.; Chávez, R. Identification and Functional Analysis of the Mycophenolic Acid Gene Cluster of Penicillium roqueforti. PLoS ONE 2016, 11, e0147047. [Google Scholar] [CrossRef]
- Regueira, T.B.; Kildegaard, K.R.; Hansen, B.G.; Mortensen, U.H.; Hertweck, C.; Nielsen, J. Molecular Basis for Mycophenolic Acid Biosynthesis in Penicillium brevicompactum. Appl. Environ. Microbiol. 2011, 77, 3035–3043. [Google Scholar] [CrossRef]
- Hansen, B.G.; Mnich, E.; Nielsen, K.F.; Nielsen, J.B.; Nielsen, M.T.; Mortensen, U.H.; Larsen, T.O.; Patil, K.R. Involvement of a Natural Fusion of a Cytochrome P450 and a Hydrolase in Mycophenolic Acid Biosynthesis. Appl. Environ. Microbiol. 2012, 78, 4908–4913. [Google Scholar] [CrossRef]
- Zhang, W.; Cao, S.; Qiu, L.; Qi, F.; Li, Z.; Yang, Y.; Huang, S.; Bai, F.; Liu, C.; Wan, X.; et al. Functional Characterization of MpaG′, the O-Methyltransferase Involved in the Biosynthesis of Mycophenolic Acid. ChemBioChem 2015, 16, 565–569. [Google Scholar] [CrossRef]
- Canonica, L.; Kroszczynski, W.; Ranzi, B.M.; Rindone, B.; Santaniello, E.; Scolastico, C. Biosynthesis of Mycophenolic Acid. J. Chem. Soc. Perkin 1 1972, 116, 2639–2643. [Google Scholar] [CrossRef]
- You, C.; Li, F.; Zhang, X.; Ma, L.; Zhang, Y.-Z.; Zhang, W.; Li, S. Structural Basis for Substrate Specificity of the Peroxisomal Acyl-CoA Hydrolase MpaH’ Involved in Mycophenolic Acid Biosynthesis. FEBS J. 2021, 288, 5768–5780. [Google Scholar] [CrossRef]
- Hansen, B.G.; Genee, H.J.; Kaas, C.S.; Nielsen, J.B.; Regueira, T.B.; Mortensen, U.H.; Frisvad, J.C.; Patil, K.R. A New Class of IMP Dehydrogenase with a Role in Self-Resistance of Mycophenolic Acid Producing Fungi. BMC Microbiol. 2011, 11, 202. [Google Scholar] [CrossRef]
- Gillot, G.; Jany, J.-L.; Dominguez-Santos, R.; Poirier, E.; Debaets, S.; Hidalgo, P.I.; Ullán, R.V.; Coton, E.; Coton, M. Genetic Basis for Mycophenolic Acid Production and Strain-Dependent Production Variability in Penicillium roqueforti. Food Microbiol. 2017, 62, 239–250. [Google Scholar] [CrossRef]
- Fontaine, K.; Hymery, N.; Lacroix, M.Z.; Puel, S.; Puel, O.; Rigalma, K.; Gaydou, V.; Coton, E.; Mounier, J. Influence of Intraspecific Variability and Abiotic Factors on Mycotoxin Production in Penicillium roqueforti. Int. J. Food Microbiol. 2015, 215, 187–193. [Google Scholar] [CrossRef]
- Rojas-Aedo, J.F.; Gil-Durán, C.; Goity, A.; Vaca, I.; Levicán, G.; Larrondo, L.F.; Chávez, R. The Developmental Regulator Pcz1 Affects the Production of Secondary Metabolites in the Filamentous Fungus Penicillium roqueforti. Microbiol. Res. 2018, 212–213, 67–74. [Google Scholar] [CrossRef]
- Torrent, C.; Gil-Durán, C.; Rojas-Aedo, J.F.; Medina, E.; Vaca, I.; Castro, P.; García-Rico, R.O.; Cotoras, M.; Mendoza, L.; Levicán, G.; et al. Role of Sfk1 Gene in the Filamentous Fungus Penicillium roqueforti. Front. Microbiol. 2017, 8, 2424. [Google Scholar] [CrossRef]
- García-Rico, R.; Chavez, R.; Fierro, F.; Martin, J. Effect of a Heterotrimeric G Protein α Subunit on Conidia Germination, Stress Response, and Roquefortine C Production in Penicillium roqueforti. Int. Microbiol. Off. J. Span. Soc. Microbiol. 2009, 12, 123–129. [Google Scholar] [CrossRef]
- Vinokurova, N.G.; Ivanushkina, N.E.; Kochkina, G.A.; Arinbasarov, M.U.; Ozerskaya, S.M. Production of Mycophenolic Acid by Fungi of the Genus Penicillium Link. Appl. Biochem. Microbiol. 2005, 41, 83–86. [Google Scholar] [CrossRef]
- Chen, M.; Wang, J.; Lin, L.; Xu, X.; Wei, W.; Shen, Y.; Wei, D. Synergistic Regulation of Metabolism by Ca2+/Reactive Oxygen Species in Penicillium brevicompactum Improves Production of Mycophenolic Acid and Investigation of the Ca2+ Channel. ACS Synth. Biol. 2022, 11, 273–285. [Google Scholar] [CrossRef]
- El-Sayed, E.-S.R.; Zaki, A.G. Unlocking the Biosynthetic Potential of Penicillium roqueforti for Hyperproduction of the Immunosuppressant Mycophenolic Acid: Gamma Radiation Mutagenesis and Response Surface Optimization of Fermentation Medium. Biotechnol. Appl. Biochem. 2023, 70, 306–317. [Google Scholar] [CrossRef]
- Rojas-Aedo, J.F.; Gil-Durán, C.; Del-Cid, A.; Valdés, N.; Álamos, P.; Vaca, I.; García-Rico, R.O.; Levicán, G.; Tello, M.; Chávez, R. The Biosynthetic Gene Cluster for Andrastin A in Penicillium roqueforti. Front. Microbiol. 2017, 8, 813. [Google Scholar] [CrossRef]
- Uchida, R.; Shiomi, K.; Inokoshi, J.; Sunazuka, T.; Tanaka, H.; Iwai, Y.; Takayanagi, H.; Omura, S. Andrastins A-C, New Protein Farnesyltransferase Inhibitors Produced by Penicillium sp. FO-3929. 2. Structure Elucidation and Biosynthesis. J. Antibiot. 1996, 49, 418–424. [Google Scholar] [CrossRef]
- Shiomi, K.; Uchida, R.; Inokoshi, J.; Tanaka, H.; Iwai, Y.; Ōmura, S. Andrastins A∼C, New Protein Farnesyltransferase Inhibitors, Produced by Penicillium sp. FO-3929. Tetrahedron Lett. 1996, 37, 1265–1268. [Google Scholar] [CrossRef]
- Rho, M.-C.; Toyoshima, M.; Hayashi, M.; Uchida, R.; Shiomi, K.; Komiyama, K.; Ōmura, S. Enhancement of Drug Accumulation by Andrastin A Produced by Penicillium sp. FO-3929 in Vincristine-Resistant KB Cells. J. Antibiot. 1998, 51, 68–72. [Google Scholar] [CrossRef]
- Fernández-Bodega, M.A.; Mauriz, E.; Gómez, A.; Martín, J.F. Proteolytic Activity, Mycotoxins and Andrastin A in Penicillium roqueforti Strains Isolated from Cabrales, Valdeón and Bejes–Tresviso Local Varieties of Blue-Veined Cheeses. Int. J. Food Microbiol. 2009, 136, 18–25. [Google Scholar] [CrossRef]
- Nielsen, K.F.; Dalsgaard, P.W.; Smedsgaard, J.; Larsen, T.O. Andrastins A−D, Penicillium roqueforti Metabolites Consistently Produced in Blue-Mold-Ripened Cheese. J. Agric. Food Chem. 2005, 53, 2908–2913. [Google Scholar] [CrossRef]
- Matsuda, Y.; Awakawa, T.; Abe, I. Reconstituted Biosynthesis of Fungal Meroterpenoid Andrastin A. Tetrahedron 2013, 69, 8199–8204. [Google Scholar] [CrossRef]
- Lo, H.-C.; Entwistle, R.; Guo, C.-J.; Ahuja, M.; Szewczyk, E.; Hung, J.-H.; Chiang, Y.-M.; Oakley, B.R.; Wang, C.C.C. Two Separate Gene Clusters Encode the Biosynthetic Pathway for the Meroterpenoids Austinol and Dehydroaustinol in Aspergillus nidulans. J. Am. Chem. Soc. 2012, 134, 4709–4720. [Google Scholar] [CrossRef]
- Matsuda, Y.; Awakawa, T.; Itoh, T.; Wakimoto, T.; Kushiro, T.; Fujii, I.; Ebizuka, Y.; Abe, I. Terretonin Biosynthesis Requires Methylation as Essential Step for Cyclization. ChemBioChem 2012, 13, 1738–1741. [Google Scholar] [CrossRef] [PubMed]
- Martín, J.F. Vacuolal and Peroxisomal Calcium Ion Transporters in Yeasts and Fungi: Key Role in the Translocation of Intermediates in the Biosynthesis of Fungal Metabolites. Genes 2022, 13, 1450. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Aguado, M.; Ullán, R.V.; Teijeira, F.; Rodríguez-Castro, R.; Martín, J.F. The Transport of Phenylacetic Acid across the Peroxisomal Membrane Is Mediated by the PaaT Protein in Penicillium chrysogenum. Appl. Microbiol. Biotechnol. 2013, 97, 3073–3084. [Google Scholar] [CrossRef] [PubMed]
- van den Berg, M.A.; Albang, R.; Albermann, K.; Badger, J.H.; Daran, J.-M.; Driessen, A.J.M.; Garcia-Estrada, C.; Fedorova, N.D.; Harris, D.M.; Heijne, W.H.M.; et al. Genome Sequencing and Analysis of the Filamentous Fungus Penicillium chrysogenum. Nat. Biotechnol. 2008, 26, 1161–1168. [Google Scholar] [CrossRef]
- Kordula, B.; Sandra, Z.; Katharina, L.; Michael, F.; Ulrich, K. Genome-Wide Chromatin Immunoprecipitation Sequencing Analysis of the Penicillium chrysogenum Velvet Protein PcVelA Identifies Methyltransferase PcLlmA as a Novel Downstream Regulator of Fungal Development. mSphere 2016, 1, e00149-16. [Google Scholar] [CrossRef]
- Dubey, M.K.; Aamir, M.; Kaushik, M.S.; Khare, S.; Meena, M.; Singh, S.; Upadhyay, R.S. PR Toxin–Biosynthesis, Genetic Regulation, Toxicological Potential, Prevention and Control Measures: Overview and Challenges. Front. Pharmacol. 2018, 9, 288. [Google Scholar] [CrossRef]
- Wei, R.-D.; Still Paul, E.; Smalley, E.B.; Schnoes, H.K.; Strong, F.M. Isolation and Partial Characterization of a Mycotoxin from Penicillium roqueforti. Appl. Microbiol. 1973, 25, 111–114. [Google Scholar] [CrossRef]
- Wei, R.; Schnoes, H.K.; Hart, P.A.; Strong, F.M. The Structure of PR Toxin, a Mycotoxin from Penicillium roqueforti. Tetrahedron 1975, 31, 109–114. [Google Scholar] [CrossRef]
- Chen, F.-C.; Chen, C.-F.; Wei, R.-D. Acute Toxicity of PR Toxin, a Mycotoxin from Penicillium roqueforti. Toxicon 1982, 20, 433–441. [Google Scholar] [CrossRef]
- Lee, Y.-H.; Fang, S.-C.; Wei, R.-D. The Effects of Penicillium roqueforti Toxin on the Activity of Rat Hepatic DNA Polymerases. Toxicology 1984, 33, 43–57. [Google Scholar] [CrossRef]
- Moule, Y.; Jemmali, M.; Rousseau, N. Mechanism of the Inhibition of Transcription by Pr Toxin, a Mycotoxin from Penicillium roqueforti. Chem. Biol. Interact. 1976, 14, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Moulé, Y.; Jemmali, M.; Darracq, N. Inhibition of Protein Synthesis by PR Toxin, a Mycotoxin from Penicillium roqueforti. FEBS Lett. 1978, 88, 341–344. [Google Scholar] [CrossRef] [PubMed]
- Ueno, Y.; Kubota, K.; Ito, T.; Nakamura, Y. Mutagenicity of Carcinogenic Mycotoxins in Salmonella Typhimurium1. Cancer Res. 1978, 38, 536–542. [Google Scholar] [PubMed]
- Polonelli, L.; Lauriola, L.; Morace, G. Preliminary Studies on the Carcinogenic Effects of P Penicillium roqueforti Toxin (PR Toxin) on Rats. Mycopathologia 1982, 78, 125–127. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.-H.; Ding, W.-H.; Wei, R.-D. Biochemical Effects of PR Toxin on Rat Liver Mitochondrial Respiration and Oxidative Phosphorylation. Arch. Biochem. Biophys. 1984, 230, 400–411. [Google Scholar] [CrossRef]
- Hymery, N.; Puel, O.; Tadrist, S.; Canlet, C.; Scouarnec, H.; Coton, E.; Coton, M. Effect of PR Toxin on THP1 and Caco-2 Cells: An in Vitro Study. World Mycotoxin J. 2017, 10, 375–386. [Google Scholar] [CrossRef]
- Chang, S.C.; Wei, Y.H.; Wei, D.L.; Chen, Y.Y.; Jong, S.C. Factors Affecting the Production of Eremofortin C and PR Toxin in Penicillium roqueforti. Appl. Environ. Microbiol. 1991, 57, 2581–2585. [Google Scholar] [CrossRef]
- Rasmussen, R.R.; Storm, I.M.L.D.; Rasmussen, P.H.; Smedsgaard, J.; Nielsen, K.F. Multi-Mycotoxin Analysis of Maize Silage by LC-MS/MS. Anal. Bioanal. Chem. 2010, 397, 765–776. [Google Scholar] [CrossRef]
- Siemens, K.; Zawistowski, J. Occurrence of PR Imine, a Metabolite of Penicillium roqueforti, in Blue Cheese. J. Food Prot. 1993, 56, 317–320. [Google Scholar] [CrossRef]
- Polonelli, L.; Morace, G.; Monache, F.D.; Samson, R.A. Studies on the PR Toxin of Penicillium roqueforti. Mycopathologia 1978, 66, 99–104. [Google Scholar] [CrossRef]
- Moulé, Y.; Moreau, S.; Bousquet, J.F. Relationships between the Chemical Structure and the Biological Properties of Some Eremophilane Compounds Related to PR Toxin. Chem. Biol. Interact. 1977, 17, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Scott, P.; Kanhere, S. Instability of PR Toxin in Blue Cheese. J. Assoc. Off. Anal. Chem. 1979, 62, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.C.; Lu, K.L.; Yeh, S.F. Secondary Metabolites Resulting from Degradation of PR Toxin by Penicillium roqueforti. Appl. Environ. Microbiol. 1993, 59, 981–986. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.-C.; Yeh, S.-F.; Li, S.-Y.; Lei, W.-Y.; Chen, M.-Y. A Novel Secondary Metabolite Relative to the Degradation of PR Toxin by Penicillium roqueforti. Curr. Microbiol. 1996, 32, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.-C.; Cheng, M.-K.; Wei, Y.-H. Production of PR-Imine, PR-Acid, and PR-Amide Relative to the Metabolism of PR Toxin by Penicillium roqueforti. Fungal Sci. 2004, 19, 39–46. [Google Scholar] [CrossRef]
- Chang, S.-C.; Ho, C.-P.; Cheng, M.-K. Isolation, Purification, and Characterization of the PR-Amide Synthetase from Penicillium roqueforti. Fung Sci. 2004, 19, 117–123. [Google Scholar]
- Chang, S.-C.; Lei, W.-Y.; Tsai, Y.-C.; Wei, Y.-H. Isolation, Purification, and Characterization of the PR Oxidase from Penicillium roqueforti. Appl. Environ. Microbiol. 1998, 64, 5012–5015. [Google Scholar] [CrossRef]
- Hohn, T.M.; Plattner, R.D. Purification and Characterization of the Sesquiterpene Cyclase Aristolochene Synthase from Penicillium roqueforti. Arch. Biochem. Biophys. 1989, 272, 137–143. [Google Scholar] [CrossRef]
- Riclea, R.; Dickschat, J.S. Identification of Intermediates in the Biosynthesis of PR Toxin by Penicillium roqueforti. Angew. Chem. Int. Ed. 2015, 54, 12167–12170. [Google Scholar] [CrossRef]
- Brock, N.L.; Dickschat, J.S. PR Toxin Biosynthesis in Penicillium roqueforti. ChemBioChem 2013, 14, 1189–1193. [Google Scholar] [CrossRef]
- Moreau, S.; Lablache-Combier, A.; Biguet, J. Production of Eremofortins A, B, and C Relative to Formation of PR Toxin by Penicillium roqueforti. Appl. Environ. Microbiol. 1980, 39, 770–776. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.-C.; Wei, Y.-H.; Liu, M.-L.; Wei, R.-D. Isolation and Some Properties of the Enzyme That Transforms Eremofortin C to PR Toxin. Appl. Environ. Microbiol. 1985, 49, 1455–1460. [Google Scholar] [CrossRef] [PubMed]
- Moreau, S.; Cacan, M.; Lablache-Combier, A. Eremofortin C, a New Metabolite Obtained from Penicillium roqueforti Cultures and from Biotransformation of PR Toxin. J. Org. Chem. 1977, 42, 2632–2634. [Google Scholar] [CrossRef] [PubMed]
- Hidalgo, P.I.; Ullán, R.V.; Albillos, S.M.; Montero, O.; Fernández-Bodega, M.Á.; García-Estrada, C.; Fernández-Aguado, M.; Martín, J.-F. Molecular Characterization of the PR-Toxin Gene Cluster in Penicillium roqueforti and Penicillium chrysogenum: Cross Talk of Secondary Metabolite Pathways. Fungal Genet. Biol. 2014, 62, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Hidalgo, P.I.; Poirier, E.; Ullán, R.V.; Piqueras, J.; Meslet-Cladière, L.; Coton, E.; Coton, M. Penicillium roqueforti PR Toxin Gene Cluster Characterization. Appl. Microbiol. Biotechnol. 2017, 101, 2043–2056. [Google Scholar] [CrossRef]
- Cane, D.E.; Wu, Z.; Proctor, R.H.; Hohn, T.M. Overexpression in Escherichia coli of Soluble Aristolochene Synthase from Penicillium roqueforti. Arch. Biochem. Biophys. 1993, 304, 415–419. [Google Scholar] [CrossRef]
- Proctor, R.H.; Hohn, T.M. Aristolochene Synthase. Isolation, Characterization, and Bacterial Expression of a Sesquiterpenoid Biosynthetic Gene (ari1) from Penicillium roqueforti. J. Biol. Chem. 1993, 268, 4543–4548. [Google Scholar] [CrossRef]
- Frisvad, J.C.; Houbraken, J.; Popma, S.; Samson, R.A. Two New Penicillium Species Penicillium buchwaldii and Penicillium spathulatum, Producing the Anticancer Compound Asperphenamate. FEMS Microbiol. Lett. 2013, 339, 77–92. [Google Scholar] [CrossRef]
- Lai, S.; Shizuri, Y.; Yamamura, S.; Kawai, K.; Furukawa, H. Three New Phenolic Metalolites from Penicillium Species. Heterocycles 1991, 32, 297–305. [Google Scholar]
- Larsen, T.O.; Breinholt, J. Dichlorodiaportin, Diaportinol, and Diaportinic Acid: Three Novel Isocoumarins from Penicillium nalgiovense. J. Nat. Prod. 1999, 62, 1182–1184. [Google Scholar] [CrossRef]
- Asiri, I.A.M.; Badr, J.M.; Youssef, D.T.A. Penicillivinacine, Antimigratory Diketopiperazine Alkaloid from the Marine-Derived Fungus Penicillium vinaceum. Phytochem. Lett. 2015, 13, 53–58. [Google Scholar] [CrossRef]
- Cui, H.; Liu, Y.; Nie, Y.; Liu, Z.; Chen, S.; Zhang, Z.; Lu, Y.; He, L.; Huang, X.; She, Z. Polyketides from the Mangrove-Derived Endophytic Fungus Nectria sp. HN001 and Their α-Glucosidase Inhibitory Activity. Mar. Drugs 2016, 14, 86. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, J.L.; Nielsen, K.F.; Sondergaard, T.E. Redirection of Pigment Biosynthesis to Isocoumarins in Fusarium. Fungal Genet. Biol. 2012, 49, 613–618. [Google Scholar] [CrossRef] [PubMed]
- Costa, J.H.; Wassano, C.I.; Angolini, C.F.F.; Scherlach, K.; Hertweck, C.; Pacheco Fill, T. Antifungal Potential of Secondary Metabolites Involved in the Interaction between Citrus Pathogens. Sci. Rep. 2019, 9, 18647. [Google Scholar] [CrossRef]
- Bertinetti, B.V.; Peña, N.I.; Cabrera, G.M. An Antifungal Tetrapeptide from the Culture of Penicillium canescens. Chem. Biodivers. 2009, 6, 1178–1184. [Google Scholar] [CrossRef]
- Salvatore, M.M.; Nicoletti, R.; DellaGreca, M.; Andolfi, A. Occurrence and Properties of Thiosilvatins. Mar. Drugs 2019, 17, 664. [Google Scholar] [CrossRef]
- Vinale, F.; Salvatore, M.M.; Nicoletti, R.; Staropoli, A.; Manganiello, G.; Venneri, T.; Borrelli, F.; DellaGreca, M.; Salvatore, F.; Andolfi, A. Identification of the Main Metabolites of a Marine-Derived Strain of Penicillium brevicompactum Using LC and GC MS Techniques. Metabolites 2020, 10, 55. [Google Scholar] [CrossRef]
- Capon, R.J.; Stewart, M.; Ratnayake, R.; Lacey, E.; Gill, J.H. Citromycetins and Bilains A–C: New Aromatic Polyketides and Diketopiperazines from Australian Marine-Derived and Terrestrial Penicillium spp. J. Nat. Prod. 2007, 70, 1746–1752. [Google Scholar] [CrossRef]
- Usami, Y.; Aoki, S.; Hara, T.; Numata, A. New Dioxopiperazine Metabolites from a Fusarium Species Separated from a Marine Alga. J. Antibiot. 2002, 55, 655–659. [Google Scholar] [CrossRef]
- Huber, E.M. Epipolythiodioxopiperazine-Based Natural Products: Building Blocks, Biosynthesis and Biological Activities. ChemBioChem 2022, 23, e202200341. [Google Scholar] [CrossRef]
- Yang, F.; Cheng, L.; Du, Y.; Xia, L.; Long, C. Functional Identification of the DHN Melanin Synthesis Gene Cluster and Its Role in UV-C Tolerance in Citrus Postharvest Pathogenic Fungus Penicillium digitatum. Fungal Biol. 2022, 126, 566–575. [Google Scholar] [CrossRef] [PubMed]
- Seekles, S.J.; Teunisse, P.P.P.; Punt, M.; van den Brule, T.; Dijksterhuis, J.; Houbraken, J.; Wösten, H.A.B.; Ram, A.F.J. Preservation Stress Resistance of Melanin Deficient Conidia from Paecilomyces variotii and Penicillium roqueforti Mutants Generated via CRISPR/Cas9 Genome Editing. Fungal Biol. Biotechnol. 2021, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Tam, E.W.T.; Chong, K.T.K.; Cai, J.J.; Tung, E.T.K.; Ngan, A.H.Y.; Lau, S.K.P.; Yuen, K.-Y. High Diversity of Polyketide Synthase Genes and the Melanin Biosynthesis Gene Cluster in Penicillium marneffei. FEBS J. 2010, 277, 3750–3758. [Google Scholar] [CrossRef] [PubMed]
- Perez-Cuesta, U.; Aparicio-Fernandez, L.; Guruceaga, X.; Martin-Souto, L.; Abad-Diaz-de-Cerio, A.; Antoran, A.; Buldain, I.; Hernando, F.L.; Ramirez-Garcia, A.; Rementeria, A. Melanin and Pyomelanin in Aspergillus fumigatus: From Its Genetics to Host Interaction. Int. Microbiol. 2020, 23, 55–63. [Google Scholar] [CrossRef]
- van der Nest, M.A.; Chávez, R.; De Vos, L.; Duong, T.A.; Gil-Durán, C.; Ferreira, M.A.; Lane, F.A.; Levicán, G.; Santana, Q.C.; Steenkamp, E.T.; et al. IMA Genome-F14. IMA Fungus 2021, 12, 5. [Google Scholar] [CrossRef]
- Coton, E.; Coton, M.; Hymery, N.; Mounier, J.; Jany, J.-L. Penicillium Roqueforti: An Overview of Its Genetics, Physiology, Metabolism and Biotechnological Applications. Fungal Biol. Rev. 2020, 34, 59–73. [Google Scholar] [CrossRef]
- Wei, Q.; Bai, J.; Yan, D.; Bao, X.; Li, W.; Liu, B.; Zhang, D.; Qi, X.; Yu, D.; Hu, Y. Genome Mining Combined Metabolic Shunting and OSMAC Strategy of an Endophytic Fungus Leads to the Production of Diverse Natural Products. Acta Pharm. Sin. B 2021, 11, 572–587. [Google Scholar] [CrossRef]
- Mao, X.-M.; Xu, W.; Li, D.; Yin, W.-B.; Chooi, Y.-H.; Li, Y.-Q.; Tang, Y.; Hu, Y. Epigenetic Genome Mining of an Endophytic Fungus Leads to the Pleiotropic Biosynthesis of Natural Products. Angew. Chem. Int. Ed. 2015, 54, 7592–7596. [Google Scholar] [CrossRef]
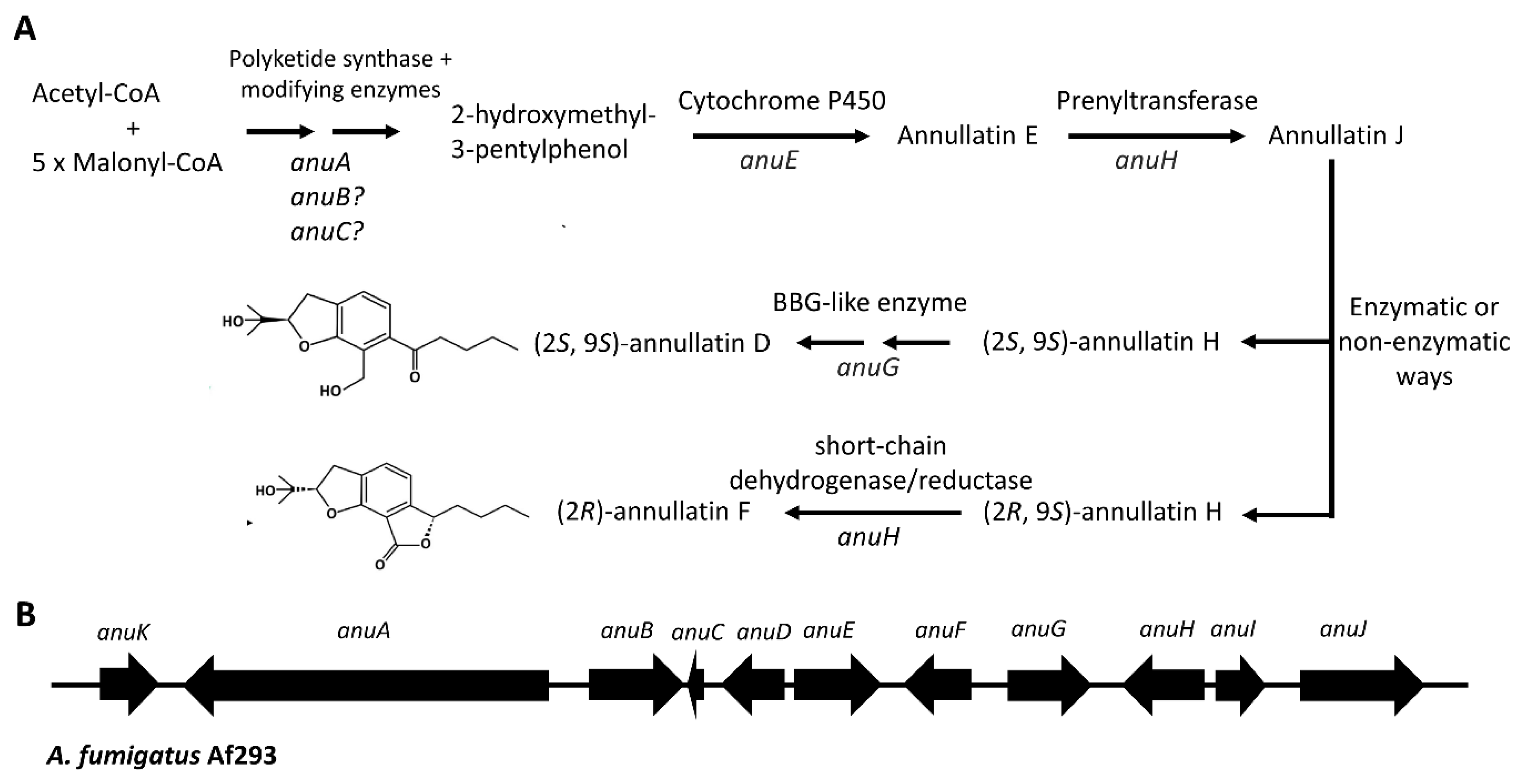
- Xiang, P.; Kemmerich, B.; Yang, L.; Li, S.-M. Biosynthesis of Annullatin D in Penicillium roqueforti Implies Oxidative Lactonization between Two Hydroxyl Groups Catalyzed by a BBE-like Enzyme. Org. Lett. 2022, 24, 6072–6077. [Google Scholar] [CrossRef]
- Asai, T.; Luo, D.; Obara, Y.; Taniguchi, T.; Monde, K.; Yamashita, K.; Oshima, Y. Dihydrobenzofurans as Cannabinoid Receptor Ligands from Cordyceps annullata, an Entomopathogenic Fungus Cultivated in the Presence of an HDAC Inhibitor. Tetrahedron Lett. 2012, 53, 2239–2243. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Metin, B. Penicillium roqueforti Secondary Metabolites: Biosynthetic Pathways, Gene Clusters, and Bioactivities. Fermentation 2023, 9, 836. https://doi.org/10.3390/fermentation9090836
Metin B. Penicillium roqueforti Secondary Metabolites: Biosynthetic Pathways, Gene Clusters, and Bioactivities. Fermentation. 2023; 9(9):836. https://doi.org/10.3390/fermentation9090836
Chicago/Turabian StyleMetin, Banu. 2023. "Penicillium roqueforti Secondary Metabolites: Biosynthetic Pathways, Gene Clusters, and Bioactivities" Fermentation 9, no. 9: 836. https://doi.org/10.3390/fermentation9090836
APA StyleMetin, B. (2023). Penicillium roqueforti Secondary Metabolites: Biosynthetic Pathways, Gene Clusters, and Bioactivities. Fermentation, 9(9), 836. https://doi.org/10.3390/fermentation9090836





