Abstract
Koumiss is a traditional fermented dairy product in Inner Mongolia pastoral areas, which is deeply loved by the local people; however, there has been little research on the characteristics of probiotics. This study comprehensively explored the properties of potential probiotics in koumiss, combining in vitro assays and whole-genome sequencing. The biochemical identification and phylogenetic tree results showed that the branches of this strain were close to Lacticaseibacillus paracasei, indicating that the strain was L. paracasei. The agar diffusion assay showed that the strain could effectively inhibit the growth of pathogenic bacteria. We have also identified the CHAP structural domain at the genomic level, which may be associated with the antibacterial activity of this strain. The strain was well tolerated in a simulated gastrointestinal environment, growing well at pH = 2.5, 0.3% bile salts, and 5% NaCl while exhibiting hydrophobicity, aggregation, and antioxidant properties. In vitro experiments and genome, levels showed that resistance (resistance genes) to the antibiotics used in this study was not present in this strain. In addition, we did not observe toxic effects in acute oral administration in mice, and no virulence genes were identified at the genomic level. Therefore, the strain has the potential for probiotic development.
1. Introduction
Koumiss is a specialty dairy product of minorities in China, fermented using natural ferments and traditional Chinese fermentation techniques. The effect of koumiss is inseparable from that of lactic acid bacteria (LAB). LAB and their metabolites in koumiss are popular among pastoralists due to their beneficial effects on human health [1]. Koumiss contains high levels of vitamins A, C, E, B2, B12, minerals, and folic acid, which strengthen the immune system and maintain blood pressure; koumiss also has effects on the kidneys, endocrine glands, intestinal system, liver, nervous system, and vascular system [2]. LAB have been closely related to human life throughout history, playing a positive role in improving sensory, safety, and quality characteristics of koumiss [3].
In recent years, with the development of LAB genome sequencing, considerable published genomic data have revealed the importance of LAB, which brings unprecedented opportunities for the development of LAB [4]. Park et al. isolated the Lacticaseibacillus S1 strain from traditional Korean fermented rice wine and used whole-genome sequencing to provide a theoretical basis for the study of alcohol resistance of Lacticaseibacillus [5]. Meanwhile, Yang et al. isolated a strain of L. helveticus CAUH18 from traditional koumiss with intestinal colonization ability for whole-genome sequencing and bioinformatics analysis, providing a basis for further studies on its molecular genetics and probiotic function [6]. However, Tarrah et al. isolated a strain of L. paracasei DTA93 from Brazilian infant feces; the in vitro characterization and whole-genome sequencing studies comprehensively reveal the potential of this strain as a probiotic [7].
In this study, to obtain potential LAB from koumiss, we collected samples from pastoral areas in Inner Mongolia, China, and a strain of L. paracasei with potential probiotic properties named XM-38 was successfully isolated and identified. Subsequently, we applied in vitro and in vivo assays and whole-genome sequencing to comprehensively evaluate the probiotic properties of the strains, laying the foundation for the subsequent development of traditional koumiss and high-quality probiotics.
2. Materials and Methods
2.1. Isolation of LAB from Koumiss
The naturally fermented koumiss was collected from the home of pastoralists in Inner Mongolia. The sample was diluted in 6 gradients (1 × 10−1–1 × 10−6) and coated in DeMan–Rogosa–Sharpe (MRS) solid medium for anaerobic culture (37 °C for 36 h). The MRS plates were screened for acceptable counts of bacteria, and a certain number of colonies were picked and placed in MRS liquid medium for anaerobic incubation (37 °C for 36 h), then cultured in MRS solid medium plates for purification by scratching. The morphology of the colonies was observed, and they were detected using Gram staining and peroxidase. Successfully detected bacteria were stored at −80 °C in 80% glycerol for later use as an isolate (bacterium) was named XM-38.
2.2. Extraction of XM-38 Supernatant
The bacterial solution was removed from the −80 °C refrigerator, inoculated into fresh MRS broth medium at a ratio of 1:1000, and cultured anaerobically at 37 °C for 24 h. Then, the bacterial solution was centrifuged at 15,000× g at 4 °C for 10 min to collect the supernatant. The sterile supernatant was obtained by filtration using a 0.22 µm pore size filter (Millipore, Bellerica, MA, USA).
2.3. In Vitro Assays of Probiotic Properties
2.3.1. Antibacterial Assay
The antimicrobial activity of XM-38 against pathogenic indicator bacteria Listeria monocytogenes C53-3, Staphylococcus aureus NCTC8325, Salmonella typhimurium S50333, Escherichia coli K88, and Shigella ATCC12022 was identified by Oxford Cup Method [8]. The normal nutrient agar medium was poured into the plates as a base layer, the additive zone was divided, and placed in sterile Oxford cups. Subsequently, 4.5 mL of semi-solid nutrient agar medium at 48–50 °C was mixed with 0.5 mL of pathogen bacteria suspension (approximately 1 × 109 CFU·mL−1 is an effective pathogenic bacteria suspension) diluted to approximately 106 CFU·mL−1 and poured into the plates. After complete solidification of the semi-solid medium, the Oxford cups were removed, and 200 μL of XM-38 supernatant was added to the wells; diffusion at 4 °C was performed for 5 h, then the wells were incubated at 37 °C for 16–24 h. The diameter of the zone of inhibition was observed and measured. All data are presented as averages of three independent experiments.
2.3.2. Toleration of Temperature
The XM-38 was inoculated into the MRS liquid medium at a ratio of 2% after it had been activated and then placed in a constant temperature incubator at 15, 25, 37, 40, and 45 °C to culture for 5 d. The tolerance of XM-38 to temperature was evaluated by measuring the absorbance at OD600 wavelength [9]. All data are presented as averages of three independent experiments.
2.3.3. Toleration of NaCl
The activated XM-38 was centrifuged at 8000× g for 10 min at 4 °C, and the precipitation was suspended in an MRS broth medium. A volume of 100 μL of the suspension was inoculated into an MRS broth medium containing various concentrations of NaCl (0%, 1%, 3%, and 5%) and cultured at 37 °C for 24 h. After inoculation, the MRS broth medium was diluted in a 10-fold gradient, and 20 μL of each was added to MRS agar to the culture at 37 °C for 24 h. The viability was calculated by colony count (number per milliliters) [10]. All data are presented as averages of three independent experiments.
2.3.4. Toleration to Acid and Bile Salt
According to the method of de Albuquerque et al. [11], 1 mL of activated XM-38 was inoculated with 10 mL MRS broth medium containing different pH values (2.5, 3.5, 4.5, or 5.5, using 1 M HCl) and different bile salt concentrations (0.1%, 0.2%, or 0.3% (w/v)) (Solarbio Biotechnology Co., Ltd., Beijing China), cultured at 37 °C. Every hour, 1 mL of bacterial solution was collected (1–4 h), serially diluted with 0.15% sterile peptone water, and cultured in MRS solid medium plates for purification by scratching. Colony counts were manually performed and expressed as log CFU·mL−1. The control group was cultured in MRS broth medium (Ph = 6.2–6.6) without bile salt.
2.3.5. Auto-Aggregation Analysis
The self-aggregating ability of XM-38 was evaluated according to the method of Kos et al. [12]. First, a single XM-38 colony was inoculated into an MRS broth medium and grown at 37 °C for 18 h. Then, colonies were centrifuged at 8000× g at 4 °C for 15 min to collect the precipitation, then washed twice with PBS, vortexed for 10 s, and adjusted to OD600 to 0.5. Then, samples were left to stand at room temperature for 5 h. The OD600 value was measured every hour. Equation (1) is used to measure the percentage of automatic aggregation.
In Equation (1), At is the absorbance at time t; t = 1, 2, 3, 4, and 5 h; A0 is the absorbance at 0 h.
2.3.6. Hydrophobicity
Following the method of Doyle and Rosenberg [13], with slight modifications, the activated XM-38 for 18 h was centrifuged at 8000× g for 15 min, the precipitation was washed twice with PBS, vortexed for 10 s, and adjusted to OD600 = 0.40 ± 0.05. The bacterial solution was mixed with xylene in equal proportions and vortexed for 5 min. The absorbance (A0) of its aqueous phase was measured immediately. After the mixture was allowed to stand at 37 °C for 1 h, the absorbance (At) of the aqueous phase was measured. Equation (2) was used to calculate the hydrophobicity of XM-38.
In Equation (2), A0 is the absorbance at 0 h; At is the absorbance after 1 h.
2.3.7. Antioxidant Properties
The antioxidant activity of XM-38 was evaluated according to the method of [14] with slight modifications and three groups. In the sample group, 1 mL of bacterial solution was mixed with 1 mL of DPPH ethanol (0.2 mmol L−1). In the control group, 1 mL of DPPH was mixed with 1 mL of normal saline. In the blank group, 1 mL of absolute ethanol mix was mixed with 1 mL of normal saline. The tubes were vortexed and placed in the dark at room temperature for 30 min. The absorbance was measured at 517 nm, and all data are presented as averages of three independent experiments. Equation (3) was used to calculate the DPPH clearance.
In Equation (3), A1 is the absorbance of the control group, and A2 is the absorbance of the sample group.
2.3.8. Drug Susceptibility Analysis
The drug susceptibility of XM-38 was evaluated according to the standard disc diffusion method of Kassim et al. [15]. The XM-38 bacterial solution (106 CFU·mL−1) was spread on the modified MRS solid medium containing 1–5% agar. The antibiotic-containing susceptibility papers were attached to each plate while maintaining an appropriate distance. The diameters of the transparent circles around the antimicrobial susceptibility discs were measured after being cultured at 37 °C for 48 h. Additionally, results were compared with information in Performance Standard for Antimicrobial Susceptibility Testing, developed by American Clinical and Laboratory Standards [16]. The antibiotics include ampicillin, gentamicin, kanamycin, streptomycin, erythromycin, clindamycin, tetracycline, and chloramphenicol.
2.3.9. Acute Oral Toxicity Test in Mice
The safety of XM-38 was evaluated according to the criteria provided by the OECD (Organization for Economic Co-operation and Development) [17]. The experimental animals were provided by Henan Skebes Biotechnology Co., Ltd. (Henan, China) (license number SCXK2020–0005). Twenty healthy Balb/c mice (ten male, ten female) have fasted for 6 h. All mice were fed 20 g·kg−1 bacterial solution three times every 4 h and fed a regular diet after the last dose; they were observed for health or death over 14 d.
2.4. Identification of XM-38
2.4.1. Whole-Genome Extraction, Sequencing, and Construction of Phylogenetic Trees
The XM-38 genome was extracted using the PureLink™ Pro 96 Genomic DNA Purification Kit (Invitrogen, Carlsbad, CA, USA). The whole genome of XM-38 was sequenced using the PacBio platform. By aligning with the 16S rRNA sequences of other lactic acid bacteria included in GenBank, a systematic purification tree was constructed using Mege7 [18] software to determine the classification of XM-38.
2.4.2. Genome Annotation
Coding genes were predicted on newly sequenced genomes using GeneMarkS [19] (Version 4.17) software (http://topaz.gatech.edu/GeneMark/, assessed on 19 December 2022). Additionally, the KEGG database (http://www.genome.jp/kegg/, assessed on 19 December 2022) was used to annotate predicted genes. The tRNA and rRNA were predicted by tRNAscan-SE [20] software (Version 1.3.1) and rRNAmmer [21] (Version 1.2) software.
2.4.3. Prediction of Bacteriocin Genes
Bacteriocins are secreted proteins. In this study, the signal peptide prediction tool SignalP [22] was used to annotate whether the protein sequence contained signal peptides. At the same time, the TMHMM [23] was used to annotate whether the protein sequence contained transmembrane regions. Finally, the proteins containing the signal peptide and no transmembrane region were screened for secretion protein.
2.4.4. Antibiotic-Resistance Gene
The amino acid sequence of the gene encoded by XM-38 was compared with the ARDB database [24] using Diamond software [25], and the gene of the target species was combined with its corresponding drug resistance function annotation information to obtain the annotation result.
2.4.5. Virulence Gene Prediction
The amino acid sequence of XM-38 was compared with the VFDB database [26] using Diamond software [25], and the gene of the target species was combined with its corresponding virulence factor functional annotation information to obtain the annotation result.
2.4.6. Statistical Analysis
The experimental data are expressed as the mean ± standard deviation (SE). The differences were analyzed using the one-way ANOVA test. p < 0.05 was considered statistically significant. All analyses were performed using the SPSS version 22 software.
3. Results
3.1. Biological Identification of XM-38
3.1.1. Antibacterial Assay
The results of the agar diffusion assay showed that XM-38 has strong antibacterial activity against Gram-negative and Gram-positive bacteria, such as Listeria monocytogenes C53-3, Staphylococcus aureus NCTC8325, Salmonella typhimurium S50333, Escherichia coli K88, and Shigella ATCC12022 (Table 1). The antibacterial activity of XM-38 against other pathogenic strains remains to be studied. LAB inhibits the growth of pathogenic bacteria mainly by lowering the pH in the environment, producing bacteriocin, and competing for nutrients [27]. In this study, a protein containing the bacteriocin CHAP domain was successfully identified from secreted proteins using the bioinformatics software SignalP and TMHMM, and the result showed that the bacteriocin had good antibacterial activity against Staphylococcus aureus [28]. In conclusion, XM-38 inhibited the growth of pathogenic bacteria, possibly related to bacteriocin.

Table 1.
The results of agar diffusion assay of XM-38 inhibiting pathogenic bacteria.
3.1.2. Toleration to Temperature and NaCl
The potential of probiotics to grow at different temperatures is important for their application in the food industry [29]. XM-38 can grow at a temperature of 15–45 °C, with the highest activity at 37 °C (Figure 1A). At the same time, XM-38 exhibited good salt tolerance to 1~5% NaCl, and the viable count ranged from 10.52 to 10.44 log CFU·mL−1 (Figure 1B). Although XM-38 was cultured in media with different salt concentrations for 24 h, the viable bacteria counts decreased slightly with the increase in salt concentration, but the viable bacteria rate was still as high as 98.5% in a 5% NaCl medium. At the same time, the colony counts were all above 10 log CFU·mL−1. Therefore, XM-38 has strong salt tolerance and can resist strong osmotic pressure in the stomach to maintain its growth characteristics.
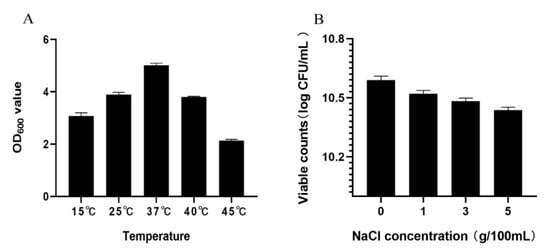
Figure 1.
Temperature and NaCl tolerance of XM-38. (A) Temperature tolerance results; (B) NaCl tolerance results. All data are shown as averages of three independent experiments. The bars represent the standard deviation.
3.1.3. Toleration to Acid and Bile Salt
XM-38 is acid- and bile-tolerant; when inoculated in an MRS medium with a low pH value and high bile salt concentration, the viable bacterial counts decreased slightly (Table 2 and Table 3). The survival rate of XM-38 was 81.29% in pH = 2.5 medium after 3 h of culture and 76.62% in bile salt-containing medium after 4 h of culture. These results indicated that the XM-38 could grow normally under these conditions while exerting the effect of probiotics. Therefore, this strain can meet the concentration requirements of probiotics.

Table 2.
Colony counts of XM-38 at different pH values and different periods (n = 3).

Table 3.
Colony counts of XM-38 were exposed to different bile salt concentrations (w/v) for different periods (n = 3).
3.1.4. Hydrophobicity and Antioxidant Properties
The hydrophobicity of LAB is closely associated with adhesion [30]. The hydrophobicity of XM-38 can reach 76.32 ± 8.68%, indicating that it has excellent hydrophobicity and adhesion. Studies have shown that oxidative stress is associated with dysbiosis of the gut flora; therefore, improving gut flora with probiotics has been widely studied [31]. XM-38 exhibits strong DPPH free radical scavenging activity at a rate of 78.29 ± 10.76%.
3.1.5. Auto-Aggregation Analysis
The auto-aggregation of probiotics is closely related to their adhesion properties [32,33]. The increased adhesion of probiotics results in prolonged colonization of the gut, resulting in improved local microbiota and immune responses [34]. In this study, we evaluated the auto-aggregation ability of XM-38 over 5 h. The results showed that the auto-aggregation rate of the strain was 10% in the first hour, which soared to 70.6% in 5 h, indicating that the strain has a good auto-aggregation ability.
3.1.6. Drug Susceptibility Analysis
Sensitivity to antibiotics is an important characteristic of probiotics [35,36]. The susceptibility and resistance of XM-38 to antibiotics were evaluated according to the minimum inhibitory concentration (MIC) values (cut-off values) specified by the EFSA [37]. In this study, the MIC values of XM-38 were all lower than the cut-off value specified by the EFSA, which indicated that this strain was sensitive to ampicillin, gentamicin, kanamycin, streptomycin, erythromycin, clindamycin, tetracycline, and chloramphenicol. At the same time, to detect antibiotic resistance genes at the gene level, this study also compared the encoded gene of XM-38 with the ARDB database. Table 4 indicated that eight antibiotic resistance genes in total were detected, but only when the identity value of the comparison was greater than the minimum identity value could it be regarded as a credible result. In conclusion, XM-38 is a safe strain but only resistant to bacitracin.

Table 4.
Antibiotic-resistance gene XM38.
3.1.7. Acute Oral Toxicity Test in Mice
After ingesting XM-38 with an LD50 > 20 g/kg, all mice did not find any adverse effects, weight loss, and death. At the same time, the encoded gene of XM-38 was compared with the VFDB database to obtain the annotation information of virulence genes. Table 5 indicates that 10 virulence-related genes were identified in total, but the consistency was less than 50%. Thus, the XM-38 genome did not contain any virulence-related genes, which proves that XM-38 is a non-toxic strain that can be further developed and utilized.

Table 5.
Virulence-related genes encoded by XM-38.
3.2. Identification of XM-38
The whole genome of XM-38 is composed of a circular chromosome of 2.91 Mb (Table 6). Based on 16S rRNA analysis, a phylogenetic tree was constructed using MEGA 7 software [18], and the results showed that XM-38 has a close homologous relationship with L. paracasei R094 (Figure 2). Biochemical tests showed that the immobile, Gram-positive, and catalase tests of XM-38 were negative. At the same time, the function of the XM-38 genome product was annotated using the KEGG database. KEGG [38,39] is a database for systematic analysis of metabolic pathways of gene products and compounds in cells and the functions of these gene products. It integrates data from genomics, chemical molecules, and biochemical systems, including KEGG pathways, KEGG drugs, KEGG diseases, KEGG modules, KEGG genes, KEGG genomes, etc. (Figure 3). The whole-genome sequence of Lacticaseibacillus paracasei has been submitted to GenBank (accession numbers: CP104699).

Table 6.
Details of the Lacticaseibacillus paracasei XM-38 genome.
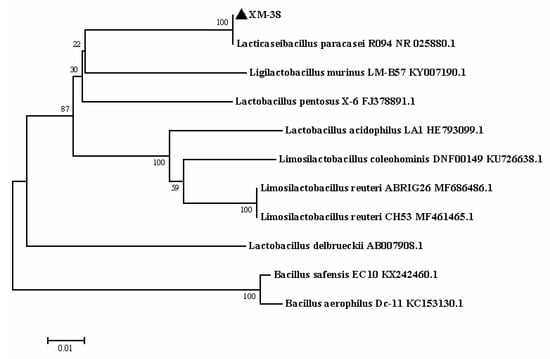
Figure 2.
Phylogenetic tree of XM-38. The phylogenetic tree was constructed using the distance-based neighbor-joining algorithm in Mega7 with 1000 bootstrap replicates (▲ indicates the XM-38) (strain name representation: strain name + GenBank number).
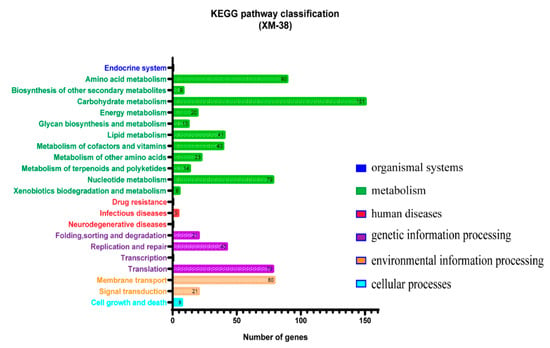
Figure 3.
Histogram of the function and quantity of XM-38 genes annotated in the KEGG database. This graph was produced using GraphPad Prism V 7.03 (GraphPad Software Inc., San Diego, CA, USA).
4. Discussion
Traditional fermented foods play an important role in the human diet. Archaeological studies have found that fermented foods came from Asia as early as 8000 BC [40]. Fermented food is the product of converting raw animal and plant materials into nutrients needed for animal survival using functional microorganisms. During food fermentation, the bioavailability of nutrients increases, and the function of probiotics and prebiotics is stimulated, thereby improving the nutritional properties and health benefits of related foods [41]. There are many types of fermented foods around the world. Koumiss is a traditional fermented dairy product in Inner Mongolia with a unique flavor. It can regulate the gastrointestinal environment, improve the absorption of nutrients, reduce the body’s lactose intolerance, enhance immunity, prevent scurvy and atherosclerosis, and help to treat tuberculosis [42]. In koumiss food, microorganisms involved in the fermentation process belong to a group of LAB [43]. Due to intensive in vivo and in vitro research, the use of LAB in treatments for various diseases has been increasing year by year [44,45,46]. Advances in biotechnology have boosted the potential to explore the functions of LAB. A combination of the in vitro characteristics and genomics of LAB can reveal the molecular mechanism behind the characteristics of LAB to discover potential probiotics. In order to qualify for probiotics, candidate microorganisms must have specific functional and safety characteristics, including an ability to produce different bacteriocins, temperature tolerance, acid resistance, bile salt resistance, and non-pathogenicity. Therefore, probiotics can be widely used in the food and health industries [45,47,48,49]. L. paracasei XM-38 exhibits broad bacteriostatic activity against pathogenic Gram-positive and Gram-negative bacteria. This feature not only balances normal intestinal flora but also prevents food spoilage during processing [50,51]. Adewale et al. isolated lactic acid bacteria with Salmonella inhibitory activity from bovine feces but did not detect the presence of bacteriocins, which is suspected to be the effect of lactic acid [52]. In addition, in this study, we screened the bacteriocin CHAP (cysteine, histidine-dependent amidohydrolase/peptidase) domain with bacteriostatic activity at the genome level. The CHAP domain is usually associated with other domains that cleave peptidoglycans, leading to bacterial lysis [53,54]. Bacteriocins are a class of protein-like substances with an antibacterial activity that is synthesized and secreted into the environment by lactic acid bacteria in the process of growth and metabolism, which have great potential for application in the research and development of natural food biological preservatives [55]. Therefore, this study found that the CHAP structural domain of XM-38 has good application prospects. It is still unclear whether this domain determines the antibacterial activity of XM-38 and needs to be further verified. The ability of probiotics to grow at different temperatures is important for industrial production [29]. It can be seen from the temperature tolerance test that XM-38 can grow well between 15 and 45 °C, and the fastest growth temperature is 37 °C. The ability to tolerate the acidic environment of the gastrointestinal tract and the secretion of bile salts in the small intestine is important for obtaining potential probiotics [29,35]. L. paracasei XM-38 can grow well in pH = 2.5 and a 0.3% bile salt environment and can also grow in 5% NaCl, which reflects the potential for clinical applications of this strain. Xu et al. isolated L. paracasei subsp L1 from sweet potato sour liquid, and it can also grow normally in a medium with pH = 2.5, 0.3% bile salt, and 5% sodium chloride, indicating that the results of this study are consistent with previous studies and have certain significance [10]. Meanwhile, Zarate et al. reported that strains with bile tolerance significantly reduced the symptoms of lactose intolerance [56]. Therefore, XM-38 may have a greater potential for development in functional dairy products. The adhesion of probiotics is critical to inhibiting other pathogens on epithelial tissues [57]. Current research shows that the hydrophobicity and auto-aggregation ability of probiotics is highly correlated with adhesion and biofilm formation, and biofilm is an important property that hinders the invasion of pathogenic bacteria [58,59]. Al Kosa et al. found that CMUL57 (L. palustris), CMUL67 (L. acidophilus), and CMUL140 (L. plantarum) were the most hydrophobic strains among the screened strains, and these strains also showed the greatest self-aggregation ability [60]. The auto-aggregation of XM-38 reached 70.6% after 5 h, indicating the potential to hinder the invasion of other pathogens. Meanwhile, the hydrophobicity of XM-38 was 76.32 ± 8.68%. Haydee et al. isolated L. paracasei CT12 from Tibios. The bacterium had good hydrophobicity (97–99%), self-aggregation (about 70%), and adhesiveness (up to 90%), showing the characteristics of probiotics [61]. Specific probiotics can exhibit antioxidant activity and reduce damage caused by oxidation, indicating that their free radical scavenging is specific [62]. L. paracasei has a strong free radical scavenging efficiency (78.29 ± 10.76%). In this study, we tested the acute oral toxicity in mice and antibiotic susceptibility to evaluate the safety of XM-38. The results showed that XM-38 had no effect on the weight and health of the mice and was sensitive to all antibiotics. Interestingly, we found that XM-38 has the unique flavor of fermented milk during the cultivation process, which can arouse consumers’ appetite. Therefore, this LAB is capable of causing changes in the flavor of koumiss. In summary, this study comprehensively and systematically evaluated the efficacy and safety of XM-38 as a good probiotic, indicating that it is a promising candidate for clinical application.
5. Conclusions
The experiment showed that L. paracasei XM-38 is a safe, non-toxic, and stable Lacticaseibacillus strain with probiotic-related genes and characteristics. The study of this strain can lay a strong foundation for the development and utilization of probiotics.
Author Contributions
H.-Z.Z., Q.-J.S., C.Y. and Y.-J.W. proposed the test idea and design; H.-Z.Z. performed the test; H.G. revised the manuscript language; C.-Y.L., X.L., Y.-X.W. and Z.-P.M. analyzed and discussed the test data; F.-X.W. and Y.-J.W. acquired the reagents and materials; H.-Z.Z. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded in whole or in part by the Inner Mongolia Natural Science Foundation (2020MS03047), the Education Department of Inner Mongolia Autonomous Region “Young scientific and technological talents in universities” project (NJYT22043), and the Introducing Talents Scientific Research Project of Inner Mongolia Agricultural University (NDYB2018-2 and NDGCC2016-22).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
The study is reported in accordance with ARRIVE guidelines (https://arriveguidelines.org, accessed on 19 December 2022); all animal experiments were carried out in accordance with the regulations of the China Laboratory Animal Administration. The experimental protocol was approved by the Animal Welfare and Research Ethics Committee of Inner Mongolia Agricultural University (approval number: 20210513-1). All authors agreed to and participated in the study.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the GenBank repository (accession numbers: CP104699), published on 1 September 2024.
Acknowledgments
The study was also supported by the Key Laboratory of Clinical Diagnosis and Treatment Technology in Animal Disease, Ministry of Agriculture, PR China, and Ruminant Animal Disease Diagnosis Center, Inner Mongolia Agricultural University.
Conflicts of Interest
The authors declare that the study was conducted without any conflicts of interest.
References
- Sudun; Wulijideligen; Arakawa, K.; Miyamoto, M.; Miyamoto, T. Interaction between lactic acid bacteria and yeasts in airag, an alcoholic fermented milk. Anim. Sci. J. 2013, 84, 66–74. [Google Scholar] [CrossRef]
- Afzaal, M.; Saeed, F.; Anjum, F.; Waris, N.; Husaain, M.; Ikram, A.; Ateeq, H.; Muhammad Anjum, F.; Suleria, H. Nutritional and ethnomedicinal scenario of koumiss: A concurrent review. Food Sci. Nutr. 2021, 9, 6421–6428. [Google Scholar] [CrossRef] [PubMed]
- Lynch, K.M.; Zannini, E.; Coffey, A.; Arendt, E.K. Lactic Acid Bacteria Exopolysaccharides in Foods and Beverages: Isolation, Properties, Characterization, and Health Benefits. Annu. Rev. Food Sci. Technol. 2018, 9, 155–176. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Huang, J.; Zhou, R. Genomics of lactic acid bacteria: Current status and potential applications. Crit. Rev. Microbiol. 2017, 43, 393–404. [Google Scholar] [CrossRef] [PubMed]
- Park, G.S.; Hong, S.J.; Jung, B.K.; Park, S.; Jin, H.; Lee, S.J.; Shin, J.H.; Lee, H.S. Whole genome sequence of lactic acid bacterium Pediococcus acidilactici strain S1. Braz. J. Microbiol. 2017, 48, 395–396. [Google Scholar] [CrossRef]
- Yang, Y.; An, H.; Zhai, Z.; Wang, G.; Li, J.; Hao, Y. Complete genome sequence of Lactobacillus helveticus CAUH18, a potential probiotic strain originated from koumiss. J. Biotechnol. 2016, 224, 18–19. [Google Scholar] [CrossRef]
- Tarrah, A.; Pakroo, S.; Corich, V.; Giacomini, A. Whole-genome sequence and comparative genome analysis of Lactobacillus paracasei DTA93, a promising probiotic lactic acid bacterium. Arch. Microbiol. 2020, 202, 1997–2003. [Google Scholar] [CrossRef]
- Zhang, J.; Cui, X.; Zhang, M.; Bai, B.; Yang, Y.; Fan, S. The antibacterial mechanism of perilla rosmarinic acid. Biotechnol. Appl. Biochem. 2021, 69, 1757–1764. [Google Scholar] [CrossRef]
- Masuda, M.; Ide, M.; Utsumi, H.; Niiro, T.; Shimamura, Y.; Murata, M. Production potency of folate, vitamin B(12), and thiamine by lactic acid bacteria isolated from Japanese pickles. Biosci. Biotechnol. Biochem. 2012, 76, 2061–2067. [Google Scholar] [CrossRef]
- Xu, Y.; Tian, Y.; Cao, Y.; Li, J.; Guo, H.; Su, Y.; Tian, Y.; Wang, C.; Wang, T.; Zhang, L. Probiotic Properties of Lactobacillus paracasei subsp. paracasei L1 and Its Growth Performance-Promotion in Chicken by Improving the Intestinal Microflora. Front. Physiol. 2019, 10, 937. [Google Scholar] [CrossRef]
- De Albuquerque, T.M.R.; Garcia, E.F.; de Oliveira Araujo, A.; Magnani, M.; Saarela, M.; de Souza, E.L. In Vitro Characterization of Lactobacillus Strains Isolated from Fruit Processing By-Products as Potential Probiotics. Probiotics Antimicrob. Proteins 2018, 10, 704–716. [Google Scholar] [CrossRef]
- Kos, B.; Suskovic, J.; Vukovic, S.; Simpraga, M.; Frece, J.; Matosic, S. Adhesion and aggregation ability of probiotic strain Lactobacillus acidophilus M92. J. Appl. Microbiol. 2003, 94, 981–987. [Google Scholar] [CrossRef]
- Doyle, R.J.; Rosenberg, M. Measurement of microbial adhesion to hydrophobic substrata. Methods Enzymol. 1995, 253, 542–550. [Google Scholar]
- Son, S.; Lewis, B.A. Free radical scavenging and antioxidative activity of caffeic acid amide and ester analogues: Structure-activity relationship. J. Agric. Food Chem. 2002, 50, 468–472. [Google Scholar] [CrossRef]
- Kassim, A.; Omuse, G.; Premji, Z.; Revathi, G. Comparison of Clinical Laboratory Standards Institute and European Committee on Antimicrobial Susceptibility Testing guidelines for the interpretation of antibiotic susceptibility at a University teaching hospital in Nairobi, Kenya: A cross-sectional study. Ann. Clin. Microbiol. Antimicrob. 2016, 15, 21. [Google Scholar] [CrossRef] [PubMed]
- Selt, F.; Hohloch, J.; Hielscher, T.; Sahm, F.; Capper, D.; Korshunov, A.; Usta, D.; Brabetz, S.; Ridinger, J.; Ecker, J.; et al. Establishment and application of a novel patient-derived KIAA1549:BRAF-driven pediatric pilocytic astrocytoma model for preclinical drug testing. Oncotarget 2017, 8, 11460–11479. [Google Scholar] [CrossRef] [PubMed]
- OECD. OECD Guidelines for the Testing of Chemicals No. 423: Acute Oral Toxicity—Acute Toxic Class Method; OECD: Paris, France, 1996. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Besemer, J.; Lomsadze, A.; Borodovsky, M. GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001, 29, 2607–2618. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Lagesen, K.; Hallin, P.; Rodland, E.A.; Staerfeldt, H.H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Pop, M. ARDB—Antibiotic Resistance Genes Database. Nucleic Acids Res. 2009, 37, D443–D447. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Chen, L.; Xiong, Z.; Sun, L.; Yang, J.; Jin, Q. VFDB 2012 update: Toward the genetic diversity and molecular evolution of bacterial virulence factors. Nucleic Acids Res. 2012, 40, D641–D645. [Google Scholar] [CrossRef]
- O’Shea, E.F.; Cotter, P.D.; Ross, R.P.; Hill, C. Strategies to improve the bacteriocin protection provided by lactic acid bacteria. Curr. Opin. Biotechnol. 2013, 24, 130–134. [Google Scholar] [CrossRef]
- Horgan, M.; O’Flynn, G.; Garry, J.; Cooney, J.; Coffey, A.; Fitzgerald, G.F.; Ross, R.P.; McAuliffe, O. Phage lysin LysK can be truncated to its CHAP domain and retain lytic activity against live antibiotic-resistant staphylococci. Appl. Environ. Microbiol. 2009, 75, 872–874. [Google Scholar] [CrossRef]
- Qureshi, N.; Gu, Q.; Li, P. Whole genome sequence analysis and in vitro probiotic characteristics of a Lactobacillus strain Lactobacillus paracasei ZFM54. J. Appl. Microbiol. 2020, 129, 422–433. [Google Scholar] [CrossRef]
- Koryszewska-Baginska, A.; Gawor, J.; Nowak, A.; Grynberg, M.; Aleksandrzak-Piekarczyk, T. Comparative genomics and functional analysis of a highly adhesive dairy Lactobacillus paracasei subsp. paracasei IBB3423 strain. Appl. Microbiol. Biotechnol. 2019, 103, 7617–7634. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, Y.; Wang, Y.; Xu, H.; Mei, X.; Yu, D.; Wang, Y.; Li, W. Antioxidant Properties of Probiotic Bacteria. Nutrients 2017, 9, 521. [Google Scholar] [CrossRef]
- Vlkova, E.; Rada, V.; Smehilova, M.; Killer, J. Auto-aggregation and co-aggregation ability in bifidobacteria and clostridia. Folia Microbiol. 2008, 53, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Valeriano, V.D.; Parungao-Balolong, M.M.; Kang, D.K. In vitro evaluation of the mucin-adhesion ability and probiotic potential of Lactobacillus mucosae LM1. J. Appl. Microbiol. 2014, 117, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.M.; Im, D.S. Screening and characterization of probiotic lactic acid bacteria isolated from Korean fermented foods. J. Microbiol. Biotechnol. 2009, 19, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Ren, D.; Li, C.; Qin, Y.; Yin, R.; Du, S.; Ye, F.; Liu, C.; Liu, H.; Wang, M.; Li, Y.; et al. In vitro evaluation of the probiotic and functional potential of Lactobacillus strains isolated from fermented food and human intestine. Anaerobe 2014, 30, 1–10. [Google Scholar] [CrossRef]
- Vizoso Pinto, M.G.; Franz, C.M.; Schillinger, U.; Holzapfel, W.H. Lactobacillus spp. with in vitro probiotic properties from human faeces and traditional fermented products. Int. J. Food Microbiol. 2006, 109, 205–214. [Google Scholar] [CrossRef]
- EFSA Panel. Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA J. 2012, 10, 2740. [Google Scholar]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Hattori, M.; Aoki-Kinoshita, K.F.; Itoh, M.; Kawashima, S.; Katayama, T.; Araki, M.; Hirakawa, M. From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Res. 2006, 34, D354–D357. [Google Scholar] [CrossRef]
- McGovern, P.E.; Zhang, J.; Tang, J.; Zhang, Z.; Hall, G.R.; Moreau, R.A.; Nunez, A.; Butrym, E.D.; Richards, M.P.; Wang, C.S.; et al. Fermented beverages of pre- and proto-historic China. Proc. Natl. Acad. Sci. USA 2004, 101, 17593–17598. [Google Scholar] [CrossRef]
- Tamang, J.P.; Shin, D.H.; Jung, S.J.; Chae, S.W. Functional Properties of Microorganisms in Fermented Foods. Front. Microbiol. 2016, 7, 578. [Google Scholar] [CrossRef]
- Cuamatzin-Garcia, L.; Rodriguez-Rugarcia, P.; El-Kassis, E.G.; Galicia, G.; Meza-Jimenez, M.L.; Banos-Lara, M.D.R.; Zaragoza-Maldonado, D.S.; Perez-Armendariz, B. Traditional Fermented Foods and Beverages from around the World and Their Health Benefits. Microorganisms 2022, 10, 1151. [Google Scholar] [CrossRef] [PubMed]
- Mathur, H.; Beresford, T.P.; Cotter, P.D. Health Benefits of Lactic Acid Bacteria (LAB) Fermentates. Nutrients 2020, 12, 1679. [Google Scholar] [CrossRef] [PubMed]
- Mokoena, M.P. Lactic Acid Bacteria and Their Bacteriocins: Classification, Biosynthesis and Applications against Uropathogens: A Mini-Review. Molecules 2017, 22, 1255. [Google Scholar] [CrossRef]
- LeBlanc, J.G.; Levit, R.; Savoy de Giori, G.; de Moreno de LeBlanc, A. Application of vitamin-producing lactic acid bacteria to treat intestinal inflammatory diseases. Appl. Microbiol. Biotechnol. 2020, 104, 3331–3337. [Google Scholar] [CrossRef] [PubMed]
- De Boeck, I.; Spacova, I.; Vanderveken, O.M.; Lebeer, S. Lactic acid bacteria as probiotics for the nose? Microb. Biotechnol. 2021, 14, 859–869. [Google Scholar] [CrossRef] [PubMed]
- Vasconcelos, F.M.; Silva, H.L.A.; Poso, S.M.V.; Barroso, M.V.; Lanzetti, M.; Rocha, R.S.; Graca, J.S.; Esmerino, E.A.; Freitas, M.Q.; Silva, M.C.; et al. Probiotic Prato cheese attenuates cigarette smoke-induced injuries in mice. Food Res. Int. 2019, 123, 697–703. [Google Scholar] [CrossRef]
- Moradi, M.; Kousheh, S.A.; Almasi, H.; Alizadeh, A.; Guimaraes, J.T.; Yilmaz, N.; Lotfi, A. Postbiotics produced by lactic acid bacteria: The next frontier in food safety. Compr. Rev. Food Sci. Food Saf. 2020, 19, 3390–3415. [Google Scholar] [CrossRef]
- Dowarah, R.; Verma, A.K.; Agarwal, N.; Singh, P.; Singh, B.R. Selection and characterization of probiotic lactic acid bacteria and its impact on growth, nutrient digestibility, health and antioxidant status in weaned piglets. PLoS ONE 2018, 13, e0192978. [Google Scholar] [CrossRef]
- Ndahetuye, J.B.; Koo, O.K.; O’Bryan, C.A.; Ricke, S.C.; Crandall, P.G. Role of lactic acid bacteria as a biosanitizer to prevent attachment of Listeria monocytogenes F6900 on deli slicer contact surfaces. J. Food Prot. 2012, 75, 1429–1436. [Google Scholar] [CrossRef]
- Liu, Z.; Xu, C.; Tian, R.; Wang, W.; Ma, J.; Gu, L.; Liu, F.; Jiang, Z.; Hou, J. Screening beneficial bacteriostatic lactic acid bacteria in the intestine and studies of bacteriostatic substances. J. Zhejiang Univ. Sci. B 2021, 22, 533–547. [Google Scholar] [CrossRef]
- Adetoye, A.; Pinloche, E.; Adeniyi, B.A.; Ayeni, F.A. Characterization and anti-salmonella activities of lactic acid bacteria isolated from cattle faeces. BMC Microbiol. 2018, 18, 96. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Rawlings, N.D. The CHAP domain: A large family of amidases including GSP amidase and peptidoglycan hydrolases. Trends Biochem. Sci. 2003, 28, 234–237. [Google Scholar] [CrossRef] [PubMed]
- Becker, S.C.; Swift, S.; Korobova, O.; Schischkova, N.; Kopylov, P.; Donovan, D.M.; Abaev, I. Lytic activity of the staphylolytic Twort phage endolysin CHAP domain is enhanced by the SH3b cell wall binding domain. FEMS Microbiol. Lett. 2015, 362, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, G.M.; Davidson, B.E.; Hillier, A.J. Novel expression system for large-scale production and purification of recombinant class IIa bacteriocins and its application to piscicolin 126. Appl. Environ. Microbiol. 2004, 70, 3292–3297. [Google Scholar] [CrossRef] [PubMed]
- Zarate, G.; Chaia, A.P.; Gonzalez, S.; Oliver, G. Viability and beta-galactosidase activity of dairy propionibacteria subjected to digestion by artificial gastric and intestinal fluids. J. Food Prot. 2000, 63, 1214–1221. [Google Scholar] [CrossRef] [PubMed]
- Ku, S.; Park, M.S.; Ji, G.E.; You, H.J. Review on Bifidobacterium bifidum BGN4: Functionality and Nutraceutical Applications as a Probiotic Microorganism. Int. J. Mol. Sci. 2016, 17, 1544. [Google Scholar] [CrossRef]
- Dunne, C.; O’Mahony, L.; Murphy, L.; Thornton, G.; Morrissey, D.; O’Halloran, S.; Feeney, M.; Flynn, S.; Fitzgerald, G.; Daly, C.; et al. In vitro selection criteria for probiotic bacteria of human origin: Correlation with in vivo findings. Am. J. Clin. Nutr. 2001, 73 (Suppl. 2), 386S–392S. [Google Scholar] [CrossRef]
- Pan, W.H.; Li, P.L.; Liu, Z. The correlation between surface hydrophobicity and adherence of Bifidobacterium strains from centenarians’ faeces. Anaerobe 2006, 12, 148–152. [Google Scholar] [CrossRef]
- Al Kassaa, I.; Hamze, M.; Hober, D.; Chihib, N.E.; Drider, D. Identification of vaginal lactobacilli with potential probiotic properties isolated from women in North Lebanon. Microb. Ecol. 2014, 67, 722–734. [Google Scholar] [CrossRef]
- Romero-Luna, H.E.; Peredo-Lovillo, A.; Hernandez-Mendoza, A.; Hernandez-Sanchez, H.; Cauich-Sanchez, P.I.; Ribas-Aparicio, R.M.; Davila-Ortiz, G. Probiotic Potential of Lactobacillus paracasei CT12 Isolated from Water Kefir Grains (Tibicos). Curr. Microbiol. 2020, 77, 2584–2592. [Google Scholar] [CrossRef]
- Kerry, R.G.; Patra, J.K.; Gouda, S.; Park, Y.; Shin, H.S.; Das, G. Benefaction of probiotics for human health: A review. J. Food Drug Anal. 2018, 26, 927–939. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).