Influence of Salinity on the Microbial Community Composition and Metabolite Profile in Kimchi
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Materials
2.2. Preparation of Kimchi
2.3. Microbial Community Analysis
2.4. Analysis of pH and Titratable Acidity
2.5. Analysis of Organic Acids
2.6. Analysis of Free Sugars
2.7. Analysis of Free Amino Acids
2.8. Statistical Analysis
3. Results and Discussion
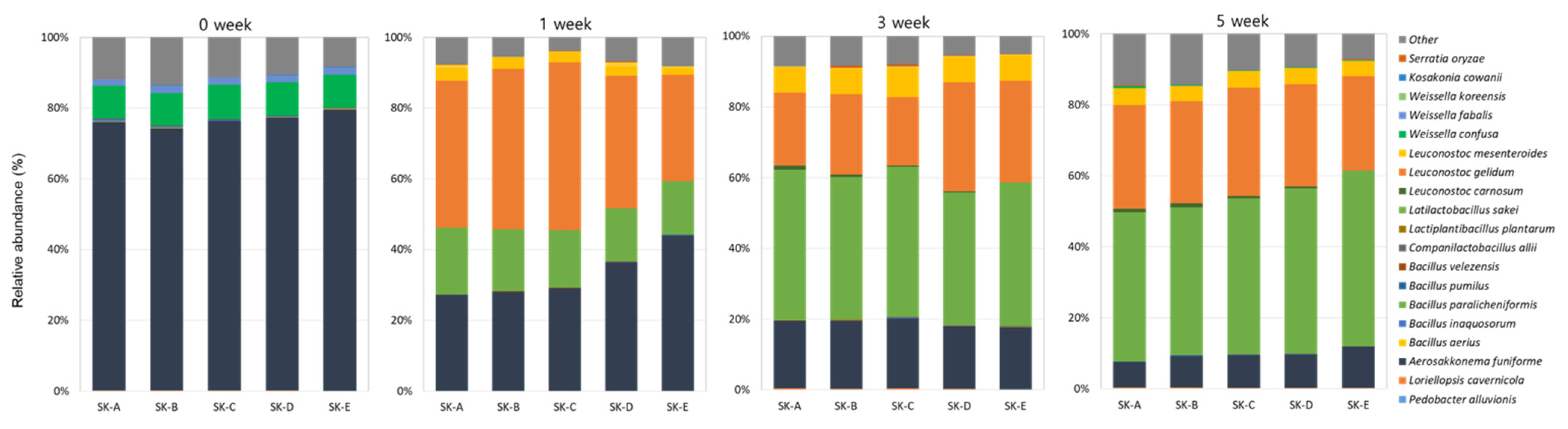
3.1. Microbial Community Analysis
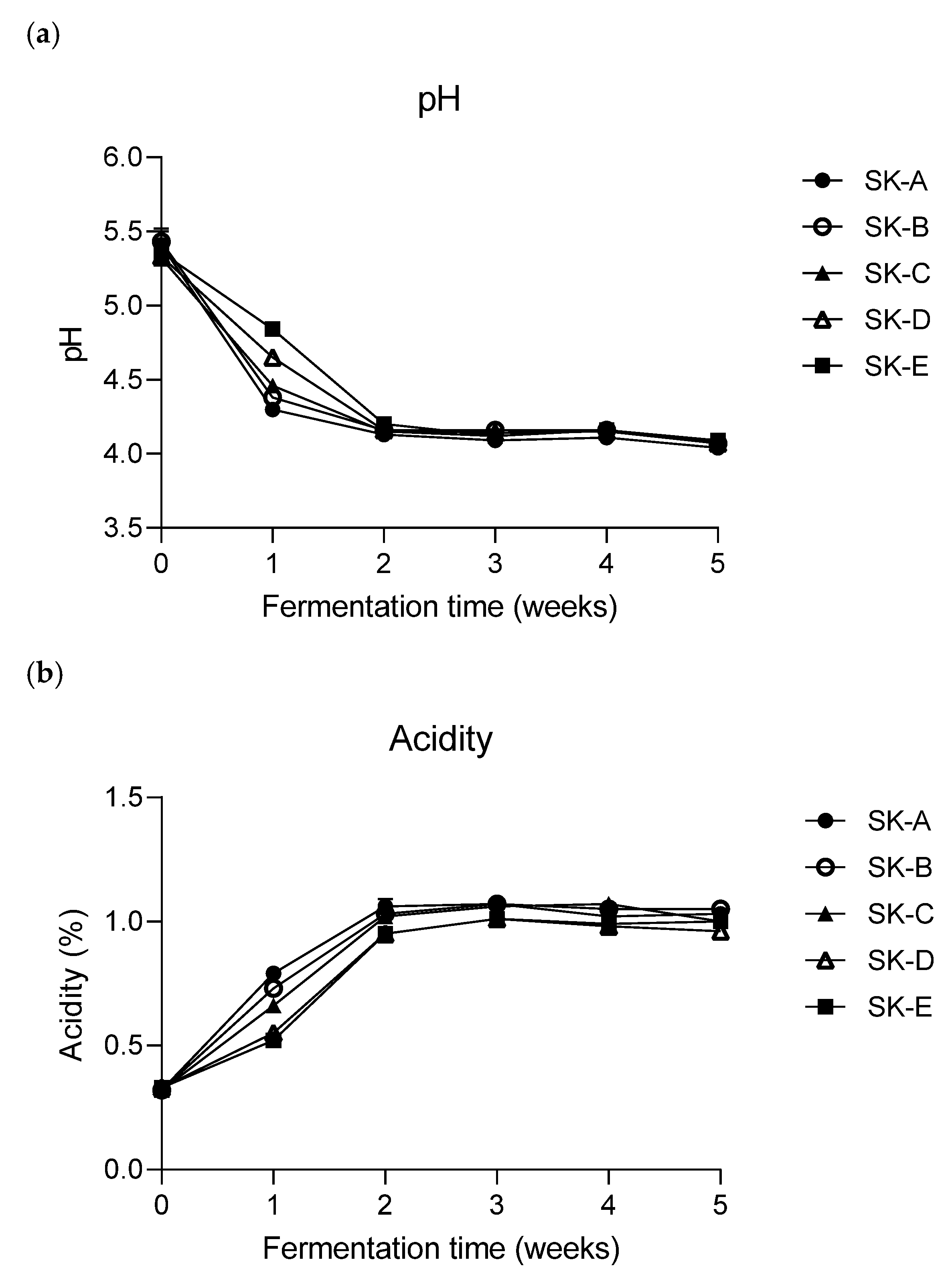
3.2. Changes in the pH and Titratable Acidity of Kimchi
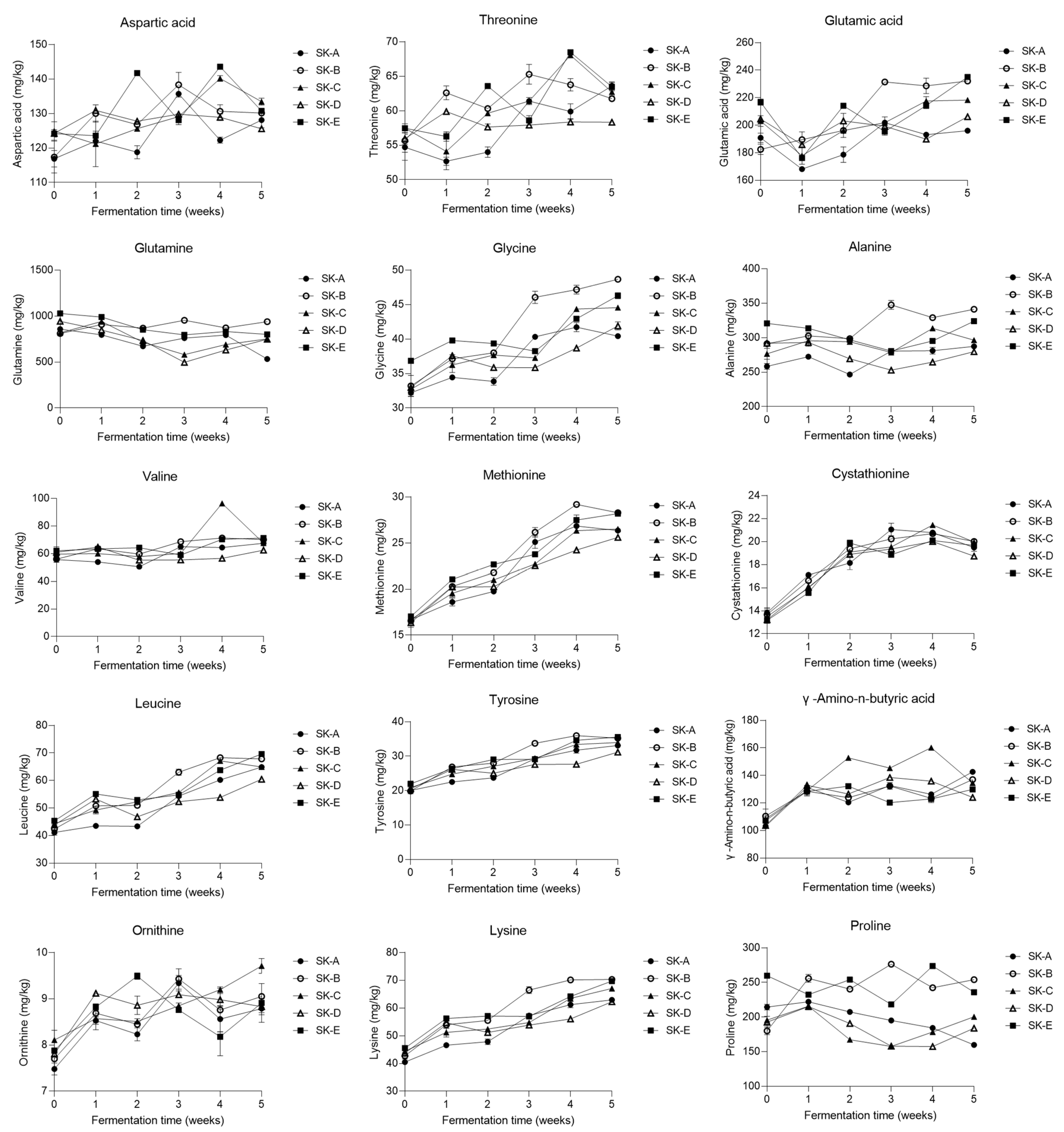
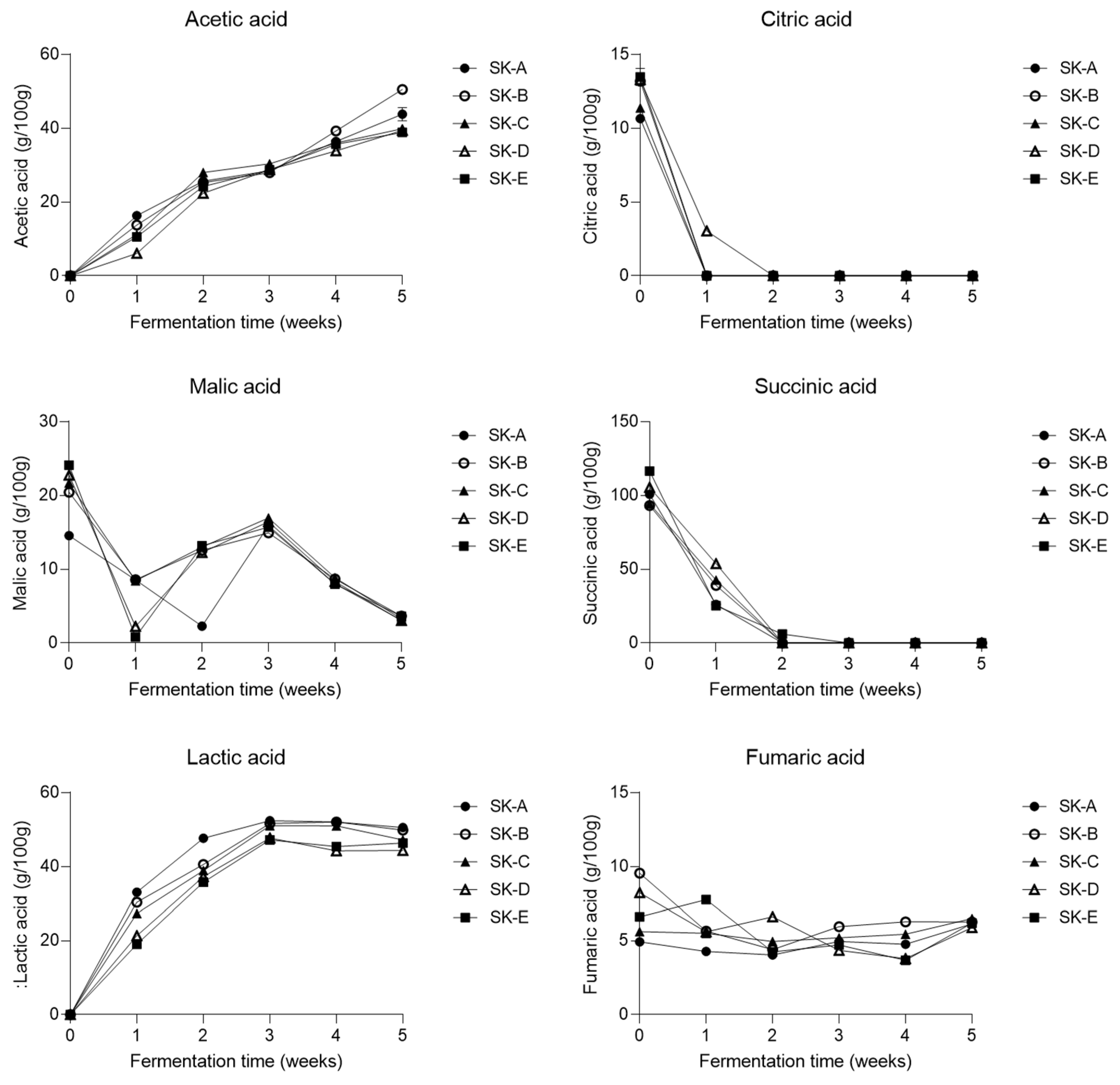
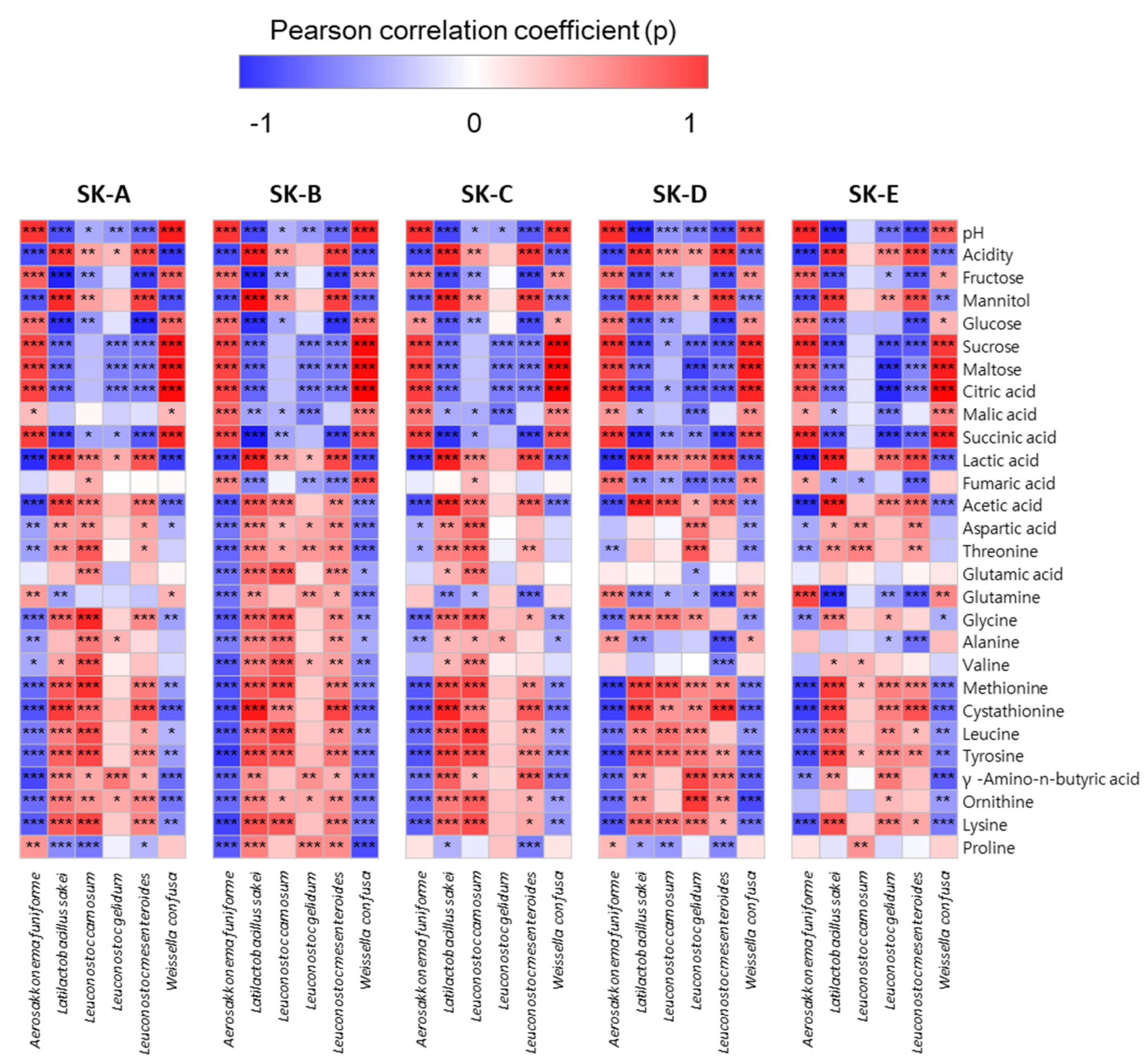
3.3. Changes in the Metabolite Profile of Kimchi
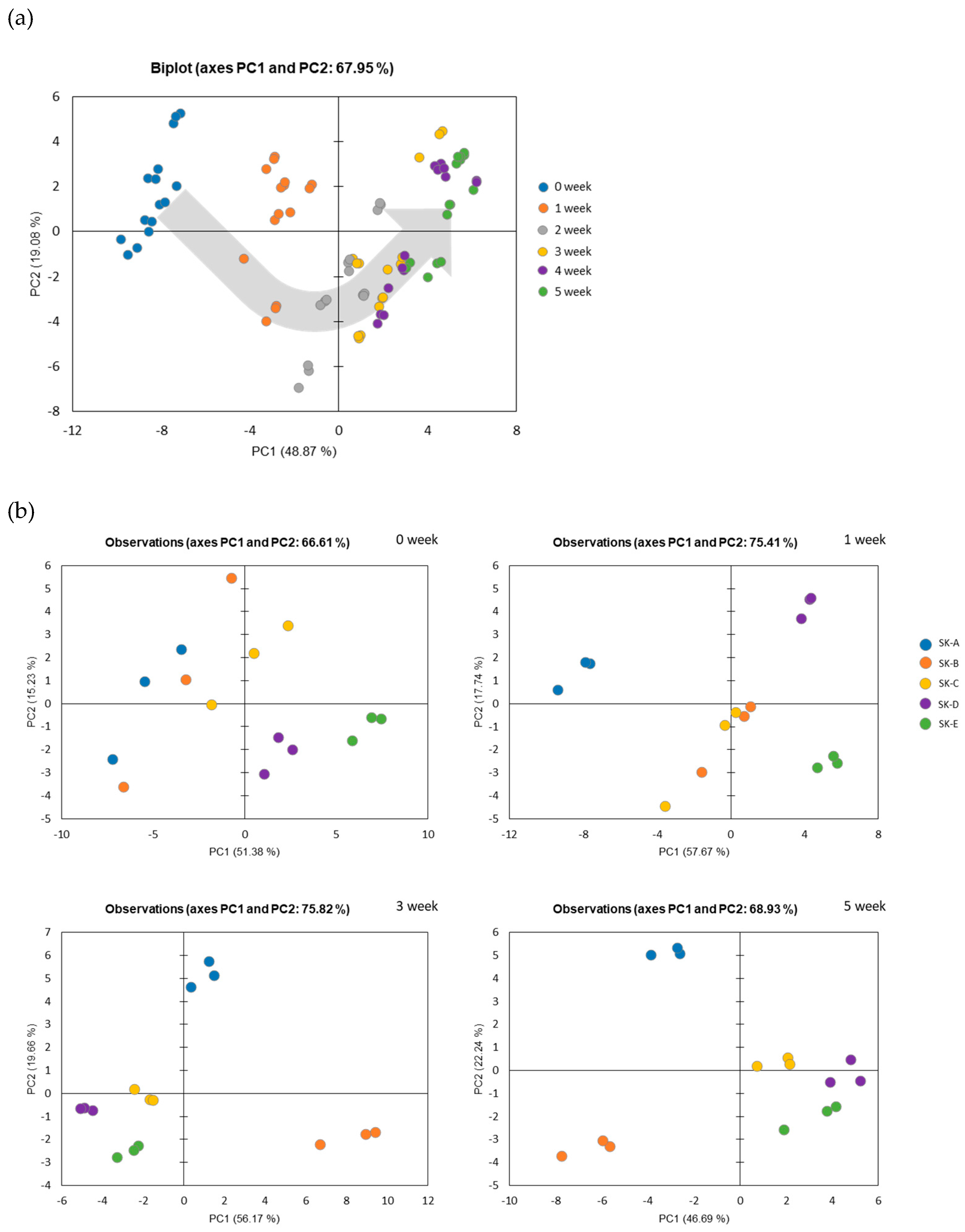
3.4. Multivariate Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jung, J.Y.; Lee, S.H.; Lee, H.J.; Seo, H.Y.; Park, W.S.; Jeon, C.O. Effects of Leuconostoc Mesenteroides Starter Cultures on Microbial Communities and Metabolites during Kimchi Fermentation. Int. J. Food Microbiol. 2012, 153, 378–387. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.H.; Lee, J.H.; Min, S.; Min, D.B. Changes of Volatile Compounds, Lactic Acid Bacteria, pH, and Headspace Gases in Kimchi, a Traditional Korean Fermented Vegetable Product. J. Food Sci. 2003, 68, 849–854. [Google Scholar] [CrossRef]
- Kim, E.J.; Seo, S.H.; Park, S.E.; Lim, Y.W.; Roh, S.W.; Son, H.S. Initial Storage of Kimchi at Room Temperature Alters Its Microbial and Metabolite Profiles. LWT. Food Sci. Technol. 2020, 134, 110160. [Google Scholar] [CrossRef]
- Song, H.S.; Whon, T.W.; Kim, J.; Lee, S.H.; Kim, J.Y.; Kim, Y.B.; Choi, H.J.; Rhee, J.K.; Roh, S.W. Microbial Niches in Raw Ingredients Determine Microbial Community Assembly During Kimchi Fermentation. Food Chem. 2020, 318, 126481. [Google Scholar] [CrossRef] [PubMed]
- Cheigh, H.S.; Park, K.Y. Biochemical, Microbiological, and Nutritional Aspects of Kimchi (Korean Fermented Vegetable Products). Crit. Rev. Food Sci. Nutr. 1994, 34, 175–203. [Google Scholar] [CrossRef]
- Lee, M.E.; Jang, J.Y.; Lee, J.H.; Park, H.W.; Choi, H.J.; Kim, T.W. Starter Cultures for Kimchi Fermentation. J. Microbiol. Biotechnol. 2015, 25, 559–568. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.H.; Lee, H.J.; Jung, J.Y.; Lee, S.H.; Seo, H.Y.; Park, W.S.; Jeon, C.O. Effects of Red Pepper Powder on Microbial Communities and Metabolites during Kimchi Fermentation. Int. J. Food Microbial. 2013, 160, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.W.; Kim, B.M.; Lee, H.J.; Jang, G.J.; Song, S.H.; Lee, J.I.; Lee, S.B.; Shim, J.M.; Lee, K.W.; Kim, J.H.; et al. Effects of Different Salt Treatments on the Fermentation Metabolites and Bacterial Profiles of Kimchi. J. Food Sci. 2017, 82, 1124–1131. [Google Scholar] [CrossRef]
- Seo, S.H.; Park, S.E.; Kim, E.J.; Lee, K.I.; Na, C.S.; Son, H.S. A GC-MS Based Metabolomics Approach to Determine the Effect of Salinity on Kimchi. Food Res. Int. 2018, 105, 492–498. [Google Scholar] [CrossRef] [PubMed]
- Bautista-Gallego, J.; Rantsiou, K.; Garrido-Fernández, A.; Cocolin, L.; Arroyo-López, F.N. Salt Reduction in Vegetable Fermentation: Reality or Desire? J. Food Sci. 2013, 78, R1095–R1100. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H. Lactic Acid Fermented Foods and Their Benefits in Asia. Food Control 1997, 8, 259–269. [Google Scholar] [CrossRef]
- Bibbins-Doming, K.; Chertow, G.M.; Coxson, P.G.; Moran, A.; Lightwood, J.M.; Plecher, M.J.; Goldman, L. Projected Effect of Dietary Salt Reduction on Future Cardiovascular Disease. N. Engl. J. Med. 2010, 362, 590–599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meneton, P.; Jeunemaitre, X.; de Wardener, H.E.; Macgregor, G.A. Links Between Dietary Salt Intake, Renal Salt Handling, Blood Pressure, and Cardiovascular Diseases. Physiol. Rev. 2005, 85, 679–715. [Google Scholar] [CrossRef] [PubMed]
- He, F.J.; Macgregor, G.A. Effect of Modest Salt Reduction on Blood Pressure: A Meta-Analysis of Randomized Trials. Implications for Public Health. J. Hum. Hypertens. 2002, 16, 761–770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, S.J.; Yang, S.Y.; Yoon, K.S. Lactic Acid Bacteria Starter in Combination With Sodium Chloride Controls Pathogenic Escherichia coli (EPEC, ETEC, and EHEC) in Kimchi. Food Microbiol. 2021, 100, 103868. [Google Scholar] [CrossRef] [PubMed]
- Hong, G.H.; Lee, S.Y.; Park, E.S.; Park, K.Y. Changes in Microbial Community by Salt Content in Kimchi During Fermentation. J. Korean Soc. Food Sci. Nutr. 2021, 50, 648–653. [Google Scholar] [CrossRef]
- Lee, K.W.; Shim, J.M.; Kim, D.W.; Yao, Z.; Kim, J.A.; Kim, H.J.; Kim, J.H. Effects of Different Types of Salts on the Growth of Lactic Acid Bacteria and Yeasts During Kimchi Fermentation. Food Sci. Biotechnol. 2018, 27, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Yun, Y.R.; Lee, J.J.; Lee, H.J.; Choi, Y.J.; Lee, J.H.; Park, S.J.; Park, S.H.; Seo, H.Y.; Min, S.G. Comparison of Quality Characteristics of Commercial Kimchi Manufactured in Korea, China, and the United States. Foods 2021, 10, 2488. [Google Scholar] [CrossRef] [PubMed]
- Park, B.; Yang, J.S.; Moon, E.W.; Seo, H.Y.; Ha, J.H. Influence of Capsaicinoids Content on the Microbial Community during Kimchi Fermentation. J. Microbiol. Biotechnol. 2019, 29, 1580–1590. [Google Scholar] [CrossRef] [PubMed]
- You, S.Y.; Yang, J.S.; Kim, S.H.; Hwang, I.M. Changes in the Physicochemical Quality Characteristics of Cabbage Kimchi With Respect to Storage Conditions. J. Food Qual. 2017, 2017, 9562981. [Google Scholar] [CrossRef] [Green Version]
- Choi, Y.J.; Young, S.J.; Lee, M.J.; Park, S.J.; Yun, Y.R.; Park, S.H.; Lee, M.A. Changes in Volatile and Non-Volatile Compounds of Model Kimchi through Fermentation by Lactic Acid Bacteria. LWT Food Sci. Technol. 2019, 105, 118–126. [Google Scholar] [CrossRef]
- Lee, M.H.; Jeong, I.H.; Jeong, S.T.; Chang, Y.H. Effect of Nuruk protease activity on the quality of anchovy sauce. Korean J. Food Sci. Technol. 2021, 53, 356–363. [Google Scholar]
- Jung, J.Y.; Lee, S.H.; Jin, H.M.; Hahn, Y.S.; Madsen, E.L.; Jeon, C.O. Metatranscriptomic Analysis of Lactic Acid Bacterial Gene Expression During Kimchi Fermentation. Int. J. Food Microbiol. 2013, 163, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Lee, H.W.; Lee, M.E.; Roh, S.W.; Kim, T.W. Mixed Starter of Lactococcus lactis and Leuconostoc citreum for Extending Kimchi Shelf-Life. J. Microbiol. 2019, 57, 479–484. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.J.; Choi, Y.J.; Lee, M.J.; Park, S.J.; Oh, S.J.; Yun, Y.R.; Min, S.G.; Seo, H.Y.; Park, S.H.; Lee, M.A. Effects of Combining Two Lactic Acid Bacteria as a Starter Culture on Model Kimchi Fermentation. Food Res. Int. 2020, 136, 109591. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.Y.; Lee, S.H.; Jeon, C.O. Kimchi Microflora: History, Current Status, and Perspectives for Industrial Kimchi Production. Appl. Microbiol. Biotechnol. 2014, 98, 2385–2393. [Google Scholar] [CrossRef] [PubMed]
- Park, S.E.; Yoo, S.A.; Seo, S.H.; Lee, K.I.; Na, C.S.; Son, H. GC-MS Based Metabolomics Approach of Kimchi for the Understanding of Lactobacillus plantarum Fermentation Characteristics. LWT Food Sci. Technol. 2016, 68, 313–321. [Google Scholar] [CrossRef]
- Ryu, J.Y.; Lee, H.S.; Rhee, H.S. Changes of Organic Acid and Volatile Flavor Compounds in Kimchi Fermented With Different Ingredients. Korean J. Food Sci. Technol. 1984, 16, 169–174. [Google Scholar]
- Shim, S.M.; Kim, J.Y.; Lee, S.M.; Park, J.B.; Oh, S.K.; Kim, Y.S. Profiling of Fermentative Metabolites in Kimchi: Volatile and Non-Volatile Organic Acids. J. Korean Soc. Appl. Biol. Chem. 2012, 55, 463–469. [Google Scholar] [CrossRef]
- Wisselink, H.W.; Weusthuis, R.A.; Eggink, G.; Hugenholtz, J.; Grobben, G.J. Mannitol Production by Lactic Acid Bacteria: A Review. Int. Dairy J. 2002, 12, 151–161. [Google Scholar] [CrossRef]
- Giuliani, A. The Application of Principal Component Analysis to Drug Discovery and Biomedical Data. Drug Discov. Today 2017, 22, 1069–1076. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Jeon, H.S.; Yoo, J.Y.; Kim, J.H. Some Important Metabolites Produced by Lactic Acid Bacteria Originated From Kimchi. Foods 2021, 10, 2148. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, M.-A.; Choi, Y.-J.; Lee, H.; Hwang, S.; Lee, H.J.; Park, S.J.; Chung, Y.B.; Yun, Y.-R.; Park, S.-H.; Min, S.; et al. Influence of Salinity on the Microbial Community Composition and Metabolite Profile in Kimchi. Fermentation 2021, 7, 308. https://doi.org/10.3390/fermentation7040308
Lee M-A, Choi Y-J, Lee H, Hwang S, Lee HJ, Park SJ, Chung YB, Yun Y-R, Park S-H, Min S, et al. Influence of Salinity on the Microbial Community Composition and Metabolite Profile in Kimchi. Fermentation. 2021; 7(4):308. https://doi.org/10.3390/fermentation7040308
Chicago/Turabian StyleLee, Mi-Ai, Yun-Jeong Choi, Hyojung Lee, Sojeong Hwang, Hye Jin Lee, Sung Jin Park, Young Bae Chung, Ye-Rang Yun, Sung-Hee Park, Sunggi Min, and et al. 2021. "Influence of Salinity on the Microbial Community Composition and Metabolite Profile in Kimchi" Fermentation 7, no. 4: 308. https://doi.org/10.3390/fermentation7040308
APA StyleLee, M.-A., Choi, Y.-J., Lee, H., Hwang, S., Lee, H. J., Park, S. J., Chung, Y. B., Yun, Y.-R., Park, S.-H., Min, S., Kwon, L.-S., & Seo, H.-Y. (2021). Influence of Salinity on the Microbial Community Composition and Metabolite Profile in Kimchi. Fermentation, 7(4), 308. https://doi.org/10.3390/fermentation7040308






