Abstract
This study was aimed at screening potential probiotic candidates to enhance the antimicrobial activity of Artemisia argyi against foodborne pathogens. Ten LAB strains were isolated from natural Artemisia argyi fermentation liquor (AAFL) and assessed for safety and antimicrobial ability. Therein, Lactiplantibacillus plantarum WLPL01, Lacticaseibacillus casei WLCA01, WLCA02, and WLCA03, and Lactobacillus harbiness WLHA01 were further evaluated for their potential probiotic properties (gastrointestinal tolerance and adhesion capacity). The results suggested that L. plantarum WLPL01 exhibited excellent properties and was, therefore, selected as the starter for A. argyi leaves fermentation. Then, L. plantarum WLPL01-fermented AAFL (AAFL-LP) was further investigated for its antimicrobial activity against foodborne pathogens. The results demonstrated that the inhibitory effect of AAFL-LP to foodborne pathogens, such as Listeria monocytogenes CMCC54007, Salmonella Typhimurium ATCC 13311, and Candida albicans ATCC 14053, was enhanced when compared to spontaneously fermented AAFL (AAFL-spontaneous). In addition, an analysis of the whole genome of L. plantarum WLPL01 revealed the presence of 13.9 kb long and 16 plantaricin-encoding loci (pln locus), and the increased antimicrobial activities of AAFL-LP might correlate with the production of bacteriocin. Our results indicate that L. plantarum WLPL01 can be used as a starter for Artemisia argyi fermentation to enhance its antimicrobial activity against foodborne pathogens.
1. Introduction
Artemisia argyi (AA) is a herbaceous plant distributed in most regions of East Asia. It is frequently used as a traditional herbal medicine for the treatment of microbial infections, osteoarthritis, inflammatory diseases, asthma, diarrhea, hepatitis, malaria, cancer, and circulatory disorders [1,2] due to its abundant bioactive compounds such as Artemisia oil, glycosides, coumarins, flavonoids, polyacetylenes, monoterpenes, triterpenes, and sesquiterpene lactones [3,4,5,6]. Recently, AA found increasing applications as a feed additive in animal production for decreasing diarrhea and regulating gastrointestinal function [7], improving the breeding environment, and increasing animal feed intake [8]. Fermentation is a traditional process in which microorganisms or enzymes can generate biological compounds, and simultaneously the metabolism of microorganisms can be altered to produce new ingredients [9,10].
Traditional or artisanal fermented foods are deemed as abundant sources of lactic acid bacteria (LAB). For instance, Lactiplantibacillus plantarum Bom 816, L. paracasei B41, and L. pentosus N3 isolated from a traditional Turkish fermented cereal beverage boza showed antimicrobial activity against Escherichia coli, Klebsiella pneumoniae, Vibrio cholerae, and Bacillus subtilis [11], and L. plantarum ULAG24 and ULAG11 from West African fermented cereals demonstrated antagonism to foodborne pathogens [12]. Gioia et al. reported that L. plantarum PCS20 was capable of effectively surviving in ground meat and performing antimicrobial activity in carnis against Clostridium [13].
On the other hand, LAB are widely used as a starter culture to control the fermentation rate, extend the shelf-life, improve functionality, ameliorate flavor, and ensure safety [14,15]. For example, the antagonistic effect of A. princeps was enhanced after fermentation with Bifidobacterium infantis K-525 [16]. Similarly, Jo’s results confirmed that Lactobacillus pentosus SMB718 isolated from Korean traditional paste has many beneficial probiotic activities, such as antimicrobial and anti-inflammatory effects, and it could promote a significantly higher amount of allyl mercaptan production when used as a starter in garlic and onion fermentation [17].
However, reports that describe the distribution of native flora, e.g., the constitution of LAB strains in AAFL, are scarce, and research on the antagonistic effect of AAFL artificially fermented by native species of LAB is even more so. In this study, LAB strains from a naturally fermented A. argyi liquor were isolated and evaluated for the probiotic properties and food safety indexes. Based on the antibiotic sensitivity and survival ability in an artificial gastrointestinal tract (GI), a native strain of L. plantarum WLPL01 was screened out. Subsequently, L. plantarum WLPL01 was used as a starter to ferment A. argyi, and the antagonistic effect of L. plantarum WLPL01-fermented AAFL (AAFL-LP) against pathogens was compared with that of AAFL-spontaneous. Moreover, the bacteriocin-encoding genes in the genome of L. plantarum WLPL01 were bioinformatically analyzed. This work might provide a fundamental basis for using A. argyi fermentation liquor as a feed additive to control pathogens in the breeding industry.
2. Materials and Methods
2.1. Isolation and Identification of Strains from AAFL
Artemisia argyi fermentation liquor (AAFL) was presented by Professor Ziniu Yu (Huazhong Agricultural University, Wuhan, China). To isolate LAB strains, an aliquot of AAFL was 10-fold diluted and plated on De Man Rogosa and Sharpe (MRS) agar (Solarbio, Beijing, China), KFS agar (Hopebiol, Qingdao, China), and M17 agar (Hopebiol, Qingdao, China). After anaerobic incubation for 48 h at 37 °C in a thermostatic anaerobic incubator chamber (Gene Science, San Diego, CA, USA), single colonies were selected and purified on an MRS agar plate. Then, the obtained strains were screened by Gram staining and catalase testing. The Gram-positive and catalase-negative isolates were further identified by PCR with 16S rDNA universal primers, 27F and 1492R [18], the amplicons were sequenced by Sangon (Sangon, Shanghai, China), and the sequencing results were compared with the NCBI database using the BLAST algorithm (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 1 October 2015) to identify species.
2.2. Phylogenetic Analysis
The genetic presence and comparison of 16S rRNA was performed by analyzing the nucleotide sequence data available at the National Center for Biotechnology Information (NCBI) database. The 16S rRNA gene sequences of related organisms were obtained from the NCBI database and compared with the sequences of our strains to establish the closest neighbor in the evolutionary tree. The neighbor-joining (NJ) phylogenetic tree with the p-distance model was constructed using the MEGA software (version 7.0, Mega Limited, Auckland, New Zealand).
2.3. Growth Curve and Acid Production
The strains were inoculated (1%, v/v) and incubated anaerobically at 37 °C, the OD600 was measured using a microplate reader (Varioskan™ LUX, Waltham, MA, USA), and the pH was monitored using a pH meter (Bell, Dalian, China).
2.4. Antimicrobial Activity Assay
The antimicrobial activity of LAB was analyzed according to our previous report [19] with some modifications. Briefly, aliquots of 100 μL (106 CFU/mL) indicator strains viz. Escherichia coli O157:H7, Staphylococcus aureus CMCC26001, Candida albicans ATCC14053, Listeria monocytogenes CMCC54007, and Salmonella Typhimurium ATCC13311 were spread on the surface of LB agar plates, separately. Then, 200 μL of LAB supernatant was loaded into an Oxford cup (outer diameter 7.8 ± 0.1 mm, inner diameter 6.0 ± 0.1 mm, and height 10.0 ± 0.1 mm), which was placed on the surface of the agar. After incubation for 10 h at 37 °C, the inhibition zone around the cup was measured as a quantitative measure of inhibition ability. All the experiments were performed in triplicate.
2.5. Determination of Antibiotic Susceptibility
Antibiotic susceptibility was tested according to the method in [20] with slight modifications. Briefly, bacterial strains were inoculated (1%, v/v) in MRS broth supplemented with antibiotic (chloramphenicol, ampicillin, ciprofloxacin, polymyxin B, tetracycline, erythromycin, or gentamycin) (Solarbio, Beijing, China) at various final concentrations (2, 4, 8, 16, 32, 64, 128, 256, 512, and 1024 μg/mL) and the OD600 was measured after a 24 h incubation period at 37 °C. All these experiments were performed in triplicate.
2.6. Detection of Biogenic Amine Production
The production of biogenic amine was tested according to the method in [21] with slight modifications. In brief, strains were incubated in the modified decarboxylase medium, which contained tryptone (0.5%, w/v), yeast extract (0.5%, w/v), NaCl (0.5%, w/v), glucose (0.1%, w/v), pyridoxal-5-phosphate (0.005%, w/v), Tween 80 (0.05%, w/v), MgSO4·7H2O (0.02%, w/v), CaCO3 (0.01%, w/v), MnSO4·4H2O (0.005%, w/v), FeSO4·7H2O (0.004%, w/v), bacteriological agar (2%, w/v), and purple bromocresol (0.006%, w/v), in which the last one was used as a pH indicator. The precursor amino acids of each biogenic amine (histidine, lysine, ornithine, and tyrosine) (Solarbio, Beijing, China) were added individually to the culture medium at a final concentration of 2%, and pH was adjusted to 5.5 ± 0.1. After incubation, obvious pink circles around the growing single colonies were denoted as the positive result of biogenic amine.
2.7. Growth Characteristics of Strains in the Presence of Bile Salts and Acid
Bile salt resistance and acid tolerance assays were carried out according to the method in [22] with minor modifications. For the bile salt resistance assay, overnight cultures of the isolates were inoculated into MRS broth (1%, v/v) with 0.15%, 0.3%, and 0.45% ox bile salts (w/v) (Solarbio, Beijing, China). After incubation at 37 °C for 24 h, the OD600 was measured. The results were expressed as the percentage of growth compared with the control (without ox bile salts). For the acid tolerance assay, bacterial cells were inoculated in MRS broth at pH 6.5, 4.5, and 3.5. Resistance was assessed in terms of colony counts and enumerated on an MRS agar plate after incubation at 37 °C for 0, 1, 3, and 12 h. All the assays were performed in triplicates.
2.8. Survival of Strains under Simulated Gastric and Intestinal Fluid
Overnight cultured cells were harvested by centrifugation (5000× g, 10 min, 4 °C), washed twice with PBS (pH 7.2), and resuspended in simulated gastric fluid (0.3% pepsin, 7 mmol/L KCl, 125 mmol/L NaCl, and 45 mmol/L NaHCO3, pH 2.5) or simulated intestinal fluid (0.1 g/L pancreatin, 3 g/L ox bile salts, 0.835 g/L KCl, 6.5 g/L NaCl, 0.22 g/L CaCl2, and 1.386 g/L NaHCO3, pH 7.5) containing 10% (w/v) skimmed milk, and incubated for 120 min at 37 °C in anaerobic condition. Viable counts were counted on an MRS agar plate before and after the simulated gastric and intestinal fluid challenges. The tests were performed in triplicate.
2.9. Adhesive Ability of Strains to Caco-2 Cells
The adhesion of strains to Caco-2 cells was evaluated according to previous report [23]. Briefly, 5 × 105 Caco-2 cells were seeded in 6-well tissue culture plates and incubated at 37℃ in a humidified 5% (v/v) CO2 atmosphere for 24 h. One milliliter of each strain suspension (1 × 108 CFU/mL in antibiotic-free DMEM) or DMEM solution (as control) was added into each well and incubated for 2 h at 37 °C. After incubation, the monolayers were washed to remove the unbound bacteria. The adherent bacteria were detached and dispersed in 1 mL of 0.05% Trypsin-EDTA, and then diluted serially and spread onto MRS agar plates for counting. The adhesion ability was expressed as the percentage of the adhesion of selected strains relative to the well-known probiotic strain Lactobacillus rhamnosus GG.
2.10. Preparation of AAFL-LP
Artemisia argyi was obtained from Qichun, Wuhan, and fermented using the traditional artisan method. Briefly, after drying and trimming, AA were cut and filtered, and then 5 g (10%, w/v) of sterile AA powders and 2.5 g (5%, w/v) of glucose were added to 50 mL of distilled water in a 100 mL conical flask (AA sterile). Then, L. plantarum WLPL01 (5 × 107 CFU/mL) or 1 mL of 0.85% NaCl solution (for spontaneous fermentation as control) was inoculated. After mixing, samples were incubated at 37 °C for 5 days, and the obtained AAFL was named AAFL-LP or AAFL-spontaneous. The pH values and viable bacterial numbers of the fermented samples were monitored.
2.11. Antimicrobial Activity of AAFL-LP by Oxford Cup Assay
To investigate the antimicrobial activity, AAFL sterile, AAFL-spontaneous, and AAFL-LP were extracted by ultrasonic extraction at 25 °C for 1 h with 80% (v/v) ethanol. After filtering through filter paper, the extracts were vacuum concentrated and freeze-dried. The antimicrobial activity of crude extracts was tested in an agar spot assay, as described above.
2.12. Genomic DNA Extraction and Genome Analysis
The genomic DNA of L. plantarum WLPL01 was extracted using the cetyltrimethyl ammonium bromide (CTAB) method, and the DNA concentration, quality, and integrity were determined using a Qubit Flurometer (Invitrogen, Waltham, MA, USA) and a NanoDrop Spectrophotometer (Thermo Scientific, Waltham, MA, USA). Sequencing libraries were generated using the TruSeq DNA Sample Preparation Kit (Illumina, San Diego, CA, USA) and the Template Prep Kit (Pacific Biosciences, Menlo Park, CA, USA). The genome sequencing was then performed by the Personal Biotechnology Company (Shanghai, China) using the Pacific Biosciences platform and the Illumina Miseq platform.
Data assembly proceeded after adapter contamination removal and data filtering using AdapterRemoval [24] and SOAPec [25]. The filtered reads were assembled using SPAdes [26] and A5-miseq [27] to construct scaffolds and contigs. The Canu [28] software was used to assemble the data obtained by Pacbio platform sequencing. Subsequently, all the assembled results were integrated to generate a complete sequence. Finally, the genome sequence was acquired after the rectification using the pilon software [29].
2.13. Identification of Bacteriocin-Encoding Genes of L. plantarum WLPL01
Different plantaricin genes were identified in whole-genome sequence of L. plantarum WLPL01 with a similarity search of the sequence using BLASTP and compared with known plantaricin genes [30]. Subsequently, the organization of plantaricin genes of the individual strain of L. plantarum was analyzed using the bacteriocin database BAGEL4 [31].
2.14. Statistical Analysis
The results were evaluated using analysis of variance (ANOVA) and Tukey test; p-values less than 0.05 were considered statistically significant. The analysis was performed with GraphPad Prism version 7.0 (GraphPad Software, Inc., La Jolla, CA, USA).
3. Results
3.1. Isolation and Identification of Strains from AAFL
Ten isolates of LAB strains from A. argyi fermentation liquid (AAFL) were identified as one strain of Lactiplantibacillus plantarum (WLPL01), three strains of Lacticaseibacillus casei (WLCA01, WLCA02, and WLCA03), one strain of Lactobacillus harbiness (WLHA01), and five strains of Lentilallactobacillus buchneri (WLBU01, WLBU02, WLBU03, WLBU04, and WLBU05). The relevant phylogenetic tree of these strains is shown in Figure 1.
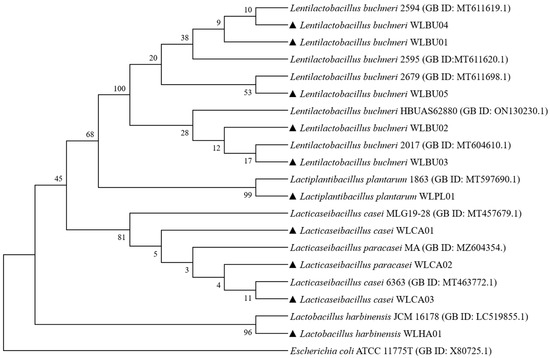
Figure 1.
Phylogenetic tree of the LAB isolates from Artemisia argyi fermentation liquor (AAFL) based on 16S rRNA gene sequences. E. coli ATCC 11775T was taken as an out-group. Bootstrap values are given at branching points. Filled upward triangles represent the LAB strains isolated in this study.
3.2. Growth Curve and Acid Production of Strains
The growth curve and pH value of the 10 isolates are shown in Figure 2. Strains were categorized into two groups: one group included one of L. plantarum strain, three of L. casei strains, and one of L. harbinesis strain, with strains entering the stationary phase at 12 h, with a final pH of 3.8; the other group included five L. buchneri strains that had a long logarithm growth phase (48 h) and final higher pH of 4.3. Among these 10 strains, L. plantarum WLPL01 showed the highest growth tendency with rapid acidification.
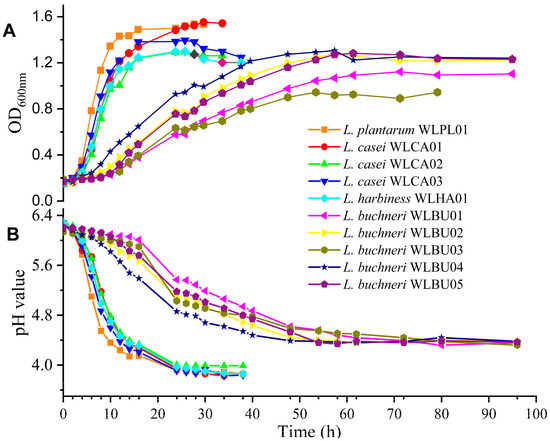
Figure 2.
The growth curves (A) and pH value (B) of the LAB strains from AAFL under anaerobic conditions in MRS broth at 37 °C.
3.3. Antibiotic Sensitivity and Biogenic Amine Test of Strains
Antibiotic resistance is a risk factor in food safety. We chose typical antibiotics, including cell wall synthesis inhibitors (ampicillin), DNA synthesis inhibitors (ciprofloxacin), and protein synthesis inhibitors (tetracycline, gentamycin, erythromycin, and chloramphenicol), to evaluate the antibiotic sensitivity of 10 strains, compared with the breakpoints set by EUCAST (2017). The results showed that one L. harbinesis (WLHA01) and two out of three L. casei strains (WLCA01 and WLCA02) were found to be resistant to gentamycin, four out of five L. buchneri (WLBU02, WLBU03, WLBU04, and WLBU05) strains showed resistance to tetracycline, and L. plantarum WLPL01, L. casei WLCA03, and L. buchneri WLBU01 showed susceptibility to all the tested antibiotics (Table 1).

Table 1.
MIC distribution of LAB strains isolated from AAFL to antibiotics.
Free from or reduction in biogenic amines is a criterion for screening LAB strains. The result showed that strains of L. buchneri, i.e., WLBU03 and WLBU05, were positive in the yielding of putrescine and cadaverine, respectively, while other strains were all negative in biogenic amine production.
3.4. Antimicrobial Activity of Strains
The antimicrobial spectrum of 10 strains was investigated using five kinds of pathogens as indicators. As shown in Figure 3, L. plantarum WLPL01, L. harbinesis WLHA01, and L. casei (WLCA01, WLCA02, and WLCA03) showed a broad spectrum against Gram-positive (L. monocytogenes and S. aureus), Gram-negative (E. coli and S. Typhimurium), and fungus (C. albicans). From these, L. plantarum demonstrated the strongest antagonistic effect. In contrast, all the five strains of L. buchneri showed no antimicrobial activity except for E. coli O157:H7 and S. Typhimurium (with a very small inhibition zone for WLBU01 and WLBU02).
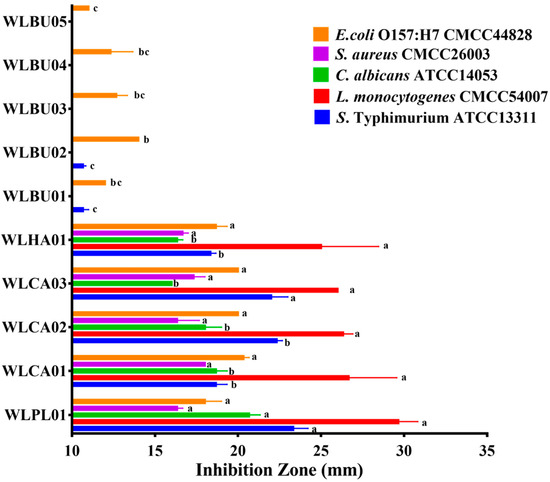
Figure 3.
Antagonistic activities of the LAB strains from AAFL against Escherichia coli O157:H7, Staphylococcus aureus CMCC26001, Candida albicans ATCC14053, Listeria monocytogenes CMCC54007, and Salmonella Typhimurium ATCC13311 in well diffusion assay. Values are presented as means ± standard deviation in triplicate. Different letters represent significant differences (p < 0.05).
3.5. Survival of Strains in Mimic GI and Adhesion to Caco-2 Cells
As probiotics, lactic acid bacteria are expected to colonize in the gut under the stress of GI condition. Therefore, based on the results of growth curves, antimicrobial properties, and biogenic amine production, five strains (L. plantarum WLPL01, L. casei WLCA01, WLCA02, WLCA03, and L. harbinesis WLHA01) were selected for further testing in an artificial GI.
In the bile salt tolerance test (Table 2), L. plantarum WLPL01, L. casei WLCA01, WLCA02, WLCA03, and L. harbinesis WLHA01 all showed a slight and nonstatistically significant decrease in survival rates, in a range of 0.15–0.45% bile salts. For low pH tolerance (Figure 4A), all five strains maintained values as high as 107 CFU/mL, even under pH 3.5 for 12 h, showing a significant growth tendency in MRS medium at pH 4.5 for 12 h (p < 0.05). To further evaluate the survival capability of the five strains, separate simulated gastric digestion and intestinal digestion tests were performed. As shown in Figure 4B, WLPL01 and WLHA01 maintained the initial level of viable counts, while the counts of all three L. casei strains increased significantly (p < 0.01) in the simulated gastric fluid for 2 h. However, L. casei WLCA01 and WLCA02 were significantly (p < 0.001) decreased after incubation with simulated intestinal fluids for 2 h, while WLPL01 and WLHA01 were still stable in the simulated intestinal fluids.

Table 2.
Tolerance of five selected LAB strains to bile salt stress. Values are presented as means ± standard deviation in triplicate.
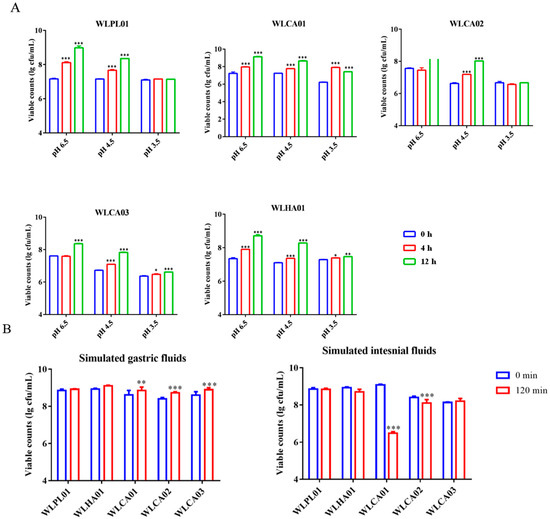
Figure 4.
Low pH tolerance (A) and survival capacity in simulated gastric and intestinal fluids (B) of the five selected LAB strains. Values are presented as means ± standard deviation in triplicate. Significant differences compared with the respective control (0 h or 0 min) are denoted as * p < 0.05; ** p < 0.01; *** p < 0.001.
Adherence on epithelial cells is considered to be an important and initial event for lactic acid bacteria to function as probiotics. Lactobacillus rhamnosus GG (LGG), as an outstanding Lactobacillus due to its excellent probiotic function, is often selected as a positive control when exploring other probiotic functions. As shown in Figure 5, the highest adhesion ratio of 82.17 ± 6.85% was obtained for WLCA03, followed by WLPL01 and WLCA01. All of them showed a weaker adhesion ability when compared with the strain of LGG, which was also used as a positive control. Its adhesion ability (%) was set as 100%.
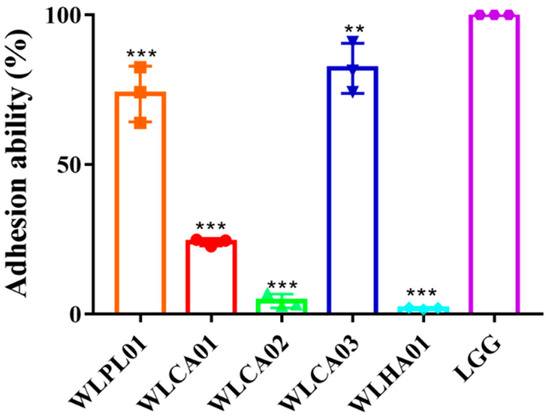
Figure 5.
The adhesion of the five selected LAB strains to Caco-2 cells. The adhesion ability was expressed as a percent compared to the LGG group. Values are presented as means ± standard deviation in triplicate. Significant differences compared with LGG group are denoted as ** p < 0.01; *** p < 0.001.
3.6. Antimicrobial Activity of L. plantarum WLPL01-Fermented AAFL (AAFL-LP) against Pathogens
Chemical and microbiological changes were monitored during the fermentation of A. argyi by L. plantarum WLPL01 (Figure 6A). Generally, viable counts of WLPL01 achieved the highest value of 8.51 ± 0.09 log CFU/mL at 48 h, while spontaneous fermentation took 96 h to reach a peak of 8.56 ± 0.03 log CFU/mL, under the condition of A. argyi supplied with an extra carbon source. For the pH, inoculation of WLPL01 resulted in a rapid decease in the pH value (final pH 3.3). All the above results suggested that fermented A. argyi might be affected by the existence of either WLPL01 or native flora of A. argyi. As shown in Figure 6B, neither AAFL-LP nor AAFL-spontaneous exhibited a significantly enhanced antagonistic ability against all of the tested pathogens. For C. albicans, both AAFL-LP and AAFL-spontaneous showed enhanced antagonistic ability; therein, AAFL-spontaneous at day 5 and AAFL-LP at day 1, 3, and 5 significantly increased the inhibition zone compared to unfermented A. argyi liquor (p < 0.05). Remarkably, AAFL-LP significantly strengthened the inhibition ability at day 3, compared with AAFL-spontaneous (p < 0.05). For L. monocytogenes and S. Typhimurium, AAFL-LP and AAFL-spontaneous also achieved a significant inhibition. Compared with AAFL-spontaneous, AAFL-LP showed a significantly higher inhibition ability against L. monocytogenes at day 3 (p < 0.001) and day 5 (p < 0.01), and S. Typhimurium exhibited a higher ability at day 5 (p < 0.05).
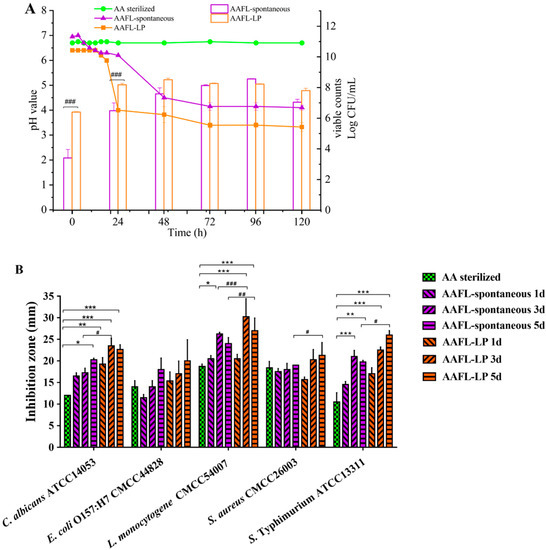
Figure 6.
Effect of fermentation on viable counts of microbial cells and pH (A), antimicrobial activity (B) of AAFL. Values are presented as means ± standard deviation in triplicate. * Denotes significant differences compared with control (AA sterilized) (* p < 0.05; ** p < 0.01, *** p < 0.001); # Denotes significant difference between AAFL-spontaneous and AAFL-LP (# p < 0.05, ## p < 0.01, ### p < 0.001).
We further used S. Typhimurium as an example to test the inhibitory effect of AAFL-LP with different concentrations. As shown in Figure 7, the viable counts of S. Typhimurium were significantly decreased (p < 0.001) when co-cultured with 25 mg/mL AAFL-LP, and no living cell was observed when co-cultured with 50 mg/mL of AAFL-LP. Moreover, the morphology of S. Typhimurium observed by SEM showed that S. Typhimurium cells were rough, severely wrinkled, and collapsed when co-cultured with 50 mg/mL of AAFL-LP.
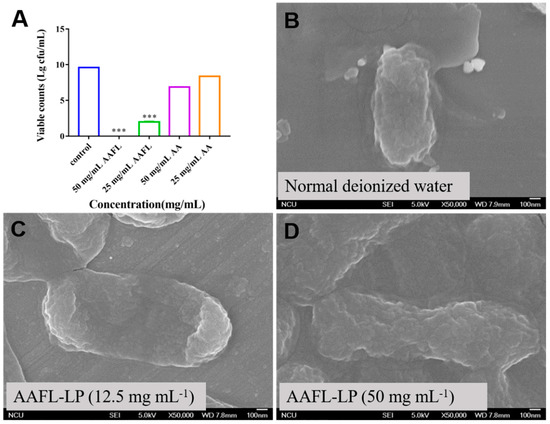
Figure 7.
In vitro antimicrobial activity of AAFL-LP against S. Typhimurium ATCC 13311 (A) and scanning electron micrographs of S. Typhimurium after 2 h in presence of normal deionized water (B), AAFL-LP (12.5 mg/mL) (C), and AAFL-LP (50 mg/mL) (D). * Denotes significant differences compared with control (*** p < 0.001).
3.7. Genome Analysis and Bacteriocin Identification of L. plantarum WLPL01
An analysis of the whole-genome sequence for the L. plantarum WLPL01 (NZ_CP122974.1) was carried out. The result (Figure 8A) showed that the genome of L. plantarum WLPL01 contains a circular chromosome, with a size of 3,142,471 bp, an average G + C content of 44.68%, containing 2946 candidate protein-coding genes (with an average size of 840 bp), among which 2942 proteins were found to be functionally categorized. The chromosomal properties of L. plantarum WLPL01 are summarized in Table 3, with L. plantarum WCFS1 (GenBank Accession no. AL935263) as a reference. In addition, the chromosome of L. plantarum WLPL01 has 147 RNA genes including 71 tRNA, 16 rRNA, and 60 ncRNA. In addition, the genome of L. plantarum WLPL01 has four plasmids, with sizes of 42,746, 38,441, 10,743, and 6349 bp, respectively, and a G + C content that ranged from 35.85% to 40.77%.
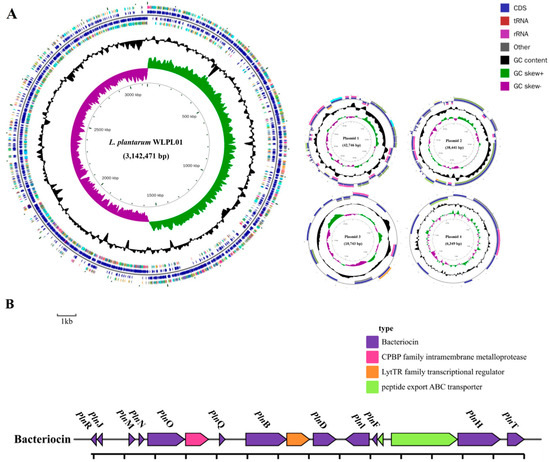
Figure 8.
Circular map of the L. plntarum WLPL01 genome (A) and genetic organization map of the plantaricins gene cluster of L. plantarum WLPL01 (B).

Table 3.
Comparison of the features of the L. plantarum WLPL01 genome with the reference genome.
The capacity of AAFL-LP against S. Typhimurium might correlate with the bacteriocin produced by L. plantarum WLPL01 during fermentation; thus, we identified the bacteriocin-encoding genes in the L. plantarum WLPL01 genome. Screening of the entire genome of L. plantarum WLPL01 revealed that the bacteriocin-encoding locus (pln locus) was located in a 13.9 kb long region organized in an operon-like structure, which consists of 16 genes (pln), including pln genes of plnJ, plnK, plnM, plnN, plnO, plnQ, plnB, plnD, plnL, plnE, plnF, plnH, and plnT, one peptide export ABC transporter, and one transcriptional regulator (Figure 8B).
4. Discussion
Considerable functional studies have been conducted on A. argyi to discover its physiochemical properties. These have been focused on its essential oil, flavone, and polysaccharide. Little is known about the function of A. argyi after fermentation or its composition and contribution to the microbial inside. In this study, according to its primary properties, the L. plantarum WLPL01 strain was screened out from 10 indigenous strains in AAFL, and its safety and potential antimicrobial capacity was tested for the fermentation of A. argyi, targeting its use as a starter culture and as a feed additive.
It has been suggested that the low diversity of LAB in grass is possibly attributed to the low water solvent carbohydrate levels in it [1]. In our work, 10 strains belonged to limited species of L. casei, L. harbinesis, L. plantarum, and L. buchneri were isolated. This is consistent with the finding of Mundt and Hammer (1968) [32], in which only two strains of L. plantarum, two of L. brevis, and one strain of L. buchneri were isolated in greens.
The safety evaluation of the selected LAB strains was a priority to enable consumer consumption [33], which included the potential to produce biogenic amines, antibiotic resistance, toxin genes, and hemolysis potential. For antibiotic resistance, Mathur and Singh proposed that, when used as a starter culture, the risk of the undesirable transfer of resistance from LAB to the host or conferment of resistance to endogenous bacteria should be avoided [34]. In this aspect, strains of L. plantarum WLPL01, L. casei WLCA03, and L. buchneri WLBU01 exhibited sensitivity to chloramphenicol, ampicillin, ciprofloxacin, tetracycline, erythromycin, and gentamycin, suggesting their advantage as a starter culture. In addition, the production of biogenic amine during fermentation might potentiate toxic effects; therefore, free or reduced biogenic amines were critical for the selection of potential LAB strains. We found that two of the five L. buchneri strains, i.e., WLBU03 and WLBU05, were positive for putrescine and cadaverine production, respectively. Similarly, W. Straub reported that eight out of eleven L. buchneri strains produced histamine in a synthetic media [35]; another study also reported that L. buchneri produced extraordinary amounts of histamine in cheese, even in very low numbers in the inoculum [36].
The biomass and acidification were important criteria in screening strains in the starter culture. It was proposed that the fast-acidifying strains are good candidates in the dairy fermentation process as primary starter organisms [37]. Our results showed that five strains: L. plantarum WLPL01, L. casei WLCA01, WLCA02, WLCA03, and L. harbinesis WLHA01, had a shorter logarithm time and faster decline in pH than the others: L. buchneri WLBU01, WLBU02, WLBU03, WLBU04, and WLBU05.
Considering the results of the growth rate, the basic safety evaluation, and the microbial inhibition test, five strains (WLPL01, WLCA01, WLCA02, WLCA03, and WLHA01) were further selected to test their abilities via a survival and adhesion in vitro assay. All strains showed a similar bile salt resistance ability, and an increased tendency for viable cell counts in pH 4.5 for 12 h, and some of them maintained a slight increase even in pH 3.5. In the simulated gastric and intestinal test, all five strains, except for WLCA01, maintained values as high as 108 CFU/mL for 2 h. Besides the basic criteria for LAB isolation, more standards need to be met for the isolation and intended use as a starter culture.
Based on the results above, L. plantarum WLPL01 was chosen as a starter culture for the fermentation of A. argyi and to further analyze the antimicrobial activates of AAFL-LP. Neither AAFL-LP nor AAFL-spontaneous maintained cell counts as high as 107–108 CFU/mL after fermentation for 5 days, with AAFL-LP decreasing in time to reach stability and maintaining a lower pH of 3.5 as compared with AAFL-spontaneous. Of the five tested pathogens, AAFL-LP significantly enhanced the antimicrobial activities of A. argyi against C. albicans and its antagonistic potential was even superior to that of AAFL-spontaneous. In the former inhibition assay of 10 LAB strains, WLPL01 also showed the best antagonistic effect against C. albicans. Similarly, the higher antimicrobial activities of AAFL-LP for L. monocytogenes and S. Typhimurium were achieved in day 5. Our findings indicated that LAB fermentation might strengthen the antagonistic abilities of A. argyi against certain pathogens and the activities might change over time. Previous studies reported the antagonistic effect of the essential oil of Artemisia species to C. albicans and L. monocytogenes [38] or the inhibitory effects of L. plantarum strains against C. albicans [39] and L. monocytogenes [40,41]. The enhanced antimicrobial activities of L. plantarum-fermented A. argyi against L. monocytogenes and C. albicans were verified in this study. In addition, Salmonella is the most common foodborne pathogen, affecting millions of people annually, sometimes with severe and fatal outcomes [42]. Examples of foods involved in outbreaks of Salmonella include eggs, poultry, and other products of animal origin (https://www.who.int/news-room/fact-sheets/detail/food-safety, accessed on 1 January 2020). In this work, we found that AAFL-LP strengthened the inhibitory ability against S. Typhimurium compared to sterile AA and AAFL-spontaneous.
Bacteriocins are antibacterial peptides produced by bacteria. They have antibacterial properties and can inhibit the growth of different microorganisms. Because of their antibacterial activity, they have attracted much interest in the food industry as natural preservatives. L. plantarum WLPL01 was identified to encode genes for two peptides, plantaricin plnJK (class IIb) and plnEF (class I), previously described for other L. plantarum strains [43]. The presence of two pln loci-encoding plantaricins from two different classes contributes to the broad inhibitory spectrum of L. plantarum [44]. Moreover, the pln locus was also found to contain plnN described in other L. plantarum strains, such as C11, WCFS1, and V90. The presence of plantaricin secretary genes plnH has also been confirmed in L. plantarum strains DHCU70 and DKP1, which are involved in the ABC transport system [45].
5. Conclusions
In conclusion, we found that AAFL-LP significantly enhanced the antimicrobial activities, effectively preventing S. Typhimurium in vitro, which might correlate with the bacteriocins produced by L. plantrum WLPL01 during fermentation. Hence, our work might provide a fundamental basis for using L. plantrum WLPL01 and AAFL-LP as feed additives to control pathogens in the breeding industry.
Author Contributions
Conceptualization, X.T. and H.W.; Methodology, H.Z.; Software, H.Z. and Y.H.; Validation, Q.W., Q.L. and L.H.; Data Curation, H.Z. and Y.H.; Writing—Original Draft Preparation, H.Z. and Y.H.; Writing—Review & Editing, X.T. and H.W.; Project Administration, X.T. and H.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by National Natural Science Foundation of China (32060030).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cai, Y.; Benno, Y.; Ogawa, M.; Kumai, S. Effect of Applying Lactic Acid Bacteria Isolated from Forage Crops on Fermentation Characteristics and Aerobic Deterioration of Silage. J. Dairy Sci. 1999, 82, 520–526. [Google Scholar] [CrossRef]
- Zimmermann-Klemd, A.M.; Reinhardt, J.K.; Morath, A.; Schamel, W.W.; Steinberger, P.; Leitner, J.; Huber, R.; Hamburger, M.; Gründemann, C. Immunosuppressive Activity of Artemisia argyi Extract and Isolated Compounds. Front. Pharmacol. 2020, 11, 402. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, H.-K.; Jeon, S.B.; Son, K.-H.; Kim, E.H.; Kang, S.K.; Sung, N.-D.; Kwon, B.-M. New sesquiterpene—Monoterpene lactone, artemisolide, isolated from Artemisia argyi. Tetrahedron Lett. 2002, 43, 6205–6208. [Google Scholar] [CrossRef]
- Bao, X.; Yuan, H.; Wang, C.; Liu, J.; Lan, M. Antitumor and immunomodulatory activities of a polysaccharide from Artemisia argyi. Carbohydr. Polym. 2013, 98, 1236–1243. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, J.; Sun, J.; Zeng, K.-W.; Cui, J.-R.; Jiang, Y.; Tu, P.-F. NO inhibitory guaianolide-derived terpenoids from Artemisia argyi. Fitoterapia 2013, 85, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-L.; Zhang, H.-J.; Chao, J.; Liu, J.-F. Essential oil of Artemisia argyi suppresses inflammatory responses by inhibiting JAK/STATs activation. J. Ethnopharmacol. 2017, 204, 107–117. [Google Scholar] [CrossRef]
- Kim, C.H.; Kim, G.-B.; Chang, M.B.; Bae, G.S.; Paik, I.K.; Kil, D.Y. Effect of dietary supplementation of Lactobacillus-fermented Artemisia princeps on growth performance, meat lipid peroxidation, and intestinal microflora in Hy-line Brown male chickens. Poult. Sci. 2017, 91, 2845–2851. [Google Scholar] [CrossRef]
- Wang, Z.; Tan, Z.; Wu, G.; Wang, L.; Qin, G.; Wang, Y.; Pang, H. Microbial community and fermentation characteristic of whole-crop wheat silage treated by lactic acid bacteria and Artemisia argyi during ensiling and aerobic exposure. Front. Microbiol. 2022, 13, 1004495. [Google Scholar] [CrossRef]
- Jiménez-López, J.; Ruiz-Medina, A.; Ortega-Barrales, P.; Llorent-Martínez, E. Phytochemical profile and antioxidant activity of caper berries (Capparis spinosa L.): Evaluation of the influence of the fermentation process. Food Chem. 2018, 250, 54–59. [Google Scholar] [CrossRef]
- Wang, L.; Lu, W.; Wu, Y.; Wu, Z. Impact of fermentation degree on phenolic compositions and bioactivities during the fermentation of guava leaves with Monascus anka and Bacillus sp. J. Funct. Foods 2018, 41, 183–190. [Google Scholar] [CrossRef]
- Petrova, P.M.; Petrov, K.K. Antimicrobial Activity of Starch-Degrading Lactobacillus Strains Isolated from Boza. Biotechnol. Biotechnol. Equip. 2011, 25, 114–116. [Google Scholar] [CrossRef]
- Oguntoyinbo, F.A.; Narbad, A. Multifunctional properties of Lactobacillus plantarum strains isolated from fermented cereal foods. J. Funct. Foods 2015, 17, 621–631. [Google Scholar] [CrossRef]
- Di Gioia, D.; Mazzola, G.; Nikodinoska, I.; Aloisio, I.; Langerholc, T.; Rossi, M.; Raimondi, S.; Melero, B.; Rovira, J. Lactic acid bacteria as protective cultures in fermented pork meat to prevent Clostridium spp. growth. Int. J. Food Microbiol. 2016, 235, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.-E.; Jang, J.-Y.; Lee, J.-H.; Park, H.-W.; Choi, H.-J.; Kim, T.-W. Starter Cultures for Kimchi Fermentation. J. Microbiol. Biotechnol. 2015, 25, 559–568. [Google Scholar] [CrossRef] [PubMed]
- Saelim, K.; Jampaphaeng, K.; Maneerat, S. Functional properties of Lactiplantibacillus plantarum S0/7 isolated fermented stinky bean (Sa Taw Dong) and its use as a starter culture. J. Funct. Foods 2017, 38, 370–377. [Google Scholar] [CrossRef]
- Bae, E.-A.; Min, S.-W.; Lee, B.; Kim, N.-J.; Baek, N.-I.; Han, E.-J.; Chung, H.-G.; Kim, N.-H. Antiasthmic effect of fermented Artemisia princeps in asthmic mice induced by ovalbumin. J. Microbiol. Biotechnol. 2007, 17, 1554–1557. [Google Scholar] [PubMed]
- Jo, Y.M.; Seo, H.; Kim, G.Y.; Cheon, S.W.; Kim, S.-A.; Park, T.S.; Hurh, B.-S.; Han, N.S. Lactobacillus pentosus SMB718 as a probiotic starter producing allyl mercaptan in garlic and onion-enriched fermentation. Food Funct. 2020, 11, 10913–10924. [Google Scholar] [CrossRef]
- Zhang, F.; Jiang, M.; Wan, C.; Chen, X.; Chen, X.; Tao, X.; Shah, N.P.; Wei, H. Screening probiotic strains for safety: Evaluation of virulence and antimicrobial susceptibility of enterococci from healthy Chinese infants. J. Dairy Sci. 2016, 99, 4282–4290. [Google Scholar] [CrossRef]
- Jiang, M.; Zhang, F.; Wan, C.; Xiong, Y.; Shah, N.P.; Wei, H.; Tao, X. Evaluation of probiotic properties of Lactobacillus plantarum WLPL04 isolated from human breast milk. J. Dairy Sci. 2016, 99, 1736–1746. [Google Scholar] [CrossRef]
- Argyri, A.A.; Zoumpopoulou, G.; Karatzas, K.A.G.; Tsakalidou, E.; Nychas, G.-J.E.; Panagou, E.Z.; Tassou, C.C. Selection of potential probiotic lactic acid bacteria from fermented olives by in vitro tests. Food Microbiol. 2013, 33, 282–291. [Google Scholar] [CrossRef]
- Moreno-Arribas, M.V.; Polo, M.C.; Jorganes, F.; Muñoz, R. Screening of biogenic amine production by lactic acid bacteria isolated from grape must and wine. Int. J. Food Microbiol. 2003, 84, 117–123. [Google Scholar] [CrossRef] [PubMed]
- García-Ruiz, A.; de Llano, D.G.; Esteban-Fernández, A.; Requena, T.; Bartolomé, B.; Moreno-Arribas, M.V. Assessment of probiotic properties in lactic acid bacteria isolated from wine. Food Microbiol. 2014, 44, 220–225. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Jeong, H.; Lee, H.; Ahn, J. Assessment of cell surface properties and adhesion potential of selected probiotic strains. Lett. Appl. Microbiol. 2009, 49, 434–442. [Google Scholar] [CrossRef]
- Lindgreen, S. AdapterRemoval: Easy cleaning of next-generation sequencing reads. BMC Res. Notes 2012, 5, 337. [Google Scholar] [CrossRef] [PubMed]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y.; et al. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. Gigascience 2012, 1, 18. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Coil, D.; Jospin, G.; Darling, A.E. A5-miseq: An updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 2014, 31, 587–589. [Google Scholar] [CrossRef]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Diep, D.B.; Straume, D.; Kjos, M.; Torres, C.; Nes, I.F. An overview of the mosaic bacteriocin pln loci from Lactobacillus plantarum. Peptides 2009, 30, 1562–1574. [Google Scholar] [CrossRef]
- Van Heel, A.J.; De Jong, A.; Montalbán-López, M.; Kok, J.; Kuipers, O.P. BAGEL3: Automated identification of genes encoding bacteriocins and (non-) bactericidal post-translationally modified peptides. Nucleic Acids Res. 2013, 41, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Mundt, J.O.; Hammer, J.L. Lactobacilli on plants. Appl. Microbiol. 1968, 16, 1326–1330. [Google Scholar] [CrossRef] [PubMed]
- Casalta, E.; Montel, M.-C. Safety assessment of dairy microorganisms: The Lactococcus genus. Int. J. Food Microbiol. 2008, 126, 271–273. [Google Scholar] [CrossRef] [PubMed]
- Mathur, S.; Singh, R. Antibiotic resistance in food lactic acid bacteria—A review. Int. J. Food Microbiol. 2005, 105, 281–295. [Google Scholar] [CrossRef] [PubMed]
- Straub, B.W.; Kicherer, M.; Schilcher, S.M.; Hammes, W.P. The formation of biogenic amines by fermentation organisms. Z. Leb. Unters. Forsch. 1995, 201, 79–82. [Google Scholar] [CrossRef]
- Nishino, N.; Hattori, H.; Wada, H.; Touno, E. Biogenic amine production in grass, maize and total mixed ration silages inoculated with Lacticaseibacillus casei or Lactobacillus buchneri. J. Appl. Microbiol. 2007, 103, 325–332. [Google Scholar] [CrossRef]
- Ayad, E.; Nashat, S.; El-Sadek, N.; Metwaly, H.; El-Soda, M. Selection of wild lactic acid bacteria isolated from traditional Egyptian dairy products according to production and technological criteria. Food Microbiol. 2004, 21, 715–725. [Google Scholar] [CrossRef]
- Abad, M.J.; Bedoya, L.M.; Apaza, L.; Bermejo, P. The Artemisia L. Genus: A Review of Bioactive Essential Oils. Molecules 2012, 17, 2542–2566. [Google Scholar] [CrossRef]
- Wynne, A.G.; Mccartney, A.L.; Brostoff, J.; Hudspith, B.N.; Gibson, G.R. An in vitro assessment of the effects of broad-spectrum antibiotics on the human gut microflora and concomitant isolation of a Lactobacillus plantarum with anti-Candida activities. Anaerobe 2004, 10, 165–169. [Google Scholar] [CrossRef]
- Ennahar, S.; Aoude-Werner, D.; Sorokine, O.; Van Dorsselaer, A.; Bringel, F.; Hubert, J.-C.; Hasselmann, C. Production of pediocin AcH by Lactobacillus plantarum WHE 92 isolated from cheese. Appl. Environ. Microbiol. 1996, 62, 4381–4387. [Google Scholar] [CrossRef]
- Reenen, V.; Dicks, L.M.; Chikindas, M.L. Isolation, purification and partial characterization of plantaricin 423, a bacteriocin produced by Lactobacillus plantarum. J. Appl. Microbiol. 1998, 84, 1131–1137. [Google Scholar] [CrossRef]
- Liss, V.; Swart, A.L.; Kehl, A.; Hermanns, N.; Zhang, Y.; Chikkaballi, D.; Böhles, N.; Deiwick, J.; Hensel, M. Salmonella enterica Remodels the Host Cell Endosomal System for Efficient Intravacuolar Nutrition. Cell Host Microbe 2017, 21, 390–402. [Google Scholar] [CrossRef]
- Goel, A.; Halami, P.M.; Tamang, J.P. Genome Analysis of Lactobacillus plantarum Isolated from Some Indian Fermented Foods for Bacteriocin Production and Probiotic Marker Genes. Front. Microbiol. 2020, 11, 40. [Google Scholar] [CrossRef]
- Tai, H.F.; Foo, H.L.; Abdul Rahim, R.; Loh, T.C.; Abdullah, M.P.; Yoshinobu, K. Molecular characterization of new organisation of plnEF and plw loci of bacteriocin genes harbour concomitantly in Lactobacillus plantarum I-UL4. Microb. Cell Fact. 2015, 14, 89. [Google Scholar] [CrossRef]
- Rizzello, C.G.; Filannino, P.; Di Cagno, R.; Calasso, M.; Gobbetti, M. Quorum-Sensing Regulation of Constitutive Plantaricin by Lactobacillus plantarum Strains under a Model System for Vegetables and Fruits. Appl. Environ. Microbiol. 2014, 80, 777–787. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).