Abstract
The growing need for industrial production of bio-based acetoin and 2,3-butanediol (2,3-BD) is due to both environmental concerns, and their widespread use in the food, pharmaceutical, and chemical industries. Acetoin is a common spice added to many foods, but also a valuable reagent in many chemical syntheses. Similarly, 2,3-BD is an indispensable chemical on the platform in the production of synthetic rubber, printing inks, perfumes, antifreeze, and fuel additives. This state-of-the-art review focuses on representatives of the genus Bacillus as prospective producers of acetoin and 2,3-BD. They have the following important advantages: non-pathogenic nature, unpretentiousness to growing conditions, and the ability to utilize a huge number of substrates (glucose, sucrose, starch, cellulose, and inulin hydrolysates), sugars from the composition of lignocellulose (cellobiose, mannose, galactose, xylose, and arabinose), as well as waste glycerol. In addition, these strains can be improved by genetic engineering, and are amenable to process optimization. Bacillus spp. are among the best acetoin producers. They also synthesize 2,3-BD in titer and yield comparable to those of the pathogenic producers. However, Bacillus spp. show relatively lower productivity, which can be increased in the course of challenging future research.
1. Introduction
The contemporary production of acetoin and 2,3-butanediol (2,3-BD) is primarily related to the use of oil and fossil fuels. However, the rising environmental problems such as global warming and greenhouse gas emissions require rapid replacement of chemical methods with environmentally friendly and sustainable biotechnologies for the synthesis of both platform chemicals [1]. More than fifteen years ago, the United States Department of Energy classified acetoin among the 30 chemical compounds, stating that its microbial synthesis needs priority scientific development [2,3,4]. Recent forecasts show that the global acetoin market will grow by 5.79% annually, and the market size of 2,3-BD will reach USD 300 million by 2030 [5,6].
Acetoin has a pleasant butter odor; therefore, it is widely applied as a taste and aroma enhancer in butter, milk, yogurt, and many other foods manufacturing [7,8]. In nature, acetoin is found in fruits (apple, blackcurrants, blackberries, and cantaloupes), wheat, vegetables (broccoli and brussels sprouts), maple syrup, or fermented products such as vinegar, butter, yogurt, and wine. This compound is responsible for the aroma of strawberries, raspberries, vanilla, butter, coconut, and coffee, and therefore, it is used as a flavoring in cosmetics and the liquids for electronic cigarettes [9]. In its optically pure form, acetoin is a precursor for the synthesis of heterocyclic compounds, as 2,3,5,6-tetramethylpyrazine, α-hydroxy ketone, liquid crystal composites, and pharmaceutical intermediates [10]. Currently, most of the industrial production of acetoin is carried out by chemical synthesis by partial reduction of diacetyl through deoxidization with zinc or other catalysts [11]. Other common approaches of acetoin production are the selective oxidation of 2, 3-butanediol [12], and the oxidation of butan-2-one (MEK), followed by basic hydrolysis and hydrogenation of acetaldehyde [13].
Mainly used in the food, pharmaceutical, polymer, and cosmetic industries, 2,3-BD is used in the production of printing ink, perfumes, plasticizers (cellulose nitrates, polyacrylates, and polyvinyl chlorides, etc.), explosives, humidifiers, and emollients [14]. It is less toxic to microbial cells than other alcohols, but at the same time, its energy content is comparable to other liquid fuels obtained by microbial synthesis. The equimolar mixture of ethanol and 2,3-BD has an energy content of 27,700 kJ/kg, and comparatively pure ethanol, 2,3-BD, and methanol contain 29,100, 27,200, and 22,100 kJ/kg, respectively [15]. Dehydration of 2,3-BD results in the formation of methyl ethyl ketone (MEK), an important solvent and fuel additive [16,17]. In addition, 2,3-BD can be converted to 1,3-butadiene, which is the building block of synthetic rubber [18]. The dehydrogenation product of 2,3-BD, diacetyl, is a valuable flavoring in food products, such as original yogurt [19]. Esterification of 2,3-BD with maleic acid yields a polymeric material (polyurethane maleamide (PUMAs) used in medicine for the manufacturing of implants for cardiovascular disease [20]. Furthermore, 2,3-BD has antiseptic properties: 0.1% solution of the compound possesses a bactericide effect towards many pathogenic bacteria [21]. Due to its low freezing point of −60 °C, 2,3-BD can be used as an antifreeze [22,23]. It is also very promising as a cryoprotectant with properties similar to those of propylene glycol, DMSO, and glycerol [24] and creates fewer osmotic changes in the cell [25].
As a primer microbial metabolite, acetoin has been used in bacterial taxonomy as a classification marker of Enterobacteriaceae since 1898, when Voges and Proskauer, German bacteriologists at the Institute for Infectious Diseases in Berlin developed the legendary test that detects the oxidation of acetylmethylcarbinol to diacetyl in the presence of a strong base. Besides Gram-negative Klebsiella pneumoniae, Salmonella, and Enterobacter aerogenes, acetoin can be synthesized by many microorganisms, including Gram-positive wild-type strains of Bacillus subtilis, Leuconostoc citrovorum, Leuc. mesenteroides, Lactococcus lactis, Lacticaseibacillus casei, and yeasts Saccharomyces carlsbergensis, Kloeckera apiculata, and Hanseniaspora guilliermondii [26,27,28,29,30,31,32,33,34]. Most of them, however, produce less than 1 g/L acetoin and until 2011 the highest titer of 41.26 g/L was achieved by B. licheniformis [35].
Biotechnological production of 2,3-BD dates back to 1906, when Harden and Walpole studied its production by Klebsiella pneumoniae [36]. In 1926, Donker noticed it as a metabolic product of Paenibacillus polymyxa (Bacillus polymixa), but the idea for the industrial production of 2,3-BD was first suggested by Fulmer et al. in 1933 [37]. Later, during World War II, the interest in the production of 2,3-BD became more significant than ever. Due to the need for synthetic rubber, the first pilot plant for the production of 2,3-BD was built, as the process was carried out by microbial fermentation of strains of K. oxytoca and P. polymyxa. However, a large-scale industrial production never began, because it turns out that it is more cost-effective to produce 1,3-butadiene from oil. In the 1970s, the interest in 2,3-BD biotechnology returned, mainly due to rising oil prices. Pilot plants were built again in the United States, and later in China, this time with the idea of converting cheap lignocellulosic waste [38,39]. However, to date, despite great scientific progress, the industrial microbial production of 2,3-BD has not been established.
2,3-BD and acetoin are products of mixed-acid fermentation performed by many bacterial species with affiliation to genera Klebsiella, Enterobacter, Serratia, Raoultella, Paenibacillus, and Bacillus [40,41,42,43,44,45]. The first four of them belong to risk group 2 (pathogenic bacteria) and are therefore not suitable for industrial fermentation due to safety rules [15]. Conversely, bacilli are Generally Regarded as Safe (GRAS) producers of acetoin and 2,3-BD, suitable for large-scale industrial bioprocesses. These species are particularly promising due to their ability to grow well in both microaerophilic and aerobic conditions, their cultural adaptability, and in particular the capability to utilize a wide range of substrates. On the other hand, D(−), or (2R, 3R) stereoisomer of 2,3-BD is more desirable by the industry, because it could be used as an antifreeze. This isomer, as well as R-acetoin, are valuable reagents in chiral syntheses. In contrast to the strains of genus Klebsiella and Enterobacter, which produce L(+) and meso-2,3-BD, many members of the genus Bacillus and P. polymyxa generate pure D(−) form [22]. However, current progress in microbial 2,3-BD production employing bacilli was mostly due to the development of bioprocess and strain engineering. That is why, in terms of 2,3-BD titer and yield, Bacillus spp. are already competitive to the “classical” pathogenic producers.
The members of genus Bacillus and Paenibacillus polymyxa (formerly B. polymyxa) are Gram-positive, motile, non-pathogenic bacteria ubiquitously spread in natural habitats. They are usually isolated from soil, water, and the rhizosphere [46,47,48], medicine plants [49], marine water [50], and even animals’ guts [51,52]. Bacilli are widely used in the industrial manufacturing of enzymes, antibiotics, and exopolysaccharides with antimicrobial, antioxidant, and anticancer activities [53]. That is why the species is commonly used in horticulture and pharmacy, the biocontrol of plant diseases, and the development of probiotics [54,55,56].
This review summarizes the vast flow of experimental data involving the GRAS producers of 2,3-BD and acetoin, being the first focused on bacilli. Here we describe the significant advance in the employment of B. subtilis [57,58], P. polymyxa [59,60,61,62], B. amyloliquefaciens [63,64], and B. licheniformis [65,66,67,68,69,70,71,72,73,74]. In addition, the potential of the quite recently reported as 2,3-BD producers B. atrophaeus, B. mojavensis, B. vallismortis, B. velesensis, B. tequilensis, and B. safensis [69,75,76] for the valorization of plant biomass and agricultural wastes was assessed. The strains’ limitations and the opportunities for their improvement by genetic engineering are also discussed.
2. Chemical Structure of Acetoin and 2,3-Butanediol and Their Stereoisomers
Acetoin (also named acetylmethylcarbinol), or 3-hydroxybutan-2-one, according to IUPAC, appears as slightly yellow liquid or crystals. It has a molecular weight of 88.11 g/mol and a boiling point of 143.0 °C. Acetoin has the molecular formula C4H8O2 and exists in two chiral enantiomers: Levo-(R-), and Dextro-(S-), shown in Figure 1 [77].

Figure 1.
Structural formulae of acetoin stereoisomers. (a) R-acetoin; (b) S-acetoin. Source: https://pubchem.ncbi.nlm.nih.gov (accessed on 3 November 2021).
Also known as 2,3-butylene glycol, dimethylene glycol, dimethyl ethylene glycol, or butane-2,3-diol (according to IUPAC), 2,3-Butanediol (2,3-BD) a is a compound that has the chemical formula C4H10O2 and a molecular weight of 90.12 g/mol and density of 0.987 g/mL. It has three isomeric forms, two of which are optically active: L(+)-2,3-BD (2S, 3S) and D(−)-2,3-BD (2R, 3R), shown in Figure 2a,c, as well as an optically inactive form meso-2,3-BD (2R, 3S, or 2S, 3R), shown at Figure 2b [77]. In all three forms, 2,3-BD is found both as a colorless and odorless liquid and in crystalline form. It is soluble in water, alcohols, ethers, and ketones. In addition, 2,3-DB has a very high boiling point (depending on the stereoisomer, it varies between 177 °C and 182 °C) and a very low freezing point (−60 °C), which determines many of its characteristics and applications.
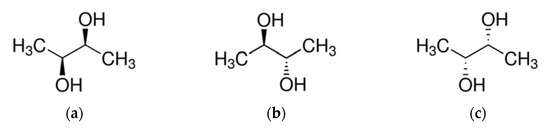
Figure 2.
Stereoisomers of 2,3-bitanediol (2,3-BD). (a) L(+)-BD; (b) meso-2,3-BD; (c) D(−)-2,3-BD Source: https://pubchem.ncbi.nlm.nih.gov (accessed on 3 November 2021).
The key for the stereoisomer’s formation is the enzyme specificity of butanediol dehydrogenase BDH, which is influenced not only by the nucleotide sequence of the responsible gene but also by factors such as carbon source, the mineral content of the medium (Ca, S, Mg), NADH/NAD+ ratio, aeration, and the growth phase of the culture, etc. [67,78].
3. Metabolic Pathway for Acetoin and 2,3-Butanediol Synthesis by Bacillus spp.
Acetoin and 2,3-BD metabolic pathway can be determined both as anabolic and catabolic. The biological role of the synthesis of these compounds is to serve as an alternative to acid production in the course of mixed-acids fermentation and thus, to prevent excessive culture acidification. The second important function of the pathway is to support NADH/NAD+ levels by the reversible transformation between acetoin and 2,3-BD, which is coupled with this redox balance. In addition, acetoin is an intermediate metabolite in amino acids biosynthesis. It, along with 2,3-BD, could be consumed by the microbial cells under starvation conditions.
In Bacillus spp. acetoin and 2,3-BD are produced from pyruvate, obtained from sugars and other carbohydrates metabolized either by the glycolytic pathway (hexoses) or via a combination with the pentose–phosphate metabolic pathway (pentoses). Both metabolites are obtained in the course of mixed-acid fermentation, which also generates other final metabolites such as lactic, acetic, succinic, and formic acids, ethanol, and glycerol. Depending on the type of producer and the cultivation conditions, these products are obtained in different proportions or do not accumulate at all. The same bacterial strain can produce predominantly 2,3-BD or acetoin as alternative final metabolites via the manipulation of process parameters.
The biosynthesis of 2,3-BD occurs through the intermediates α-acetolactate and acetoin, under the action of three key enzymes: (i) α-acetolactate synthase I/II/III (EC 2.2.1.6), which small and large subunits in bacilli are encoded by three genes: ilvH, ilvB, alsS; (ii) α-acetolactate decarboxylase (EC 4.1.1.5), encoded by alsD and catalyzing the conversion of α-acetolactate to acetoin; and (iii) 2,3-butanediol dehydrogenase (BDH), which displays multiple enzyme activities in bacilli, but its primary function is to reduce acetoin to 2,3-BD. Depending on the BDH type, R-acetoin can be reduced into meso-2,3-BD by R,S-BDH or D(−)2,3-BD by R,R-BDH, whereas S-acetoin could be converted into meso-2,3-BD by R,S-BDH, and L-2,3-BD by S,S-BDH.
In the presence of α-acetolactate and under aerobic conditions, acetoin is spontaneously decarboxylated to diacetyl (without enzymatic reaction), which under the action of diacetyl reductase (DAR) is irreversibly converted to acetoin. The secondary function of BDH in Bacillus spp. is to act as a stereospecific DAR.
According to the KEGG database [79], the conversion of pyruvate to 2,3-BD in Bacillus spp. can be presented by two reference pathways; the first, of B. subtilis subsp. subtilis 168 (Figure 3), and the second, of B. licheniformis ATCC 14580T (Figure 3 and Figure 4). The main difference between them is in bdh genes. B. subtilis genome contains bdhA (1041 base pairs) encoding (R,R)-butanediol dehydrogenase (346 amino acids), which produces D(−)2,3-BD from R-acetoin (EC 1.1.1.4). BdhA also displays DAR activity (EC 1.1.1.303) for diacetyl conversion to R-acetoin.
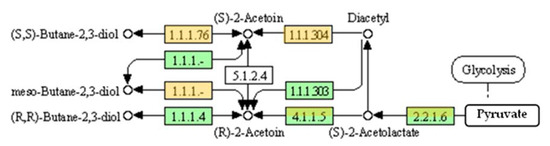
Figure 3.
Scheme of enzymes responsible for 2,3-BD production by B. subtilis subsp. subtilis 168 (green) and B. licheniformis ATCC 14580T (orange). Genes encoding the enzymes with EC 4.1.1.5 and EC 2.2.1.6 presented in both genomes, while the genes for the hypothetical enzyme with EC 5.1.2.4 were absent. Source: https://www.genome.jp/kegg-bin/show_pathway?bsu00650; https://www.genome.jp/pathway/bld00650, 4 December 2021.
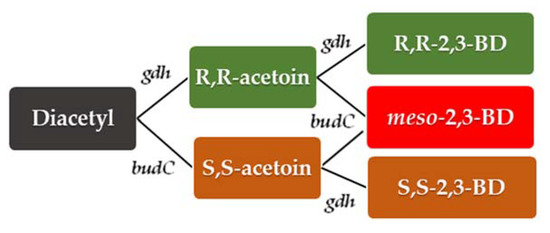
Figure 4.
The genetic basis of alternative pathways of diacetyl conversion into 2,3-BD stereoisomers in B. licheniformis. The enzyme activities of BudC (EC 1.1.1.304 and EC 1.1.176), and Gdh (EC 1.1.1.4) were proven for B. licheniformis MW3 [45].
B. licheniformis ATCC 14580T contains meso-BDH encoded by budC (EC 1.1.1-, 260 amino acids) with S-acetoin forming DAR stereo-specificity (EC 1.1.1.304). Figure 3 shows the conversion of the spontaneously formed diacetyl to S-acetoin, which, in turn, is reduced to (S,S)-2,3-BD by the same BDH BudC (EC 1.1.1.76). Although the gene encoding an enzyme with EC 1.1.1.4 has not been determined as active in the type strain ATCC 14580T, it was identified in B. licheniformis strain MW3 by Ge et al. [45]. This enzyme, encoded by gdh gene forms R-acetoin from diacetyl, and then D(−)2,3-BD stereoisomer (Figure 4), but also meso-2,3-BD from S-acetoin. Acetoin and 2,3-BD stereoisomers interconversion and the versatile catalytic activities of BDH are typical for Bacillus spp. However, this phenomenon is also the main obstacle to the successful production of 2,3-BD stereoisomers in pure form.
The analyses of Bacillus spp. draft genomes by the metabolic model of Rapid Annotations using Subsystems Technology (RAST) by ModelSEED v2.3 [80,81] allow for building a complex 2,3-BD synthesis model (Figure 5).
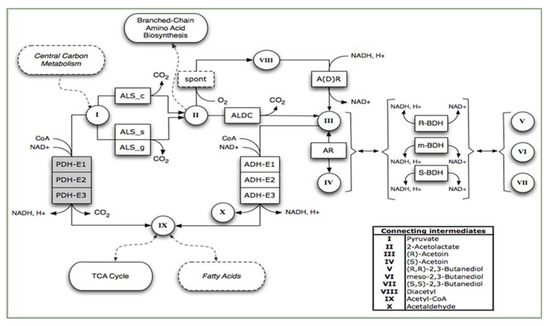
Figure 5.
RAST model of the metabolic network for acetoin and 2,3-BD synthesis in Bacillus spp. Designations: PDH-E1, PDH-E2, PDH-E3, pyruvate dehydrogenase multienzyme complex, [85]; ALS_c, ALS_s, ALS_g, acetoin dehydrogenase enzyme system; spont, spontaneous reaction; ADLC, α-acetolactate decarboxylase, A(D)R, acetoin dehydrogenase, AR, acetoin racemase; BDH, 2,3-butanediol dehydrogenase.
The enzyme acetoin racemase (EC 5.1.2.4) was postulated previously by Volloch [82], or the responsible gene was not found in Bacillus spp. genomes. The conversion of R-acetoin in acetaldehyde is performed by the acetoin dehydrogenase enzyme system ADH ES (EC 2.3.1.190) [83]. The step from pyruvate to Acetyl-CoA is carried out by the pyruvate dehydrogenase multienzyme complex (PDH) encoded by the pdhABCD operon [84]. Acetoin biosynthesis pathway genes alsD and alsS are organized in an operon, and the expression is regulated by AlsR, and encoded by alsR gene. This is a LysR-type transcriptional activator that switches “on” the operon in conditions of high medium acidification (especially in the presence of acetate). The genomes of both B. subtilis and B. licheniformis contain acoABCL operon, which is responsible for acetoin and 2,3-BD degradation. This operon is regulated by acoR gene with sigmaL-dependent promoter. Its transcription is repressed by glucose and activated by acetoin [85].
4. Natural Producers of Acetoin and 2,3-BD
4.1. B. licheniformis
Numerous attempts have been made over time to optimize the content of the medium and the cultivation conditions of B. licheniformis. The strains and bioprocesses that reached high titer, yields, and productivities of 2,3-BD are presented in Table 1; those of acetoin are shown in Table 2.

Table 1.
Natural producers of 2,3-butanediol belonging to genera Bacillus and Paenibacillus.

Table 2.
Natural producers of acetoin belonging to genera Bacillus and Paenibacillus.
As top acetoin and 2,3-BD producers among bacteria, native bacilli are selected just by ordinary enriching and screening procedure. However, as it was mentioned above, the main problem of mixed-acid fermentation is the synthesis of undesirable by-products, which reduces the carbon flux to acetoin and 2,3-BD. In addition to the target metabolites, the strains of B. licheniformis also produce acetic acid and ethanol [65], lactate, formic acid, and glycerol [86,87]. In 1993, Romano et al. [33] managed to improve the yield of acetoin produced by B. licheniformis MEL09, varying one factor at a time and using orthogonal array tests. In this way, after medium composition and culture conditions’ improvement, the maximum acetoin concentration achieved 41.26 g/L, and had a 41.26% glucose conversion efficiency (84.39% of theoretical). Then, a series of experiments engaging thermophilic B. licheniformis strains were performed by Li et al. in 2013 and 2014 [70,73,88]. At optimal temperature 50 °C and pH 7.0, with a two-stage agitation speed control strategy, 115.7 g/L of 2,3-BD was obtained from glucose by fed-batch with productivity 2.4 g/L/h and yield 94% of the theoretical value. The same authors used B. licheniformis strain ATCC 14,580 for efficient 2,3-BD synthesis from fructose (at 50 °C), and for simultaneous saccharification and fermentation of inulin, receiving 103.0 g/L 2,3-BD in 30 h with a productivity of 3.4 g/L/h [88]. Recent trials of Song et al. [75] showed that among 4000 Bacillus candidates isolated by pre-treatment and enrichment, the thermophilic B. licheniformis GSC3102 produced the highest 2,3-BD titer of 92.0 g/L. However, it was accompanied by accumulation of the toxic formate; the amount was dependent on the nitrogen source and reached up to 29.1 g/L [75].
The highest yield and productivity of B. licheniformis to date was reported by Jurchescu et al. [71]. Fed-batch cultivation of the strain DSM 8785 with substrate glucose resulted in 144.7 g/L 2,3-BD, yielding 0.4 g/g, with a productivity of 1.14 g/L/h. A very similar titer was recently obtained by Tsigoriyna et al. [67]. By applying the Plackett–Burman design and RSM through CCD, a complex optimization of medium and process parameters for 2,3-BD production by B. licheniformis strain 24 was performed. The highest concentration of 2,3-BD was 138.8 g/L, the productivity-1.16 g/L/h, and the yield—close to the theoretical—0.479 g/g. The most important factors affecting the process were the amounts of yeast extract, tryptone, K2HPO4, and MgSO4, as well as the aeration rate. Comparison with the results of Jurchescu et al. [71] reveals that the values of 2,3-BD around 140 g/L are probably the highest that can be obtained using the species B. licheniformis and indicate maximal natural capabilities of the species as a 2,3-BD producer.
4.2. B. subtilis
The natural strains of B. subtilis, which produce acetoin, have been isolated from soil, distiller’s grains, and agricultural controlled residues, spirits or vinegar factories, and even from food as natto. Zhang et al. [7], after optimization of several process parameters, efficiently produced acetoin by B. subtilis strain JNA 3-10 (42.2 g/L), but the targeted product was mixed with 2,3-BD (15.8 g/L). A similar titer, but from a cheaper substrate, was obtained by B. subtilis CICC 10,025 [97]. RSM was applied to determine the optimal levels of acid-treated molasses and soybean meal hydrolysate for acetoin production, thus obtaining 35.4 g/L acetoin in 56 h (Table 2). Tian et al. [2] also showed that the carbon and nitrogen source and agitation play a crucial role in acetoin production. By the use of B. subtilis strain SF4-3, from 150 g/L glucose, the authors obtained 48.9 g/L acetoin (with a yield corresponding to 79.90% of the theoretical conversion). High acetoin production was achieved by Dai et al. [98]. New marine B. subtilis strain DL01 demonstrated high sugar tolerance and relatively low oxygen requirement. From 210 g/L glucose, 76 g/L acetoin (60.9 g/L R-form) was received, with a productivity of 1 g/L/h and a yield of 0.421 g/g. From the cheaper molasses (200 g/L sugar), the strain produced a smaller amount of acetoin (61.15 g/L) [98].
4.3. B. amyloliquefaciens
As a close relative to B. subtilis, B. amyloliquefaciens is able to produce an enantiomer pure form of acetoin and 2,3-BD. Noticing that the lower agitation speeds favored 2,3-BD accumulation, while the higher agitation speeds favored reverse transformation of 2,3-BD to acetoin, Zhang et al. (2013) proposed a two-stage agitation speed control (350 rpm for 24 h, then-500 rpm) for acetoin production [64]. Following this strategy, through the use of B. amyloliquefaciens FMME044, the authors achieved yield of 51.2 g/L acetoin [64].
A record-high concentration of acetoin obtained from this species was achieved by Maina et al. [5]. The new approach of variation of the volumetric oxygen transfer for either acetoin or 2,3-BD production was applied. Thus, when the volumetric oxygen transfer coefficient kL a value was 64 h−1, 55.2 g/L D(−)2,3-BD and 47.4 g/L acetoin were received; when kL a value was 200 h−1, the acetoin reached 65.9 g/L with a productivity of 1.57 g/L/h. The same effect was observed when cane sugar with very high polarity (VHP) was used as a substrate. Increasing the value of kL a to 104 h−1, led to acetoin titer of 25.6 g/L, while the kL a at 49 h−1, led to an increase in 2,3-BD yield. High titer of 2,3-BD, 92.3 g/L has been achieved by Yang et al. [45] by process parameters optimization (agitation and pH) [63].
4.4. Other 2,3-BD and Acetoin Producers
4.4.1. Bacillus spp.
The interest in the selection of other GRAS microorganisms for the synthesis of 2,3-BD and acetoin is due to the need to reduce the cost of bioprocesses and in the hope that hitherto unknown bacilli will be effective producers. Thus, the species B. atrophaeus, B. mojavensis, B. vallismortis, B. pumilus, B. velezensis, B. toyonensis, and B. safensis are considered as particularly promising, because they can use the sugars falling into the lignocellulose content and may be applied in biomass valorization. For the development of economical production of 2,3-BD, the screening procedures included tests of the utilization of xylose, fructose, sucrose, or corn steep liquor [75]. For instance, B. vallismortis B-14891 converts 14 different substrates that can be obtained from residual biomass sources to 2,3-BD [76]. Recently, Petrova et al. [69] added three new species (B. velezensis, B. toyonensis, and B. safensis) to the group of 2,3-BD producers. They all displayed cellulolytic activity but also degraded arabinoxylan, arabinan, galactomannan, xyloglucan, xylan, and galactan (included in hemicellulose), as well as starch and fructans. B. velezensis 5RB was able to convert nine explored substrates to acetoin and D(−)2,3-BD, as relatively high 2,3-BD titer was obtained from glucose (18.1 g/L), cellobiose (14.8 g/L), mannose (13.8 g/L), and inulin (11.7 g/L), but low from xylose (2.6 g/L) and galactose (0.6 g/L), differing from B. toyonensis 11RA that yielded 4.48 g/L 2,3-BD from galactose [69]. However, the sugar that is most difficult to convert to acetoin and 2,3-DB is xylose. Of the bacilli studied, only two strains convert xylose to significant amounts of the target products-B. vallismortis B-14891 [76], and B. velezensis 5RB [69].
4.4.2. Paenibacillus spp.
In addition to glucose and cellulosic hydrolysates, P. polymyxa is able to ferment crude inulin to almost pure D(−)-stereoisomer of 2,3-BD [61,62,93,94,95,96]. A good natural producer of acetoin was the strain CS107 isolated by Zhang et al. [100], yielding 55.3 g/L by precise oxygen supply control. Dai et al. [93] observed the whole cellular redox status and oxygen availability as a tool to enhance 2,3-BD production by P. polymyxa. The neccessarry high NADH/NAD+ ratio was reached by decreasing agitation speed and adding ascorbic acid (vitamin C), which elevated the 2,3-BD titer 71.71 g/L. Okonkwo et al. [60] determined 50 g/L of 2,3-BD as a toxic threshold on P. polymyxa DSM 365, but after optimization of the medium content by Box–Behnken design received 68.5 g/L 2,3-BD [61]. These authors stated as problematic the synthesis of exopolysaccharide (EPS) by the used strain. Indeed, P. polymyxa, as well as B. licheniformis, produce high amounts of EPS along with 2,3-BD fermentation, especially when high substrate concentration or fed-batch fermentation mode is performed. EPS can reach 4.97 g/L for P. polymyxa [61] and 12.6 g/L for B. licheniformis [53].
P. brasilensis PB24 produces D(−), and meso-2,3-BD through activity of S-2,3-butanediol dehydrogenase encoded by butA, presented in its genome [96]. The obtained 2,3-BD by P. brasilensis was 27 g/L without any optimization. Regarding the great similarity of the genomes (including 2,3-BD operons) of P. brasilensis, P. terrae, and P. peoriae, we could expect the emergence of new producers belonging to the little-studied species of the genus Paenibacillus.
5. Engineered Producers of 2,3-BD and Acetoin
5.1. Engineered Producers of 2,3-BD
The genetic manipulations of bacilli for 2,3-BD and acetoin production have two main purposes: (i) to obtain enantiopure stereoisomers; and (ii) to enhance the fermentation process for higher titer, yield, and productivity of the target metabolite.
The first strategy is usually performed by eliminating specific bdh genes (Table 3).

Table 3.
Engineered producers of 2,3-butanediol belonging to genera Bacillus and Paenibacillus.
Knowing that B. licheniformis MW3 produces a mix of meso- and D(−)2,3-BD, Ge et al. [45] consecutively determined and eliminated the genes encoding the stereospecific 2,3-BD dehydrogenases. The engineered MW3ΔbudC strain produced 123.7 g/L D(−)2,3-BD with 99.4% purity, while MW3Δgdh produced 90.1 g/L meso-2,3-BD, which was 99.2% pure. The same approach was applied to B. licheniformis strain WX-02. Qi et al. [101] deleted budC, while Qui et al. [102] eliminated gdh gene, but also acoR gene (which encodes the activator of acoABCL operon). The recombinant WX-02ΔgdhΔacoR produced 99% pure meso-2,3-BD in a higher amount than WX-02ΔbudC.
In P. polymyxa ZJ-9, by elimination of dudA gene (encoding DAR) by homologous recombination [106], D(−)2,3-BD stereoisomer was produced to purity of 99%.
The main approach to enhance 2,3-BD production is the redistribution of the carbon flux towards 2,3-BD branch by NADH/NAD+ ratio increase. This goal was successively accomplished by Yang et al. [104] using B. amyloliquefaciens B10-127 with crude glycerol as a substrate. The manipulation of carbon flux was conducted as an extra copy of an NADH/NAD+ regeneration system was introduced into the strain by overproduction of two enzymes: glycerol dehydrogenase and acetoin reductase. This co-overexpression accelerated the NADH/NAD+ regeneration rate and, finally, caused a decrease in by-products acetoin and acids. In addition, the transcriptional regulator alsR was cloned under PbdhA promoter control, which also increased the carbon flux towards the 2,3-BD(−)branch. These genetic manipulations, accompanied by three-stage dissolved oxygen control and two-stage pH control, led to a significant increase in 2,3-BD titer (102.3 g/L).
Employing the same strain B. amyloliquefaciens B10-127 for glucose fermentation, Yang et al. [105] increased NADH/NAD+ ratio using another approach, by the complex effect of the over-production of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and NADH-dependent BDH. NAD+-dependent GAPDH-catalyzed reaction is coupled with reduction of NAD+ to NADH, while the conversion of acetoin to 2,3-BD by BDH is concomitant with oxidation of NADH to NAD+. That is why GAPDH excess increased 2,3-BD by 12.7%, and decreased acetoin yield by 44.3%. By its side, BDH excess increased 2,3-BD yield by 16.6%. Moreover, the fermentation was accelerated, and the overall process time was reduced. Thus, the final fed-batch fermentation yielded 132.9 g/L 2,3-BD in 45 h.
The desired increased NADH/NAD+ ratio for higher 2,3-BD titer was achieved by the cloning of NAD+-dependent formate dehydrogenase (fdh) gene from Candida boidinii in P. polymyxa strain ZJ-9 [107]. The recombinant P. polymyxa strain XG-1 reached 51.3 g/L 2,3-BD.
Białkowska et al. [103] tried to intensify the production of 2,3-BD B. subtilis by homologous overexpression of bdhA and heterologous expression of vhb gene encoding bacterial hemoglobin in Vitreoscilla stercoraria. The recombinant LOCK 1086 did not display higher productivity in flasks, however, the fed-batch processes were successful and reached 75.73 g/L 2,3-BD concentration.
5.2. Engineered Producers of Acetoin
When the goal is to produce the maximum amount of acetoin, the redox balance of the cell should be shifted exactly the opposite i.e., to NADH/NAD+ ratio reduction. In this way, Bao et al. [108] developed B. subtilis 168 derivate for high acetoin production with NAD+ regeneration system through homologous co-expression of 2,3-BDH and NADH oxidase. This decreased intracellular NADH concentration and NADH/NAD+ ratio and in the batch process, the recombinant produced 91.8 g/L acetoin. Another attempt of manipulation of the NADH levels in B. subtilis for high acetoin yield was performed by Zhang et al. [109] by deletion of bdhA and moderate overexpression of yodC gene encoding NADH oxidase. Thus, 2,3-BD decreased by 92.3%, while acetoin productivity reached 0.639 g/L/h (Table 4).

Table 4.
Engineered producers of acetoin belonging to genera Bacillus and Paenibacillus.
The deletion of (R,R)-2,3-BD dehydrogenase gene bdhA of B. subtilis 168 enhanced four-fold the acetoin synthesis by the recombinant variant reaching 24.6 g/L [110]. The amount of 2.4 g/L of meso-2,3-BD from 100 g/L glucose indicated that the bdhA gene was the only gene responsible for (2R,3R)-2,3-BD synthesis. A similar approach was applied by Liu et al. [111] for high acetoin production by engineered B. licheniformis WX-02. Further, two- and three-genes deletion mutants were developed, as the highest acetoin was obtained by the recombinant WX-02ΔbudCΔacoR. In optimized fed-batch fermentation, by the use of the latter, 110.04 g/L 2,3-BD was accumulated and then converted to 78.79 g/L acetoin.
For enhanced acetoin production from lignocellulosic sugars, Zhang et al. [8] used recombinant B. subtilis 168 ARSRCPΔacoAΔbdhA strain, which was able to use xylose. This strain was engineered to produce acetoin from glucose, xylose, and arabinose mixtures by cloning of araE, xylA, and xylB encoding sugar transport protein, xylose isomerase, and xylulokinase in E. coli. The recombinant strain simultaneously utilized glucose, xylose, and arabinose, and produced 62.2 g/L acetoin from mixed sugars (fed-batch process), and 11.2 g/L acetoin from lignocellulosic hydrolysate fermentation in flasks. Zhang et al. [112] reported that moderate enhancement of alsR expression under PbdhA promoter control significantly improves the acetoin production in B. subtilis. This was due to the two-fold increase in the activities of α-acetolactate synthase (ALS) and α-acetolactate decarboxylase and yielded 41.5 g/L of acetoin.
Some B. subtilis acetoin overproducers were selected after chemical or UV mutagenesis. The mutants fulfilled the requirements, although the genetic basis of these properties was not elucidated. Such a mutant is TH-49, which reached a yield of 56.9 g/L acetoin in 100 L fermentation of glucose [114]. Other such examples are B. subtilis JNA-UD(−)6 that derives from UV-light and diethyl sulfate-treated JNA 3-10, which was selected by a deficiency in BDH [115], and IPE5-4-UD(−)4, which acquired the ability to ferment simultaneously glucose and xylose to high acetoin concentrations, converting lignocellulose hydrolysates [116].
B. amyloliquefaciens E-11 mutant was selected after adaptive evolution by acetoin stress of parental strain FMME044. In comparison with the parent, the accumulated mutations resulted in a higher unsaturated fatty acid content and a lower unsaturated fatty acid content. E-11 was able to resist 80 g/L acetoin and possessed a 2,5-fold higher growth rate compared to its parent [117].
6. Substrates
An important advantage of bacilli is the wide range of substrates they can convert. Because substrate costs account for the largest share of production costs, bioprocesses based on renewable, non-food, and cheaper sources are being sought. A decade ago, Ji et al. [22] classified the cheap alternative substrates for 2,3-BD production as two types: non-cellulosic and lignocellulosic. In the first category fall the raw material or hydrolysates of plants containing sucrose (sugar cane and beet), starch, and inulin (chicory and Jerusalem artichoke).
The first alternative substrates employed for 2,3-BD production were starch and inulin. B. licheniformis NCIMB 8059 produced 2,3-BD and acetoin with a productivity of 0.58 g/L/h from cornstarch hydrolysate [68]. The one-step fermentation of raw inulin extract from Jerusalem artichoke tubers by P. polymyxa ZJ-9 was developed to produce R,R-2,3-BD [92], while Li et al. [88] performed SSF process for 2,3-BD synthesis with thermophilic B. licheniformis ATCC 14,580 (with productivity 3.4 g/L/h).
A total of two recent reports revealed the high potential of sucrose for 2,3-BD production by B. subtilis. Wang et al. [58] fermented sucrose using B. subtilis strain SC13 and reached 132.4 g/L 2,3-BD, with a productivity of 2.45 g/L/h, and yield of 0.45 g/g in fed-batch process. Feng et al. [118] developed four combinations of heterologous sucrose utilization pathways and introduced them into two different B. subtilis strains for successful conversion of sucrose and sugar cane juice into 2,3-BD.
Both raw and decolored molasses are widely employed in 2,3-BD fermentations by all bacilli. B. subtilis strains CICC10025 [97], engineered 1A1 [119], and DL01 [98] yielded between 35 and 65 g/L 2,3-BD from molasses. B. amyloliquefaciens TUL 322 also fermented molasses to 25.3 g/L 2,3-BD but this strain needed some glucose feeding for better results [120]. A higher concentration (75 g/L) was obtained by B. subtilis strain TUL 322 [121], in a medium containing sugar beet molasses in a course of a fed-batch process with four glucose additions.
A series of experiments devoted to apple pomace as a substrate were performed, but for effective 2,3-BD production, glucose addition was needed. B. subtilis TUL 322, with substrate apple pomace hydrolysate, needed three glucose feedings to produce 51 g/L 2,3-BD [121]. The more potent B. licheniformis NCIMB 8059 yielded 113 g/L 2,3-BD at the same conditions [89]. In a medium based on the apple pomace hydrolysate with three feedings with sucrose, the recombinant B. subtilis LOCK 1086 produced 51.53 g/L 2,3-BD (with a productivity of 0.43 g/L/h) [103].
The techno-economical evaluation of 2,3-BD production by Koutinas et al. [122] showed that among the three most promising substrates for the industrial production of 2,3-BD, glycerol is the most profitable, followed by molasses and sucrose. Although the highest yields are obtained from sucrose and molasses, the minimum selling price of 2,3-BD produced from glycerol is lower, because of the low glycerol price.
A substrate combination of molasses (15%) with biodiesel-derived glycerol (80%) appeared to be very promising for 2,3-BD production by B. amyloliquefaciens. Beet molasses as a co-substrate stimulated 2,3-BD production and reduced the duration of fermentation to reach the productivity of 0.87 g/L/h. From this substrate, a very high titer of 2,3-BD (102.3 g/L) was obtained by an engineered B. amyloliquefaciens strain [104]. Hence, other new-isolated strains that can convert glycerol as sole carbon source into 2,3-BD, such as B. subtilis 35 [69], may be also promising for 2,3-BD synthesis in future experiments.
Cellulose and lignocellulose containing plant biomass is a renewable, abundant, environmentally friendly, sustainable carbon feedstock for microbial biorefinery. Agro-industrial residues typically contain a mix of both, for example, corncob, cereals, and potato wastes contain cellulose and residual starch; sugar beet, sugar cane, fruits, and vegetables are rich in sucrose and fructans. The wheat straw, barley hull, rice husks, oat, ray, and sorghum straw, corn stover, sugarcane bagasse, and wood are composed of 30–50% cellulose, 16–28% hemicellulose, and 6–12% lignin [69]. Several processes for 2,3-BD and acetoin production using hydrolysates have been reported. The thermophilic strain B. licheniformis X10 effectively converted lignocellulose hydrolysates to 2,3-BD [73]. Batch fermentation with corn stover hydrolyzate as a substrate reached a titer of 74.0 g/L 2,3-BD, with a productivity of 2.1 g/L/h. Furthermore, X10 had a high tolerance to the toxic compounds of lignocellulose hydrolysis such as furfural and vanillin, and the fermentation by-products such as formic and acetic acids.
B. licheniformis mutants, WX-02ΔbudC and WX-02ΔgldA, were engaged in optically pure 2,3-BD production from Miscanthus floridulus (silver grass) hydrolysate [123]. This application of the recombinants is promising because in the course of fed-batch fermentation, 32.2 g/L D(−)2,3-BD and 48.5 g/L meso-2,3-BD were obtained, respectively.
From alkali-pretreated corncob, the thermotolerant (52 °C) acetoin producer B. subtilis IPE5-4 produced 22.76 g/L acetoin, with a yield of 0.46 g/g and productivity 0.38 g/L/h [116]. This is by far the highest acetoin yield from lignocellulosic biomass in a process of simultaneous (enzymatic) saccharification and fermentation.
Another species that could utilize the lignocellulose sugars is P. polymyxa. Adlakha et al. [94] and Ma et al. [95] employed natural P. polymyxa strains and lignocellulose hydrolysate as a substrate, but applied different process performances. However, both teams obtained a similar yield of 2,3-BD (0.3 g/g sugar). Some Bacillus strains can directly degrade cellulose, hemicelluloses, and lignin owing to the action of their glycoside-hydrolase enzymes [124,125]. Among them, the plant-associated rhizobacteria (B. amyloliquefaciens, B. velezensis, B. siamensis), belonging to “B. amyloliquefaciens operational group” [126,127] contain the richest genetic pool of cellulases and xylanases [69]. B. subtilis BY-4 isolated from Tibetan’s pig gut [128], B. subtilis UMC7 from termites [129], or B. paralicheniformis strains from the Red Sea [130], are a source of powerful endoglucanases.
Hence, the attempts to develop bioprocesses for direct microbial conversion of plant biomass into 2,3-BD and acetoin by genetic improvement of strains are worthy and prospective. For example, Liu et al. [131] cloned and expressed several clostridial genes encoding cellobiohydrolases in B. subtilis strains. Later, the genes encoding cellulases cel8A and cel48S of Acetivibrio thermocellus were cloned and expressed in 2,3-BD producers B. licheniformis 24 and B. velesensis 5RB [132]. However, despite successful expression and high cellulase activity, the accumulation of fermentable sugars was not observed in these studies.
7. Media and Process Parameters for 2,3-BD and Acetoin Production by Bacilli
7.1. Medium Content
In addition to the carbon source, the nitrogen source is crucially important for the microbial synthesis of high amounts of 2,3-BD and acetoin. Tryptone, peptone, yeast extract, corn steep liquor (or powder), casamino acids, urea, and ammonium salts are known to highly improve the yield [9,22,60,61]. That was why Häßler et al. [62] added up to 60 g/L yeast extract in the medium for 2,3-BD synthesis by P. polymyxa. Later, Okonkwo et al. [61] assessed the impact of tryptone and yeast extract and showed that concentrations above 5 g/L may not engender a further increase in 2,3-BD production by this species. However, concerning B. licheniformis, the estimated optimal values were 13.38 g/L yeast extract and 6.41 g/L tryptone [67]. Corn steep liquor was frequently applied as an inexpensive organic nitrogen source [104], however, its addition is less suitable than yeast extract for 2,3-BD yield by B. licheniformis [67,75]. Taking into account the high price of yeast extract, it should be replaced by raw materials in industrial-scale fermentation [11,28]. Xiao et al. [97] estimated that molasses and soybean meal hydrolysate are more productive than pure sucrose and yeast extract plus peptone in acetoin fermentation by B. subtilis.
Other medium supplements affecting 2,3-BD production are phosphate and acetate [9,67]. Fed-batch processes by feeding glucose and acetate at a ratio of 0.35 mol acetate per mol glucose at constant pH 6.8 for P. polymyxa were found to the enhancement of 2,3-BD [133]. Fe2+, Mn2+, K+, and Mg2+ affect the enzymes activity and their stereospecificity [134]. The highly productive media also contain vitamins (biotin), and trace elements, supplied by the following salts: MgSO4, KH2PO4, K2HPO4, FeSO4, CuSO4, NaMoO4, MnCl2, 0.1 ZnSO4, Co(NO3)2, and CaCl2 [61].
7.2. Process Parameters
A total of three modes of process performance are applied for bacilli: flask-batch, batch, and fed-batch, which can overcome the inhibition of the process by the high substrate concentrations. The most influencing process parameters are temperature, pH of the media, aeration rate, and agitation speed, as the last two are crucial for acetoin/2,3-BD production ratio and are an object of optimization most frequently (Table 5).

Table 5.
Process parameters used for 2,3-BD production by strains belonging to genera Bacillus and Paenibacillus.
7.2.1. Temperature
The optimal temperature for acetoin/2,3-BD production is strain-dependent, although the species-specific characteristics should also be taken into account. For most producers of B. subtilis and B. amyloliquefaciens, the optimal temperature is usually around 37 °C. In most cases, using producers of these two species, this temperature is accepted to be optimal without being an object of optimization. In one of the few optimizations performed, Yang et al. [63] reported that the optimal temperature of B. amyloliquefaciens strain B10-127 for 2,3-BD production is 37 °C, using an experimental range from 25 to 40 °C. At 40 °C, the obtained titer 2,3-BD is slightly lower than the maximum (57.4 vs. 59.1 g/L at 37 °C). Similarly, in the studied range 25–40 °C, the strain B. subtilis JNA 3-10 produced acetoin with maximum titer and yield at 37 °C [7]. However, detailed monitoring of the effect of the temperature revealed that some strains of B. subtilis and B. amyloliquefaciens possess a higher temperature optimum for 2,3-BD and acetoin production. For example, the strain B. amyloliquefaciens 18025 produced 2,3-BD with a maximum titer, yield, and productivity at 40 °C, when the range between 27 and 44 °C was tested [78]. Likewise, when the fermentations were performed in the range between 20 °C and 50 °C, B. subtilis 168 displayed the highest conversion rate of 2,3-BD to acetoin at 40 °C [108]. When B. licheniformis is employed, the optimal temperature for 2,3-BD or acetoin production varies in a wide range from 30 to 50 °C, depending on the specificity of the strain used [70], (Table 6).

Table 6.
Process parameters used for acetoin production by strains belonging to genera Bacillus and Paenibacillus.
When glucose to acetoin is converted by the strain B. licheniformis MEL09, it was shown that temperature has no significant effect on the process between 34 °C and 40 °C [35]. However, the temperature appeared to be a significant factor in starch hydrolysate conversion into 2,3-BD by B. licheniformis NCIMB 8059. In this case, a maximum 2,3-BD concentration was obtained at 37 °C [68].
The temperature was also the object of optimization of glucose fermentation to 2,3-BD by B. licheniformis 24 [67]. The statistical analysis of the experimental results showed that a maximum 2,3-BD titer should be obtained at 37.8 °C. However, higher temperatures accelerate glucose consumption rate and 2,3-BD productivity, respectively. Thus, maximum productivity can be obtained at temperatures ≥ 39 °C [67]. On the other hand, there are many thermophilic B. licheniformis strains among the best 2,3-BD producers. Strains such as B. licheniformis ATCC 14,580 [88], 10-1-A [70], X10 [73], and BL1 [135], all have a temperature optimum of 50 °C for 2,3-BD production. Curiously, the highest 2,3-BD concentration obtained to date by Bacillus spp. (144.7 g/L from glucose) is received by B. licheniformis DSM 8785 at 30 °C [12].
Using P. polymyxa DSM 365, Okonkwo et al. [61] optimized the temperature as a factor in the Box–Behnken design during the process of glucose conversion into 2,3-BD. The optimal value appeared to be 35 °C. Significant quantities of acetoin from glucose [100] and 2,3-BD from corn stover hydrolysate [95] were received by the cultivation of P. polymyxa strains at 37 °C. Dias et al. [96] cultivated P. brasilensis at 32 °C for 2,3-BD production from glucose.
7.2.2. pH
When bacilli are employed for 2,3-BD production, a proper pH control taking into account their distinguishing characteristics should comply. The opposite of Enterobacteriaceae producers, in acetoin/2,3-BD fermentation, Bacillus spp. form organic acids in relatively low quantities. Thus, when pH is not controlled, its changes are limited and follow subsequent periods of decrease and increase. Hereof, depending on the specificity of the strain and its by-products formation, in many fermentations, the pH is not necessary to be maintained during the process, which is an important advantage of the Bacillus producers. In these cases, only initial pH is determined, as its value is the object of optimization and maintenance during the process. In such optimization for acetoin production from glucose by B. licheniformis MEL09, in the tested range of initial pH from 6.0 to 8.5, the optimal initial pH was determined to be 6.5 [35]. The same was the optimal initial pH for 2,3-BD production from glucose by B. amyloliquefaciens B 10-127 [63]. In other studies, when fermentations without pH control have been conducted, the most frequently used initial pH was 6.5 [98], or 7.0 [2,97] for acetoin production, and 6.5 [103,105] for 2,3-BD production, with no species specificity.
In pH-controlled fermentation, the optimal control values of pH are strain-dependent and if the maximum concentration of the product is intended, they varied from pH 6.0 to 7.0. At pH ≥ 7.0 the metabolic pathway shifts to acids production and the yields of both acetoin and 2,3-BD sharply decreased, while at pH ≤ 6.0 the substrate conversion was slow, reflecting on reduced productivity [9,67,86]. Raspoet et al. investigated the effect of pH on ten B. licheniformis strains and found out that eight of them produced a maximum 2,3-BD from glucose at pH 6.0, and the other two produced equal amounts at pH 6.0 and 6.5 [86]. The maximum 2,3-BD titer, yield, and productivity obtained by B. licheniformis 10-1-A from glucose were at pH 7.0. The highest acetoin titer also was achieved at pH 7.0 [70].
It remains unclear whether pH has an impact on the 2,3-BD/acetoin production ratio or not. Bao et al. [108] investigated the process of 2,3-BD conversion to acetoin using an engineered strain of B. subtilis. The authors found out that BDH (acetoin reductase/butanediol dehydrogenase), possessing reversible activities, has different pH optima for oxidation (pH 8.5) and reduction (pH 6.5). Thus, the strain showed the highest conversion rate of 2,3-BD to acetoin at pH 8.5 [108]. This effect of pH on the acetoin reductase activity was confirmed later for B. amyloliquefaciens B10-127 [104]. However, in the process of sugar conversion to acetoin and 2,3-BD, the highest amounts of both products were obtained at equal [70], or close [78] values of pH. Using B. amyloliquefaciens 18,025 in the process of bioconversion of VHP cane sugar, Maina et al. [78] noticed that at a certain aeration mode (kL a = 49 h−1) maximum D(−)2,3-BD was formed at pH 6.0 (32.3 g/L), but the highest acetoin titer was reached at pH 6.4 (8.5 g/L). However, at higher levels of aeration, when acetoin is the main product, the highest titer, yield, and productivity of acetoin was observed at pH 6.0. This indicates that pH and aeration are most likely interdependent parameters.
7.2.3. Aeration
Aeration is the crucial parameter determining the acetoin/2,3-BD production ratio. In the same manner as all producers of 2,3-BD, Bacillus spp. can obtain energy by two different pathways, fermentation and respiration [22], and metabolites formation depends on the relative activity of both pathways [78]. The high oxygen supply favors respiration, and in conditions with an excess of oxygen, the carbon flux is mainly channeled toward cell mass formation [136], resulting in a low yield of fermented products. On the opposite, in conditions with a lack of oxygen, cell mass is insufficient, which results in slow carbon consumption and low productivity.
Thus, for effective production of acetoin and 2,3-BD, both pathways should work simultaneously, in a proportion determined by the product specificity. Because the reduction of acetoin to 2,3-BD by BDH requires simultaneous oxidation of NADH to NAD+, NADH availability is essential for 2,3-BD formation. Under conditions of oxygen excess, through the oxidative phosphorylation more NADH is converted to NAD+, which hampers 2,3-BD production at the expense of acetoin accumulation [78]. Thus, acetoin appears to be a more aerobic product than 2,3-BD. Using B. subtilis culture, Moes et al. [90] found that, at a level of dissolved oxygen (DO) more than 100 ppb, acetoin is mainly accumulated, and vice versa, when DO is below 100 ppb–2,3-BD production is predominant. The required DO level can be obtained by manipulation of aeration flow rate or agitation speed. On the other hand, oxygen demand depends on the specificity of the producer, hence, aeration is generally an object of optimization to ensure effective production.
To provide optimal aeration, the most usual optimized process parameters are aeration flow rate and agitation speed (Table 5 and Table 6). Wang et al. [58] optimized both aeration flow rate and agitation speed in batch processes of sucrose utilization by B. subtilis CS13. They found that maximum acetoin and 2,3-BD are produced at aeration rates of 2 vvm and 1 vvm, respectively. For both products, the optimal was agitation speed of 500 rpm [58]. In the process of glucose conversion by B. amyloliquefaciens FMME044, using an aeration rate of 1 vvm at different agitation speeds was found that 300 rpm is optimal for 2,3-BD, and 400 rpm–for acetoin production [64]. Similarly, in glucose conversion by P. polymyxa CS107 at 0.5 vvm, 400 rpm is the optimal agitation for 2,3-BD, and 500 rpm–for acetoin production [100].
It is clear from all indicated optimizations so far, that acetoin production is maximized at higher aerations level than 2,3-BD for any organism used. However, in the course of fermentation, DO level is not constant. As a rule, DO sharply drops in the first few hours and slowly increases when the culture is exhausted [67]. This also affects the production of acetoin and 2,3-BD. On the other hand, when maximum 2,3-BD production is desired, at the beginning of the process more oxygen is needed for biomass formation, followed by a period with less oxygen for product formation.
Based on these presumptions, for an additional increase in product formation, two-stage or three-stage aeration strategies are applied. For 2,3-BD production from glucose by strain B. licheniformis 10-1-A, using aeration of 1 vvm, agitation rate was kept at 400 rpm for the first 10 h, and 200 rpm from then to the end of the process. Thus, 115.7 g/L 2,3-BD was obtained [70]. Yang et al. [104] used a three-stage aeration strategy for 2,3-BD production from a mix of glycerol and molasses by B. amyloliquefaciens B10-127. Agitation was kept consistently at 350 rpm (0–10 h), 400 rpm (5–22 h), and 350 rpm (22–88 h). Song et al. [75] applied two-stage agitation control for 2,3-BD production from glucose by B. licheniformis GSC3102. At this aeration strategy, the agitation rate was changed from 500 to 400 rpm when acetoin concentration passes over 10 g/L. To obtain maximum acetoin concentration from sugarcane molasses by B. subtilis DL01, Dai et al. [98] used three-stage aeration–agitation rate of 200 rpm (0–6 h), 300 rpm (6–24 h), and 350 rpm (after 24 h). The maximum obtained acetoin was 61.2 g/L.
Optimizations of aeration and agitation rates have the disadvantage that they apply only to the specific apparatus. Therefore, to obtain more information about the process, control of DO levels (%) or kL a (volumetric oxygen transfer coefficient, h−1) can be applied. As it was mentioned, during acetoin and 2,3-BD production from bakery waste hydrolysate, Maina et al. [5] found that for optimal acetoin formation, kL a = 203 h−1 (corresponding to 1 vvm and 600 rpm), while the optimal for 2,3-BD production is a far lower value of kL a-64 h−1 (corresponding to 1 vvm and 300 rpm). Similarly, in the process of sugarcane molasses conversion, for optimal acetoin production, kL a = 104 h−1, and for 2,3-BD production, kL a = 30 h−1 [78].
7.3. By-Products Reduction Strategies
The techno-economic evaluation of the production of acetoin and 2,3-BD reveals that efficiency of fermentation is the most important factor in the industry, followed by the process of separation and purification [137,138]. Industrial feasibility depends on the high titer of the target metabolite and the reduced concentration of the by-products. Because acetoin is a precursor to 2,3-BD, both metabolites present in all processes performed with bacilli. As already noted above, the directing of the production towards 2,3-BD or acetoin can be conducted by fine-tuning the process parameters, with aeration and agitation being of the utmost importance. EPS synthesis was successively diminished by optimization of the medium content [61]. As for strategies to reduce other by-products such as acetate, lactate, and succinate, this is often performed through genetic engineering improvement of Bacillus spp. producers. For example, Yang et al. achieved significant reduction of by-products formed by B. amyloliquefaciens B10-127 after over-expression of glyceraldehyde-3-phosphate dehydrogenase and NADH-dependent 2,3-BDH. The simultaneous action of these enzymes redistributed glucose flux, decreasing molar yields of unwanted by-products acetoin, lactate, and succinate by 80.8%, 33.3% and 39.5%, compared to the parent strain [105]. To decrease the accumulation of acetate and L-lactate by B. subtilis 168, Fu et al. disrupted both pta (coding phosphate acetyltransferase) and ldh (coding L-lactate dehydrogenase) genes in the genome of the strain [139]. The absence of pta gene resulted in less acetate and succinate accumulation. The deletion of ldh gene did not affect the metabolic flux in 2,3-BD pathway directly, but increased the NADH/NAD+ ratio, which is the key factor for 2,3-BD production. The deletion of ldh gene in B. licheniformis BL1 fully eliminated the lactate production [135]. The deletion of acoA (encoding acetoin dehydrogenase E1 component) in B. subtilis reduced acetate synthesis and increased acetoin concentration [11,139].
8. Conclusions
The two most important factors influencing the successful application of the microbial process on an industrial scale are: the cost of the carbon source in the composition of the fermentation medium, and the stability and safety of the microbial producer. In both respects, the GRAS producers of acetoin and 2,3-BD belonging to Bacillus and Penibacillus genera are promising. In addition to their non-pathogenic nature, another important property of bacilli is that they can synthesize pure stereoisomers of 2,3-BD and acetoin that can be used in chiral chemical syntheses. Another advantage of bacilli is their ability to convert raw, cheap, and waste substrates, including plant biomass, which makes the processes cost-effective and a good basis for the development of biotechnology. Finally, Bacillus spp. are highly tolerant to 2,3BD and acetoin, which allows greater production. Advances in mathematical and statistical methods for modeling and analyzing processes have led to the development of highly efficient fermentations, almost reaching the limits of process optimization. Therefore, a further impetus to increase the production of 2,3-BD and acetoin will be devoted to the genetic improvement of the strains through genetic and metabolic engineering.
Author Contributions
Conceptualization, K.P.; writing—original draft preparation, review and editing, K.P. and P.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Scientific Fund, Ministry of Education and Science, Republic of Bulgaria, grant number DN 17/1.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lee, J.H.; Lee, D.Y.; Lee, S.K.; Kim, H.R.; Chun, Y.; Yoo, H.Y.; Kwak, H.S.; Park, C.; Lee, J.H.; Kim, S.W. Development of 2,3-Butanediol Production Process from Klebsiella aerogenes ATCC 29007 Using Extracted Sugars of Chlorella pyrenoidosa and Biodiesel-Derived Crude Glycerol. Processes 2021, 9, 517. [Google Scholar] [CrossRef]
- Tian, Y.; Fan, Y.; Liu, J.; Zhao, X.; Chen, W. Effect of nitrogen, carbon sources and agitation speed on acetoin production of Bacillus subtilis SF4-3. Electron. J. Biotechnol. 2016, 19, 41–49. [Google Scholar] [CrossRef]
- Gao, C.; Zhang, L.; Xie, Y.; Hu, C.; Zhang, Y.; Li, L.; Wang, Y.; Ma, C.; Xu, P. Production of (3S)-acetoin from diacetyl by using stereoselective NADPH-dependent carbonyl reductase and glucose dehydrogenase. Bioresour. Technol. 2013, 137, 111–115. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Chen, F.; Sun, M.; Gao, H.; Guo, Z.; Lin, H.; Chen, J.; Jin, W.; Yang, Y.; Zhang, L.; et al. Efficient (3S)-Acetoin and (2S,3S)-2,3-Butanediol Production from meso-2,3-Butanediol Using Whole-Cell Biocatalysis. Molecules 2018, 23, 691. [Google Scholar] [CrossRef] [PubMed]
- Maina, S.; Schneider, R.; Alexandri, M.; Papapostolou, H.; Nychas, G.-J.; Koutinas, A.; Venus, J. Volumetric oxygen transfer coefficient as fermentation control parameter to manipulate the production of either acetoin or D(−)2,3-butanediol using bakery waste. Bioresour. Technol. 2021, 335, 125155. [Google Scholar] [CrossRef] [PubMed]
- Global Acetoin Market 2018–2023. Available online: https://menafn.com/1099300202/Global-Acetoin-Market-2018–2023 (accessed on 3 November 2021).
- Zhang, X.; Yang, T.-W.; Lin, Q.; Xu, M.-J.; Xia, H.-F.; Xu, Z.-H.; Li, H.-Z.; Rao, Z.-M. Isolation and identification of an acetoin high production bacterium that can reverse transform 2,3-butanediol to acetoin at the decline phase of fermentation. World J. Microbiol. Biotechnol. 2011, 27, 2785–2790. [Google Scholar] [CrossRef]
- Zhang, B.; Li, X.-L.; Fu, J.; Li, N.; Wang, Z.; Tang, Y.-J.; Chen, T. Production of Acetoin through Simultaneous Utilization of Glucose, Xylose, and Arabinose by Engineered Bacillus subtilis. PLoS ONE 2016, 11, e0159298. [Google Scholar] [CrossRef]
- Maina, S.; Prabhu, A.A.; Vivek, N.; Vlysidis, A.; Koutinas, A.; Kumar, V. Prospects on bio-based 2,3-butanediol and acetoin production: Recent progress and advances. Biotechnol. Adv. 2021, 107783. [Google Scholar] [CrossRef]
- Cui, X.; Zhao, X.; Liu, D. A novel route for the flexible preparation of hydrocarbon jet fuels from biomass-based platform chemicals: A case of using furfural and 2,3- butanediol as feedstocks. Green Chem. 2018, 20, 2018–2026. [Google Scholar] [CrossRef]
- Xiao, Z.; Xu, P. Acetoin Metabolism in Bacteria. Crit. Rev. Microbiol. 2007, 33, 127–140. [Google Scholar] [CrossRef]
- Hilmi, A.; Belgsir, E.M.; Leger, J.M.; Lamy, C. Electrocatalytic oxidation of aliphatic diols Part V. Electro-oxidation of butanediols on platinum-based electrodes. J. Electroanal. Chem. 1997, 435, 69–75. [Google Scholar] [CrossRef]
- Kochius, S.; Paetzold, M.; Scholz, A.; Merkens, H.; Vogel, A.; Ansorge-Schumacher, M.; Hollmann, F.; Schrader, J.; Holtmann, D. Enantioselective enzymatic synthesis of the α-hydroxy ketone (R)-acetoin from meso-2,3-butanediol. J. Mol. Catal. Enzym. 2014, 103, 61–66. [Google Scholar] [CrossRef]
- Garg, S.K.; Jain, A. Fermentative production of 2,3-butanediol: A review. Bioresour. Technol. 1995, 51, 103–109. [Google Scholar] [CrossRef]
- Celinska, E.; Grajek, W. Biotechnological production of 2,3-butanediol current state and prospects. Biotechnol. Adv. 2009, 27, 715–725. [Google Scholar] [CrossRef] [PubMed]
- Song, D.; Cho, S.-Y.; Vu, T.-T.; Duong, H.-P.-Y.; Kim, E. Dehydration of 2,3-Butanediol to 1,3-Butadiene and Methyl Ethyl Ketone: Modeling, Numerical Analysis and Validation Using Pilot-Scale Reactor Data. Catalysts 2021, 11, 999. [Google Scholar] [CrossRef]
- Yu, E.K.; Saddler, J.N. Power of solvent production by Klebsiella pneumoniae grown on sugars present in wood hemicelluloses. Biotechnol. Lett. 1982, 4, 121–126. [Google Scholar] [CrossRef]
- Hazeena, S.H.; Sindhu, R.; Pandey, A.; Binod, P. Lignocellulosic bio-refinery approach for microbial 2,3-Butanediol production. Bioresour. Technol. 2020, 302, 122873. [Google Scholar] [CrossRef] [PubMed]
- Petrova, P.; Ivanov, I.; Tsigoriyna, L.; Valcheva, N.; Vasileva, E.; Parvanova-Mancheva, T.; Arsov, A.; Petrov, K. Traditional Bulgarian Dairy Products: Ethnic Foods with Health Benefits. Microorganisms 2021, 9, 480. [Google Scholar] [CrossRef] [PubMed]
- Petrini, P.; De Ponti, S.; Farea, S.; Tanzi, M.C. Polyurethane-maleamides for cardiovascular applications: Synthesis and properties. J. Mater. Sci.-Mater. Med. 1999, 10, 711–714. [Google Scholar] [CrossRef] [PubMed]
- Bartowsky, E.J.; Henschke, P.A. The ‘buttery’ attribute of wine-diacetyl-desirability, spoilage and beyond. Int. J. Food Microbiol. 2004, 96, 235–252. [Google Scholar] [CrossRef]
- Ji, X.J.; Huang, H.; Ouyang, P.K. Microbial 2,3-butanediol production: A state of the art review. Biotechnol. Adv. 2011, 29, 351–364. [Google Scholar] [CrossRef] [PubMed]
- Syu, M.J. Biological production of 2,3-butanediol. Appl. Microbiol. Biotechnol. 2001, 55, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Anchordoguy, T.J.; Cecchini, C.A.; Crowe, J.H.; Crowe, L.M. Insights into the cryoprotective mechanism of dimethyl sulfoxide for phospholipid bilayers. Cryobiology 1991, 28, 467–473. [Google Scholar] [CrossRef]
- Armitage., W.J.; Juss, B.K. Osmotic response of mammalian cells: Effects of permeating cryoprotectants on nonsolvent volume. J. Cell. Physiol. 1996, 168, 532–538. [Google Scholar] [CrossRef]
- Petrov, K.; Petrova, P. High production of 2,3-butanediol from glycerol by Klebsiella pneumoniae G31. Appl. Microbiol. Biotechnol. 2009, 84, 659–665. [Google Scholar] [CrossRef] [PubMed]
- Levante, A.; Bancalari, E.; Tambassi, M.; Lazzi, C.; Neviani, E.; Gatti, M. Phenotypic Diversity of Lactobacillus casei Group Isolates as a Selection Criterion for Use as Secondary Adjunct Starters. Microorganisms 2020, 8, 128. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Lu, J.R. Strategies for enhancing fermentative production of acetoin: A review. Biotechnol. Adv. 2014, 32, 492–503. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.Y.; Lee, S.H.; Lee, H.J.; Seo, H.Y.; Park, W.S.; Jeon, C.O. Effects of Leuconostoc mesenteroides starter cultures on microbial communities and metabolites during kimchi fermentation. Int. J. Food Microbiol. 2012, 53, 378–387. [Google Scholar] [CrossRef] [PubMed]
- Laroute, V.; Tormo, H.; Couderc, C.; Mercier-Bonin, M.; Le Bourgeois, P.; Cocaign-Bousquet, M.; Daveran-Mingot, M.-L. From Genome to Phenotype: An Integrative Approach to Evaluate the Biodiversity of Lactococcus lactis. Microorganisms 2017, 5, 27. [Google Scholar] [CrossRef]
- Cañas, A.; Owens, J.D. Acetoin production in growing Leuconostoc mesenteroides. World J. Microbiol. Biotechnol. 1999, 62, 339–344. [Google Scholar] [CrossRef]
- Humia, B.V.; Santos, K.S.; Barbosa, A.M.; Sawata, M.; Mendonça, M.d.C.; Padilha, F.F. Beer Molecules and Its Sensory and Biological Properties: A Review. Molecules 2019, 24, 1568. [Google Scholar] [CrossRef] [PubMed]
- Romano, P.; Suzzi, G.; Zironi, R.; Comi, G. Biometric study of acetoin production in Hanseniaspora guilliermondii and Kloeckera apiculata. Appl. Environ. Microbiol. 1993, 59, 1838–1841. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, R.M.; Cavalheiro, D.; Ninow, J.L.; Furigo, A.J. Optimization of acetoin production by Hanseniaspora guilliermondii using experimental design. Braz. J. Chem. Eng. 2002, 19, 181–186. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, S.; Yong, Y.-C.; Ji, Z.; Ma, X.; Xu, Z.; Chen, S. Efficient production of acetoin by the newly isolated Bacillus licheniformis strain MEL09. Process Biochem. 2011, 46, 390–394. [Google Scholar] [CrossRef]
- Harden, A.; Walpole, G.S. 2,3-butylene glycol fermentation by Aerobacter aerogenes. Proc. R. Soc. 1906, 77, 399–405. [Google Scholar]
- Fulmer, E.I.; Christensen, L.M.; Kendall, A.R. Production of 2,3-butylene glycol by fermentation. Ind. Eng. Chem. 1933, 25, 798–800. [Google Scholar] [CrossRef]
- Voloch, M.; Jansen, N.B.; Ladish, M.R.; Tsao, G.T.; Narayan, R.; Rodwell, V.W. 2,3-butanediol. In Comprehensive Biotechnology: The Principles, Applications, and Regulations of Biotechnology in Industry, Agriculture, and Medicine, 1st ed.; Blanch, H.W., Drew, S., Wang, D.I.C., Eds.; Pergamon Press: Oxford, UK, 1985; pp. 933–944. [Google Scholar]
- Li, Z.J.; Ji, X.J.; Kan, S.L.; Qiao, H.Q.; Jiang, M.; Lu, D.; Wang, J.; Huang, H.; Jia, H.; Ouyuang, P.; et al. Past, present, and future industrial biotechnology in China. Adv. Biochem. Eng. Biotechnol. 2010, 122, 1–42. [Google Scholar] [CrossRef]
- Petrov, K.; Petrova, P. Isolation and molecular identification of Klebsiella pneumoniae strains, producing diols from glycerol. Biotechnol. Biotechnol. Equip. 2009, 23, 814–817. [Google Scholar] [CrossRef][Green Version]
- Tsvetanova, F.; Petrova, P.; Petrov, K. 2,3-butanediol production from starch by engineered Klebsiella pneumoniae G31-A. Appl. Microbiol. Biotechnol. 2014, 98, 2441–2451. [Google Scholar] [CrossRef] [PubMed]
- Petrov, K.; Petrova, P. Enhanced production of 2,3-butanediol from glycerol by forced pH fluctuations. Appl. Microbiol. Biotechnol. 2010, 87, 943–949. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Guo, W.; Shi, L.; Yu, Y.; Zhang, C.; Yang, H. Characterization of acetoin production in a budC gene disrupted mutant of Serratia marcescens G12. J. Ind. Microbiol. Biotechnol. 2014, 41, 1267–1274. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Yang, H.H.; Feng, X.H.; Li, S.; Xu, H. A 2,3-butanediol dehydrogenase from Paenibacillus polymyxa ZJ-9 for mainly producing R,R-2,3-butanediol: Purification, characterization and cloning. J. Basic Microbiol. 2013, 53, 733–741. [Google Scholar] [CrossRef]
- Ge, Y.; Li, K.; Li, L.; Gao, C.; Zhang, L.; Ma, C.; Xu, P. Contracted but effective production of enantiopure 2,3-butanediol by thermophilic and GRAS Bacillus licheniformis. Green Chem. 2016, 18, 4693–4703. [Google Scholar] [CrossRef]
- Banoon, S.; Ali, Z.; Salih, T. Antibiotic resistance profile of local thermophilic Bacillus licheniformis isolated from Maysan province soil. Comun. Sci. 2020, 11, e3291. [Google Scholar] [CrossRef]
- Šovljanski, O.; Pezo, L.; Stanojev, J.; Bajac, B.; Kovač, S.; Tóth, E.; Ristić, I.; Tomić, A.; Ranitović, A.; Cvetković, D.; et al. Comprehensive Profiling of Microbiologically Induced CaCO3 Precipitation by Ureolytic Bacillus Isolates from Alkaline Soils. Microorganisms 2021, 9, 1691. [Google Scholar] [CrossRef]
- Mahdi, I.; Fahsi, N.; Hafidi, M.; Allaoui, A.; Biskri, L. Plant Growth Enhancement using Rhizospheric Halotolerant Phosphate Solubilizing Bacterium Bacillus licheniformis QA1 and Enterobacter asburiae QF11 Isolated from Chenopodium quinoa Willd. Microorganisms 2020, 8, 948. [Google Scholar] [CrossRef] [PubMed]
- Vinothkanna, A.; Sathiyanarayanan, G.; Balaji, P.; Mathivanan, K.; Pugazhendhi, A.; Ma, Y.; Sekar, S.; Thirumurugan, R. Structural characterization, functional and biological activities of an exopolysaccharide produced by probiotic Bacillus licheniformis AG-06 from Indian polyherbal fermented traditional medicine. Int. J. Biol. Macromol. 2021, 174, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Hirad, A.; Bahkali, A.; Khiyami, M.; Ahmed, M.; Santhapa, C.; Elgorban, A.; Al-Sum, B. Bioactivity of Marine Bacillus licheniformis Ksawd3 Isolated from Arabian Gulf, Saudi Arabia. J. Pure Appl. Microbiol. 2014, 8, 607–613. [Google Scholar]
- Biswas, J.K.; Banerjee, A.; Sarkar, B.; Sarkar, D.; Sarkar, S.K.; Rai, M.; Vithanage, M. Exploration of an Extracellular Polymeric Substance from Earthworm Gut Bacterium (Bacillus licheniformis) for Bioflocculation and Heavy Metal Removal Potential. Appl. Sci. 2020, 10, 349. [Google Scholar] [CrossRef]
- Wang, S.; Hou, Q.; Guo, Q.; Zhang, J.; Sun, Y.; Wei, H.; Shen, L. Isolation and Characterization of a Deoxynivalenol-Degrading Bacterium Bacillus licheniformis YB9 with the Capability of Modulating Intestinal Microbial Flora of Mice. Toxins 2020, 12, 184. [Google Scholar] [CrossRef] [PubMed]
- Petrova, P.; Arsov, A.; Ivanov, I.; Tsigoriyna, L.; Petrov, K. New Exopolysaccharides Produced by Bacillus licheniformis 24 Display Substrate-Dependent Content and Antioxidant Activity. Microorganisms 2021, 9, 2127. [Google Scholar] [CrossRef] [PubMed]
- Ilyas, N.; Mumtaz, K.; Akhtar, N.; Yasmin, H.; Sayyed, R.Z.; Khan, W.; Enshasy, H.A.E.; Dailin, D.J.; Elsayed, E.A.; Ali, Z. Exopolysaccharides Producing Bacteria for the Amelioration of Drought Stress in Wheat. Sustainability 2020, 12, 8876. [Google Scholar] [CrossRef]
- Kwon, J.-H.; Won, S.-J.; Moon, J.-H.; Lee, U.; Park, Y.-S.; Maung, C.E.H.; Ajuna, H.B.; Ahn, Y.S. Bacillus licheniformis PR2 Controls Fungal Diseases and Increases Production of Jujube Fruit under Field Conditions. Horticulturae 2021, 7, 49. [Google Scholar] [CrossRef]
- Matei, M.-C.; Andrei, S.M.; Buza, V.; Cernea, M.S.; Dumitras, D.A.; Neagu, D.; Rafa, H.; Popovici, C.P.; Szakacs, A.R.; Catinean, A.; et al. Natural Endotoxemia in Dogs—A Hidden Condition That Can Be Treated with a Potential Probiotic Containing Bacillus subtilis, Bacillus licheniformis, and Pediococcus acidilactici: A Study Model. Animals 2021, 11, 1367. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Qin, J.; Gao, C.; Hua, D.; Ma, C.; Li, L.; Wang, Y.; Xu, P. Production of (2S,3S)-2,3-butanediol and (3S)-acetoin from glucose using resting cells of Klebsiella pneumonia and Bacillus subtilis. Bioresour. Technol. 2011, 102, 10741–10744. [Google Scholar] [CrossRef]
- Wang, D.; Oh, B.R.; Lee, S.; Kim, D.-H.; Joe, M.-H. Process optimization for mass production of 2,3-butanediol by Bacillus subtilis CS13. Biotechnol. Biofuels 2021, 14, 15. [Google Scholar] [CrossRef] [PubMed]
- Marwoto, B.; Nakashimada, Y.; Kakizono, T.; Nishio, N. Metabolic analysis of acetate accumulation during xylose consumption by Paenibacillus polymyxa. Appl. Microbiol. Biotechnol. 2004, 64, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Okonkwo, C.C.; Ujor, V.; Ezeji, T.C. Investigation of relationship between 2,3-butanediol toxicity and production during growth of Paenibacillus polymyxa. New Biotechnol. 2017, 34, 23–31. [Google Scholar] [CrossRef]
- Okonkwo, C.C.; Ujor, V.C.; Mishra, P.K.; Ezeji, T.C. Process Development for Enhanced 2,3-Butanediol Production by Paenibacillus polymyxa DSM 365. Fermentation 2017, 3, 18. [Google Scholar] [CrossRef]
- Häßler, T.; Schieder, D.; Pfaller, R.; Faulstich, M.; Sieber, V. Enhanced fed-batch fermentation of 2,3-butanediol by Paenibacillus polymyxa DSM 365, Bioresour. Technol. 2012, 124, 237–244. [Google Scholar] [CrossRef]
- Yang, T.; Rao, Z.; Zhang, X.; Lin, Q.; Xia, H.; Xu, Z.; Yang, S. Production of 2,3-butanediol from glucose by GRAS microorganism Bacillus amyloliquefaciens. J. Basic Microbiol. 2011, 51, 650–658. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, S.; Liu, L.; Wu, J. Acetoin production enhanced by manipulating carbon flux in a newly isolated Bacillus amyloliquefaciens. Bioresour. Technol. 2013, 130, 256–260. [Google Scholar] [CrossRef] [PubMed]
- Nilegaonkar, S.; Bhosale, S.; Kshirsagar, D.; Kapadi, A. Production of 2,3-butanediol from glucose by Bacillus licheniformis. World J. Microbiol. Biotechnol. 1992, 8, 378–381. [Google Scholar] [CrossRef] [PubMed]
- Nilegaonkar, S.S.; Bhosale, S.B.; Dandage, C.N.; Kapadi, A.H. Potential of Bacillus licheniformis for the production of 2,3-butanediol. J. Ferment. Bioeng. 1996, 82, 408–410. [Google Scholar] [CrossRef]
- Tsigoriyna, L.; Ganchev, D.; Petrova, P.; Petrov, K. Highly Efficient 2,3-Butanediol Production by Bacillus licheniformis via Complex Optimization of Nutritional and Technological Parameters. Fermentation 2021, 7, 118. [Google Scholar] [CrossRef]
- Perego, P.; Converti, A.; del Borghi, M. Effects of temperature, inoculum size and starch hydrolyzate concentration on butanediol production by Bacillus licheniformis. Bioresour. Technol. 2003, 89, 125–131. [Google Scholar] [CrossRef]
- Petrova, P.; Petlichka, S.; Petrov, K. New Bacillus spp. with potential for 2,3-butanediol production from biomass. J. Biosci. Bioeng. 2020, 130, 20–28. [Google Scholar] [CrossRef]
- Li, L.; Zhang, L.; Li, K.; Wang, Y.; Gao, C.; Han, B.; Ma, C.; Xu, P. A newly isolated Bacillus licheniformis strain thermophilically produces 2,3-butanediol, a platform and fuel bio-chemical. Biotechnol. Biofuels 2013, 6, 123. [Google Scholar] [CrossRef]
- Jurchescu, I.M.; Hamann, J.; Zhou, X.; Ortmann, T.; Kuenz, A.; Prusse, U.; Lang, S. Enhanced 2,3-butanediol production in fed batch cultures of free and immobilized Bacillus licheniformis DSM 8785. Appl. Microbiol. Biotechnol. 2013, 97, 6715–6723. [Google Scholar] [CrossRef] [PubMed]
- Lü, C.; Ge, Y.; Cao, M.; Guo, X.; Liu, P.; Gao, C.; Xu, P.; Ma, C. Metabolic Engineering of Bacillus licheniformis for Production of Acetoin. Front. Bioeng. Biotechnol. 2020, 8, 125. [Google Scholar] [CrossRef]
- Li, L.X.; Li, K.; Wang, K.; Chen, C.; Gao, C.; Ma, C.Q.; Xu, P. Efficient production of 2,3-butanediol from corn stover hydrolysate by using a thermophilic Bacillus licheniformis strain. Bioresour. Technol. 2014, 170, 256–261. [Google Scholar] [CrossRef]
- Rebecchi, S.; Pinelli, D.; Zanaroli, G.; Fava, F.; Frascari, D. Effect of oxygen mass transfer rate on the production of 2,3-butanediol from glucose and agro-industrial byproducts by Bacillus licheniformis ATCC 9789. Biotechnol. Biofuels 2018, 11, 145. [Google Scholar] [CrossRef] [PubMed]
- Song, C.W.; Rathnasingh, C.; Park, J.M.; Lee, J.; Song, H. Isolation and evaluation of Bacillus strains for industrial production of 2,3-butanediol. J. Microbiol. Biotechnol. 2018, 28, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Kallbach, M.; Horn, S.; Kuenz, A.; Prusse, U. Screening of novel bacteria for the 2,3-butanediol production. Appl. Microbiol. Biotechnol. 2017, 101, 1025–1033. [Google Scholar] [CrossRef]
- PubChem Database at the National Institutes of Health (NIH). Available online: https://pubchem.ncbi.nlm.nih.gov (accessed on 3 November 2021).
- Maina, S.; Mallouchos, A.; Nychas, G.-J.E.; Freire, D.M.; de Castro, A.M.; Papanikolaou, S.; Kookos, I.K.; Koutinas, A. Bioprocess development for (2R,3R)-butanediol and acetoin production using very high polarity cane sugar and sugarcane molasses by a Bacillus amyloliquefaciens strain. J. Chem. Technol. Biotechnol. 2019, 94, 2167–2177. [Google Scholar] [CrossRef]
- KEGG BRITE Database. Available online: https://www.genome.jp/kegg/brite.html (accessed on 6 November 2021).
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef] [PubMed]
- Petrova, P.; Velikova, P.; Petrov, K. Genome sequence of Bacillus velezensis 5RB, an overproducer of 2,3-butanediol. Microbiol. Resour. Announc. 2019, 8, e01475-18. [Google Scholar] [CrossRef]
- Voloch, M.; Ladish, M.R.; Rodwell, V.W.; Tsao, G.T. Reduction of acetoin to 2,3-butanediol in Klebsiella pneumoniae: A new model. Biotechnol. Bioeng. 1983, 25, 173–183. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, Y.; Yang, D.C.; Zhang, J.L. Expression and functional analysis of three nicosulfuron-degrading enzymes from Bacillus subtilis YB1. J. Environ. Sci. Health B 2018, 53, 476–485. [Google Scholar] [CrossRef]
- Gao, H.; Jiang, X.; Pogliano, K.; Aronson, A.I. The E1-beta and E2 subunits of the Bacillus subtilis pyruvate dehydrogenase complex are involved in regulation of sporulation. J. Bacteriol. 2002, 184, 2780–2788. [Google Scholar] [CrossRef] [PubMed]
- Thanh, T.N.; Jurgen, B.; Bauch, M.; Liebeke, M.; Lalk, M.; Ehrenreich, A.; Evers, S.; Maurer, K.H.; Antelmann, H.; Ernst, F.; et al. Regulation of acetoin and 2,3-butanediol utilization in Bacillus licheniformis. Appl. Microbiol. Biotechnol. 2010, 87, 2227–2235. [Google Scholar] [CrossRef]
- Raspoet, D.; Pot, B.; De Deyn, D.; De Vos, P.; Kersters, K.; De Ley, J. Differentiation between 2,3-Butanediol Producing Bacillus licheniformis and B. polymyxa Strains by Fermentation Product Profiles and Whole-Cell Protein Electrophoretic Patterns. Syst. Appl. Microbiol. 1991, 14, 1–7. [Google Scholar] [CrossRef]
- Shariati, P.; Mitchell, W.J.; Boyd, A.; Priest, F.G. Anaerobic metabolism in Bacillus licheniformis NClB 6346. Microbiology 1995, 141, 1117–1124. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Chen, C.; Li, K.; Wang, Y.; Gao, C.; Ma, C.; Xua, P. Efficient Simultaneous Saccharification and Fermentation of Inulin to 2,3-Butanediol by Thermophilic Bacillus licheniformis ATCC 14580. Appl. Environ. Microbiol. 2014, 80, 6458–6464. [Google Scholar] [CrossRef]
- Białkowska, A.M.; Gromek, E.; Krysiak, J.; Sikora, B.; Kalinowska, H.; Jędrzejczak-Krzepkowska, M.; Celina Kubik, C.; Lang, S.; Schütt, F.; Turkiewicz, M. Application of enzymatic apple pomace hydrolysate to production of 2,3-butanediol by alkaliphilic Bacillus licheniformis NCIMB 8059. J. Ind. Microbiol. Biotechnol. 2015, 42, 1609–1621. [Google Scholar] [CrossRef] [PubMed]
- Moes, J.; Griot, M.; Keller, J.; Heinzle, E.; Dunn, I.J.; Bourne, J.R. A microbial culture with oxygen-sensitive product distribution as a potential tool for characterizing bioreactor oxygen-transport. Biotechnol. Bioeng. 1985, 27, 482–489. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.W.; Rao, Z.M.; Zhang, X.; Xu, M.J.; Xu, Z.H.; Yang, S.T. Fermentation of biodiesel-derived glycerol by Bacillus amyloliquefaciens: Effects of co-substrates on 2,3-butanediol production. Appl. Microbiol. Biotechnol. 2013, 97, 7651–7658. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Xu, H.; Li, Q.-J.; Feng, X.-H.; Li, S. Optimization of medium for one-step fermentation of inulin extract from Jerusalem artichoke tubers using Paenibacillus polymyxa ZJ-9 to produce R,R-2,3-butanediol. Bioresour. Technol. 2010, 101, 7076–7082. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.-J.; Cheng, J.-S.; Liang, Y.-Q.; Jiang, T.; Yuan, Y.-J. Regulation of extracellular oxidoreduction potential enhanced (R,R)-2,3-butanediol production by Paenibacillus polymyxa CJX518. Bioresour. Technol. 2014, 167, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Adlakha, N.; Yazdani, S.S. Efficient production of (R,R)-2,3-butanediol from cellulosic hydrolysate using Paenibacillus polymyxa ICGEB2008. J. Ind. Microbiol. Biotechnol. 2015, 42, 21–28. [Google Scholar] [CrossRef]
- Ma, K.; He, M.; You, H.; Pan, L.; Wang, Z.; Wang, Y.; Hu, G.; Cui, Y.; Maeda, T. Improvement of (R,R)-2,3-butanediol production from corn stover hydrolysate by cell recycling continuous fermentation. Chem. Eng. J. 2018, 332, 361–369. [Google Scholar] [CrossRef]
- Dias, B.D.C.; Lima, M.E.D.N.V.; Vollú, R.E.; da Mota, F.F.; da Silva, A.J.R.; de Castro, A.M.; Freire, D.M.G.; Seldin, L. 2,3-Butanediol production by the non-pathogenic bacterium Paenibacillus brasilensis. Appl. Microbiol. Biotechnol. 2018, 102, 8773–8782. [Google Scholar] [CrossRef]
- Xiao, Z.J.; Liu, P.H.; Qin, J.Y.; Xu, P. Statistical optimization of medium components for enhanced acetoin production from molasses and soybean meal hydrolysate. Appl. Microbiol. Biotechnol. 2007, 74, 61–68. [Google Scholar] [CrossRef]
- Dai, J.Y.; Cheng, L.; He, Q.F.; Xiu, Z.L. High acetoin production by a newly isolated marine Bacillus subtilis strain with low requirement of oxygen supply. Process Biochem. 2015, 50, 1730–1734. [Google Scholar] [CrossRef]
- Xu, P.; Xiao, Z.J.; Du, Y.; Wei, Z. Acetoin High-Yield Bacillus pumilus Strain. U.S. Patent US 8,158,402 B2, 17 April 2012. [Google Scholar]
- Zhang, L.; Chen, S.; Xie, H.; Tian, Y.; Hu, K. Efficient acetoin production by optimization of medium components and oxygen supply control using a newly isolated Paenibacillus polymyxa CS107. J. Chem. Technol. Biotechnol. 2012, 87, 1551–1557. [Google Scholar] [CrossRef]
- Qi, G.; Kang, Y.; Li, L.; Xiao, A.; Zhang, S.; Wen, Z.; Xu, D.; Chen, S. Deletion of meso-2,3-butanediol dehydrogenase gene budC for enhanced D(−)2,3-butanediol production in Bacillus licheniformis. Biotechnol. Biofuels 2014, 7, 16. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Zhang, J.; Li, L.; Wen, Z.; Nomura, C.T.; Wu, S.; Chen, S. Engineering Bacillus licheniformis for the production of meso-2,3-butanediol. Biotechnol. Biofuels 2016, 9, 117. [Google Scholar] [CrossRef]
- Białkowska, A.M.; Jędrzejczak-Krzepkowska, M.; Gromek, E.; Krysiak, J.; Sikora, B.; Kalinowska, H.; Kubik, C.; Schütt, F.; Turkiewicz, M. Effects of genetic modifications and fermentation conditions on 2,3-butanediol production by alkaliphilic Bacillus subtilis. Appl. Microbiol. Biotechnol. 2016, 100, 2663–2676. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Rao, Z.; Zhang, X.; Xu, M.; Xu, Z.; Yang, S.-T. Enhanced 2,3-butanediol production from biodiesel-derived glycerol by engineering of cofactor regeneration and manipulating carbon flux in Bacillus amyloliquefaciens. Microb. Cell Fact. 2015, 14, 122. [Google Scholar] [CrossRef]
- Yang, T.; Rao, Z.; Zhang, X.; Xu, M.; Xu, Z.; Yang, S.T. Improved production of 2,3-butanediol in Bacillus amyloliquefaciens by over-expression of glyceraldehyde-3-phosphate dehydrogenase and 2,3-butanediol dehydrogenase. PLoS ONE 2013, 8, e76149. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Cao, C.; Jiang, R.; Xu, H.; Xue, F.; Huang, W.; Ni, H.; Gao, J. Production of R,R-2,3-butanediol of ultra-high optical purity from Paenibacillus polymyxa ZJ-9 using homologous recombination. Bioresour. Technol. 2018, 261, 272–278. [Google Scholar] [CrossRef]
- Zhang, L.; Xu, Y.; Gao, J.; Xu, H.; Cao, C.; Xue, F.; Ding, G.; Peng, Y. Introduction of the exogenous NADH coenzyme regeneration system and its influence on intracellular metabolic flux of Paenibacillus polymyxa. Bioresour. Technol. 2016, 201, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Bao, T.; Zhang, X.; Rao, Z.; Zhao, X.; Zhang, R.; Yang, T.; Xu, Z.; Yang, S. Efficient Whole-Cell Biocatalyst for Acetoin Production with NAD+ Regeneration System through Homologous Co-Expression of 2,3-Butanediol Dehydrogenase and NADH Oxidase in Engineered Bacillus subtilis. PLoS ONE 2019, 9, e102951. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, R.; Bao, T.; Rao, Z.; Yang, T.; Xu, M.; Xu, Z.; Li, H.; Yang, S. The rebalanced pathway significantly enhances acetoin production by disruption of acetoin reductase gene and moderate-expression of a new water-forming NADH oxidase in Bacillus subtilis. Metab. Eng. 2014, 23, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhao, X.; Zhang, J.; Zhao, C.; Liu, J.; Tian, Y.; Yang, L. Effect of deletion of 2, 3-butanediol dehydrogenase gene (bdhA) on acetoin production of Bacillus subtilis. Prep. Biochem. Biotechnol. 2017, 47, 761–767. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wei, X.; Yu, W.; Wen, Z.; Chen, S. Enhancement of acetoin production from Bacillus licheniformis by 2,3-butanediol conversion strategy: Metabolic engineering and fermentation control. Process Biochem. 2017, 57, 35–42. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, R.; Bao, T.; Yang, T.; Xu, M.; Li, H.; Xu, Z.; Rao, Z. Moderate expression of the transcriptional regulator ALsR enhances acetoin production by Bacillus subtilis. J. Ind. Microbiol. Biotechnol. 2013, 40, 1067–1076. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Fu, J.; Zhang, X.; Chen, T. Metabolic engineering of Bacillus subtilis for enhanced production of acetoin. Biotechnol. Lett. 2012, 34, 1877–1885. [Google Scholar] [CrossRef]
- Xu, H.; Jia, S.; Liu, J. Production of acetoin by Bacillus subtilis TH-49. In Proceedings of the 2011 International Conference on Consumer Electronics, Communications and Networks (CECNet), Xianning, China, 11–13 March 2011; pp. 1524–1527. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, R.; Yang, T.; Zhang, J.; Xu, M.; Li, H.; Xu, Z.; Rao, Z. Mutation breeding of acetoin high producing Bacillus subtilis blocked in 2,3-butanediol dehydrogenase. World J. Microbiol. Biotechnol. 2013, 29, 1783–1789. [Google Scholar] [CrossRef]
- Jia, X.; Peng, X.; Liu, Y.; Han, Y. Conversion of cellulose and hemicellulose of biomass simultaneously to acetoin by thermophilic simultaneous saccharification and fermentation. Biotechnol. Biofuels 2017, 10, 232. [Google Scholar] [CrossRef]
- Luo, Q.; Wu, J.; Wu, M. Enhanced acetoin production by Bacillus amyloliquefaciens through improved acetoin tolerance. Process Biochem. 2014, 49, 1223–1230. [Google Scholar] [CrossRef]
- Feng, J.; Gu, Y.; Yan, P.-F.; Song, C.; Wang, Y. Recruiting Energy-Conserving Sucrose Utilization Pathways for Enhanced 2,3-Butanediol Production in Bacillus subtilis. ACS Sustain. Chem. Eng. 2017, 5, 11221–11225. [Google Scholar] [CrossRef]
- Deshmukh, A.N.; Nipanikar-Gokhale, P.; Jain, R. Engineering of Bacillus subtilis for the Production of 2,3-Butanediol from Sugarcane Molasses. Appl. Biochem. Biotechnol. 2016, 179, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Sikora, B.; Kubik, C.; Kalinowska, H.; Gromek, E.; Białkowska, A.; Jędrzejczak-Krzepkowska, M.; Schüett, F.; Turkiewicz, M. Application of byproducts from food processing for production of 2,3-butanediol using Bacillus amyloliquefaciens TUL 308. Prep. Biochem. Biotechnol. 2016, 46, 610–619. [Google Scholar] [CrossRef]
- Białkowska, A. Strategies for efficient and economical 2,3-butanediol production: New trends in this field. World J. Microbiol. Biotechnol. 2016, 32, 200. [Google Scholar] [CrossRef]
- Koutinas, A.A.; Yepez, B.; Kopsahelis, N.; Freire, D.M.G.; de Castro, A.M.; Papanikolaou, S.; Kookos, I.K. Techno-economic evaluation of a complete bioprocess for 2,3-butanediol production from renewable resources. Bioresour. Technol. 2016, 204, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Huang, H.; Chen, S.; Qi, G. Production of optically pure 2,3-butanediol from Miscanthus floridulus hydrolysate using engineered Bacillus licheniformis strains. World J. Microbiol. Biotechnol. 2018, 34, 66. [Google Scholar] [CrossRef]
- Khelil, O.; Choubane, S.; Cheba, B.A. Polyphenols content of spent coffee grounds subjected to physico-chemical pretreatments influences lignocellulolytic enzymes production by Bacillus sp. R2. Bioresour. Technol. 2016, 211, 769e773. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Tanabe, S.H.; Xie, C.; Honda, D.; Sun, J.; Ai, L. Bacillus ligniniphilus sp. nov.; an alkaliphilic and halotolerant bacterium isolated from sediments of the South China Sea. Int. J. Syst. Evol. Microbiol. 2014, 64, 1712e1717. [Google Scholar] [CrossRef]
- Borriss, R.; Chen, X.H.; Rueckert, C.; Blom, J.; Becker, A.; Baumgarth, B.; Fan, B.; Pukall, R.; Schumann, P.; Spröer, C.; et al. Relationship of Bacillus amyloliquefaciens clades associated with strains DSM 7 T and FZB42 T: A proposal for Bacillus amyloliquefaciens subsp. nov. and Bacillus amyloliquefaciens subsp. plantarum subsp. nov. based on complete genome sequence comparisons. Int. J. Syst. Evol. Microbiol. 2011, 61, 1786e1801. [Google Scholar] [CrossRef]
- Chen, L.; Gu, W.; Xu, H.-Y.; Yang, G.-L.; Shan, X.-F.; Chen, G.; Kang, Y.-H.; Wang, C.-F.; Qian, A.-D. Comparative genome analysis of Bacillus velezensis reveals a potential for degrading lignocellulosic biomass. 3 Biotech 2018, 8, 253. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Yang, W.; Meng, F.; Ji, S.; Xin, H.; Cao, B. Characterization of an acidic cellulase produced by Bacillus subtilis BY-4 isolated from gastrointestinal tract of Tibetan pig. J. Taiwan Inst. Chem. Eng. 2015, 56, 67–72. [Google Scholar] [CrossRef]
- Wei, K.S.C.; Teoh, T.C.; Koshy, P.; Salmah, I.; Zainudin, A. Cloning, expression and characterization of the endoglucanase gene from Bacillus subtilis UMC7 isolated from the gut of the indigenous termite Macrotermes malaccensis in Escherichia coli. Electron. J. Biotechnol. 2015, 18, 103–109. [Google Scholar] [CrossRef]
- Fatani, S.; Saito, Y.; Alarawi, M.; Gojobori, T.; Mineta, K. Genome sequencing and identification of cellulase genes in Bacillus paralicheniformis strains from the Red Sea. BMC Microbiol. 2021, 21, 254. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.M.; Xin, X.J.; Li, C.X.; Xu, J.H.; Bao, J. Cloning of thermostable cellulase genes of Clostridium thermocellum and their secretive expression in Bacillus subtilis. Appl. Biochem. Biotechnol. 2012, 166, 652–662. [Google Scholar] [CrossRef] [PubMed]
- Arsov, A.; Petrov, K.; Petrova, P. Enhanced Activity by Genetic Complementarity: Heterologous Secretion of Clostridial Cellulases by Bacillus licheniformis and Bacillus velezensis. Molecules 2021, 26, 5625. [Google Scholar] [CrossRef]
- Nakashimada, Y.; Marwoto, B.; Kashiwamura, T.; Kakizono, T.; Nishio, N. Enhanced 2,3-butanediol production by addition of acetic acid in Paenibacillus polymyxa. J. Biosci. Bioeng. 2000, 90, 661–664. [Google Scholar] [CrossRef]
- Deshmukh, A.; Mistry, S.; Yewale, T.; Mahajan, D.; Jain, R. Production of 2,3-butanediol from Sugarcane Molasses Using Bacillus subtilis. Int. J. Adv. Biotechnol. Res. 2015, 6, 66–79. Available online: https://www.researchgate.net/publication/273453911_Production_of_2_3-Butanediol_from_Sugarcane_Molasses_Using_Bacillus_subtilis (accessed on 9 December 2021).
- Wang, Q.; Chen, T.; Zhao, X.; Chamu, J. Metabolic engineering of thermophilic Bacillus licheniformis for chiral pure D(−)2,3-butanediol production. Biotechnol. Bioeng. 2012, 109, 1610–1621. [Google Scholar] [CrossRef]
- Jansen, N.B.; Flickinger, M.C.; Tsao, G.T. Production of 2,3-butanediol from D(−)xylose by Klebsiella oxytoca ATCC 8724. Biotechnol. Bioeng. 1984, 26, 362–369. [Google Scholar] [CrossRef] [PubMed]
- Hakizimana, O.; Emmanuel Matabaro, E.; Lee, B.H. The current strategies and parameters for the enhanced microbial production of 2,3-butanediol. Biotechnol. Rep. 2020, 25, e00397. [Google Scholar] [CrossRef]
- Harvianto, G.R.; Haider, J.; Hong, J.; Long, N.V.D.; Shim, J.-J.; Cho, M.H.; Kim, W.K.; Lee, M. Purification of 2,3-butanediol from fermentation broth: Process development and techno-economic analysis. Biotechnol. Biofuels 2018, 11, 18. [Google Scholar] [CrossRef]
- Fu, J.; Huo, G.; Feng, L.; Mao, Y.; Wang, Z.; Ma, H.; Chen, T.; Zhao, X. Metabolic engineering of Bacillus subtilis for chiral pure meso-2,3-butanediol production. Biotechnol. Biofuels 2016, 9, 90. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).