Abstract
Cardiovascular disease (CVD) has become the leading cause of death, and it is critical to develop new functional foods to prevent intravascular thrombosis, the key cause of CVD. Fermented soy-based food is a good choice because of its native fibrinolytic enzyme (FE) activity. In this study, a strain that can produce a new type of fibrinolytic enzyme was selected from Chinese Douchi and identified as Bacillus licheniformis SFD-Y5 by molecular biology experiments and physiological and biochemical experiments. Single factor experiments combined with statistical experiments, including Plackett–Burman experiment, steepest ascent experiment and RSM (Box–Behnken design), were used to optimize the fermentation of FE by B. licheniformis SFD-Y5. The final FE activity was 2434.45 ± 28.49 IU/mL under optimal conditions, which is the highest FE activity produced by wild B. licheniformis so far. Further studies showed that Y5 FE is a serine metalloproteinase with good stability at alkaline pHs (pH 8.0–11.0). The results of our study could lay a foundation for the future production, molecular modification and further application in functional foods of Y5 FE.
1. Introduction
According to a report by the World Health Organization (WHO) in 2023, cardiovascular disease (CVD) accounts for most noncommunicable disease (NCD) deaths in the world—nearly 17.9 million people every year [1]. Intravascular thrombosis, the accumulation of fibrin in the blood vessels under abnormal conditions, is one of the major reasons for CVD [2]. For CVD patients, it is critical to rapidly dissolve blood clots and reestablish blood flow [3]. Therapies, surgical operations, anti-coagulants, antiplatelet drugs and thrombolytic agents have been used commonly in clinic [4]. However, the disadvantages of these methods, such as high price and adverse side effects, including hair loss, gastrointestinal bleeding, hypersensitivity reactions, esophagitis, liver injury, etc., have encouraged researchers to investigate new methods to cure or prevent CVD [5,6,7].
Nowadays, functional foods with fibrinolytic enzyme activity have attracted much attention in order to find new, safe and cost-effective thrombolytic agents [8]. Among them, traditional fermented soy-based foods, which are widely consumed in Asia, are found to be a good choice. Nattokinase (NK), discovered in the traditional Japanese fermented soy-based food, natto, is an alkaline serine protease with strong fibrinolytic and thrombolytic activity [9]. In addition, NK can enhance fibrinolysis and the release of an endogenous plasminogen activator as well as reduce carotid plaque area and common carotid artery media thickness in patients with hyperlipidemia by direct oral administration [5,9]. At present, NK is mainly applied as a functional food ingredient, and its potential application as a drug is being investigated [10]. Other similar fibrinolytic enzymes have also been found in Chinese traditional fermented soy-based foods such as Douchi [11], Dajang [12] and Chinese soybean paste [13]; Korean food such as Chungkook-Jang [14], Doen-Jang [15] and soy sauce [16]; Thailand food such as Thou nao [17]; and Vietnamese traditional fermented soybean paste such as Miso and Green Chili [18]. Further studies showed that the fibrinolytic enzymes in these fermented soy-based foods were produced by the microorganisms which took part in the production process of the foods [2]. By far, most of the reported microorganisms belong to Bacillus, especially Bacillus subtilis, which is the most common fibrinolytic enzyme-producing strain. Compared to Bacillus subtilis, there are few reports about fibrinolytic enzymes from B. licheniformis that have also been conferred GRAS (Generally Recognized as Safe) status by the Food and Drug Administration and are suitable for protease production [19].
Douchi is a typical and popular traditional soybean-based fermented food, with a history of more than 3000 years [20]. According to the types of microorganisms used in the production process, douchi can be divided into four types: Aspergillus type, Mucor type, Rhizopus type and bacterial type. Among them, bacterial Douchi, especially the douchi in Shandong Province, is similar to natto in many aspects such as raw materials, production process and food components [21]. Therefore, it is reasonable to screen new fibrinolytic enzyme-producing strains from bacterial Douchi.
In this study, a Bacillus licheniformis Y5 that can produce a new fibrinolytic enzyme and exhibited high fibrinolytic enzyme activity was isolated from Shandong Douchi. Various nutritional components and fermentation parameters were systematically examined to optimize conditions for FE production through single factor experiment, Plackett–Burman (P-B) design and response surface methodology (RSM). The effects of different protease inhibitors and metal ions on the FEA of Y5 FE, as well as the optimal pH, temperature, pH stability and thermal stability were examined. The results can lay a foundation for the production, molecular modification and further application of Y5 fibrinolytic enzyme.
2. Materials and Methods
2.1. Chemicals and Reagents
The PrimeSTAR® MaxDNA polymerase, DNA marker and other molecular biological reagents were purchased from Takara Biological Co., Ltd. (Shiga, Japan). Fibrinogen (bovine), urokinase and thrombin were obtained from Shanghai Jingke Biological Co., Ltd. (Shanghai, China). PMSF, EDTA, pepstatin A, and DL-dithiothreitol (DTT) were purchased from Yuanye Bio-Technology Co., Ltd. (Shanghai, China). All of the other chemicals used in the study were analytical grade and purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China).
2.2. Bacterial Diversity Analysis of Douchi Sample
In this study, 10 homemade douchi samples were collected from 5 cities in Shandong Province. The samples were collected from different parts of douchi and stored in sterile bag at −80 °C until testing. For bacterial diversity analysis, 2 g of each sample was mixed and microbial DNA was extracted using OMEGA Soil DNA Kit (M5636-02) (Omega Bio-Tek, Norcross, GA, USA), following the manufacturer’s instructions. Then, the bacteria 16S rRNA genes V3-V4 regions were amplified with the primers 338F (5′-ACTCC-TACGGGAGGCAGCA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) by PCR. PCR amplicons were purified with Vazyme VAHTSTM DNA Clean Beads (Vazyme, Nanjing, China) and quantified using the Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA, USA). After the individual quantification step, amplicons were pooled in equal amounts, and pair-end 2 × 250 bp sequencing was performed using the Illumina MiSeq platform with MiSeq Reagent Kit v3 at Shanghai Personal Biotechnology Co., Ltd. (Shanghai, China). The species composition analysis was conducted with Krona software.
2.3. Isolation of FE-Producing Bacteria
Five grams of douchi was mixed with 25 mL sterilized normal saline, cultured at 37 °C, 200 r/min for 30 min and heated at 95 °C for 5 min. Then, the supernatant was collected and tenfold serial dilutions of the suspension were prepared with sterilized normal saline until a final dilution of 10−5-fold was achieved. Next, 200 µL of each sample was plated onto the skim milk medium with 1.8% agar and incubated at 37 °C for 18 h. Then, the strains with bigger transparent zone were separated, purified and cultured in LB medium at 37 °C for 72 h. Cultures were centrifuged at 8000 r/min for 10 min, and 10 µL of the supernatant was used to detect the fibrinolytic enzyme activity. The strains with higher fibrinolytic enzyme activity were used for further experimentation.
2.4. Identification of FE-Producing Bacteria
The physiological and biochemical identification of FE-producing bacteria were determined by Bacillus identification kit (HBI G14, Hopebio, China) and API system (API 50CHB, bio Mérieux, France). The species-level identification of FE-producing bacteria was detected by comparing their 16S rDNA sequence with those in the GenBank database through BLAST (NCBI). The fibrinolytic enzyme gene identification of the strains was detected by PCR with the genome as the template and the primers of fibrinolytic enzymes reported in the previous study, including primers F1 and R1 for Bacillus subtilis [22], primers F2 and R2 for Bacillus licheniformis [23] and primers F3 and R3 for Bacillus Bacillus velezensis [24] (Table 1). The isolated strain was saved at −80 °C in 25% (v/v) glycerin.

Table 1.
Primers designed for amplification of the fibrinolytic enzyme gene.
2.5. Measurement of Fibrinolytic Enzyme Activity
The fibrinolytic enzyme activity was detected by the method of Astrup and Mullerz with urokinase as standard [25]. The preparation method of fibrin plate was according to the report of Li et al. [26].
2.6. Optimization of the Culture Medium and Fermentation Parameters for Y5 FE Production
2.6.1. The Effect of Culture Medium on Y5 FE Production
Single experiments were designed to optimize the culture medium of SFD-Y5, including carbon source, nitrogen source and metal ions as the base of basic fermentation medium with a single factor changed at one time. Different carbon sources (glucose, fructose, xylose, maltose, sucrose, soluble starch, glycerol, maltodextrin, malt extract, soybean oligosaccharides, dextrose mother liquid and high fructose corn syrups) at 20 g/L were individually added to the basal fermentation medium to evaluate the effect of carbon source on Y5 FE production. Different nitrogen sources (20 g/L yeast extract powder (A), 20 g/L tryptone (B), 20 g/L soy peptone (C), 20 g/L bean cake powder (D), 20 g/L soy isolate protein (E), 20 g/L ammonium citrate (F), 1%A + 1%B, 1%A + 1%C, 1%A + 1%D, 1%A + 1%E, 1%A + 1%F, 1%A + 1%corn steep liquor (G)) were individually added to the basal fermentation medium to evaluate the effect of nitrogen source on Y5 FE production. Different concentrations of NaCl (0, 0.05, 0.10, 0.30, 0.50 and 1.0%), CaCl2 (0, 0.1, 0.2, 0.5, 0.75 and 1.0‰) and MgSO4 (0, 0.1, 0.25, 0.5, 0.75 and 1.0‰) were added to the basal fermentation medium to evaluate the effect of metal ions on Y5 FE production. Moreover, the optimal concentration of carbon source and nitrogen source were also evaluated.
2.6.2. The Effect of Fermentation Parameters on Y5 FE Production
Single experiments were carried out to evaluate the effect of fermentation parameters on Y5, including inoculum size (1, 5, 8, 10 and 15%), temperature (28, 32, 37, 42 and 47 °C), rotate speed (150, 175, 200, 225 and 250 r/min) and medium volume (20, 30, 40, 50 and 60 mL).
2.6.3. Plackett–Burman, Steepest Ascent Experiment and Response Surface Experiment
Plackett–Burman experimental design was carried out to determine the three most important factors that influence the yield of Y5 MFE. Then, steepest ascent experiment was carried out to determine the optimal conditions of the three factors. At last, a 3-level Box–Behnken design (BBD) with 3 factors was carried out to optimize the effects of glucose, yeast extract powder and CaCl2 on the production of Y5 fibrinolytic enzyme.
2.7. The Physiological-Biochemical Characteristic of Y5 FE
Ammonium sulfate was added to the supernatant of fermentation broth until 60% saturation was achieved. Then, the liquid was left at 4 °C overnight and centrifuged at 12,000× g and 4 °C for 40 min. The precipitation was dissolved in Tris-HCl buffer (pH 7.5) and centrifuged at 12,000× g and 4 °C for 20 min. Then, the supernatant was dialyzed to remove excessive ions in dialysis bags (3 kDa), condensed and lyophilized to obtain crude Y5 FE powder for further physiological-biochemical characteristic experiments.
2.7.1. The Effects of Protease Inhibitors on Y5 FE
The effects of various protease inhibitors, including pepstatin A (1 and 5 mM), EDTA (1 and 5 mM) and PMSF (1, 5 and 10 mM), on the fibrinolytic enzyme activity of the crude FE were examined according to the method of Salunke et al. with some modifications [27]. The crude FE with inhibitors were incubated at 37 °C for 30 min. Then, samples were evaluated for fibrinolytic enzyme activity. The crude FE without inhibitors was used as control and regarded as 100%.
2.7.2. The Effects of Metal Ions on Y5 FE
The effects of metal ions including K+, Ca2+, Cu2+, Mg2+, Fe2+, Na, Zn2+, Co2+ and Mn2+ at final concentrations of 5 mM and 10 mM on the fibrinolytic enzyme activity of the crude FE were examined according to the report of Salunke et al. with some modifications [27]. The crude FE without metal ions was used as control and taken as 100%.
2.7.3. The Optimal pH and pH Stability of Y5 FE
Y5 FE activity was detected in a pH range of 3.0–11.0 at a 1.0 interval. The pH stability of Y5 FE was investigated by incubating the enzyme solution at 37 °C for 37 min at a pH range of 6.0–11.0. The highest Y5 FE activity was regarded as 100%.
2.7.4. The Optimal Temperature and Thermal Stability of Y5 FE
Y5 FE activity was detected in a temperature range of 22–62 °C at a 5 °C interval. The highest FE activity was regarded as 100%. The thermal stability was measured by incubating the Y5 FE at 4, 32, 37, 42, 47, 52, 57 and 62 °C for 30 min and the FE incubating at 4 °C for 30 min was regarded as 100%.
2.8. Statistical Analysis
Data from all the experiments in this paper were analyzed by one-way ANOVA using the SPSS software (version 26, SPSS Inc., Chicago, IL, USA) with one variable being procedure treatment. All assays were carried out in triplicate. Results were reported as mean ± standard deviation (SD). Differences were considered significant if p < 0.05 with the Tukey test.
3. Results and Discussion
3.1. Bacterial Diversity Analysis of Douchi Sample
Douchi is one of the traditional natural fermented soybean-based foods in China, whose production process is similar to that of natto. In order to screen FE-producing bacteria more efficiently, the Illumina MiSeq method was used to analyze the bacterial diversity of mixed Douchi samples. According to our statistical analysis, it was found that there were about four phyla, six classes, seven orders, seven families and eight genera. Based on the data, Korna software was used to analyze the species composition of bacteria in the douchi samples. As is shown in Figure 1, about 99% of the bacteria in Douchi belonged to the genus Bacillus. This result is similar to previous reports that most fibrinolytic enzyme-producing strains found in douchi belonged to Bacillus [22,23,24]. However, the species composition was different from the reports about Aspergillus type douchi [28], the main genus of which was Staphylococcus and Corynebacterium. The differences may be because of the different natural production process and the differences of the main microorganisms that took part in the process.
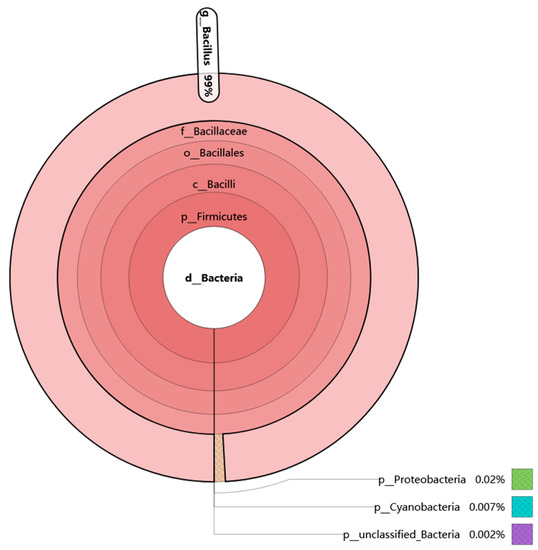
Figure 1.
Bacterial biodiversity analysis of douchi based on Krona analysis. In the figure, the circles represent the five classification levels of phylum, class, order, family and genus from the inside to the outside. The size of the fan reflects the relative abundance of different classification units and gives specific values.
3.2. Isolation and Identification of FE-Producing Bacteria
3.2.1. Isolation of FE-Producing Bacteria
Through primary and secondary screening, seven strains were found to form large transparent zones (diameter > 12.0 mm) in fibrin plates, which indicated high fibrinolytic enzyme activity (Table 2). Among the strains, P6, ZY and Y6 were identified as Bacillus subtilis; B4, D1 and X1 were identified as Bacillus velezensis; and Y5 was identified as Bacillus licheniformis by 16S rDNA sequencing. There have been reports that all three kinds of strains could produce fibrinolytic enzymes. In order to investigate whether the strains have the ability to produce a new kind of fibrinolytic enzyme, PCR was carried out with the reported fibrinolytic enzyme gene as primers and the genome of seven strains as templates [22,23,24]. As can be seen in Figure 2, the fibrinolytic enzyme of Y5 is different from the previous fibrinolytic enzymes of Bacillus velezensis, Bacillus subtilis and Bacillus licheniformis, which indicates a new fibrinolytic enzyme. Therefore, Y5 was chosen for the further identification and further study.

Table 2.
The diameter product of the transparent zone and 16S rDNA identification of the fibrinolytic enzyme-producing strains.
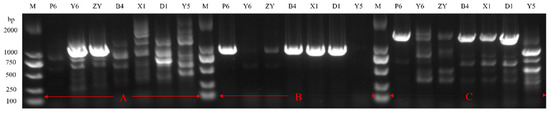
Figure 2.
The PCR amplification of genes coding fibrinolytic enzymes. The primer from the FE of (A) B. subtilis, (B) B. licheniformis, and (C) B. velezensis.
3.2.2. Further Identification of Y5
After being cultured on LB medium for 18 h at 37 °C, the colonies of Y5 were round, milky white, opaque and with a smooth surface on the edge (Figure 3B). Based on simple Gram staining and electron micrograph, the Y5 cell was rod-shaped, G+, peritrichate, with a length of 1.5–3.0 μm and a width of 0.4–0.8 μm, which is consistent with the previous reports about the morphology of Bacillus licheniformis. Moreover, the 16S rDNA sequence of Y5 was compared to all sequences in the GenBank for species identification. It was found that the best match (91% homology) was to that of Bacillus licheniformis strain ATCC 14,580 (NR 074923.1). A phylogenetic tree was constructed using the neighbor-joining method (Figure 3A), and Y5 was identified as Bacillus licheniformis using phylogenetic analysis. However, sometimes Bacillus licheniformis, Bacillus subtilis, Bacillus velezensis and Bacillus amyloliquefaciens are easily confused when tested only by 16S rDNA analysis. Two kits, HBI G14 and API 50 CHB, were used to carry out physiological and biochemical identification of Y5. The results of Table 3 show that Y5 is a typical Bacillus licheniformis, which is also consistent with the previous reports about Bacillus licheniformis. Bacillus licheniformis Y5 was deposited in the China Center for Type Culture Collection (CCTCC No:M2020796).
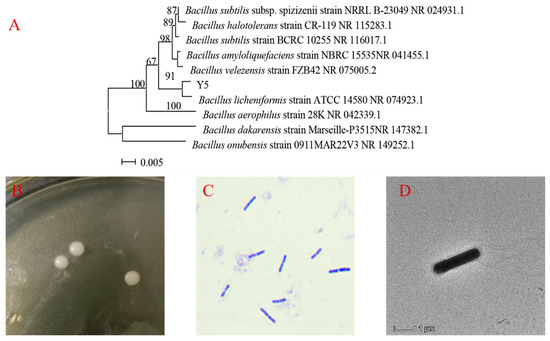
Figure 3.
The further identification of Y5. (A) The phylogenetic position of Y5 based on 16S rDNA analysis, (B) the colony morphology of Y5, (C) Gram staining of Y5, (D) electron micrograph image of Y5.

Table 3.
Physiological and biochemical characteristics of Y5 by HBI G14 Kit and API 50 CHB.
3.3. Optimization of the Culture Medium and Fermentation Parameters for Y5 FE Production
3.3.1. The Effects of Fermentation Medium Components on Y5 FE Production
The effects of fermentation medium components, including carbon sources, nitrogen sources and metal ions, on Y5 FE production were determined. Carbon sources can provide energy as well as the carbon skeleton of microbial cell and microbial metabolites [5]. Nitrogen sources can provide nitrogen for bacterial proteins, enzymes, nucleic acids and other biological macromolecules that the microorganism need [29]. Metal ions also play an important role in the growth and reproduction of microorganisms and the activity and yield of enzymes, such as calcium, chlorine, sodium and zinc. The metal ions can help to ensure the microorganism is in a normal osmotic pressure and redox potential environment. Moreover, some metal ions are also important components of enzymes and participate in enzymatic reactions as activators, while some metal ions may inactivate the enzyme molecule by binding to the thiol group of the enzyme, destroying its disulfide bond, or replace the native metal ions [30,31]. Therefore, it is important to select the appropriate carbon source, nitrogen source and metal ions.
In this study, it was found that xylose was the most conductive for Y5 FE production, followed by glucose (Figure 4A). The result was consistent with the previous reports of Xie et al. [32] and Singh et al. [33]: xylose could promote the synthesis of Nattokinase and cellulase. That may be because xylose can relieve the inhibition effect of the repressor protein on the operon and promote the synthesis of FE [34]. However, compared to xylose, glucose has a wider source and much lower cost, which is more suitable for large-scale production of Y5 FE. Further studies showed that 30 g/L of glucose is the optimal carbon source concentration for FE production (Figure 4B). The optimal nitrogen source for enzyme production is yeast extract and the optimal concentration is 20 g/L, which is in accordance with the result on FE production by Xanthomonas oryzae IND3 [35]. Moreover, the results confirmed that a complex nitrogen source addition could enhance the production of FE (Figure 4C,D). Among the metal ions, it was found that the addition of 0.2 g/L CaCl2 and 1.0 g/L NaCl could enhance the FE production (Figure 4E,F). Ca2+ has been reported to play an important role in the activity of NK, while Na+ may help the strain to sustain a normal osmotic pressure [36].
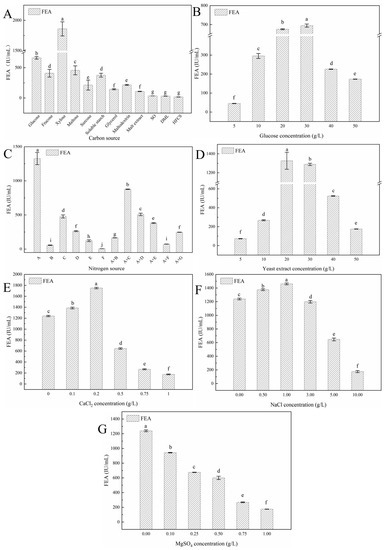
Figure 4.
Effects of fermentation medium components on Y5 FE production: (A) carbon source, (B) glucose concentration, (C) nitrogen source, (D) yeast extract concentration, (E) CaCl2 concentration, (F) NaCl concentration, and (G) MgSO4. In the figures, FE refers to fibrinolytic enzyme; FEA, fibrinolytic enzyme activity; SO, soybean oligosaccharides; DML, dextrose mother liquid; HFCS, high fructose corn syrup. (A) refers to 20 g/L of yeast extract, (B) refers to 20 g/L of tryptone, (C) refers to 20 g/L of soy peptone, (D) refers to 20 g/L of bean cake powder, (E) refers to 20 g/L of soy isolate protein, (F) refers to 20 g/L of ammonium citrate, (G) refers to 20 g/L of corn steep liquor, A + X refers to 10 g/L of yeast extract and 10 g/L of X (X = B, C, D, F, E, G). Different lowercase letters indicate significant (p < 0.05) differences in means according to Tukey’s HSD test. All assays in the figure were carried out in triplicate (n = 3).
3.3.2. The Effects of Fermentation Parameters on Y5 FE Production
Single-factor experiments were carried out to evaluate the effect of fermentation parameters including inoculum size, temperature, loading volume and rotate speed on Y5 FE production. Inoculum size, as one of critical factors for microorganism fermentation, has a significant influence on Y5 FE production. As shown in Figure 5A, 10% is the optimal inoculum size for Y5, which is different from 2% for NK production of Bacillus subtilis subsp. natto and 5% for FE production of Aspergillus niger W-47 [37]. That may because of the differences of strain species, fermentation state and the FE characteristics [37]. Temperature could not only affect microorganism metabolism but also influence the oxygen concentration and mass transfer rate during the fermentation process [10]. The production of Y5 FE increased significantly at the temperature of 28–37 °C, reached the highest value at 37 °C, then gradually decreased with the increase of temperature (Figure 5B). The results show that 37 °C is the optimal fermentation temperature of Y5, which is consistent with the previous reports about most Bacillus strains as well as the fermentation temperature of most fermented soy-based food [2].
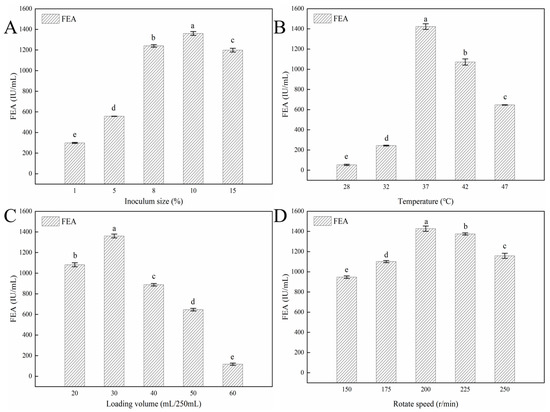
Figure 5.
Effects of fermentation conditions on Y5 FE production: (A) inoculum size, (B) temperature, (C) loading volume, and (D) rotate speed. Different lowercase letters indicate significant (p < 0.05) differences in means according to Tukey’s HSD test. All assays in the figure were carried out in triplicate (n = 3).
Rotation speed and loading volume influenced FE production by affecting the oxygen concentration, which is critical for aerobic microorganisms. In this study, FE production was highest when the rotation speed was 200 r/min and the loading volume was 30 mL in the 250 mL flask, which is similar to previous reports [37]. Bacillus licheniformis, as an aerobic microorganism, needs a large amount of dissolved oxygen for its growth and FE production. In liquid fermentation, the higher the rotation speed, the smaller the liquid volume in the flask, and the larger the amount of dissolved oxygen in the liquid broth. However, if the loading volume is too small, the water in the medium will seriously evaporate, which will result in the increase of osmotic pressure, decrease of the mass transfer efficiency and reduction of FE production. Similarly, the extensive rotation speed will damage the bacterial cells and affect the synthesis of FE, according to previous reports about FE production by Bacillus subtilis WTC016 [37].
3.3.3. Optimization of Y5 FE Production by Statistical Experiments
In order to select the most significant variables for Y5 FE production, eight variables including glucose, yeast extract, NaCl, CaCl2, inoculum size, temperature, loading volume and rotation speed, were tested and identified by the Plackett–Burman experiment. As shown in Table 4a,b, glucose, yeast extract and CaCl2 were the top three parameters that influence Y5 FE production. Next, a steepest ascent experiment was carried out to confirm the optimal concentration range of the three parameters and the results are shown in Table 5. It was found that FEA was 1742.63 ± 43.24 IU/mL, about two folds of the original FEA, when 25 g/L of glucose, 25 g/L of yeast extract and 0.3 g/L of CaCl2 were added in the fermentation medium.

Table 4.
(a) Design and results for Plackett–Burman experiment. (b) Statistical analysis in the Plackett–Burman design.

Table 5.
The design and results for steepest ascent experiment.
Based on the results, a Box–Behnken experiment was designed to optimize the production of Y5 FE. The experimental design and results from the analysis of variances and effect estimates for the main factors are shown in Table 6. The p-value of the model was less than 0.1 while the p-value for lack of fit was 0.2049 (not significant) and the coefficient R2 (0.9767) was larger than the R2Adj (0.9467), indicating the model is good and could exactly reflect the relationship of every factor. The simulated multiple regression equation is as follows:
where FEA is the fibrinolytic enzyme activity (U/g), and A, B and C are glucose, yeast extract and CaCl2.
FEA = 2082.77 + 144.16A + 29.06B + 7.46C − 112.26AB + 124.21AC − 160.76BC − 246.57A2 − 176.22B2 − 529.98C2,

Table 6.
(a) Design and results for Box–Behnken experiment. (b) ANOVA for response surface test.
The interactive effects of variables on FEA are reflected by the three-dimensional (3D) surface curves and contour plot (Figure 6). The optimal levels for FEA are glucose 28.48 g/L, yeast extract 24.24 g/L and CaCl2 0.35 g/L. The realistic FEA is 2434.45 ± 28.49 IU/mL at the optimal conditions which is close to the predicted FEA (2373.53 IU/mL), indicating the accuracy and reliability of the developed model.
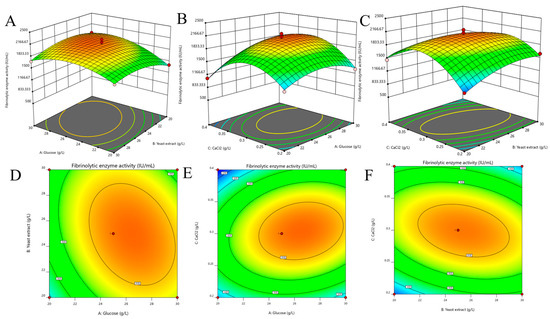
Figure 6.
Three-dimensional (3D) surface graph and contour plot of fibrinolytic enzyme production for every two factors. (A,D): for glucose concentration and yeast extract concentration; (B,E): for glucose concentration and CaCl2 concentration; (C,F): yeast extract concentration and CaCl2 concentration.
Statistical experimental design plays important roles in fermentation optimization for industrial production. Among the statistical tools, P-B experiment and steepest ascent experiment combined with RSM are common methods to investigate the key variables, study the interactive effects of variables and determine the optimal concentration of them for target products [2]. These methods have been used to optimize the FE production by Bacillus subtilis [38] and Bacillus amyloliquefaciens [21]. In this study, after optimization by these methods, FE produced by Y5 achieved 2434.45 ± 28.49 IU/mL, about three folds higher than non-optimized medium. As far as know, this is the highest fibrinolytic enzyme activity produced by wild Bacillus licheniformis.
3.4. The Physiological-Biochemical Characteristic of Y5 FE
3.4.1. The Effects of Protease Inhibitors and Metal Ions on Y5 FE
The effects of protease inhibitors on the fibrinolytic enzyme activity of crude Y5 FE are shown in Table 7. The results show that Y5 FE could not be influenced by pepstatin A (1–5 mM) and DTT (1–5 mM). However, it is significantly inhibited by EDTA (1–5 mM) and PMSF (1–10 mM). The type and active sites of protease could be investigated by protease inhibitors. For example, PMSF, pepstatin A, DTT and EDTA could inhibit the activity of serine, aspartate, cysteine and metal protease, respectively [39]. The results in Table 7 show that Y5 FE is a type of serine metalloprotease, which is different from the previous reports about FE produced by Bacillus licheniformis CH 3-17 [23], Bacillus licheniformis CH 3-17 [40] and Bacillus licheniformis RO3 [41], indicating that Y5 FE may be a new type of FE.

Table 7.
Effect of inhibitors on Y5 fibrinolytic enzyme activity (FEA).
As presented in Table 8, the activity of Y5 FE increased significantly in the presence of Zn2+ but decreased significantly in the presence of Cu2+ and Fe2+. The results are consistent with the effects of protease inhibitors, indicating Y5 FE is a metalloprotease.

Table 8.
Effect of metal ions on Y5 fibrinolytic enzyme activity (FEA).
3.4.2. The Optimal Temperatures and Thermal Stability of Y5 FE
Crude Y5 FE exhibited the highest activity at 42 °C and relatively stable enzymatic activity in the range of 32–47 °C (Figure 7A,B). However, the residual activity of Y5 FE decreased significantly when it was incubated at above 47 °C for 30 min and was only about 35% at 62 °C, indicating poor thermal stability of Y5 FE. The result is consistent with previous reports of NK and similar FE, almost all of which were inactivated at high temperature [5]. Generally speaking, in the production process of functional foods and food ingredients, heating treatment is necessary. Therefore, improvement in the thermal stability of Y5 FE needs further study for the future application in functional foods.
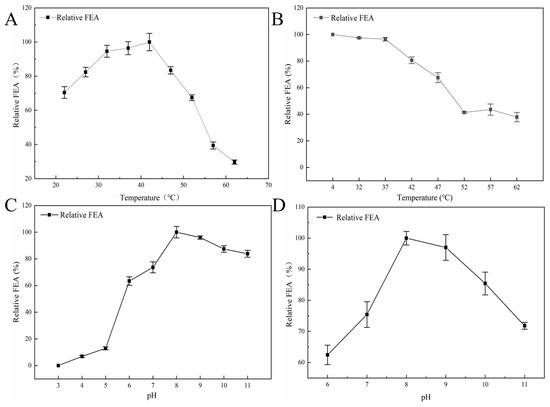
Figure 7.
The optimal temperature, pH and stability of Y5 FE: (A) optimal temperature, (B) thermal stability, (C) optimal pH, (D) stability of Y5 FE under different pH. All assays in the figure were carried out in triplicate (n = 3).
3.4.3. The Optimal pH and pH Stability of Y5 FE
The optimal pH of crude Y5 FE was 8.0 (Figure 7C), which is consistent with the optimal pH of most serine proteases produced by Bacillus [2,4]. Most (96%) of the activity of Y5 FE was maintained at pH 9.0, while about 74% of the activity remained at pH 7.0. Moreover, Y5 FE was stable at alkaline pHs (pH 8.0–11.0), and relatively stable at neutral pH (pH 6.0–7.0), but was unstable at acidic pHs (pH 3.0–5.0). The physiological pH of human is about 7.5, which is very close to the optimal pH of Y5 FE, indicating that Y5 FE may have a good fibrinolytic effect in the human body [8]. However, the in vivo effect of Y5 FE still needs further investigation.
4. Conclusions
In this study, a strain exhibited high FE activity was screened from the Chinese traditional fermented soy-based food, Douchi, and identified as Bacillus licheniformis SFD-Y5. Further study showed that the coding gene of Y5 FE was different from the previous reports, indicated a new type of FE. A remarkable FEA (2434.45 ± 28.49 IU/mL) was achieved under optimal fermentation medium and fermentation parameters which are glucose 28.48 g/L, yeast extract 24.24 g/L, CaCl2 0.35 g/L, NaCl 1.00 g/L, pH 7.0, 37 °C, inoculum size 10%, loading volume 30 mL/250 mL, rotate speed 200 r/min and fermentation for 72 h. As far as we know, this is the highest FEA produced by wild Bacillus licheniformis. The activity of Y5 FE is significantly inhibited by EDTA and PMSF, suggesting that it is a serine metalloproteinase. Importantly, our results revealed that Y5 FE has highest activity at 42 °C and pH 8.0 but has poor thermal stability. The results of our study could lay a foundation for the future production, molecular modification and further application of Y5 FE in functional foods.
Author Contributions
Conceptualization, N.Z. and M.Y.; methodology, M.Y.; software, C.M.; validation, X.X., L.L. and Q.W.; formal analysis, Y.Y.; investigation, X.B.; resources, X.B. and Y.X.; data curation, C.M.; writing—original draft preparation, M.Y.; writing—review and editing, M.Y.; visualization, C.M.; supervision, N.Z. and Y.T.; project administration, Y.T.; funding acquisition, N.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (32072258), the National Key Research and Development Program of China (2021YFD2100902-3) and Graduate research and innovation project of Harbin University of Commerce (YJSCX2020-636HSD).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data sharing not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- World Health Organization. Noncommunicable Diseases: Mortality; WHO: Geneva, Switzerland, 2023.
- Yao, M.; Yang, Y.; Fan, J.; Ma, C.; Liu, X.; Wang, Y.; Wang, B.; Sun, Z.; McClements, D.J.; Zhang, J.; et al. Production, purification, and functional properties of microbial fibrinolytic enzymes produced by microorganism obtained from soy-based fermented foods: Developments and challenges. Crit. Rev. Food Sci. Nutr. 2022, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.S.; Sabu, A. Fibrinolytic Enzymes for Thrombolytic Therapy [M]//LABROU N. In Therapeutic Enzymes: Function and Clinical Implications; Springer: Berlin/Heidelberg, Germany, 2019; pp. 345–381. [Google Scholar]
- Sharma, C.; Osmolovskiy, A.; Singh, R. Microbial Fibrinolytic Enzymes as Anti-Thrombotics: Production, Characterisation and Prodigious Biopharmaceutical Applications. Pharmaceutics 2021, 13, 1880. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tang, X.; Chen, L.; Xu, X.; Li, J. Characterization of a Nattokinase from the Newly Isolated Bile Salt-Resistant Bacillus mojavensis LY-06. Foods 2022, 11, 2403. [Google Scholar] [CrossRef] [PubMed]
- Kajihara, Y. Dabigatran-induced esophagitis. Clevel. Clin. J. Med. 2019, 86, 303–304. [Google Scholar] [CrossRef] [PubMed]
- Barzkar, N.; Jahromi, S.T.; Vianello, F. Marine Microbial Fibrinolytic Enzymes: An Overview of Source, Production, Biochemical Properties and Thrombolytic Activity. Mar. Drugs 2022, 20, 46. [Google Scholar] [CrossRef] [PubMed]
- Pinontoan, R.; Elvina; Sanjaya, A.; Jo, J. Fibrinolytic characteristics of Bacillus subtilis G8 isolated from natto. Biosci. Microbiota Food Health 2021, 40, 144–149. [Google Scholar] [CrossRef] [PubMed]
- Sumi, H.; Hamada, H.; Tsushima, H.; Mihara, H.; Muraki, H. A novel fibrinolytic enzyme (nattokinase) in the vegetable cheese Natto; a typical and popular soybean food in the Japanese diet. Experienti 1987, 43, 1110–1111. [Google Scholar] [CrossRef]
- Peng, Y.; Yang, X.; Zhang, Y. Microbial fibrinolytic enzymes: An overview of source, production, properties, and thrombolytic activity in vivo. Appl. Microbiol. Biotechnol. 2005, 69, 126–132. [Google Scholar] [CrossRef]
- Man, L.L.; Xiang, D.J.; Zhang, C.L. Strain Screening from Traditional Fermented Soybean Foods and Induction of Nattokinase Production in Bacillus subtilis MX-6. Probiotics Antimicrob. Proteins 2019, 11, 283–294. [Google Scholar] [CrossRef]
- Liu, X.L.; Kopparapu, N.K.; Zheng, H.C.; Katrolia, P.; Deng, Y.P.; Zheng, X.Q. Purification and characterization of a fibrinolytic enzyme from the food-grade fungus, Neurospora sitophila. J. Mol. Catal. B Enzym. 2016, 134, 98–104. [Google Scholar] [CrossRef]
- Wei, X.; Luo, M.; Xie, Y.; Yang, L.; Li, H.; Xu, L.; Liu, H. Strain screening, fermentation, separation, and encapsulation for production of nattokinase functional food. Appl. Biochem. Biotechnol. 2012, 168, 1753–1764. [Google Scholar] [CrossRef]
- Jeong, S.-J.; Heo, K.; Park, J.Y.; Lee, K.W.; Park, J.-Y.; Joo, S.H.; Kim, J.H. Characterization of AprE176, a fibrinolytic enzyme from Bacillus subtilis HK176. J. Microbiol. Biotechnol. 2015, 25, 89–97. [Google Scholar] [CrossRef]
- Yao, Z.; Liu, X.; Shim, J.M.; Lee, K.W.; Kim, H.-J.; Kim, J.H. Properties of a Fibrinolytic Enzyme Secreted by Bacillus amyloliquefaciens RSB34, Isolated from Doenjang. J. Microbiol. Biotechnol. 2017, 27, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.K.; Bae, D.H.; Kwon, T.J.; Lee, S.B.; Lee, H.H.; Park, J.H.; Heo, S.; Johnson, M.G. Purification and characterization of a fibrinolytic enzyme from Bacillus sp. KDO-13 isolated from soybean paste. J. Microbiol. Biotechnol. 2001, 11, 845–852. [Google Scholar]
- Han, X.; Shiwa, Y.; Itoh, M.; Suzuki, T.; Yoshikawa, H.; Nakagawa, T.; Nagano, H. Molecular cloning and sequence analysis of an extracellular protease from four Bacillus subtilis strains. Biosci. Biotechnol. Biochem. 2013, 77, 870–873. [Google Scholar] [CrossRef] [PubMed]
- Huy, D.; Hao, P.A.; Hung, P.V. Screening and identification of Bacillus sp. isolated from traditional Vietnamese soybean-fermented products for high fibrinolytic enzyme production. Int. Food Res. J. 2016, 23, 326–331. [Google Scholar]
- Sewalt, V.; Shanahan, D.; Gregg, L.; La Marta, J.; Carrillo, R. The Generally Recognized as Safe (GRAS) Process for Industrial Microbial Enzymes. Ind. Biotechnol. 2016, 12, 295–302. [Google Scholar] [CrossRef]
- Liu, L.; Chen, X.; Hao, L.; Zhang, G.; Jin, Z.; Li, C.; Yang, Y.; Rao, J.; Chen, B. Traditional fermented soybean products: Processing, flavor formation, nutritional and biological activities. Crit. Rev. Food Sci. Nutr. 2022, 62, 1971–1989. [Google Scholar] [CrossRef]
- Zhang, X.; Yun, L.-J.; Peng, L.-B.; Lu, Y.; Ma, K.-P.; Tang, F. Optimization of Douchi fibrinolytic enzyme production by statistical experimental methods. J. Huazhong Univ. Sci. Technol. Med. Sci. 2013, 33, 153–158. [Google Scholar] [CrossRef]
- Nakamura, T.; Yamagata, Y.; Ichishima, E. Nucleotide Sequence of the Subtilisin NAT Gene, aprN, of Bacillus subtilis (natto). Biosci. Biotechnol. Biochem. 1992, 56, 1869–1871. [Google Scholar] [CrossRef]
- Jo, H.-D.; Kwon, G.-H.; Park, J.-Y.; Cha, J.; Song, Y.-S.; Kim, J.H. Cloning and overexpression of aprE3-17 encoding the major fibrinolytic protease of Bacillus licheniformis CH 3-17. Biotechnol. Bioprocess Eng. 2011, 16, 352–359. [Google Scholar] [CrossRef]
- Yang, H.; Liu, Y.; Ning, Y.; Wang, C.; Zhang, X.; Weng, P.; Wu, Z. Characterization of an Intracellular Alkaline Serine Protease from Bacillus velezensis SW5 with Fibrinolytic Activity. Curr. Microbiol. 2020, 77, 1610–1621. [Google Scholar] [CrossRef] [PubMed]
- Astrup, T.; Müllertz, S. The Fibrin Plate Method for Estimating Fibrinolytic Activity. Arch. Biochem. Biophys. 1952, 40, 346–351. [Google Scholar] [CrossRef]
- Li, G.; Liu, X.; Cong, S.; Deng, Y.; Zheng, X. A novel serine protease with anticoagulant and fibrinolytic activities from the fruiting bodies of mushroom Agrocybe aegerita. Int. J. Biol. Macromol. 2021, 168, 631–639. [Google Scholar] [CrossRef] [PubMed]
- Salunke, A.S.; Nile, S.H.; Kharat, A.S. A comparative study on fibrinolytic enzymes extracted from six Bacillus spp. isolated from fruit-vegetable waste biomass. Food Biosci. 2022, 50, 102149. [Google Scholar] [CrossRef]
- Yang, L.; Yang, H.-L.; Tu, Z.-C.; Wang, X.-L. High-Throughput Sequencing of Microbial Community Diversity and Dynamics during Douchi Fermentation. PLoS ONE 2016, 11, e0168166. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Chen, G.; Pan, S.; Zeng, J.; Liang, Z. Cost-effective fibrinolytic enzyme production by Bacillus subtilis WR350 using medium supplemented with corn steep powder and sucrose. Sci. Rep. 2019, 9, 6824. [Google Scholar] [CrossRef]
- Yao, Z.; Kim, J.A.; Kim, J.H. Characterization of a Fibrinolytic Enzyme Secreted by Bacillus velezensis BS2 Isolated from Sea Squirt Jeotgal. J. Microbiol. Biotechnol. 2019, 29, 347–356. [Google Scholar] [CrossRef]
- Xin, X.; Ambati, R.R.; Cai, Z.; Lei, B. Purification and characterization of fibrinolytic enzyme from a bacterium isolated from soil. 3 Biotech 2018, 8, 90. [Google Scholar] [CrossRef]
- Qiuling, X. Optimization of Conditions in Liqiuid Fermentation to Produce Nattokinase and Research about Metabolic Regulation of Nattokinase Biosynthesis. Ph.D. Thesis, South China University of Technology, Guangzhou, China, 1999. [Google Scholar]
- Singh, K.; Richa, K.; Bose, H.; Karthik, L.; Kumar, G.; Rao, K.V.B. Statistical media optimization and cellulase production from marine Bacillus VITRKHB. 3 Biotech 2014, 4, 591–598. [Google Scholar] [CrossRef]
- Li, Y.; Liu, X.; Zhang, L.; Ding, Z.; Xu, S.; Gu, Z.; Shi, G. Transcriptional Changes in the Xylose Operon in Bacillus licheniformis and Their Use in Fermentation Optimization. Int. J. Mol. Sci. 2019, 20, 4615. [Google Scholar] [CrossRef]
- Vijayaraghavan, P.; Arasu, M.V.; Rajan, R.A.; Al-Dhabi, N. Enhanced production of fibrinolytic enzyme by a new Xanthomonas oryzae IND3 using low-cost culture medium by response surface methodology. Saudi J. Biol. Sci. 2019, 26, 217–224. [Google Scholar] [CrossRef]
- Li, D.; Hou, L.; Hu, M.; Gao, Y.; Tian, Z.; Fan, B.; Li, S.; Wang, F. Recent Advances in Nattokinase-Enriched Fermented Soybean Foods: A Review. Foods 2022, 11, 1867. [Google Scholar] [CrossRef] [PubMed]
- Ju, S.; Cao, Z.; Wong, C.; Liu, Y.; Foda, M.F.; Zhang, Z.; Li, J. Isolation and Optimal Fermentation Condition of the Bacillus subtilis Subsp. natto Strain WTC016 for Nattokinase Production. Ermentation 2019, 5, 92. [Google Scholar] [CrossRef]
- Li, T.; Zhan, C.; Guo, G.; Liu, Z.; Hao, N.; Ouyang, P. Tofu processing wastewater as a low-cost substrate for high activity nattokinase production using Bacillus subtilis. BMC Biotechnol. 2021, 21, 57. [Google Scholar] [CrossRef]
- Zhang, Y.; Xia, Y.; Lai, P.F.-H.; Liu, X.; Xiong, Z.; Liu, J.; Ai, L. Fermentation conditions of serine/alkaline milk-clotting enzyme production by newly isolated Bacillus licheniformis BL312. Ann. Microbiol. 2019, 69, 1289–1300. [Google Scholar] [CrossRef]
- KyHwang, K.-J.; Choi, K.-H.; Kim, M.-J.; Park, C.-S.; Cha, J. Purification and characterization of a new fibrinolytic enzyme of Bacillus licheniformis KJ-31, isolated from Korean traditional Jeot-gal. J. Microbiol. Biotechnol. 2007, 17, 1469–1476. [Google Scholar]
- Afifah, D.N.; Rustanti, N.; Anjani, G.; Syah, D.; Yanti; Suhartono, M.T. Proteomics study of extracellular fibrinolytic proteases from Bacillus licheniformis RO3 and Bacillus pumilus 2.g isolated from Indonesian fermented food. IOP Conf. Ser. Earth Environ. Sci. 2017, 55, 012025. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).